Abstract
OBJECTIVE
Previous work from the HLA-B27 transgenic rat model of ankylosing spondylitis (AS) suggests that macrophages develop an intracellular stress response called the Unfolded Protein Response (UPR) and as a result secrete increased cytokines in response to Toll like receptor agonists such as lipopolysaccharide (LPS). Our objective was to determine if macrophages from AS patients also undergo an UPR and secrete increased cytokines/chemokines in response to LPS.
METHODS
Peripheral blood monocytes isolated from 10 AS patients and healthy controls were differentiated in vitro with M-CSF. Select samples were treated with IFN-γ to up-regulate MHC class I (HLA-B) expression prior to stimulation with LPS for 3h (for RNA collection), or 8–24h (for supernatant collection). UPR induction was assessed by expression of ERdj4, BiP and CHOP mRNA.
RESULTS
Although IFN-γ treatment up-regulated HLA-B expression 2-fold(p=0.0094), neither IFN-γ nor LPS enhanced BiP or CHOP expression substantially (<1.3 fold). ERdj4 expression increased weakly but insignificantly in IFN-γ+LPS AS samples (2.2 fold, p=0.31). In response to LPS, AS macrophages secreted more CXCL9, IL-10, IL-12p70, IL-23, and TNF-α than controls(p ≤ 0.025)The most striking difference was observed for IL-23 (median AS patient 265 pg/mL vs. control 9 pg/mL, p=0.0007). We did not detect significant differences in IL-6, IL-8, or IFN-β production.
CONCLUSIONS
Greater IL-23 production by AS patient macrophages in response to LPS provides further support for the development of Th17/IL23-directed therapy. Since significant UPR induction was not detected in AS patient macrophages, the relationship between UPR and inflammatory cytokine production remains unclear.
Ankylosing spondylitis (AS) is an insidious spinal inflammatory disease that affects young adults and incurs a high rate of disability (1, 2). Inflammation can also involve the eyes, skin, gut, peripheral joints and tendinous insertions (entheses). AS is a complex genetic disease, with the presence of the MHC class I allele HLA-B27 conferring up to 40% of genetic risk; this allele is found in 90–95% of AS patients but only 8–9% of the Caucasian population (3–6). HLA-B27/human β2 microglobulin-expressing rats (B27-Tg) develop a spondyloarthritis-like disease involving gut inflammation and joint swelling that mimics human disease and highlights the importance of HLA-B27 as a causative factor (7). However, almost 40 years after the strong linkage to HLA-B27 was discovered, it is still not clear how this allele predisposes to disease (8). Although the physiologic role of MHC class I is to present intracellular peptides to CD8+ T cells, depletion of CD8+ T cells in the B27-Tg rat does not impact disease (9).
One hypothesis for HLA-B27-related pathogenesis stems from the observation that HLA-B27 displays an unusual propensity to fold slowly and misfold during its biosynthesis (10, 11). HLA-B27 forms aberrant dimers that can be recognized by leukocytes on the cell surface and that accumulate in the endoplasmic reticulum (ER) (12–14). Accumulation of misfolded protein in the ER triggers a conserved intracellular stress response, the Unfolded Protein Response (UPR), that decreases protein production, enhances size and folding capacity of the ER, and degrades excess ER protein. The UPR profoundly alters cellular metabolism predominantly through the induction of gene expression (15). Evidence for HLA-B27 induction of the UPR, including up-regulation of binding immunoglobulin protein (BiP/GRP78), CCAAT/-enhancer-binding protein homologous protein (CHOP), and UPR-dependent splicing of the X-box binding protein-1 (XBP1) transcription factor mRNA, has been detected in cells transfected with HLA-B27 as well as in bone marrow macrophages from diseased B27-Tg animals (16). In the animal model, the magnitude of UPR induction correlated strongly with HLA-B27 expression; in pre-morbid animals, visualization of the UPR required acute up-regulation of MHC class I by inflammatory cytokines such as TNF-α and IFN (16, 17). The clinical relevance of these observations is supported by BiP over-expression in knee synovial fluid mononuclear cells from HLA-B27 positive spondyloarthropathy patients (18). However, the relationship between HLA-B27 triggered UPR and disease remains unclear: in B27-Tg rats with intermediate HLA-B27 expression, additional human β2 microglobulin induces arthritis and spondylitis in the absence of intestinal inflammation, while simultaneously reducing B27 misfolding and BiP expression evident in conconavalin A treated spleen cells.(19).
Our previous studies suggested a link between HLA-B27, the UPR, and innate immune responses: macrophages stimulated with pharmacologic UPR inducers(e.g. tunicamycin)respond to Toll-like receptor (TLR) agonists such as lipopolysaccharide (LPS/endotoxin) with increased production of inflammatory cytokines, in particular IFN-β and IL-23 (20, 21). Bone marrow macrophages from diseased animals expressing UPR target genes also expressed type I IFN regulated genes (16). B27+ macrophages from transgenic rats stimulated in vitro with IFN-γ (to induce the UPR) produce increased IFN-β and IL-23 in response to LPS. IL-23 drives IL-17 production from pathogenic Th17 T cells and is implicated in multiple autoimmune and inflammatory diseases (22). Involvement of the IL-23/Th-17 axis in AS pathogenesis is further supported by studies in the B27-Tg rat colon, colonic biopsies from AS patients, and AS peripheral blood mononuclear cells (PBMC) (21, 23, 24).
Macrophages pre-dominate in biopsies from early spondyloarthritis inflammatory lesions (25). Also, macrophages appear to be more susceptible to UPR induction than splenocytes in the B27-Tg rat model (17). For this study, we examined peripheral blood derived macrophages from AS patients to determine if they undergo an UPR and whether UPR induction correlates with increased cytokine/chemokine responses to LPS. Despite a 2-fold up-regulation of HLA-B by IFN-γ treatment, we did not observe significant UPR induction as measured by BiP, CHOP, and ERdj4 (an XBP1 gene target) expression. Except for IL-12 secretion, IFN-γ treatment minimized differences between patients and controls. In response to LPS alone, even in the absence of obvious UPR induction, AS patient macrophages produced strikingly greater levels of IL-23 (p=0.0007) than controls. These results have implications for AS pathogenesis and further support the development and application of anti-IL23 therapeutics to AS.
PATIENTS AND METHODS
Study Subjects
Nine patients meeting modified New York criteria for AS were included in the study along with one HLA-B27+ patient with thoracic syndesmophytes and sacroiliac pain (26). None of the patients were related to each other. In one AS patient sample, we were unable to collect culture supernatants. In another AS patient sample, we only had enough cells to collect RNA for no stimulation and IFN-γ conditions only (final numbers per group are indicated in figures and tables). Control subjects were unrelated, and had no personal or family history of AS. For subject characteristics see Table 1. All subjects were recruited from within the University of Wisconsin Hospitals and Clinics. Informed consent was obtained prior to participation and the study protocol was approved by the UW Health Sciences Institutional Review Board.
Table 1.
Ankylosing Spondylitis patient and control characteristics
| AS patient(n=10) | Control (n=10) | |
|---|---|---|
| Age median (range) years: | 47 (18–58) | 45 (28–57) |
| Sex, %male: | 90 | 90 |
| Disease duration median (range) years: | 15 (2–32) | NA |
| HLA-B27, number positive: | 9 | 0 |
| BASDAI, median (range): | 3.2 (0–5.4) | N.A. |
| Arthritis meds: | ||
| NSAIDs: | 6 | 1* |
| Methotrexate: | 2 | 0 |
| TNF-blockers: | 5 | 0 |
| Other: | sulfasalazine (1) | allopurinol (1) |
| glucosamine (1) | glucosamine (1) | |
| fosamax (1) | ||
1 Control was taking 325 mg aspirin for history of myocardial infarction. Not included above were 3 other controls taking anti-platelet doses of aspirin (81 mg).
NA: not applicable, ND: not determined, BASDAI: Bath ankylosing spondylitis disease activity score, NSAIDS: non-steroidal anti-inflammatories, TNF: Tumor necrosis factor. 6 patients were on disease modifying anti-rheumatic drugs (DMARDS: Methotrexate and/or TNF blockers).
Macrophage derivation and in vitro treatments
Whole blood was obtained by venipuncture and collected in 3 × 10 mL ACD tubes (acid citrate dextrose, BD Biosciences, San Jose, CA). Samples were processed within 18h of collection. Samples were diluted with PBS, underlaid with lymphocyte separation medium (Mediatech, Menassas, VA) and centrifuged to obtain PBMC buffy coats. Cells were washed twice at 900 rpm to decrease platelets. Monocytes were purified by negative selection using MACS columns from Miltenyi Biotech (Auburn, CA) according to the manufacturer’s instructions. Purity was 87–92% CD 14 positive by flow cytometry. Monocytes were plated in serum free RPMI in 6-well dishes for RNA analysis (350,000–500,000/well) and 24-well dishes (150,000–250,000)/well) for cytokine analysis. RNA and cytokine results were normalized (described below) to account for cell number differences. For further purification, non-adherent cells were removed after 2h. The adherent cells were then cultured with RPMI 1640 containing 10% Fetal Bovine Serum, penicillin-streptomycin, and L-glutamine (HyClone, Logan, UT). Cultures were supplemented with 20 ng/mL recombinant human M-CSF (R&D Systems, Minneapolis, MN), replaced on days 2 and 4. On day 5, 1000U/mL IFN-γ (PeproTech, Rocky Hill, NJ) was added to select cultures for 24h prior to stimulation on day 6 with either 10 ng/mL or 100 ng/mL LPS (S. enteriditus, Sigma, St. Louis, MO). IFN-γ and M-CSF contained less than 0.01 EU endotoxin detected by limulus assay (GenScript, Piscataway, NJ). For the tunicamycin (Sigma, St. Louis, MO) assay, macrophages were stimulated with 0.1–1 μg/mL for 4–16h starting on day 5. Cells obtained after 5 days of culture were relatively homogeneous, large, and had a “fried egg” appearance. By flow cytometry, cells were >99% CD11b+, 94–99% HLA-DR+, and expressed macrophage markers CD163 and CD206 (80–87% and 86–88% respectively). IFN-γ and LPS treatments affected viability (assessed by propidium iodide and trypan blue exclusion) by the end of day 7 compared to no stimulation as follows: LPS 10 ng/mL 73+/−7%, LPS 100 ng/mL 87+/−7%, IFN-γ 92+/−11%, IFN-γ + LPS 10 ng/mL 60+/−12%, IFN-γ + LPS 100 ng/mL 55+/−5%.
HLA-B, HLA-B27, and UPR gene expression
RNA was obtained following 24h of IFN-γ treatment plus 3h LPS stimulation (in select samples)on day 6 by resuspending the cells in TriZol (Invitrogen, Carlsbad, CA), and processing according to the manufacturer’s instructions. RNA was treated with DNase (Invitrogen), and reverse transcribed to cDNA using reverse transcriptase (Promega, Fitchburg, WI) with random primers (Promega) according to standard protocols. Relative expression of HLA-B, and the UPR-regulated genes BiP, CHOP, and ERdj4 were determined by quantitative real time polymerase chain reaction (PCR) using a MyiQ or iCycler (Bio-Rad, Hercules, CA) with normalization to 18S rRNA expression. HLA-B27 specific primers have been previously described by Bon et al.(27). Other primers were designed using Beacon Design 7.0 software (Premier Biosoft, Palo Alto, CA); primer sequences are available upon request from the corresponding author.
Cytokine/Chemokine secretion
Cell culture supernatants were collected at 8h (0.5 mL from the 1.5 mL culture) and at 24h: at 24h, all media was removed. Two time points were selected because of production kinetics for early and late peaking inflammatory mediators (e.g. IFN-β and CXCL9). Cells were lysed in 100 μL SDS lysis buffer and total protein quantified by BCA assay (Pierce, Rockford, IL) to account for differences in cell numbers/well. Supernatants were frozen at −80°C until analysis by Luminex assay (Millipore, Billerica, MA) measured on a Luminex 100 (Austin, TX). Multiplex analytes included IFN-γ, IFN-α2, IL-1β, IL-10, IL-12 (p70), IL-15, IL-1α, IL-6, IL-8, IP-10, MCP-1, TNF-α, and VEGF. CXCL9 and IL-23 Luminex assays were run individually. IFN-β production was quantified by colorimetric ELISA (PBL, Piscataway, NJ) read on a 354 Multiskan Ascent plate reader (Thermo Labsystems, Beverly, MA). Initially we screened 6 patient and 4 control samples for the 15 cytokines and chemokines listed above. For analytes where we observed a p<0.15 by student’s T-test comparing controls and patients within a treatment group (e.g. LPS 10 ng/mL, IFN-γ+LPS 10 ng/mLetc.) we completed evaluation of the other 3 patient and 6 control samples. Duplicate samples were used to normalize between Luminex runs. Thus we measured CXCL-9, IFN-β, IL-6, IL-8, IL-10, IL-12 (p70), IL-23, and TNF-α in 9 AS patient and 10 control subjects. Only data from the complete analysis of 8 chemokines and cytokines in 19 subjects is presented below.
Statistical Analysis
UPR-regulated gene expression and up-regulation of HLA-B and HLA-B27 were analyzed using Linear Mixed Effects (LME) models. These models included a random effect for subject and fixed effects for group (patient vs. control), IFN-γ, LPS 10 ng/mL, and LPS 100 ng/mL plus two-way interactions between group and all other terms and between IFN-γ and the two LPS doses. An LME model (also called a repeated measures ANOVA) is similar to ANOVA except that at least one additional error term is included. Subject was included as a random effect in the LME model, to model within-subject correlation among all the measurements taken on a particular subject. The UPR responses were transformed to the log scale before analysis to obtain approximately normally distributed residuals. For BiP and HLA-B, the significant main effect of IFN-γ in the complete model was further investigated by examining individual treatment comparisons. Correlations were performed using log-transformed data. Response to tunicamycin was also analyzed using an LME model with fixed effects for dose and time and a random effect for blood sample. Responses were transformed to the log scale before analysis. Wilcoxon Rank Sum tests were used to assess the differences in cytokine and chemokine production between patient and control groups for all the responses under each of the treatment conditions at both 8 and 24 hours.
RESULTS
Up-regulation of HLA-B
Previous examination of GM-CSF-derived macrophages from AS patients by microarray did not show evidence of an UPR (28). However, HLA-B up-regulation was also not detected in that study. In the B27-Tg rat model, up-regulation of HLA-B27 appears to play a critical role in visualizing the UPR; in pre-morbid animals, acute up-regulation of HLA-B27 expression by inflammatory cytokines was essential, and the magnitude of UPR induction strongly correlated with the fold-increase in HLA-B27 expression (16, 17). To further examine the role of the UPR in AS, we derived macrophages from PBMC with either GM-CSF or M-CSF and stimulated with 100–1000U/mL TNF-α, IFN-α or IFN-γ for 24–48h prior to analyzing surface expression of HLA-B by flow cytometry (data not shown). Derivation with M-CSF and 24h stimulation with 1000U/mL IFN-γ yielded the greatest increase in HLA-B (1.82 ± 0.12 fold) with the least impact on viability as determined by propidium iodide exclusion (+11% ± 30%). These conditions were used for the following studies.
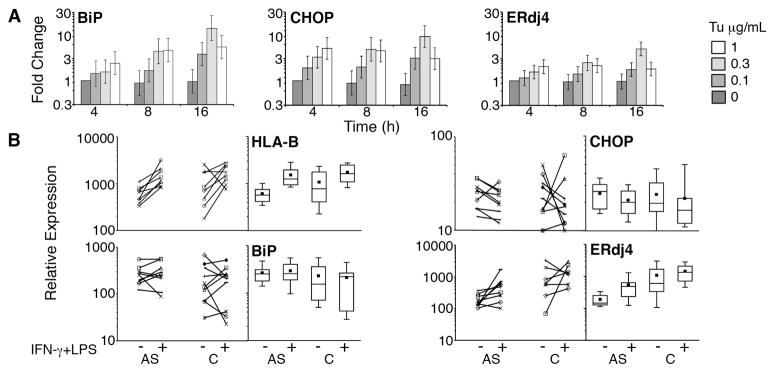
HLA-B expression was analyzed in macrophages from AS patients and controls by quantitative real time PCR (Table 2). AS patient and control macrophages responded similarly to treatment and only IFN-γ had a significant impact on mean expression (p<0.0001). In pair-wise tests combining patients and controls, IFN-γ, IFN-γ+LPS 10 ng/mL and IFN-γ+LPS 100 ng/mL were all significantly increased over no stimulation (NS), (p 0.02). In the AS patients, HLA-B expression increased from NS to IFN-γ an average of 2.21(range 1.09–5.95, p=0.0063);from NS to IFN-γ + LPS 10 ng/mL the increase was 1.83-fold (0.34–4.98, p=0.02);for NS to IFN-γ + LPS 100 ng/mL an increase of 2.44(1.3–6.75, p=0.0027)was observed. In this last group, all AS patients up-regulated HLA-B expression (Figure 1A).
Table 2.
HLA-B expression following in vitro treatment with IFN-γ or LPS*
| Treatment | Patient (mean, 95% CI) | Control (mean, 95% CI) |
|---|---|---|
| No stimulation | 559 (356, 875) | 770 (492, 1204) |
| LPS 10 ng/mL | 664 (413, 1065) | 1551 (969, 2479) |
| LPS 100 ng/mL | 596 (380, 933) | 855 (546, 1336) |
| IFN-γ | 1249 (799, 1949) | 1239 (793, 1933) |
| IFN-γ+LPS 10 ng/mL | 1071 (668, 1713) | 1911 (1195, 3052) |
| IFN-γ+LPS 100 ng/mL | 1375 (881, 2145) | 1577 (1010, 2459) |
Means were derived from log transformed expression values (see Methods). Patients did not respond differently than controls to treatment (p=0.119). The only significant effect was by IFN-γ treatment, which increased the response for all treatments (IFN-γ only or IFN-γ+LPS) for both patients and controls (p<0.0001). N=7–8 AS patients and controls per group.
Figure 1. HLA-B and UPR-regulated gene expression.
A) Human macrophages were treated with various doses of tunicamycin (Tu μg/mL) for the times indicated. Gene expression was normalized to 4h untreated controls to yield fold change. Data was combined from 4 independent experiments with bars representing geometric means and vertical bars showing +/− standard errors of the mean. The geometric mean best represents the typical fold change but downweights large fold changes. B) Analysis of AS patients (AS) and controls (C): individual data points with lines connecting unstimulated and stimulated samples are on the left, and box plots showing median, 25th percentile, 75th percentile (boxes), means (black squares) and 10th and 90th percentiles (bars), are on the right. Subject macrophages were unstimulated (−), or treated with IFN-γ for 24h followed by 100 ng/mL LPS (+) for 3h. Relative gene expression was determined by quantitative PCR with normalization to 18S rRNA. HLA-B expression represents data from 8 patients and 8 controls. P=0.0006 for upregulation of HLA-B in combined patient and control samples. CHOP and BiP expression represent data from 9 patients and 10 controls. ERdj4 expression shows data from 9 patients and 7 controls.
Others have demonstrated increased HLA-B27 expression on patient PBMC compared to healthy HLA-B27+ controls, in the absence of overall increased HLA class I expression(29). Thus we specifically examined HLA-B27 expression in patient samples. Using a linear effects model, HLA-B27 behaved similarly to HLA-B, in that only IFN-γ treatment was significant (p=0.0005). IFN-γ induced a mean of 2.24-fold increase (range 0.86–7.8, p=0.016), IFN-γ + LPS 10 ng/mL 3.74-fold (0.61–10.4, p=0.063) and IFN-γ+LPS 100 ng/mL 1.96-fold (1.32–11.23, p=0.016). These ranges are slightly larger compared to HLA-B, consistent with previously published data(29).
UPR gene expression
Part of our hypothesis predicted that IFN-γ would induce the UPR by acutely up-regulating HLA-B expression in AS patient-derived macrophages. We chose to study the expression of CHOP, ERdj4 and BiP, as these gene targets are robustly up-regulated by the UPR, and relatively specific for the three major identified signaling pathways of the UPR initiated by activation of the ER-resident proteins protein kinase receptor-like ER kinase (PERK), inositol requiring 1 (IRE-1), and activating transcription factor 6 (ATF6) respectively (15, 30, 31). In previous studies in the transgenic rat model, 20–24h IFN-γ had been sufficient to observe UPR induction and a further 3h of LPS for additional XBP1 splicing(16, 20). As seen in Figure 1A, these gene products are sensitive to change following treatment with very low doses of tunicamycin. All tunicamycin doses showed significant elevation over no stimulation (p-values ranged from 0.048 to <0.001) with the exception of ERdj4 at 0.1 μg/mL (p=0.094). The increase from 4h to 16h was significant for BiP (p=0.0044) All other effects of increased time were not significant (p-values of 0.057 to 0.88).
Contrary to our prediction, in vitro stimulation with IFN-γ and LPS had minimal impact on mean UPR-regulated gene expression, even under conditions of the greatest HLA-B up-regulation (IFN-γ + 100 ng/mL LPS, Figure 1). AS patient mean gene expression was not significantly elevated compared to controls in any treatment groups. There was no statistical difference between the responses of patients and controls to the various treatments (Table 3). Combining patients and controls, the only significant difference compared to no treatment was seen in response to IFN+10 ng/mL LPS, where BiP and CHOP decreased (fold changes of 0.68 and 0.7 respectively, p<0.02). ERdj4 showed a trend towards differential regulation in AS patient and control macrophages: In the IFN + LPS treatment groups, control macrophages had unchanged or slightly decreased mean ERdj4 expression, whereas the AS patient macrophages tended to up-regulate ERdj4 (a significant patient-IFN interaction, p=0.03). However, ERdj4 mean expression in AS patient-derived macrophages remained below control mean expression. In AS patient macrophages there was no significant correlation between fold up-regulation of HLA-B and UPR-regulated gene expression.
Table 3.
UPR-regulated genes following in vitro treatment with IFN-γ or LPS
| Gene | Treatment | Patient (mean, 95% CI) | Control (mean, 95% CI) |
|---|---|---|---|
| BiP | No stimulation | 262 (136, 499) | 150 (76, 299) |
| LPS 10 ng/mL | 197 (95, 400) | 205 (99, 414) | |
| LPS 100 ng/mL | 259 (131, 501) | 185 (93, 360) | |
| IFN-γ | 206 (106, 399) | 115 (57, 223) | |
| IFN-γ+LPS 10 | 204 (98, 414) | 69 (30, 147) | |
| IFN-γ+LPS 100 | 261 (133, 505) | 133 (67, 258) | |
| CHOP | No stimulation | 15.4 (9, 23.9) | 11.1 (5.8, 18.2) |
| LPS 10 ng/mL | 8.1 (3, 15.1) | 10.5 (4.7, 18.4) | |
| LPS 100 ng/mL | 10.9 (5.5, 18.2) | 7 (2.6, 12.9) | |
| IFN-γ | 11.7 (6.3, 19) | 7.3 (3, 13.1) | |
| IFN-γ+LPS 10 | 7.9 (2.8, 14.9) | 4.5 (0.4, 10.1) | |
| IFN-γ+LPS 100 | 10.9 (5.5, 18.2) | 9.2 (4.4, 15.7) | |
| ERdj4 | No stimulation | 185 (83, 397) | 593 (268, 1295) |
| LPS 10 ng/mL | 110 (40, 275) | 644 (248, 1651) | |
| LPS 100 ng/mL | 214 (94, 476) | 651 (282, 1486) | |
| IFN-γ | 151 (67, 327) | 284 (131, 604) | |
| IFN-γ+LPS 10 | 213 (83, 520) | 237 (94, 579) | |
| IFN-γ+LPS 100 | 391 (175, 860) | 486 (219, 1064) |
N = 10 patients and controls for no stimulation and IFN-γ, 7 patients and controls for LPS 10 ng/mL and IFN-γ + LPS 10, and 9 patients and 9–10 controls for LPS 100 ng/mL and IFN-γ +LPS 100 ng/mL. Mean expression derives from log-transformed data (see Methods).
We further analyzed the AS patients according to whether they were treated with DMARDs (6 DMARD+ and 4 DMARD−). Few cases were significant: in response to IFN-γ, BiP expression changed by 0.64-fold in the DMARD+ and 1.16-fold in DMARD− (p=0.038). In response to IFN-γ+100 ng/mL LPS, CHOP changed 0.71-fold in DMARD+ vs. 0.93-fold in DMARD− (p=0.016) and ERdj4 changed 1.31-fold in the DMARD+ vs. 4.83-fold in the DMARD− (p=0.032). Thus, in select cases, DMARD− patients tended to show greater increase in gene induction or less of a decrease as compared to the DMARD+. Our study was most likely underpowered to detect other differences between DMARD groups.
Cytokine/Chemokine production
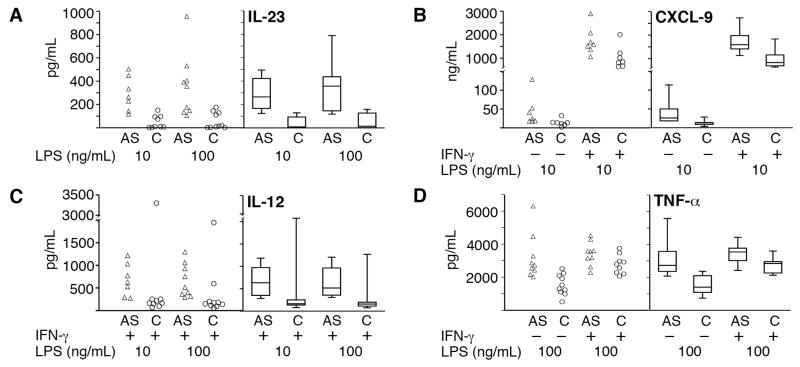
The evaluation of inflammatory mediator production was based upon previously described differences between AS patients and controls as well as studies examining the regulation of inflammation by the UPR. We had previously described a marked synergy between the UPR and LPS induction of both IFN-β and IL-23, in the context of both pharmacologic UPR induction and HLA-B27+ Tg rat macrophages undergoing an UPR (20, 21). We hypothesized that IFN-γ-treated AS patient macrophages would produce excess IFN-β and IL-23 in response to LPS stimulation. In addition to IFN-β and IL-23, we also examined macrophage production of CXCL-9, IL-6, IL-8, IL-10, IL-12 (p70), and TNF-α in 9 AS patients and 10 controls (Table 4).
Table 4.
Cytokine and chemokine production by AS patients and controls
| Mediator | Patient | Control | |||
|---|---|---|---|---|---|
| Pg/mL | Time | Stimulation | median(25%,75%) | median(25%,75%) | p-value |
| CXCL9** | 24h | no stimulation | 0.06 (0,0.14) | 0.47(0.29,0.64) | 0.1832 |
| LPS 10 ng/mL | 40.6 (22.3,154.5) | 12.6 (9.4,13.4) | 0.0041* | ||
| LPS 100 ng/mL | 27.7 (23.4,71.3) | 31.9 (16.5,35.1) | 0.5457 | ||
| IFN-γ | 1796 (1661,2026) | 1091 (1.4,2269) | 0.885 | ||
| IFN-γ+LPS 10 | 1593 (1447,1884) | 832 (731,1104) | 0.0175* | ||
| IFN-γ+LPS 100 | 1650 (1263,2031) | 833 (603,1677) | 0.1615 | ||
| IFN-β | 8h | no stimulation | 0 | 0 | NA |
| LPS 10 ng/mL | 76 (0,132) | 0 (0,65) | 0.3507 | ||
| LPS 100 ng/mL | 51 (0,148) | 0 (0,78) | 0.4530 | ||
| IFN-γ | 0 | 0 | NA | ||
| IFN-γ+LPS 10 | 522 (302,1087) | 362 (274,460) | 0.7577 | ||
| IFN-γ+LPS 100 | 627 (225,1181) | 301 (260,377) | 0.7802 | ||
| IL-6 | 8h | no stimulation | 6 (3,8) | 7 (6,22) | 0.4568 |
| LPS 10 ng/mL | 2413 (1211,3569) | 3012 (2811,3771) | 0.6065 | ||
| LPS 100 ng/mL | 2305 (1616,3440) | 2972 (2192,3713) | 0.6965 | ||
| IFN-γ | 19 (14,26) | 7 (3,12) | 0.2000 | ||
| IFN-γ+LPS 10 | 8904 (6456,12302) | 5600 (4698,7336) | 0.1142 | ||
| IFN-γ+LPS 100 | 8037 (5606,13858) | 5161 (3228,8601) | 0.0831 | ||
| IL-10 | 24h | no stimulation | 34 (20,47) | 8 (8,37) | 0.2000 |
| LPS 10 ng/mL | 2240 (1523,4869) | 486 (383,1153) | 0.0229* | ||
| LPS 100 ng/mL | 2126 (1438,2749) | 682 (471,1725) | 0.0653 | ||
| IL-12p70 | 8h | no stimulation | 0 | 0 | NA |
| LPS 10 ng/mL | 3 (1,4) | 0 (0,0) | 0.0203§ | ||
| LPS 100 ng/mL | 2 (0,5) | 0 (0,1) | 0.0929 | ||
| IFN-γ | 0 | 0 | NA | ||
| IFN-γ+LPS 10 | 634 (411,908) | 165 (150,250) | 0.0110* | ||
| IFN-γ+LPS 100 | 516 (369, 931) | 158 (118,194) | 0.0101* | ||
| IL-23 | 24h | no stimulation | 0 | 0 | NA |
| LPS 10 ng/mL | 265 (188,393) | 9 (6,92) | 0.0007* | ||
| LPS 100 ng/mL | 357 (149,405) | 15 (3,122) | 0.0022* | ||
| TNF-α | 8h | no stimulation | 3 (2,4) | 3 (2,13) | 0.8571 |
| LPS 10 ng/mL | 2667 (2260,2719) | 1393 (1094,2397) | 0.0562 | ||
| LPS 100 ng/mL | 2732 (2431,3284) | 1410 (1128,2053) | 0.0016* | ||
| IFN-γ | 10(7,16) | 4 (2,13) | 0.8571 | ||
| IFN-γ+LPS10 | 3077 (2952,3663) | 2806 (2625,3193) | 0.0556 | ||
| IFN-γ+LPS100 | 3545 (3156,3604) | 2671 (2212,2949) | 0.0408* | ||
| 24h | no stimulation | 7 (5,7) | 3 (2,6) | 0.2938 | |
| LPS 10 ng/mL | 2430 (2352,2550) | 1617 (1224,1739) | 0.0068* | ||
| LPS 100 ng/mL | 2620 (1964,2957) | 1536 (1157,2213) | 0.0177* |
Significant p-values;
CXCL9 is ng/mL and all other cytokines are pg/mL. The number of subjects for chemokine/cytokine experiments were 4–5 for unstimulated and IFN-γ only, 7 patients and 7–9 controls for LPS 10 ng/mL and IFN-γ+LPS 10 ng/mL, and 9 patients and 9–10 controls for LPS 100 ng/mL and IFN-γ+LPS 100 ng/mL. NA: not applicable.
Values at detection threshold for assay. No significant differences were detected for IL-8 production (data not shown).
We observed statistically significant increases in CXCL9, IL-10, IL-12 (p70), IL-23, and TNF-α production by AS patient macrophages compared to controls (Table 4 and Figure 2). We did not detect significant differences in production of IL-6, IL-8 or IFN-β. Apart from IL-12 (p70), these differences were primarily found in the LPS only treatment groups, where there was no HLA-B up-regulation or UPR gene induction. Contrary to our prediction, IFN-γ pre-treatment minimized differences for all inflammatory mediators tested except IL-12 (p70) (Table 4 and data not shown). Median differences between AS patients and controls for IL-10, IL-12 (p70), and CXCL9 ranged from 1.9 to 4.6 fold. Statistical differences in IL-12 production were only detected at 8h, but not 24h (data not shown). Several in vitro treatments revealed differences between AS patients and control macrophages in TNF-α production, although p-values reflected relatively less variability rather than profound differences in median production (< 2 fold). The most striking AS patient-control differences were observed for median IL-23 production following 24h LPS stimulation(see Table 4 and Figure 2): AS patient macrophages produced greater than a log-fold more IL-23 compared to controls (median AS production of 265–357 pg/mL vs. control 9–15 pg/mL, p=0.0022–0.0007). In our initial screen of 4 controls and 6 patients, no differences in IL-23 production were observed for IFN-γ pre-treated cells, and 8h LPS stimulation was comparable to 24h (median AS production of 213–341 pg/mL vs. control 24–64 pg/mL).
Figure 2. Increased IL-23, CXCL9, IL-12(p70) and TNF-α production in AS patient macrophages.
Individual sample data points are on the left and box plots showing median, 25th and 75th percentiles (boxes) and 10th and 90th percentiles (error bars) are on the right. A) IL-23: AS patient (AS, open triangles) or control samples (C, open circles) were treated with 10 or 100 ng/mL LPS for 24h. See Table 3 for associated p-values and numbers per group. B) CXCL-9: AS patient and control samples were left untreated (−) or treated with IFN-γ (+) for 24h, and then stimulated with 10 ng/mL LPS for another 24h. C) IL-12 (p70): All samples were pre-treated with IFN-γ for 24h and then stimulated with either 10 or 100 ng/mL LPS for another 8h. The outlier points for IL-12 production in response to both doses of LPS were from the same control subject. D) TNF-α: Macrophages were left untreated (−) or treated with IFN-γ (+) for 24h and then stimulated with 100 ng/mL LPS for 8h.
DISCUSSION
Based on the B27-Tg rat spondyloarthritis model we hypothesized that increased HLA-B27 expression and misfolding during inflammation induces a UPR that renders innate immune cells (macrophages and dendritic cells) more pro-inflammatory and hyper-responsive to bacterial TLR agonists. UPR-driven cells secrete excessive IL-23 (Th17 activation) and type I IFN (20, 21). However, the transgenic rat highly over-expresses HLA-B27 and thus HLA-B27 misfolding effects would most likely be exaggerated. It is important to assess the UPR-TLR inflammatory model in human disease, where there are at most 2 copies of the HLA-B27 gene.
In this study we did not detect significantly increased UPR-regulated gene expression in AS patient macrophages relative to controls at baseline, nor did we observe significant up-regulation of classic UPR target genes following in vitro IFN-γ and LPS stimulation. Recently, LPS was shown to activate IRE-1 dependent XBP1 splicing via the induction of an NADPH oxidase while suppressing activation of the PERK and ATF6 arms of the UPR (32). In this report, 10 ng/mL LPS also tended to decrease mean UPR gene expression in AS patients. However, we did not detect increased UPR gene expression following IFN-γ stimulation even in the absence of the additional LPS treatment. The significance of the increase in ERdj4 mRNA following IFN-γ+LPS treatment in AS patients is unclear, since mean expression remained below that observed in control subject macrophages.
The simplest explanation for the difference in our findings and the B27-Tg animal studies lies in the magnitude of HLA-B up-regulation. In the report describing the correlation of UPR magnitude and HLA-B27 expression in transgenic rat macrophages, IFN-γ treatment up-regulated HLA-B27 four to ten fold; although even a two fold up-regulation of HLA-B27 by TNF-α treatment yielded a two-fold induction of BiP expression (17). The mean two to three-fold HLA-B27 induction we observed may not have been sufficient to detect UPR induction in patient samples, particularly given patient to patient variability. Our small sample sizes may have been underpowered to detect subtle upregulation of gene expression. In addition, the representative UPR target genes we chose (BiP, CHOP, ERdj4) may not be the most sensitive indicators of an HLA-B27 UPR, although they are widely utilized to detect the UPR (15, 32). Alternatively, the UPR observed in the B27-Tg rats may depend upon dramatic HLA-B27 over-expression related to multiple transgene copy numbers, and thus be less relevant to human disease. However, the increased BiP expression found in the synovial cells from spondyloarthritis patients would argue against this possibility (18). Another possibility is that factors in addition to IFN-γ activation and HLA-B27 up-regulation are required to induce a UPR in AS macrophages. Our in vitro system may have been too simplistic compared to the in vivo inflammatory milieu. Future examination of inflamed patient tissues may be more revealing. Related to all these factors, our results do not rule out a role for the UPR in pathogenesis.
The study by Tran et al. showing worse arthropathy in B27-Tg rats where the UPR has been modulated by β2 microglobulin throws into question the relationship between B27-related UPR and joint disease(19). Multiple non-UPR hypotheses have sought to explain the striking contribution of HLA-B27 to genetic risk (8). For instance, expanded numbers of natural killer and CD4 T cells that recognize cell surface HLA-B27 dimers have been identified in the circulation of AS patients (14). Other efforts have focused on specific antigen presentation by HLA-B27 (33). With some variability across studies, HLA-B27 has also been shown to alter survival and persistence of intracellular organisms through unclear mechanisms (8, 34). Ultimately, our data suggest that non-UPR, cytokine modulatory mechanisms may contribute towards a pro-inflammatory diathesis and that non-UPR hypotheses bear greater exploration.
The tendency of IFN-γ to minimize differences in inflammatory mediator production is consistent with our previous data showing a global decrease in IFN-γ regulated gene expression in AS patient macrophages that could be recovered with exogenous IFN-γ treatment (28). This study and others have highlighted a relative deficit in the production of Th1 cytokines such as IFN-γ by AS patient cells (28, 35, 36). However, the IL-12 (p70) data (Table 4) suggests that under certain infectious or inflammatory conditions where IFN-γ is produced, AS patients could potentially mount an even more robust Th1 response than controls. Our findings of increased CXCL9, TNF-α and IL-10 in AS patient macrophages corroborate previous studies of patient samples (37, 38). Increased CXCL9, CXCL10/IP-10 and IL-12/23p40 are present in synovial fluid from patients with spondyloarthropathy (37, 39, 40). Reports describing serum and PBMC from AS patients have demonstrated variable increases in TNF-α, IL-1β, IL-6, IL-8, VEGF, and IL-10(38). What is unique to this study is removal of monocytes from the context of patient inflammation and medication. The in vitro derivation and stimulation conditions are controlled; thus the increased cytokine/chemokine production most likely reflects an intrinsic, genetically determined property of AS macrophages.
The most striking finding from this study is that even in the absence of an obvious UPR, AS macrophages produced significantly more IL-23 in response to LPS alone. The one known HLA-B27 negative patient produced IL-23 close to the median AS patient level (335 pg/mL in response to 10 ng/mL LPS and 405 with 100 ng/mL LPS). The increased IL-23 was much more robust than the differences observed for TNF-α. ER stress induced CHOP has recently been shown to directly regulate IL-23 expression(41). Although UPR and IL-23 production have also been closely associated in the B27-Tg rat model, our results suggest the potential for excess IL-23 production by patient macrophages in the absence of an overt UPR; in this study mean CHOP expression actually decreased following IFN-γ treatment and early during the LPS stimulation period (21). In the recent report by Martinon et al., LPS stimulation decreased pharmacologically induced UPR target gene expression (BiP, CHOP, ERdj4) out to 9h(32). In the present study we did not collect RNA after 24h of LPS, and thus cannot absolutely rule out late occurrence of an LPS-induced UPR.
Although this study is underpowered to assess the impact of HLA-B27 positivity in patients, our results support the hypothesis that AS macrophages produce an overabundance of IL-23 in response to TLR4 agonists in the infectious environment, thus predisposing to the development of inflammatory lesions. The IL-23/Th17 axis has become increasing recognized as a key component of anti-bacterial immunity, thus explaining genetic pressures to maintain risk alleles for a debilitating arthritic condition over the centuries (42). Given the responses we observed to LPS, it will be important to determine whether macrophages from AS patients produce excess IL-23 in response to colonic and infectious organisms. The finding of robust IL-23 expression in subclinical gut inflammation from AS patients suggests this may be the case, however more defined studies will be helpful in elucidating pathogenesis(24).
Our IL-23 results extend and support other recent studies examining the IL-23-Th17 axis in AS. Polymorphisms in the IL-23R were identified in genome wide association studies as susceptibility alleles for AS (43). Increased IL-23 and IL-17 levels have been observed in the serum and cultured PBMC supernatants from AS patients with active disease (23, 44). AS patients also have a higher proportion of IL-17 producing CD4+ T cells in circulation (23).
Anti-IL12p40 agents (e.g. ustekinumab), that block both IL-12-and IL-23-mediated effects, are highly efficacious in psoriasis, more than etanercept (a TNF-blocker) (45). In this study, the overproduction of IL-23 by AS patient macrophages was much more impressive than that observed for TNF-α, even though TNF-blockers are currently the therapy of choice for AS (46). The evidence from this study and others, implicating IL-23 expression in AS pathogenesis provides strong rationale for development of IL-23 blocking agents as treatment for AS.
Acknowledgments
Grants and other support: 1UL1RR025011 (CTSA), funding from U.W. Graduate School, 1UL1RR02501(KL2), AI081045 (K08).
References
- 1.Sieper J, Braun J, Rudwaleit M, Boonen A, Zink A. Ankylosing spondylitis: an overview. Ann Rheum Dis. 2002;61(Suppl 3):iii8–18. doi: 10.1136/ard.61.suppl_3.iii8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boonen A, Severens JL. Ankylosing spondylitis: what is the cost to society, and can it be reduced? Best Pract Res Clin Rheumatol. 2002;16(4):691–705. doi: 10.1053/berh.2002.0244. [DOI] [PubMed] [Google Scholar]
- 3.Reveille JD. The genetic basis of ankylosing spondylitis. Curr Opin Rheumatol. 2006;18(4):332–41. doi: 10.1097/01.bor.0000231899.81677.04. [DOI] [PubMed] [Google Scholar]
- 4.Braun J, Bollow M, Remlinger G, Eggens U, Rudwaleit M, Distler A, et al. Prevalence of spondylarthropathies in HLA-B27 positive and negative blood donors. Arthritis Rheum. 1998;41(1):58–67. doi: 10.1002/1529-0131(199801)41:1<58::AID-ART8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 5.van der Linden SM, Valkenburg HA, de Jongh BM, Cats A. The risk of developing ankylosing spondylitis in HLA-B27 positive individuals. A comparison of relatives of spondylitis patients with the general population. Arthritis Rheum. 1984;27(3):241–9. doi: 10.1002/art.1780270301. [DOI] [PubMed] [Google Scholar]
- 6.Brown MA. Genetics and the pathogenesis of ankylosing spondylitis. Curr Opin Rheumatol. 2009;21(4):318–23. doi: 10.1097/bor.0b013e32832b3795. [DOI] [PubMed] [Google Scholar]
- 7.Hammer RE, Maika SD, Richardson JA, Tang JP, Taurog JD. Spontaneous inflammatory disease in transgenic rats expressing HLA-B27 and human beta 2m: an animal model of HLA-B27-associated human disorders. Cell. 1990;63(5):1099–112. doi: 10.1016/0092-8674(90)90512-d. [DOI] [PubMed] [Google Scholar]
- 8.Smith JA, Marker-Hermann E, Colbert RA. Pathogenesis of ankylosing spondylitis: current concepts. Best Pract Res Clin Rheumatol. 2006;20(3):571–91. doi: 10.1016/j.berh.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 9.May E, Dorris ML, Satumtira N, Iqbal I, Rehman MI, Lightfoot E, et al. CD8 alpha beta T cells are not essential to the pathogenesis of arthritis or colitis in HLA-B27 transgenic rats. J Immunol. 2003;170(2):1099–105. doi: 10.4049/jimmunol.170.2.1099. [DOI] [PubMed] [Google Scholar]
- 10.Mear JP, Schreiber KL, Munz C, Zhu X, Stevanovic S, Rammensee HG, et al. Misfolding of HLA-B27 as a result of its B pocket suggests a novel mechanism for its role in susceptibility to spondyloarthropathies. J Immunol. 1999;163(12):6665–70. [PubMed] [Google Scholar]
- 11.Dangoria NS, DeLay ML, Kingsbury DJ, Mear JP, Uchanska-Ziegler B, Ziegler A, et al. HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. J Biol Chem. 2002;277(26):23459–68. doi: 10.1074/jbc.M110336200. [DOI] [PubMed] [Google Scholar]
- 12.Allen RL, O’Callaghan CA, McMichael AJ, Bowness P. Cutting edge: HLA-B27 can form a novel beta 2-microglobulin-free heavy chain homodimer structure. J Immunol. 1999;162(9):5045–8. [PubMed] [Google Scholar]
- 13.Kollnberger S, Bird LA, Roddis M, Hacquard-Bouder C, Kubagawa H, Bodmer HC, et al. HLA-B27 heavy chain homodimers are expressed in HLA-B27 transgenic rodent models of spondyloarthritis and are ligands for paired Ig-like receptors. J Immunol. 2004;173(3):1699–710. doi: 10.4049/jimmunol.173.3.1699. [DOI] [PubMed] [Google Scholar]
- 14.Chan AT, Kollnberger SD, Wedderburn LR, Bowness P. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondylarthritis. Arthritis Rheum. 2005;52(11):3586–95. doi: 10.1002/art.21395. [DOI] [PubMed] [Google Scholar]
- 15.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 16.Turner MJ, Sowders DP, DeLay ML, Mohapatra R, Bai S, Smith JA, et al. HLA-B27 misfolding in transgenic rats is associated with activation of the unfolded protein response. J Immunol. 2005;175(4):2438–48. doi: 10.4049/jimmunol.175.4.2438. [DOI] [PubMed] [Google Scholar]
- 17.Turner MJ, Delay ML, Bai S, Klenk E, Colbert RA. HLA-B27 up-regulation causes accumulation of misfolded heavy chains and correlates with the magnitude of the unfolded protein response in transgenic rats: Implications for the pathogenesis of spondylarthritis-like disease. Arthritis Rheum. 2007;56(1):215–23. doi: 10.1002/art.22295. [DOI] [PubMed] [Google Scholar]
- 18.Gu J, Rihl M, Marker-Hermann E, Baeten D, Kuipers JG, Song YW, et al. Clues to pathogenesis of spondyloarthropathy derived from synovial fluid mononuclear cell gene expression profiles. J Rheumatol. 2002;29(10):2159–64. [PubMed] [Google Scholar]
- 19.Tran TM, Dorris ML, Satumtira N, Richardson JA, Hammer RE, Shang J, et al. Additional human beta2-microglobulin curbs HLA-B27 misfolding and promotes arthritis and spondylitis without colitis in male HLA-B27-transgenic rats. Arthritis Rheum. 2006;54(4):1317–27. doi: 10.1002/art.21740. [DOI] [PubMed] [Google Scholar]
- 20.Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008;38(5):1194–203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60(9):2633–43. doi: 10.1002/art.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8(4):345–50. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- 23.Shen H, Goodall JC, Hill Gaston JS. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 2009;60(6):1647–56. doi: 10.1002/art.24568. [DOI] [PubMed] [Google Scholar]
- 24.Ciccia F, Bombardieri M, Principato A, Giardina A, Tripodo C, Porcasi R, et al. Overexpression of interleukin-23, but not interleukin-17, as an immunologic signature of subclinical intestinal inflammation in ankylosing spondylitis. Arthritis Rheum. 2009;60(4):955–65. doi: 10.1002/art.24389. [DOI] [PubMed] [Google Scholar]
- 25.Bollow M, Fischer T, Reisshauer H, Backhaus M, Sieper J, Hamm B, et al. Quantitative analyses of sacroiliac biopsies in spondyloarthropathies: T cells and macrophages predominate in early and active sacroiliitis-cellularity correlates with the degree of enhancement detected by magnetic resonance imaging. Ann Rheum Dis. 2000;59(2):135–40. doi: 10.1136/ard.59.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27(4):361–8. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 27.Bon MA, van Oeveren-Dybicz A, van den Bergh FA. Genotyping of HLA-B27 by real-time PCR without hybridization probes. Clin Chem. 2000;46(7):1000–2. [PubMed] [Google Scholar]
- 28.Smith JA, Barnes MD, Hong D, DeLay ML, Inman RD, Colbert RA. Gene expression analysis of macrophages derived from ankylosing spondylitis patients reveals interferon-gamma dysregulation. Arthritis Rheum. 2008;58(6):1640–9. doi: 10.1002/art.23512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cauli A, Dessole G, Fiorillo MT, Vacca A, Mameli A, Bitti P, et al. Increased level of HLA-B27 expression in ankylosing spondylitis patients compared with healthy HLA-B27-positive subjects: a possible further susceptibility factor for the development of disease. Rheumatology (Oxford) 2002;41(12):1375–9. doi: 10.1093/rheumatology/41.12.1375. [DOI] [PubMed] [Google Scholar]
- 30.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33(1):75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 32.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–8. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Castro JA. HLA-B27-bound peptide repertoires: their nature, origin and pathogenetic relevance. Adv Exp Med Biol. 2009;649:196–209. doi: 10.1007/978-1-4419-0298-6_14. [DOI] [PubMed] [Google Scholar]
- 34.Kim TH, Uhm WS, Inman RD. Pathogenesis of ankylosing spondylitis and reactive arthritis. Curr Opin Rheumatol. 2005;17(4):400–5. doi: 10.1097/01.bor.0000163447.44037.c4. [DOI] [PubMed] [Google Scholar]
- 35.Rudwaleit M, Siegert S, Yin Z, Eick J, Thiel A, Radbruch A, et al. Low T cell production of TNFalpha and IFNgamma in ankylosing spondylitis: its relation to HLA-B27 and influence of the TNF-308 gene polymorphism. Ann Rheum Dis. 2001;60(1):36–42. doi: 10.1136/ard.60.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brand JM, Neustock P, Kruse A, Alvarez-Ossorio L, Schnabel A, Kirchner H. Stimulation of whole blood cultures in patients with ankylosing spondylitis by a mitogen derived from Mycoplasma arthritidis (MAS) and other mitogens. Rheumatol Int. 1997;16(5):207–11. doi: 10.1007/BF01330297. [DOI] [PubMed] [Google Scholar]
- 37.Loos T, Dekeyzer L, Struyf S, Schutyser E, Gijsbers K, Gouwy M, et al. TLR ligands and cytokines induce CXCR3 ligands in endothelial cells: enhanced CXCL9 in autoimmune arthritis. Lab Invest. 2006;86(9):902–16. doi: 10.1038/labinvest.3700453. [DOI] [PubMed] [Google Scholar]
- 38.Keller C, Webb A, Davis J. Cytokines in the seronegative spondyloarthropathies and their modification by TNF blockade: a brief report and literature review. Ann Rheum Dis. 2003;62(12):1128–32. doi: 10.1136/ard.2003.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wendling D, Cedoz JP, Racadot E. Serum and synovial fluid levels of p40 IL12/23 in spondyloarthropathy patients. Clin Rheumatol. 2009;28(2):187–90. doi: 10.1007/s10067-008-1011-0. [DOI] [PubMed] [Google Scholar]
- 40.Proost P, Struyf S, Loos T, Gouwy M, Schutyser E, Conings R, et al. Coexpression and interaction of CXCL 10 and CD 26 in mesenchymal cells by synergising inflammatory cytokines: CXCL 8 and CXCL 10 are discriminative markers for autoimmune arthropathies. Arthritis Res Ther. 2006;8(4):R107. doi: 10.1186/ar1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci U S A. 2010;107(41):17698–703. doi: 10.1073/pnas.1011736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper AM. IL-17 and anti-bacterial immunity: protection versus tissue damage. Eur J Immunol. 2009;39(3):649–52. doi: 10.1002/eji.200839090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reveille JD, Sims AM, Danoy P, Evans DM, Leo P, Pointon JJ, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42(2):123–7. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Lin Z, Wei Q, Jiang Y, Gu J. Expression of IL-23 and IL-17 and effect of IL-23 on IL-17 production in ankylosing spondylitis. Rheumatol Int. 2009;29(11):1343–7. doi: 10.1007/s00296-009-0883-x. [DOI] [PubMed] [Google Scholar]
- 45.Griffiths CE, Strober BE, van de Kerkhof P, Ho V, Fidelus-Gort R, Yeilding N, et al. Comparison of ustekinumab and etanercept for moderate-to-severe psoriasis. N Engl J Med. 2010;362(2):118–28. doi: 10.1056/NEJMoa0810652. [DOI] [PubMed] [Google Scholar]
- 46.Heldmann F, Dybowski F, Saracbasi-Zender E, Fendler C, Braun J. Update on Biologic Therapy in the Management of Axial Spondyloarthritis. Curr Rheumatol Rep. 2010 doi: 10.1007/s11926-010-0125-y. [DOI] [PubMed] [Google Scholar]