Abstract
Quantum dots (QDs) are highly fluorescent and photostable, making them excellent tools for imaging. When using these QDs in cells and animals, however, intracellular biothiols (e.g., glutathione and cysteine) can degrade the QD monolayer compromising function. Here, we describe a label-free method to quantify the intracellular stability of monolayers on QD surfaces that couples laser desorption/ionization mass spectrometry (LDI-MS) with inductively coupled plasma mass spectrometry (ICP-MS). Using this new approach we have demonstrated that QD monolayer stability is correlated with both QD particle size and monolayer structure, with proper choice of both particle size and ligand structure required for intracellular stability.
Introduction
Bright and photostable quantum dots (QDs) with size-tunable fluorescence emission properties are widely employed tools1,2,3 for imaging cellular structures4,5,6 and events7,8. The small diameters and tailorable surface functionalities of QDs have resulted in their widespread use in whole animal9 and cellular imaging1,2,3 applications. For these applications QDs are functionalized with hydrophilic monolayers to increase their water solubility10,11,12, and with functional groups for targeting and biomolecular recognition11,12. QDs with heterobifunctional thiol monolayers11,12 provide much smaller hydrodynamic diameters (~4–10 nm) than other surface modification methods such as amphiphilic polymer encapsulation1, an important issue for both cellular12 and in vivo applications9 where larger particle size generates issues in terms of uptake and excretion. These functionalized thiol-based monolayers minimize non-specific binding and improve colloidal stability11,12. For example, monolayers with dihydrolipoic acid anchor groups, poly(ethylene glycol) (PEG) spacers, and streptavidin or fluorescent dye end groups have been used for single molecule tracking and sensing inside cells12. Improvements to the colloidal and pH stability of these functionalized QDs have been made by tuning the monolayer structure. For example, Mattoussi et al. showed that monolayers with multidentate thiolate anchor groups are more stable under a variety of environmental conditions than monolayers with monothiolate anchor groups13.
Effective use of functionalized QDs for imaging and sensing in living cells and animals, however, requires an accurate assessment of monolayer stability. Monolayer degradation will compromise the QD utility; biogenic thiols such as cysteine and glutathione (GSH) with intracellular concentrations as high as 10 mM can displace thiolate-bound ligands from nanoparticle surfaces14,15,16,17. The resultant loss of the protective monolayer coating results in QD aggregation in cells resulting in toxicity18. Moreover, if the QD monolayer contains a targeting antibody or ligand for the purpose of tracking a given protein/receptor7,8, monolayer loss will diminish or eliminate targeting, providing misleading or irreproducible results. Finally, monolayer displacement modifes QD optical properties; QDs coated with GSH or cysteine have shown reduced blinking19 and enhanced emission intensity19,20,21, making monolayer stability an important issue for quantitative applications.
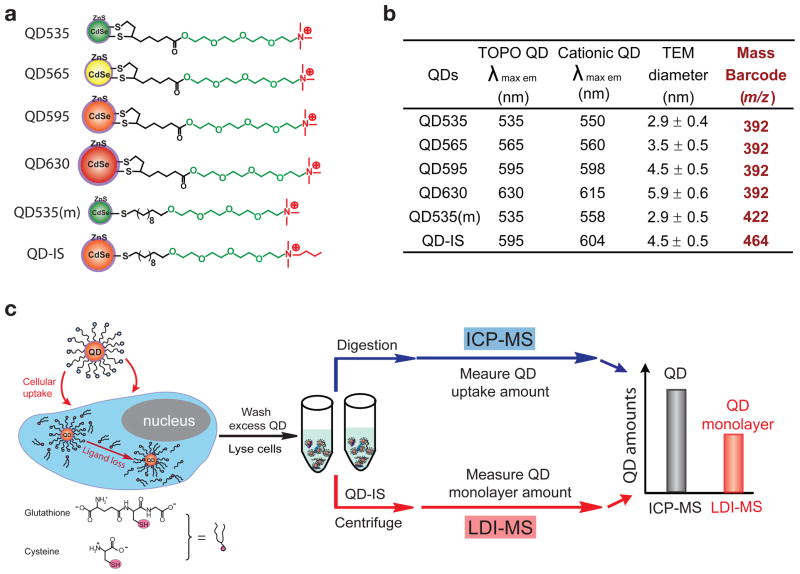
Clearly monolayer stability is required for effective applications of QDs in biology. To date, however, the intracellular stability of QD monolayers has not been quantified due to inadequate measurement tools. Here, we describe a new method based on the combination of laser desorption/ionization mass spectrometry (LDI-MS) and inductively coupled plasma mass spectrometry (ICP-MS) that enables QD monolayer stability to be quantitatively assessed inside cells (Fig. 1c). We demonstrate the utility of this new approach by comparing the stability of four cationic CdSe/ZnS QDs (Fig. 1a) with the same dithiolate monolayer but different particle sizes/colors, with core diameters ranging from 2.9 nm to 5.9 nm (Fig. 1b and Supplementary Fig. S1) and hydrodynamic diameters of 8 nm to 18 nm (Supplementary Fig. S2). We also compare the stability of QDs functionalized with dithiolate and monothiolate groups. In this method, the total amount of the QDs taken up by the cells is first quantified using ICP-MS. In a parallel measurement, monolayer amount on QD surface is measured by LDI-MS. The difference between the QD amount as determined by ICP-MS and monolayer amount as determined by LDI-MS provides the amount of monolayer released from the QD, providing quantitation of intracellular monolayer stability.
Figure 1. Quantification of QD stability in cells using Integrated ICP-MS and LDI-MS.
a, Structural illustrations of the six monolayer-protected CdSe/ZnS QDs used in this study. Four QDs have the same dithiolate monolayer (QD535, QD565, QD595, and QD630), while the QD535(m) has a monothiolate monolayer but the same core size as QD535. QD-IS was used as the internal standard for MS quantitation. b, Fluorescence emission maxima and particle sizes of QDs (Supplementary Fig. S1-3). The “mass barcode” is the mass-to-charge ratio (m/z) of the ligand ion used for quantitation of the QD monolayer. c, Parallel measurement of total QD uptake and monolayer amounts inside cells using ICP-MS and LDI-MS, respectively. The difference between the values obtained by ICP-MS and LDI-MS represents the amount of monolayer released from the QDs inside the cells.
Results and discussion
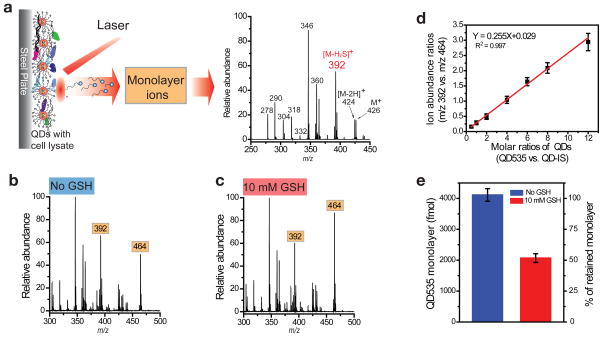
Previously, we demonstrated that thiolate-bound monolayers on gold nanoparticles (AuNPs) can be desorbed and ionized by UV laser irradiation and then readily detected by MS, even in the presence of cellular constituents22,23,24. Monolayer ions and their fragments are generated during the LDI process and act as “mass barcodes” that can be used for NP identification and quantitation22. QD cores absorb UV laser energy in similar fashion, enabling the monolayer ligands to likewise be efficiently desorbed and ionized without the addition of matrices used in MALDI-MS (see Supplementary Fig. S4 for typical LDI mass spectra of pure QDs). For example, spiking QD595 into cell lysate and then analyzing it via LDI-MS gives rise to a spectrum (Fig. 2a) in which the monolayer ligand (m/z 426) and its fragment ions (e.g., m/z 392) can be readily observed. Similar spectra were obtained for the other QDs spiked into the cell lysate (Supplementary Fig. S5). The absence of matrix allows us to see only ligands attached to the QD and not free ligands in cellular material (Supplementary Fig. S6), a key requirement for assessing stability. The ion signals for the monolayer ligand and/or fragments (e.g., m/z 392) can therefore be along with an appropriate internal standard to determine the amount of ligands attached to the QDs in complex cellular samples.
Figure 2. LDI-MS measurements of monolayer amounts on QDs.
a, The LDI-MS process and a representative LDI mass spectrum of QD595 (400 fmol) spiked into cell lysate. The fragment ion ([M-H2S]+) ion at m/z 392 was chosen as “mass barcode” for quantitation of QD595. b, c, LDI mass spectra of QD535 (m/z 392) and the internal standard QD-IS (m/z 464) in cell lysate (b) and cell lysate containing 10 mM GSH (c) after incubation at 37 °C for 3 h. QD535 (4000 fmol) was spiked into the cell lysate before incubation, and 800 fmol of QD-IS was added just prior to analysis. d, Calibration curve: Ion abundance ratios from QD535 (m/z 392) and QD-IS (m/z 464) for QD535 (400 – 9600 fmol) and QD-IS(800 fmol). e, Remaining monolayer of QD535 without (blue) and with (red) the addition of GSH to the cell lysate, showing substantial monolayer degradation with the presence of GSH in lysate.
The ability of the LDI-MS to measure the amounts of QD monolayers was first validated using cell lysate with known amounts of QDs spiked. As an example, 4000 fmol of QD535 was spiked into HeLa cell lysate with and without GSH added (Fig. 2b and c). After incubating this QD in cell lysate for 3 h, the solutions were mixed with internal standard QD-IS (800 fmol), and analyzed by LDI-MS (Fig. 2b). The “mass barcode” abundance ratio of QD535 (m/z 392) relative to QD-IS (m/z 464) is 1.34 ± 0.07. Applying the calibration curve (Fig. 2d), in which the “mass barcode” ratios are correlated with monolayer concentration ratios, QD535 monolayer amount is determined as 4100 ± 200 fmol (blue column in Fig. 2e). Note that we defined that one QD particle has one intact surface monolayer. Consequently, on the calibration curve the QD concentration and the monolayer concentration are the same. Considering that 4000 fmol of QD535 was initially spiked into lysate, this result indicates that no monolayer release was observed, and the recovery rate is 103 ± 5%. For comparison, the ion abundance ratio of QD535 in cell lysate to which 10 mM GSH was added is 0.69 ± 0.05 (Fig. 2c), which corresponds to 2100 ± 200 fmol of monolayer (red column in Fig. 2e). Therefore, this result indicates that only 53 ± 5% of the original monolayer of QD535 remains after exposure to 10 mM GSH. The same experiments were also done for the other five QDs, and in each case GSH causes monolayer release (Supplementary Fig. S7).
Determination of the monolayer stability of QD in living cells was obtained using QDs with positively-charged monolayers (see Supplementary Fig. S2a for QD zeta potentials) designed to facilitate uptake22. Confocal microscopy was used to visualize the cellular uptake of QDs (Supplementary Fig. S8). Our previous work22 and others25 have shown that the cellular uptake of cationic nanoparticles can occur through endocytosis, consistent with our observations. Confocal microscopy images (Supplementary Fig. S8) of the QDs used in this study demonstrate that different sized QDs with the same surface functionality have the same intracellular distribution and are mostly found in lysosomes after 24 h incubation. Previous studies have shown that biogenic thiols like cysteine and cysteamine are actively accumulated in lysosomes to concentrations as high as tens of mM26,27,28, indicating a possible mechanism for QD monolayer release. Compared with the lysate studies, measuring intracellular particle stability introduces an additional complication in assessing monolayer stability, as cellular uptake of QDs will vary as a function of particle size29 and monolayer structure22. To address this issue, the total amount of the QDs taken up by the cells was first quantified using ICP-MS. Total Cd concentrations were measured and then converted to particle amounts (Equation 4 in Supplementary Information). In parallel, a fraction of the same sample was analyzed by LDI-MS to determine the monolayer amounts remaining on the QD surface (calibration curves are shown in Supplementary Fig. S9). The difference between the QD particle amount in cells as determined by ICP-MS and monolayer amount left on QD surface as determined by LDI-MS provides the amount of monolayer released from the QD (Fig. 1c).
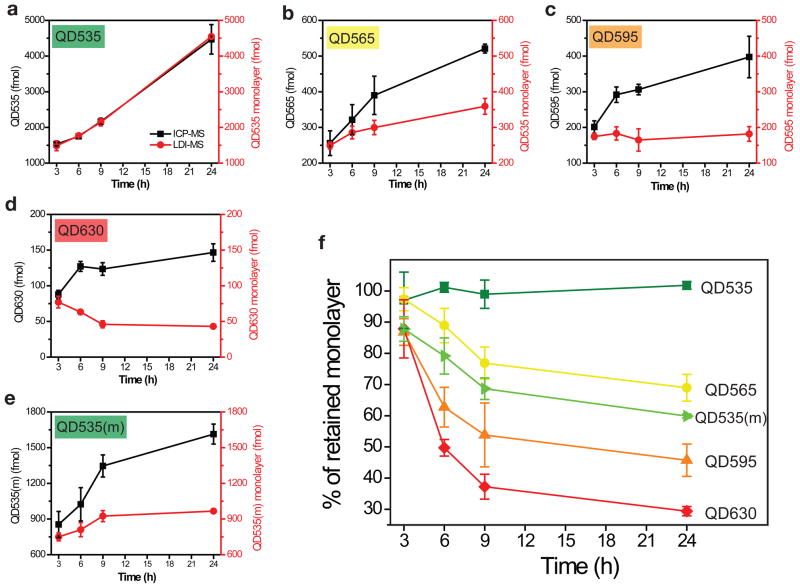
For our studies, each of the five QDs (QD535, QD565, QD595, QD630, and QD535(m)) was incubated with HeLa cells for 3 h, 6 h, 9 h, and 24 h. No cell morphology changes were observed after incubation. At each time point, the cells were lysed, and then analyzed for Cd (ICP-MS) and monolayers (LDI-MS). Representative LDI mass spectra at each time point are shown in Supplementary Fig. S10, with the overall results are summarized in Fig. 3a–e. For each QD, uptake of QD particles as determined by ICP-MS increases with time (Figure 3a–e [black curves]). The monolayer amount as determined by LDI-MS (Fig. 3a–e [red curves]), however, varies over time depending on both QD particle size and the monolayer structure.
Figure 3. Measurement of monolayer release from QDs in cells.
a–e, ICP-MS and LDI-MS measurements of QD535 (a), QD565 (b), QD595 (c), QD630 (d), and QD535(m) (e) after uptake by HeLa cells at varying time points (3 h, 6 h, 9 h, and 24 h). The difference between the values obtained by ICP-MS and LDI-MS represents the amount of monolayer released from the QDs. Incubation concentrations for QD535, QD565, QD595, and QD630 are 120 nM and for QD535(m) is 240 nM. f, Percentages of the monolayer remaining on the QDs upon uptake into HeLa cells. The results were calculated by comparing the LDI-MS and ICP-MS results shown in a–e.
From the results in Fig. 3a–d, it is clear that monolayer stability decreases as QD particle size increases. If the monolayer percentages remaining on the QDs are plotted as a function of time (Fig. 3f), only the smallest QD with the dithiolate monolayer (QD535) is stable for up to 24 h, with all the larger particles showing monolayer losses of 31–71%. Control experiments (Supplementary Fig. S11) verify that all of these QDs are stable in cell culture media and cell lysate, indicating that monolayer release happens only inside living cells, presumably through displacement with biogenic thiols. The size dependent monolayer stability is most likely related to nanoparticle curvature and surface coverage, as experimental and theoretical studies have shown that larger nanoparticles with flatter surfaces lead to less surface coverage than smaller more strongly curved nanoparticles30,31,32. Consequently, there are more free surface atoms (e.g., Zn for the QDs in this study) on larger particles, enabling greater accessibility for biogenic GSH and/or other thiols. Similar phenomena were reported by Mattoussi and coworkers for AuNPs, observing that 5 nm AuNPs are more resistant to decomposition by sodium cyanide than 10 or 15 nm AuNPs with the same dithiolate monolayer33.
Our results indicate that the monolayer anchor groups (monothiol vs. dithiol) also affect QD monolayer stability. Previous work has shown that dithiolate ligands enhance the colloidal and pH stability of gold NPs and QDs relative to monothiolate groups11,33. Our experiments provide a quantitative measure of this increased stability in live cells by comparing mono- and dithiol QDs (QD535 and QD535(m)) with the same core diameter (Fig. 3a and e). These studies confirm that the dithiolate anchor group provides significantly greater intracellular monolayer stability than the monothiolate group: after 24 h 40% of the monothiolate-bound monolayer is lost while complete monolayer retention is observed with the dithiolate monolayer (Fig. 3f). Note that QD535 and QD535(m) have 5 and 11 carbons in the hydrophobic alkane chain, respectively. For a better comparison, another QD535(m) with 5 carbons in the hydrophobic part was also synthesized, however, the shorter alkane chain together with monothiol anchor group produced unstable QDs. These results further highlight the importance of the anchor groups and ligand hydrophobicityon QD stability.
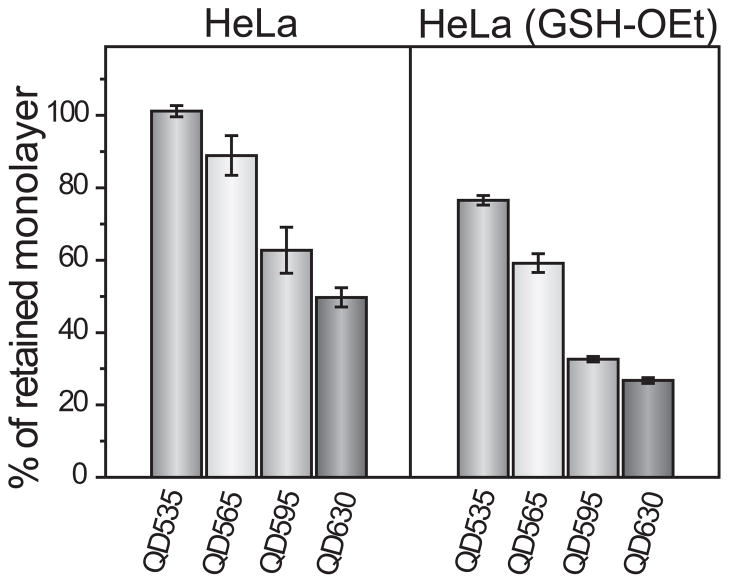
As the most abundant thiol species in the cells, GSH is also one of the most promising in situ triggers for the intracellular release of QD monolayers. The role of GSH in intracellular monolayer degradation was further established by incubating HeLa cells with 10 mM glutathione monoethyl ester (GSH-OEt)16, increasing the intracellular GSH concentration by 150% (Supplementary Fig. S13), without changing other cellular properties. As a neutral molecule, GSH-OEt efficiently penetrates cell membranes, including sub-cellular lysosome membranes, and rapidly hydrolyzes to generate GSH34, thus offering a simple method to increase GSH concentrations both in the cytosol and lysosome. It is clear that increased cellular concentrations of GSH leads to increased monolayer release (Fig. 4). For example, the retained monolayer on QD595 decreases from 63 ± 6% in the untreated HeLa cells to 33 ± 3% in the GSH-OEt treated cells. The manipulation of GSH concentrations in living cells provides a viable means of controlling monolayer stability.
Figure 4. QD monolayer stability as a function of intracellular GSH concentration.
A comparison of QD monolayer stability in two different cellular environments. HeLa (GSH-OEt) refers to HeLa cells treated with 10 mM glutathione monoethyl ester (GSH-OEt) for 1 h prior to incubation with the QDs. All QDs were incubated at 120 nM for 6 h.
In summary, we have developed a quantitative tool for measuring of QD monolayer stability in cells using a combination of LDI-MS and ICP-MS. Using this method, we find that monolayer stability decreases as QD particle size increases. The smallest QD studied, (2.9 nm core diameter), shows excellent stability in cells for up to 24 h. In contrast, the largest QD (5.9 nm core diameter) loses ~70% of its monolayer over the same 24 h. Similar differences in stability were observed when a dithiolate monolayer is replaced by a monothiolate monolayer, with monolayer stability decreasing by 40%. Taken together, these studies show that proper choice of both particle size and monolayer structure is critical for live-cell imaging applications. Our study indicates several strategies to create QDs with superior monolayer stability by choosing relatively smaller QDs and using bidentate ligands, e.g., dithiolate. Given the importance of Qds for live cell imaging, our studies likewise demonstrate the continuing need for more stable QD functionalization strategies.
Methods
QD synthesis
The TOPO capped CdSe/ZnS core-shell QDs were prepared according to reported procedures35. After that, ligand place-exchange reactions35 were used to obtain the QDs shown in Fig. 1a (see Supplementary Information for details).
Cell culture and cellular uptake of QDs
HeLa cells (60,000 cells/well) were grown on a 24-well plate in low-glucose Dulbecco’s modified Eagle’s medium (DMEM; glucose (1.0 g L−1)) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (100 I.U./ml penicillin and 100 μg/ml streptomycin). Cultures were maintained at 37 °C under constant saturated humidity with 5% CO2. After 24 h of plating, the cells were washed five times with cold phosphate buffer saline (PBS). Then, 500 μL of media containing QD (120 nM for QD535, QD565, QD595, and QD630; 240 nM for QD535(m)) was added. Following a certain culture time (3 h, 6 h, 9 h, or 24 h), the cells were washed five times with cold PBS to remove extra QDs and lysed for 10–30 min with a lysis buffer (200 μL; Genlantis, USA). Each sample was prepared in six replicates, which were split with half being separately analyzed by ICP-MS and LDI-MS. To prepare cell lysates, HeLa cells (60,000 cells/well) were grown on a 24-well plate for 24 h, and lysed with 200 μL of lysis buffer. To increase the intracellular concentration of GSH, 200 μL of media containing 10 mM glutathione ethyl ester (Sigma, USA) was incubated with HeLa cells for 1 h, and then replaced by media containing QD for a further 6 h incubation. The intracellular concentration of total GSH was measured using the glutathione detection kit (BioVision, USA).
ICP-MS sample preparation and measurements
After cellular uptake, the lysed cells were digested with 0.5 mL of fresh aqua regia (highly corrosive and must be use with extreme caution!) for 10 minutes. The digested samples were diluted into 10 mL with de-ionized water. A series of cadmium standard solutions (10, 5, 2, 1, 0.5, 0.2, 0 ppb) were prepared before each experiment. Each cadmium standard solution also contained 5% aqua regia. The cadmium standard solutions and QD sample solutions were measured on an Elan 6100 ICP mass spectrometer (PerkinElmer SCIEX, USA). The instrument was operated with 1100 W RF power, and the nebulizer Ar flow rate was optimized around 0.9–1.1 L/min.
LDI-MS sample preparation and measurements
After cellular uptake, the lysed cells were mixed for 15 min with QD-IS (typically 400 or 800 fmol) as the internal standard. Following ultracentrifugation at 60,000 rpm (~139,000 g) for 20 min, the QDs taken up by cells and the internal standard were collected as part of precipitate and washed with 60% acetonitrile/40% water. Later, the samples were transferred onto a MALDI target for LDI-MS analysis without adding any organic matrix. External calibration curves were generated before sample analyses. Each QD with the QD-IS at different ratios were spiked into cell lysate and vortexed for 15 min. The QD mixtures were collected by the centrifugation, washed, and analyzed by LDI-MS. The intensity ratios of the “mass barcode” ions for QDs and QD-IS were determined and plotted against the QD concentration ratios to generate a calibration curve (Supplementary Fig. S9). The QD monolayer amounts were then determined by using the internal standard and comparing to the calibration curve. To measure monolayer stability in cell lysate (Fig. 2), 4000 fmol of QD535 was spiked into HeLa cell lysate (200 μL) with and without added 10 mM GSH. After incubating this QD in cell lysate at 37 °C for 3 h, 800 fmol of QD-IS was added. Then, the mixture was immediately centrifuged, and the resulting precipitate was subjected to LDI-MS analysis. All of the LDI-MS measurements were carried on a Bruker Autoflex III MALDI-TOF mass spectrometer (Billerica, MA, USA). All mass spectra were acquired in the reflectron mode and represent an average of 500 laser shots at a repetition frequency at 100 Hz. The accelerating voltage was set to 19 kV. The laser power was optimized in the range of 50–90 % for each sample. The Bruker software (FlexAnalysis Version 3.3) was used for data analysis. Each sample was measured 10 times.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the NIH (Grants R21 ES017871-01 and GM077173-05) and through the Center for Hierarchical Manufacturing (NSF Grant DMI-0531171). We thank Prof. Julian F. Tyson for access to the ICP-MS instrumentation. We also thank Brian Creran for assistance with TEM.
Footnotes
Author contributions
Z.J.Z., V.M.R., and R.W.V. conceived and designed the experiments. Z.J.Z., Y.C.Y., R.T., B.Y., and J.T. performed the experiments. All the authors analyzed and discussed the data. Z.J.Z. wrote the paper. V.M.R. and R.W.V. revised the paper.
Additional information
The authors declare no competing financial interests. Supplementary information that accompanies this paper is available online.
References
- 1.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 2.Zrazhevskiy P, Sena M, Gao X. Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chem Soc Rev. 2010;39:4326–4354. doi: 10.1039/b915139g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan WCW, Nie SM. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 5.Smith AM, Duan HW, Mohs AM, Nie SM. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv Drug Deliver Rev. 2008;60:1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alivisatos AP, Gu WW, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 7.Dahan M, et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 8.Lidke DS, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 9.Choi HS, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith AM, Duan H, Rhyner MN, Ruan G, Nie S. A systematic examination of surface coatings on the optical and chemical properties of semiconductor quantum dots. Phys Chem Chem Phys. 2006;8:3895–3903. doi: 10.1039/b606572b. [DOI] [PubMed] [Google Scholar]
- 11.Susumu K, et al. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J Am Chem Soc. 2007;129:13987–13996. doi: 10.1021/ja0749744. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, et al. Compact biocompatible quantum dots functionalized for cellular imaging. J Am Chem Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart MH, et al. Multidentate poly(ethylene glycol) ligands provide colloidal stability to semiconductor and metallic nanocrystals in extreme conditions. J Am Chem Soc. 2010;132:9804–9813. doi: 10.1021/ja102898d. [DOI] [PubMed] [Google Scholar]
- 14.Li D, et al. Glutathione-mediated release of functional plasmid DNA from positively charged quantum dots. Biomaterials. 2008;29:2776–2782. doi: 10.1016/j.biomaterials.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Han G, et al. Controlled recovery of the transcription of nanoparticle-bound DNA by intracellular concentrations of glutathione. Bioconjugate Chem. 2005;16:1356–1359. doi: 10.1021/bc050173j. [DOI] [PubMed] [Google Scholar]
- 16.Hong R, et al. Glutathione-mediated delivery and release using monolayer protected nanoparticle carriers. J Am Chem Soc. 2006;128:1078–1079. doi: 10.1021/ja056726i. [DOI] [PubMed] [Google Scholar]
- 17.Chompoosor A, Han G, Rotello VM. Charge dependence of ligand release and monolayer stability of gold nanoparticles by biogenic thiols. Bioconjugate Chem. 2008;19:1342–1345. doi: 10.1021/bc8000694. [DOI] [PubMed] [Google Scholar]
- 18.Pace HE, Lesher EK, Ranville JF. Influence of stability on the acute toxicity of CdSe/ZnS nanocrystals to Daphnia magna. Environ Toxicol Chem. 2010;29:1338–1344. doi: 10.1002/etc.168. [DOI] [PubMed] [Google Scholar]
- 19.Li-Shishido S, Watanabe TM, Tada H, Higuchi H, Ohuchi N. Reduction in nonfluorescence state of quantum dots on an immunofluorescence staining. Biochem Biophys Res Commun. 2006;351:7–13. doi: 10.1016/j.bbrc.2006.09.159. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F, Ali Z, Amin F, Riedinger A, Parak W. In vitro and intracellular sensing by using the photoluminescence of quantum dots. Anal Bioanal Chem. 2010;397:935–942. doi: 10.1007/s00216-010-3609-8. [DOI] [PubMed] [Google Scholar]
- 21.Park C, Yoon T. H l-Cysteine-induced photoluminescence enhancement of CdSe/ZnSe quantum dots in aqueous solution. Colloids Surf B Biointerfaces. 2010;75:472–477. doi: 10.1016/j.colsurfb.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Zhu ZJ, Ghosh PS, Miranda OR, Vachet RW, Rotello VM. Multiplexed screening of cellular uptake of gold nanoparticles using laser desorption/ionization mass spectrometry. J Am Chem Soc. 2008;130:14139–14143. doi: 10.1021/ja805392f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu ZJ, Rotello VM, Vachet RW. Engineered nanoparticle surfaces for improved mass spectrometric analyses. Analyst. 2009;134:2183–2188. doi: 10.1039/b910428c. [DOI] [PubMed] [Google Scholar]
- 24.Yan B, et al. Laser desorption/ionization mass spectrometry analysis of monolayer-protected gold nanoparticles. Anal Bioanal Chem. 2010;396:1025–1035. doi: 10.1007/s00216-009-3250-6. [DOI] [PubMed] [Google Scholar]
- 25.Al-Hajaj NA, et al. Short Ligands Affect Modes of QD Uptake and Elimination in Human Cells. ACS Nano. 2011;5:4909–4918. doi: 10.1021/nn201009w. [DOI] [PubMed] [Google Scholar]
- 26.Pisoni RL, Acker TL, Lisowski KM, Lemons RM, Thoene JG. A cysteine-specific lysosomal transport system provides a major route for the delivery of thiol to human fibroblast lysosomes: possible role in supporting lysosomal proteolysis. J Cell Biol. 1990;110:327–335. doi: 10.1083/jcb.110.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pisoni RL, Park GY, Velilla VQ, Thoene JG. Detection and characterization of a transport-system mediating cysteamine entry into human fibroblast lysosomes -specificity for aminoethylthiol and aminoethylsulfide derivatives. J Biol Chem. 1995;270:1179–1184. doi: 10.1074/jbc.270.3.1179. [DOI] [PubMed] [Google Scholar]
- 28.Krepela E, Prochazka J, Karova B. Regulation of cathepsin B activity by cysteine and related thiols. Biol Chem. 1999;380:541–551. doi: 10.1515/BC.1999.069. [DOI] [PubMed] [Google Scholar]
- 29.Chithrani BD, Ghazani AA, Chan WCW. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 30.Hostetler MJ, et al. Alkanethiolate gold cluster molecules with core diameters from 1.5 to 5.2 nm: core and monolayer properties as a function of core size. Langmuir. 1998;14:17–30. [Google Scholar]
- 31.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. The role radius of rurvature plays in thiolated oligonucleotide loading on gold nanoparticles. ACS Nano. 2009;3:418–424. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olmos-Asar JA, Rapallo A, Mariscal MM. Development of a semiempirical potential for simulations of thiol-gold interfaces. Application to thiol-protected gold nanoparticles. Phys Chem Chem Phys. 2011;13:6500–6506. doi: 10.1039/c0cp02921a. [DOI] [PubMed] [Google Scholar]
- 33.Mei BC, et al. Effects of ligand coordination number and surface curvature on the stability of gold nanoparticles in aqueous solutions. Langmuir. 2009;25:10604–10611. doi: 10.1021/la901423z. [DOI] [PubMed] [Google Scholar]
- 34.Puri RN, Meister A. Transport of glutathione, as γ-glutamylcysteinylglycyl ester, into liver and kidney. Proc Natl Acad Sci U S A. 1983;80:5258–5260. doi: 10.1073/pnas.80.17.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh YC, et al. Synthesis of cationic quantum dots via a two-step ligand exchange process. Chem Commun. 2011;47:3069–3071. doi: 10.1039/c0cc04975a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.