Abstract
The vibriocidal titer assay can be used to detect antibodies against Vibrio cholerae in serum samples, serving as an indicator of prior infection and potential protection against cholera. The assay can be utilized in research and clinical settings to test the effectiveness of vaccines, and also in epidemiological studies relevant to cholera transmission and surveillance. This unit outlines the steps involved in conducting an easily interpreted colorimetric vibriocidal titer assay with a relatively short turnaround time for results of around 8 hours, with final result observations in 24 hours. The assay can also be easily scaled up or down to accommodate as many or as few serum samples available and is not V. cholerae strain specific.
Keywords: Vibrio cholerae, Vibriocidal Titer Assay, Serum, Antibody Titer
Introduction
The vibriocidal titer assay is the gold standard indicator of protection against Vibrio cholerae. It is a relatively simple assay that can be used to determine if a person has developed antibodies against V. cholerae, either by exposure to infectious V. cholerae or as a result of oral cholera vaccine administration. It is a quantitative assay that allows you to determine the titer of a person's serum with visually interpreted results (purple color formation) within 24 hours. The simplicity of the assay provides for easy adjustment to accommodate as many or as few serum samples as available, and can be modified to test the serum sample(s) against any V. cholerae strain.
The protocol described here requires heat-inactivated serum samples, an exogenous source of complement, live V. cholerae cultures, and a chromogenic tetrazolium salt (neotetrazolium chloride). Briefly, the V. cholerae cultures are grown to mid-log phase and incubated with an exogenous source of complement. Following incubation, the cells are added to a serially diluted sample of the serum(s), and incubated, allowing V. cholerae antibodies which may be present in the serum sample(s) in combination with complement to lyse the live bacteria. The chromogenic salt, neotetrazolium chloride (NTC), is added to all the samples, but is only reduced by live cells, indicated by the color change to purple. Cells that have been lysed by the complement through the specific binding action of the antibodies in the serum, will not be able to reduce the neotetrazolium chloride and will therefore not change in color. The titer of the serum is then calculated as the reciprocal of the highest dilution of the serum that did not result in purple color detection, or neotetrazolium chloride reduction.
Basic Protocol – Colorimetric Determination of vibriocidal activity of antibodies or sera samples
The purpose of this protocol is to provide a simple and visual colorimetric assay to determine whether or not a patient is carrying antibodies against Vibrio cholerae and to quantitate the vibriocidal activity (or titer) of the patient's serum. This protocol takes advantage of the dye salt, NTC, which, in the presence of succinic acid, is reduced by live cells resulting in the formation of a visually obvious purple color.
Briefly, Vibrio cholerae is cultured to mid-log phase and incubated with exogenous complement proteins and a dilution series of the heat-inactivated serum. This will allow for the antibodies in the serum to kill the living Vibrio cholerae cells through complement-mediated cell lysis. The cultures are allowed to recover if they have not lysed, and are then grown overnight at room temperature in the presence of the NTC. If the cells were not lysed because of no vibriocidal activity in the serum, the NTC will be reduced and a purple color will develop. The titer of the serum can then be determined based on the lowest concentration of the serum that resulted in cell lysis, or no color development.
Materials
Overnight Cultures
Glass Culture Tubes (Fisher Cat. #14-961-32)
1× PBS (ice cold) (see recipe)
2 ml Microfuge Tubes (USA Scientific Cat. #1620-2700)
20-200 μl Multichannel Pipettor (optional) (Rainin Model #L12-200)
50 ml Pipettor Solution Trough (Fisher Cat. #S505200) (only if using multichannel pipettor)
Spectrophotometer (Milton Roy Model #Genesys5)
Luria-Bertani Broth (see recipe)
Spectrophotometer Cuvettes (Sarstedt #67.742)
Ice Bucket with Ice
Guinea Pig Complement Serum (Sigma #S1639) (see recipe)
Humidified Chamber with Lid
Neotetrazolium Chloride/Sodium Succinate Solution (see recipe)
96-well U-bottom Plates (Falcon Cat. #353911)
Lids for U-bottom Plates (Falcon Cat. #353913)
Microfuge (Eppendorf Model #5415D)
- Grow overnight cultures of the Vibrio cholerae strain that the sera/antibody is to be tested against in Luria-Bertani broth.It is a good idea to test against both V. cholerae Ogawa and Inaba serotypes.
- Subculture the strains in fresh media the next morning at a dilution of 1 to 50, and grow to an OD600=0.9 under aerobic conditions.For example, add 100 μl of overnight culture to 5000 μl of fresh Luria-Bertani broth. This is necessary to ensure a healthy growing population of cells.
Harvest 1 ml of the culture by centrifugation, remove the liquid media and resuspend the cell pellet in an equal volume of cold 1× PBS.
- Mix the following in chilled 2.0 ml microfuge tubes and let sit on ice for at least 20 minutes:
- 1 volume PBS washed cells
- 7 volumes cold 1× PBS
- 2 volumes cold guinea pig complement serum (GPC)
The actual volumes used to prepare this sample will depend on how many antibody/serum samples will be tested with each strain. Generally, to test one antibody/serum sample against one strain, three rows (12-wells across) of the 96-well plate will be needed: one row for the test antibody/serum sample, one row for a positive control (if available) and one row for a negative control (if available). Therefore, in order to ensure enough sample is prepared, it is a good idea to calculate enough sample volume for 13 wells. Since 25 μl of this sample will be needed for each well of one row, 13 wells × 25 μl × 3 rows (975 μl) of sample volume should be prepared. - While chilling the cell/GPC sample on ice, thaw out the antibody or serum sample on ice.The serum sample should be heat-inactivated by heating at 56°C for 30 minutes and then placing it back in the ice until ready to dilute.
- Prepare a serial dilution of the antibody/serum sample across the 12 wells, one row per antibody/serum sample.Add 50 μl of the antibody/serum sample to well 1, and 25 μl of cold 1× PBS to the rest of the 11 wells. Pipette 25 μl of the antibody/serum sample in the first well, and add it to the second well and mix thoroughly. Pipette 25 μl of the mixture from well 2 and add it to the third well and mix thoroughly. Repeat down the row to the final well. After mixing the sample in the final well (well 12 of row 1), remove 25 μl and discard. You should now have 25 μl of antibody/serum sample at a two-fold serial dilution across the row. Although the serial dilutions can be performed by pipetting the samples into the subsequent wells individually, using 8 channels of a 12-channel multichannel pipettor would be more convenient and would also provide for more uniformity.
- Add 25 μl of the cell/GPC sample to each of the wells.The 25 μl of cell/GPC can be pipetted individually to each well, but a 12-channel multichannel pipettor is more convenient.
Cover the 96-well plate with the lid and incubate at 37°C for 1 hour.
- After the 1 hour incubation, add 100 μl of fresh Luria-Bertani broth to all the wells.This can be done one well at a time, but a multichannel pipettor is more convenient.
- Place the uncovered plate in a humidified chamber and close the lid of the chamber.To assemble a humidified chamber, use any container that has a lid and can hold a 96-well plate with ample room. Line the bottom of the container with a paper towel and add water until the paper towel is completely soaked. Sufficient water should be added so that there is a small amount of extra water on the bottom of the container that hasn't soaked into the paper towel.
Incubate this humidified chamber at 37°C for 2 hours without shaking.
- Remove the plate from the humidified chamber and add 25 μl of the neotetrazolium chloride/sodium succinate (NTC/SS) solution to each well.This can be done one well at a time, but a multichannel pipettor is more convenient.
- Place the plate back into the humidified chamber and wrap the humidified chamber in aluminum foil to protect it from light.Incubate the light protected humidified chamber overnight at room temperature and observe color development the following day (Figure 6A.3.1).Note: color development is indicative of live cells, which means the antibody/serum DID NOT cause the cells to lyse.Keep the plate covered inside a humidified chamber to prevent the samples from drying out.Due to the light sensitivity of the neotetrazolium chloride, the humidified chamber should be protected from light by wrapping the chamber in aluminum foil.The titer is determined as the reciprocal of the highest dilution of the antibody/serum sample that resulted in cell lysis. Cell lysis will result in no purple color development.
Alternate Protocol 1 – Colony counting method to determine titer
An alternative method to the colorimetric vibriocidal assay is to conduct a colony count to determine the titer. This protocol essentially follows the same steps as the colorimetric assay, but rather than adding the NTC/SS reagent, samples are transferred to microfuge tubes, concentrated, and the cell suspension plated on rich medium agar plates (i.e. Luria-Bertani agar plates). The plates are incubated at 37°C overnight and the presence of viable cells determined for each dilution the following day by visual inspection for colony growth.
Materials
Overnight Cultures
Glass Culture Tubes (Fisher Cat. #14-961-32)
1× PBS (ice cold) (see recipe)
500 μl Microfuge Tubes (USA Scientific Cat. #1605-0000)
2 ml Microfuge Tubes (USA Scientific Cat. #1620-2700)
20-200 μl Multichannel Pipettor (optional) (Rainin Model #L12-200)
50 ml Pipettor Solution Trough (Fisher Cat. #S505200) (only if using multichannel pipettor)
Spectrophotometer (Milton Roy Model #Genesys 5)
Luria-Bertani Broth (see recipe)
Spectrophotometer Cuvettes (Sarstedt #67.742)
Ice Bucket with Ice
Guinea Pig Complement Serum (Sigma #S1639) (see recipe)
Humidified Chamber with Lid
96-well U-bottom Plates (Falcon Cat. #353911)
Lids for U-bottom Plates (Falcon Cat. #353913)
Microfuge (Eppendorf Model #5415D)
Luria-Bertani Agar Plates (12 plates for each strain) (see recipe)
Can reduce to 6 plates instead of 12 (see Sub-protocol steps a through g)
Prepare the cell samples with GPC and antibody/serum sample as described in the Basic Protocol, steps 1 through 11.
After incubation, transfer the contents of each well to individual rich media agar plates (i.e. Luria-Bertani agar plates).
Sub-protocol
To reduce the number of plates from 12 plates per antibody/serum sample to 6, transfer the contents of each well to individual 650 μl microfuge tubes.
Centrifuge the samples in a microfuge for 2 mins at maximum speed to pellet the cells.
Remove 100 μl of the supernatant and resuspend the cell pellet in the remaining supernatant.
Divide one agar plate in half by drawing a line down the middle of the bottom of the plate.
Apply the first resuspended cell pellet (approximately 50 μl volume) to one half of the plate and spread the sample to cover that half of the plate.
Apply the next resuspended cell pellet in the dilution series to the other half of the plate and repeat the spreading.
Repeat sub-protocol step f for the remaining wells.
3. Incubate plates at 37°C overnight and observe the following day for colony growth.
- 4. Determine the titer of the antibody/serum sample by identifying the highest dilution to give no colony growth on the plate.The titer is determined as in the Basic Protocol by expressing the titer as the reciprocal of the highest dilution of the antibody/serum to yield no growth.
Reagents and Solutions
1× PBS (pH7.2)
Sodium chloride (Fisher Cat. #S271-500) – 8.0 g
Potassium chloride (Fisher Cat. #P217-500) – 0.2 g
Sodium phosphate (Fisher Cat. #S373-500) – 1.44 g
Potassium phosphate (Fisher Cat. #P285-500) – 0.24 g
Double-distilled water – 800 ml
Adjust to pH7.2 with HCl (Fisher Cat. #A114S-212)
Bring up volume to 1000 ml with double-distilled water
Autoclave to sterilize
Luria-Bertani Broth
Tryptone (BD Cat. #211705) – 10 g
Yeast extract (BD Cat. #212750) – 5 g
Sodium chloride (Fisher Cat. #S271-500) – 5 g
Double-distilled water – up to 1000 ml
Autoclave to sterilize
Luria-Bertani Agar Plates
Luria-Bertani broth – 1000 ml
Agar (BD Cat. #214030) – 15 g
Autoclave to sterilize
Guinea Pig Complement Serum
Guinea pig complement serum (Sigma Cat. #S1639) – one bottle
Ice cold sterile double-distilled water – 5.0 ml
Neotetrazolium Chloride/Sodium Succinate Solution – 1000 μl
10% (w/v) Neotetrazolium chloride – 10 μl
27% (w/v) Sodium succinate – 90 μl
Sterile double-distilled water – up to 1000 μl
-
10% (w/v) Neotetrazolium Chloride
Neotetrazolium chloride (MP Biomedicals Cat. # 102436) – 0.1 g
100% Methanol – 1.0 ml
-
27% (w/v) Sodium Succinate
Sodium succinate (MP Biomedicals Cat. # 102436) – 13.5 g
Sterile double-distilled water – 50.0 ml
Filter sterilize through 0.22 μm filter (Millipore Cat. #SLGV033RS)
Commentary
Background Information
The vibriodcial titer assay is the benchmark serologic assay that demonstrates the capacity of an individual's immune system to afford protection against cholera. The first vibriocidal titer assay was reported by Finkelstein in 1962 (Finkelstein 1962), using an agar plate based protocol. Although this assay was reported to be highly reproducible (Finkelstein 1962), it was very labor intensive and time-consuming because titer determination required manual enumeration of bacterial colonies. In 1968, Benenson et al developed a microtechnique (Benenson et al. 1968) to determine vibriocidal antibody responses in epidemiological studies and hospital admitted patients. Rather than requiring the tedious colony counting method described by Finkelstein, this assay involved the cessation of the complement-mediated lysis and support for the surviving Vibrio cholerae cells with the addition of a liquid broth. The theory behind this assay is that if the Vibrio cholerae cells were not lysed by the complement due to the absence of Vibrio-specific antibody in the serum, then the surviving bacteria would multiply and be easily detected by observing for growth, indicated by increased turbidity in the reaction tube. The major drawback to this method is the relatively few number of samples that can be processed at any given time.
In 1971, Holmgren et al revisited the original vibriocidal titer assay proposed by Finkelstein, and developed an agar-plaque assay in an effort to alleviate the tedious enumeration of bacterial colonies (Holmgren et al. 1971). This assay involved pouring plates with live Vibrio bacteria in the agar and applying drops of serially diluted serum and complement mixtures to the surface of the solidified bacteria-embedded plates. Serum samples positive for vibriocidal antibodies would result in plaque formation at the dilution with an effective concentration of the serum. Ultimately, the microtechnique developed by Benenson et al was favored (Attridge et al. 2000; Attridge et al. 2002; Chernyak et al. 2002) and was eventually refined by Boutonnier et al (Boutonnier et al. 2003).
The method developed by Boutonnier et al (Boutonnier et al. 2003) has been successfully applied to not only serologic samples for epidemiological studies, but also to clinical research studies for vaccine development (Yang et al. 2007; Dharmasena et al. 2009). This unit essentially uses Boutonnier's protocol with a few modifications to make the protocol more user friendly. First, the exclusion of serum incubation in the 96-well microtiter plates decreases the overall processing time by up to 16 hours. Second, because of the limited small volume that the wells of the 96-well microtiter plate can hold, a decrease in the amount of LB broth added in step 9 (Basic Protocol) allows for easier handling without the concern for spillover and contamination from adjacent wells. Third, 1% neotetrazolium choloride (final concentration) used in step 12 (Basic Protocol) was found to enhance the level of color development. Finally, because of the relatively fragile nature of the classical O1 biotype bacterial cells as compared to the El Tor O1 biotype, incubation of the plate in the humidified chamber after addition of the neotetrazolium chloride/succinic acid solution and initial room temperature incubation (Basic Protocol – step 15), the humidified chamber was left at room temperature overnight and protected from light. These small modifications enhanced the color development and the overall ease of use of the vibriocidal titer assay.
Critical Parameters and Troubleshooting
Vibrio cholerae is a biosafety level 2 (BSL2) organism, and therefore should be handled and disposed of according to the safety precautions outlined by your institution's Environmental Health and Safety Office.
Although the protocol is straightforward, some key considerations must be kept in mind to ensure clean reproducible results. The first of these considerations is to ensure the subculture is healthy and growing well. Since the optical density only measures the turbidity of the culture, dead cells and cellular debris will also contribute to the optical density reading. Therefore by starting a fresh subculture of the cells and growing the culture to an optical density of 0.9, ensures that the bacteria are healthy and metabolically active. This will ensure that any negative readings (i.e. clear wells) are the result of vibriocidal activity of the serum, and not because the cells were not growing optimally prior to the start of the assay.
The second consideration is the source of the complement used in the assay. Although there are several vendors that can supply complement in the form of guinea pig sera (i.e. Sigma, Cappel, and BioMerieux), a study in 2003 by Boutonnier et al. (Boutonnier et al. 2003), found that between the three suppliers, the guinea pig complement from Sigma was best for the microtiter plate assay due to the consistency in the enzyme activity it demonstrated.
A third consideration is the light sensitivity of the NTC salt. To alleviate this problem, it is recommended to make the NTC/SS solution fresh for each assay. Stock solutions of both NTC and SS can be prepared separately ahead of time. If stocks are made, protect the NTC solution from light (wrapped in foil or dark tube) and store at 4°C. Since only small volumes are needed, it is not necessary to prepare large amounts of the 10% stock solution; 1 ml of stock should suffice for repeated tests. The sodium succinate (SS) stock solution can be prepared well in advance, filter sterilized and stored at room temperature indefinitely.
Anticipated Results
The colorimetric assay will result in the development of a purple color due to the reduction of the NTC salt, which can only be reduced by live cells. Therefore cells which have lysed (i.e. by the serum antibodies and the complement-mediated pathway), will not be able to reduce the NTC and the well will remain a clear color. However, after incubation with the serum and complement cocktail, if no Vibrio cholerae antibodies are present in the serum, the cells will not be lysed and will remain metabolically active and will reduce the NTC salt, resulting in purple color development. The concentration of the antibody in the serum required to cause bacterial cell lysis is called the titer of the serum and can be determined using this colorimetric vibriocidal assay. Therefore, the results will vary according to the individual titers of the different patients. A PBS blank can be run as a negative control, in which PBS is substituted for the serum sample. This will result in no complement-mediated lysis of any of the Vibrio cholerae cells and therefore all the wells will become purple, due to NTC reduction. If a patient's serum sample was positive for Vibrio cholerae antibodies, some of the wells across the serial dilution would remain clear, while the remainder of the wells down the serial dilution would be purple. The titer of the patient's antibody would be determined as the reciprocal of the dilution of the last clear well. For example, if a patient's serum resulted in the first four wells of the serial dilution remaining clear (dilutions of undiluted, 1:2, 1:4 and 1:8) with the remaining wells turning purple, the titer of that patient's serum would be 8, the reciprocal of the highest dilution.
The same principle can be applied to the alternate protocol and the titer determined the same way. In this alternative protocol however, the titer is determined by the presence or growth of colonies the following day, rather than the development of a purple color, in which lysis of the bacterial cells will result in no colonies on the rich media agar plates the following day.
Time Considerations
The protocol can be performed and concluded in a single day (∼8 hrs), with final observations being made the following day. In the Basic Protocol (step 2), the time required to grow the subculture to an OD600 of approximately 0.9 must be empirically determined. However, from a typical overnight culture, a 1:50 dilution will take approximately 4 hours to reach an OD600 of approximately 0.9.
The remainder of the Basic Protocol should take around four hours, including the two incubation periods of one hour and two hours in steps 9 and 11, respectively, in the Basic Protocol. During those four hours, 1 hour should be planned to prepare the culture, make the serial dilutions and aliquot the various volumes into the wells of the 96-well plate. As it is stated in the Basic Protocol step 6a, the use of a 12-channel multichannel pipettor is not necessary, but would increase the speed and efficiency with which the dilutions and aliquots are made. Overall, this vibriocidal assay should take around 8 hours to complete, but can require up to an hour or two more, depending on how long the subculture takes to reach to the proper optical density.
If the alternative protocol of colony counting method to determine antibody titer is used, additional time will be required following the two-hour incubation for plating of the samples on rich media agar plates. Spreading a small volume of 50 μl on an agar plate requires around 2 minutes a sample for a total of 12 minutes for all the samples of one dilution series (i.e. one strain).
Figure 1.
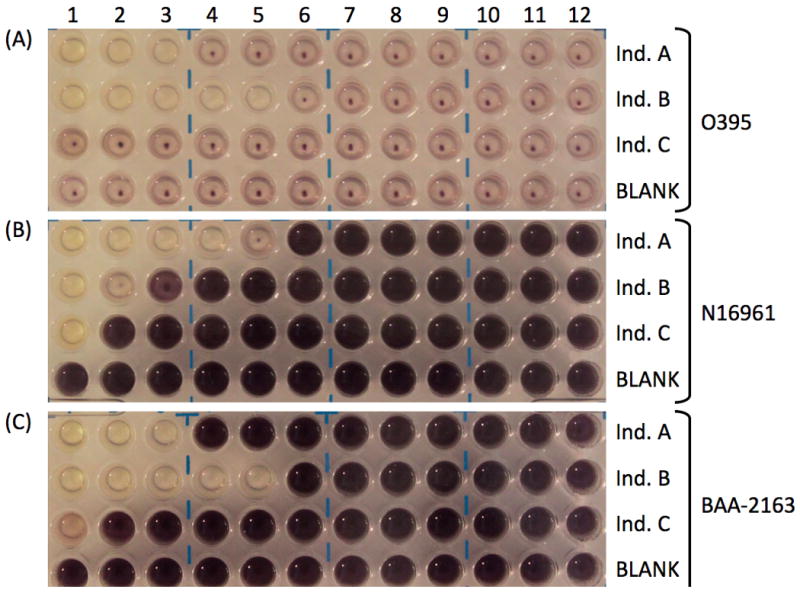
Sample plates of positive and negative vibriocidal assay results for three different Vibrio cholerae strains (A) O1 classical biotype wild-type strain O395, (B) O1 El Tor biotype wild-type strain N16961, and (C) O1 El Tor biotype strain BAA-2163 (representative strain from the 2010 Haitian cholera outbreak). For each strain, three serum samples were tested, individual A (top row – Ind. A), individual B (second row – Ind. B) and individual C (third row – Ind. C) along with one no serum negative control, BLANK (bottom row). Serial dilutions were performed from left to right (column 1 is undiluted; column 12 is 1:2048). Titers were determined as the reciprocal of the last clear well (due to cell lysis) for each row, resulting in no reduction of neotetrazolium chloride (NTC), which gives the characteristic purple coloration. For example, for (A) O1 classical biotype wild-type strain O395, the last clear well for individual A is in column 3 (dilution of 1:4) and so the titer would be 4. For individual B, the last clear well is in column 5 (dilution of 1:16), and so the titer is 16. Individual C does not have any clear wells, indicating that this serum sample does not contain any antibodies against V. cholerae. As expected the bottom row (BLANK) did not have any clear wells. Note that when testing serum samples against El Tor biotype strains, the color development is much more dramatic (B and C) than when testing against classical biotype strains (A). Classical O395 (A) showed slight purple color formation and settling of the reduced NTC, while both El Tor strains (B and C) produced a darker purple color, but still showed evidence of reduced NTC settling. This may reflect the hardiness of the strain being tested, as the longer the cells are alive and metabolizing, the more NTC will be reduced, resulting in stronger color development, as observed in the El Tor strains (B and C).
Acknowledgments
This work was supported by NIAID grants AI025096 and AI039654.
Literature Cited
- Attridge SR, Johansson C, Trach DD, Qadri F, Svennerholm AM. Sensitive microplate assay for detection of bactericidal antibodies to Vibrio cholerae O139. Clin Diagn Lab Immunol. 2002;9:383–387. doi: 10.1128/CDLI.9.2.383-387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attridge SR, Qadri F, Albert MJ, Manning PA. Susceptibility of Vibrio cholerae O139 to antibody-dependent, complement-mediated bacteriolysis. Clin Diagn Lab Immunol. 2000;7:444–450. doi: 10.1128/cdli.7.3.444-450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenson A, Saad A, Mosley WH. Serological studies in cholera: 2. The vibriocidal antibody response of cholera patients determined by a microtechnique. Bull World Health Organ. 1968;38:277–285. [PMC free article] [PubMed] [Google Scholar]
- Boutonnier A, Dassy B, Duménil R, Guénolé A, Ratsitorahina M, Migliani R, Fournier JM. A simple and convenient microtiter plate assay for the detection of bactericidal antibodies to Vibrio cholerae O1 and Vibrio cholerae O139. J Microbiol Methods. 2003;55:745–753. doi: 10.1016/j.mimet.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Chernyak A, Kondo S, Wade TK, Meeks MD, Alzari PM, Fournier JM, Taylor RK, Kovac P, Wade WF. Induction of protective immunity by synthetic Vibrio cholerae hexasaccharide derived from V. cholerae O1 Ogawa lipopolysaccharide bound to a protein carrier. J Infect Dis. 2002;185:950–962. doi: 10.1086/339583. [DOI] [PubMed] [Google Scholar]
- Dharmasena MN, Krebs SJ, Taylor RK. Characterization of a novel protective monoclonal antibody that recognizes an epitope common to Vibrio cholerae Ogawa and Inaba serotypes. Microbiol. 2009;155:2353–2364. doi: 10.1099/mic.0.025726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RA. Vibriocidal antibody inhibition (VAI) analysis: a technique for the identification of the predominant vibriocidal antibodies in serum and for the detection and identification of Vibrio cholerae antigens. J Immunol. 1962;89:264–271. [Google Scholar]
- Holmgren J, Svennerholm AM, Ouchterlony O. Quantitation of vibriocidal antibodies using agar plaque techniques. Acta Pathol Microbiol Scand B. 1971;79:708–714. [PubMed] [Google Scholar]
- Yang JS, Kim HJ, Yun CH, Kang SS, Im J, Kim HS, Han SH. A semi-automated vibriocidal assay for improved measurement of cholera vaccine-induced immune responses. J Microbiol Methods. 2007;71:141–146. doi: 10.1016/j.mimet.2007.08.009. [DOI] [PubMed] [Google Scholar]


