Abstract
The T-cell receptor (TCR) consists of a TCRαβ heterodimer, a TCRζ homodimer, and CD3γε and CD3δε heterodimers. The precise mechanism of T-cell triggering following TCR ligand engagement remains elusive. Previous studies reported that the cytoplasmic tail of CD3ε binds to the plasma membrane through a basic residue-rich stretch (BRS) and proposed that dissociation from the membrane is required for phosphorylation thereof. In this report we show that BRS motifs within the cytoplasmic tail of TCRζ mediate association with the plasma membrane and that TCR engagement results in TCRζ dissociation from the membrane. This dissociation requires phosphorylation of the TCRζ immunoreceptor tyrosine-based activation motifs by lymphocyte cell-specificprotein tyrosine kinase (Lck) but not ζ-chain–associated protein kinase 70 binding. Mutations of the TCRζ BRS motifs that disrupt this membrane association attenuate proximal and distal responses induced by TCR engagement. These mutations appear to alter the localization of TCRζ with respect to Lck as well as the mobility of the TCR complex. This study reveals that tyrosine phosphorylation of the TCRζ cytoplasmic domain regulates its association with the plasma membrane and highlights the functional importance of TCRζ BRS motifs.
T-cell triggering is initiated by the interaction of T-cell receptor (TCR) with a cognate peptide presented by a major histocompatibility complex protein (pMHC). The TCR complex consists of eight transmembrane proteins, comprising TCRαβ, CD3εγ, CD3εδ heterodimers, and a TCRζ homodimer (1). The heterodimeric TCRαβ chains are responsible for ligand engagement but have a short intracellular domain with no known signaling motifs. Instead, TCRαβ is reliant on the additional six transmembrane peptides in the TCR complex, which contain intracellular immunoreceptor tyrosine-based activation motifs (ITAMs), to convey an intracellular signal. The ligation of the TCR complex results in the phosphorylation of two tyrosine residues within these ITAMs by the lymphocyte cell-specific protein tyrosine kinase (Lck). Consequently, the phosphorylated TCR ITAMs recruit and activate ζ-chain–associated protein kinase 70 (Zap70) through the association between a doubly phosphorylated ITAM and tandem SH2 domains on Zap70, resulting in the initiation of the TCR signaling cascade. Because molecular events are ultimately required for the initiation of most adaptive immune responses, extensive research has focused on the mechanisms of TCR signaling (2, 3).
There are several nonmutually exclusive models to explain how an extracellular interaction between TCR and pMHC conveys an intracellular phosphorylation signal, a process referred to as “TCR triggering,” but the exact mechanism is controversial (2, 3). Part of the difficulty in determining the molecular mechanism of TCR triggering is the lack of knowledge surrounding the changes in structure of the TCR complex upon ligation and the contribution of different motifs in the TCR complex (4). Apart from the ITAMs, the cytoplasmic portions of the TCR complex contains two other conserved motifs: a proline-rich region (PRR) (5) and regions enriched in positively charged residues, also termed “basic rich stretch” (BRS) motifs (6). The PRR is present only on the CD3ε chain and has been reported to bind the SH3 domain of an adaptor protein Nck upon TCR engagement (5, 7). Functional studies show that this motif is not required for TCR triggering but is involved in the regulation of TCR expression levels in the thymus (8, 9).
There is one BRS motif in the membrane proximal part of CD3ε, but there are at least three in separate locations on the TCRζ (10, 11). Several studies have suggested that the BRS motifs promote close association of the CD3ε and TCRζ cytoplasmic domains with membranes, through interactions with negatively charged phospholipids (6, 10–14). Furthermore, in the case of CD3ε, NMR analysis suggests that the tyrosine residues in the ITAM motif are buried in the membrane interior (6). It has been proposed that this association protects CD3ε and TCRζ ITAMs from tyrosine phosphorylation and that TCR engagement enhances phosphorylation by somehow inducing dissociation of these ITAMs from the membrane (6, 14–16).
Previous studies looking at membrane association of the TCRζ cytoplasmic tail (TCRζcyt) have been performed in artificial membrane systems using purified peptide fragments of TCRζcyt, and questions have been raised about their interpretation (15). In this study, we investigated the interaction of TCRζcyt of native TCRs with the plasma membrane in T cells and examined the effect of TCR engagement on this association. We show that BRS motifs mediate association of the TCRζcyt with the plasma membrane in the resting state and that the TCRζcyt dissociates from the membrane upon TCR engagement. This dissociation requires ITAM phosphorylation by Lck but not Zap70 association. Mutations of the BRS motifs attenuate TCR signaling and alter spatial localization and mobility of TCR at the plasma membrane. Our results suggest that BRS motifs mediate the functionally important association of TCRζcyt with the plasma membrane and that this association could influence TCR signaling by changing the membrane environment and/or the distribution of the TCR.
Results
Cytoplasmic Tail of TCRζ Associates with the Plasma Membrane Through Its BRS Motifs.
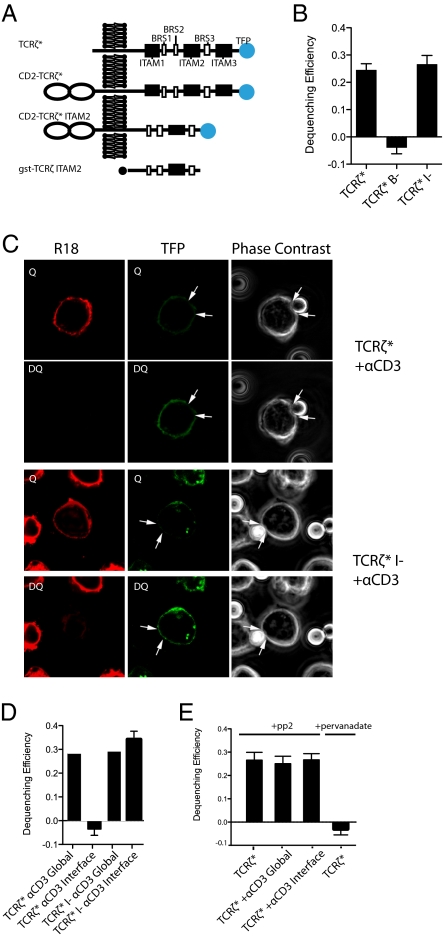
The TCRζ chain consists of a short extracellular domain (nine amino acids), a transmembrane region, and a long cytoplasmic tail containing three ITAMs and three BRS motifs (Fig. 1A and Fig. S1). A previously reported imaging approach, FRET (6), was used to examine the proximity of TCRζcyt to the plasma membrane in resting cells. In brief, a construct encoding TCRζ fused at its C terminus to teal fluorescent protein (TFP) (Fig. 1A) was retrovirally transduced into the BW5147-DC115.1 T-cell hybridoma (BW5δ cells) lacking endogenous TCRζ (17). As a result of the transduction, the surface expression of CD3ε was increased (Fig S2), indicating assembly of the transduced TFP-tagged TCRζ (TCRζ*) into the TCR/CD3 complex. To measure the proximity of the TCRζ TFP tag to the plasma membrane, the latter was labeled with the hydrophobic dye octadecyl rhodamine B chloride (R18), which acts as the FRET acceptor for the TCRζ* donor (6). FRET efficiency was determined by measuring the recovery of the donor fluorescence (dequenching) after photobleaching of the acceptor (R18) (18). There was a recovery of donor TCRζ* fluorescence after R18 bleaching (Fig. 1B and Fig. S3), suggesting that the TFP portion of TCRζ* is in close proximity to the plasma membrane. The FRET efficiency of TCRζ* was ∼0.25, consistent with the C-terminal end of the cytosolic tail of TCRζ* being closely associated with the plasma membrane in at least ∼25% of TCRζ* molecules.
Fig. 1.
Association of TCRζcyt with the plasma membrane requires BRS motifs, and anti-CD3–induced dissociation of TCRζcyt from the plasma membrane requires ITAM phosphorylation. (A) Schematic depiction of the TCRζ constructs used in this study. TCRζ* is full-length mouse TCRζ with TFP added at the C terminus. The ITAMs and BRS motifs are indicated by black and white boxes, respectively. The precise sequences are given in Fig. S1. CD2-TCRζ* is a chimeric protein in which the TCRζ* ectodomain has been replaced by the rat CD2 ectodomain. CD2-TCRζ* ITAM2: CD2-TCRζ* in which ITAM1 and ITAM3 have been deleted. (B–E) FRET efficiency measured by donor dequenching. Constructs used include TCRζ fused to C-terminal TFP (TCRζ*) and two mutant forms of TCRζ* lacking either BRS motifs (TCRζ*B−) or ITAMs (TCRζ*I−). R18 (acceptor) was bleached by a 543-nm laser (full power) for 60–80 s. Bar graphs in B, D, and E show mean dequenching efficiency from multiple (n = 30) cells calculated using the equation E = (DQ − Q)/DQ. Representative images of data summarized in B are shown in Fig. S3. In cells stimulated with anti-CD3–coated beads (D and E) the dequenching efficiency within the bead–cell interface (interface) and outside the interface (global) are shown. C shows representative images of data summarized in D. DQ, fluorescent images after R18 bleaching (donor dequenching); Q, fluorescent images before R18 bleaching (donor quenching). White arrows show the boundaries of the contact interface between a T cell and an anti-CD3–coated bead. In E TCRζ*-expressing cells were treated with pervanadate (10 μM) or PP2 (100 μM), with or without anti-CD3 beads. Representative images of data summarized in E are shown in Fig S4.
The previously reported association of the cytoplasmic tail of CD3ε with the plasma membrane was dependent on a BRS motif (6). TCRζ contains three BRS motifs in its cytoplasmic tail (Fig. 1A and Fig. S1). To investigate the role of these BRS motifs on membrane association, all positively charged residues in all three BRS motifs were mutated to alanine (TCRζ*B−) (Fig. S1). Unlike wild-type TCRζ*, TCRζ*B− mutants showed no recovery or dequenching of TFP after R18 photobleaching, indicating an absence of FRET (Fig. 1B and Fig. S3). In contrast, mutation of the tyrosine residues within the ITAMs to phenylalanine (TCRζ*I−) has no effect on dequenching efficiency (Fig. 1B and Fig. S3). This result suggests that the BRS motifs but not the ITAM tyrosine residues are required for the association of the TCRζcyt with the plasma membrane.
TCRζcyt Dissociates from the Plasma Membrane After TCR Engagement.
Having shown that in unstimulated T cells TCRζcyt is associated with the plasma membrane, we next investigated the effect of TCR engagement on this membrane association. We stimulated the cells with anti-CD3–coated beads and measured FRET between the plasma membrane and TCRζ*.
Although the dequenching efficiency in areas outside the bead-binding interface was similar to that in unstimulated cells (Fig. 1 C and D), it was greatly reduced in the contact area between the antibody-coated bead and the T cell (arrowheads in Fig. 1C). This observation suggests that the C-terminal end of TCRζcyt dissociates from the plasma membrane after TCR engagement.
Dissociation of TCRζcyt from the Plasma Membrane Requires ITAM Phosphorylation but Not Zap70 Binding.
In principle, the dissociation of TCRζcyt from the plasma membrane upon TCR ligation could be the direct consequence of TCR engagement or a secondary effect of TCRζ ITAM phosphorylation and/or binding of ZAP70/Lck. To explore these possibilities, we investigated the effect of disrupting TCRζ ITAM phosphorylation using two approaches. First, tyrosine residues within ITAMs were mutated to phenylalanine (TCRζ* I−) to prevent phosphorylation, and T cells reconstituted with TCRζ* I− were coincubated with anti-CD3 beads. Unlike wild-type TCRζ*, TCRζ* I− mutants did not dissociate from the plasma membrane upon TCR ligation because there was no change in dequenching efficiency (Fig. 1 C and D). Second, we inhibited Lck activity using the Src-kinase inhibitor PP2 (19). Incubation of T cells with PP2 also inhibited dissociation of TCRζ from the membrane upon TCR ligation (Fig. 1E and Fig. S4). Furthermore, treatment of T cells with the tyrosine phosphatase inhibitor pervanadate induced dissociation of TCRζcyt from the plasma membrane, suggesting that TCRζcyt phosphorylation is sufficient to induce TCRζcyt dissociation even in the absence of TCR engagement (Fig. 1E and Fig. S4). Taken together, these results suggest that the observed dissociation of TCRζcyt from the plasma membrane upon TCR engagement is a consequence of TCRζ ITAM phosphorylation by Lck.
Because TCRζ ITAM phosphorylation also is required for Zap70 binding to TCRζ, we investigated whether Zap70 is required for the dissociation of TCRζcyt from the plasma membrane. Human TCRζ* constructs were retrovirally transduced into a Jurkat cell line deficient in Zap70 (p116), and TCRζcyt association with the membrane was measured as above by FRET. In the absence of TCR ligation, significant dequenching of TCRζ* was observed following R18 bleaching, indicating association with the membrane (Fig. S5). Dequenching efficiency was reduced greatly following TCR engagement with anti-CD3 beads (Fig. S5), indicating dissociation of TCRζcyt from the membrane even in the absence of ZAP70. Finally, comparable changes in dequenching efficiency were observed in p116 cells that had been reconstituted with Zap70 (Fig. S5), confirming that Zap70 has no detectable effect on the TCR ligand-induced dissociation of TCRζcyt from the plasma membrane.
Collectively these results suggest that dissociation of the TCRζcyt from the membrane is dependent on TCR ligand-induced phosphorylation of TCRζ ITAMs but not on Zap70 binding.
Mutation of BRS Motifs Abrogates TCR-Induced IL-2 Production.
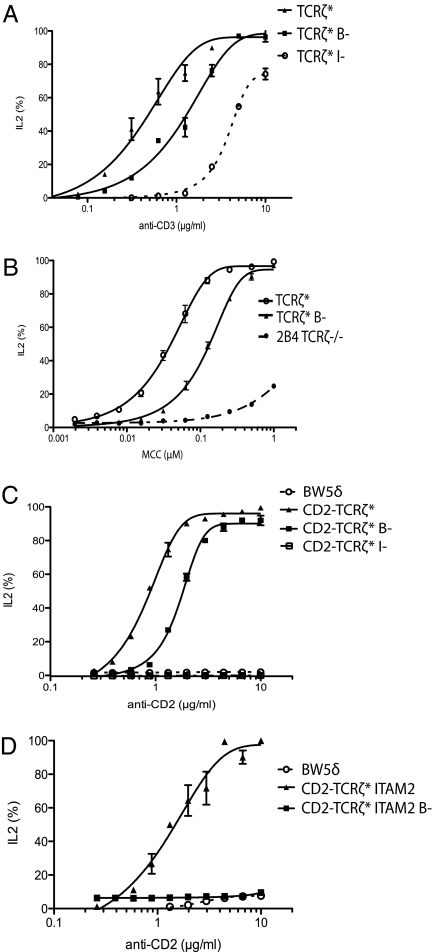
We next investigated the functional significance of BRS-mediated association of TCRζcyt with the plasma membrane. T cells expressing TCRζ in which the BRS motifs had been eliminated (TCRζ* B−) were stimulated with surface-immobilized anti-CD3 antibody or CHO cells expressing cognate pMHC (MCC in association with I-Ek) (20), and IL-2 secretion at 24 h was measured. T cells expressing TCRζ* B− displayed a modest reduction in IL-2 secretion in comparison with wild-type TCRζ* when stimulated with antibody (Fig. 2A). Similar results were obtained using T cells expressing the 2B4 TCR stimulated with cells expressing cognate pMHC (Fig. 2B).
Fig. 2.
Mutation of TCRζ BRS motifs inhibits ligand-induced IL-2 production. TCRζ-deficient BW5δ (A, C, and D) or 2B4 (B) T-cell hybridomas expressing matched levels of the indicated TCRζ constructs were stimulated by plate-bound immobilized antibodies (A, C, and D) or MCC-pulsed I-Ek–expressing CHO cells (B), and IL-2 release was measured after 24 h by ELISA. The representative results shown are the mean of experiments performed at least three times independently. IL-2 secretion was normalized to the maximal secreted amount. (A) Anti-CD3 induced IL-2 secretion. (B) pMHC-induced IL-2 secretion. (C) Anti-CD2–induced IL-2 secretion by untransduced BW5δ cells or BW5δ cells expressing the indicated CD2-TCRζ* construct. (D) Anti-CD2–induced IL-2 secretion by untransduced BW5δ cells or BW5δ cells expressing CD2-TCRζ*ITAM2 with or without (B−) intact BRS motifs.
Investigation of the functional role of TCR/CD3 cytoplasmic motifs is complicated by the fact that six cytoplasmic domains comprising 10 ITAMs are involved in signaling in a TCR/CD3 complex, resulting in significant functional redundancy (17). This redundancy is illustrated by the observation that IL-2 production can be stimulated by TCR engagement even when all TCRζ ITAMs have been disabled by tyrosine-to-phenylalanine mutations (Fig. 2A), presumably because ITAMs in the CD3 subunits are able to substitute for the eliminated TCRζ ITAMs.
To eliminate redundancy and facilitate more detailed analysis of the role of BRS motifs in TCRζ-mediated signaling, we generated a chimeric TCRζ protein comprising the extracellular portion of rat CD2 fused to the transmembrane and cytoplasmic domains of mouse TCRζ tagged with TFP (CD2-TCRζ*; Fig. 1A). Stimulation of T cells expressing this chimera with immobilized anti-CD2 antibody resulted in IL-2 production (Fig. 2C), which was eliminated completely when all ITAM tyrosine residues had been mutated to phenylalanine (Fig. 2C). In this system mutation of the BRS residues also reduced but did not eliminate IL-2 production (Fig. 2C).
To simplify the system further, we used a shortened CD2-TCRζ chimera from which ITAM1 and ITAM3 had been deleted (CD2-TCRζ*ITAM2; Fig. 1A). We reasoned that plasma membrane interactions mediated by the BRS motifs were most likely to affect access to ITAM2, because it is flanked by these motifs. FRET analysis of this chimeric protein showed the same BRS-mediated membrane association of the cytoplasmic domain as observed with native TCRζ* (Fig. S6). T cells that expressed CD2-TCRζ*ITAM2 secreted IL-2 upon stimulation with immobilized anti-CD2 (Fig. 2D). Strikingly, mutation of BRS motifs in this chimera completely abolished IL-2 secretion (Fig. 2D). Taken together, these results suggest that BRS motifs in the TCRζ are required for optimal IL-2 production, consistent with an important role in TCR signaling.
Mutation of BRS Motifs Abrogates Early TCR Signaling.
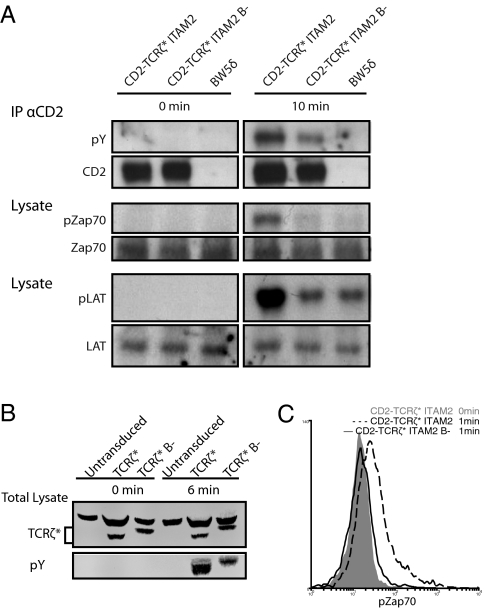
IL-2 is a relatively late consequence of TCR engagement and signaling. To investigate the role of TCRζ BRS motifs in early signaling, we examined the effect of BRS mutants in more proximal events, including TCRζ, Zap70, and linker of activated T cells (LAT) phosphorylation (Fig. 3). Mutation of the BRS motifs had no detectable effect on baseline phosphorylation of CD2-TCRζ*ITAM2 (Fig. 3A; 0 min). Treatment of T cells expressing the CD2-TCRζ*ITAM2 chimeras with pervanadate, which stimulates TCR signaling by inhibiting tyrosine phosphatases, resulted in an increase in TCRζ, Zap70, and LAT phosphorylation (Fig. 3A). Importantly, phosphorylation was reduced when the BRS motifs were mutated, suggesting that BRS motifs are required for or are involved in the early steps of TCR signaling. Similar results were obtained with native TCR, using 2B4 TCRζ−/− T-cell hybridoma reconstituted with the TCRζ* (Fig. 3B). Similar results were also obtained using Zap70 phosphorylation as a readout in T cells expressing CD2-TCRζ*ITAM2 stimulated with immobilized anti-CD2 antibody (Fig. 3C).
Fig. 3.
Mutation of BRS motifs abrogates early TCR signaling. TCRζ-deficient BW5δ (A) or 2B4 (B) cells expressing the indicated TCRζ constructs were analyzed before or after incubation with pervanadate (10 μM) at 37 oC. Immunoprecipitation was performed with anti-CD2 (OX34)–coated beads and blotted with anti-pY (4G10) antibody. Blots were stripped and reprobed with OX34. (A) Total cell lysate was used to measure pZap70 (pY319) and pLAT (pY119) and then was stripped and reprobed for total Zap70 and total LAT, respectively. (B) Total cell lysate was blotted with anti-pY (4G10) antibody and rabbit polyclonal anti-TCRζ. (C) BW5δ cells expressing matching levels of the indicated constructs were stimulated by plate-bound anti-CD2 (OX54) for 1 min. These cells then were fixed, permeabilized, and stained with a directly conjugated anti-pZap70 (pY319) antibody.
One possible trivial explanation for these results is that mutation of the BRS motifs affected the overall structure of the TCRζ cytoplasmic domain, thereby disrupting the ability of Lck to bind to and phosphorylate it. Such an effect of BRS mutations has been reported for CD3ε (21). To examine this possibility, we performed an in vitro Lck kinase assay in the absence of membranes (Materials and Methods) (Fig. S7). Under nonsaturated conditions, the ability of Lck to phosphorylate CD2-TCRζ*ITAM2 in vitro was unaffected by mutation of the BRS motifs (Fig. S8).
These results are consistent with the analysis of IL-2 secretion and suggest that BRS motifs play an important positive role in proximal TCR signaling.
Mutation of the BRS Motifs in TCRζ Alters the Mobility and Distribution of TCR/CD3.
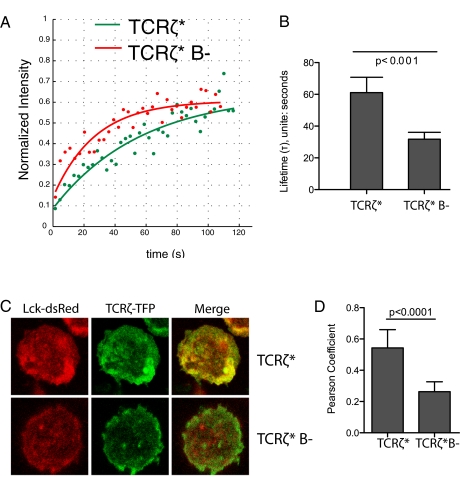
Our finding that mutation of TCRζ BRS motifs disrupts membrane association and early TCR signaling raises the question as to how these processes are linked. One possibility is suggested by the safety on/off model (14, 16), which proposes that membrane association prevents ITAM phosphorylation by inhibiting its accessibility to Lck. This model cannot account for our results, because it predicts that mutation of BRS motifs should enhance TCRζ phosphorylation, whereas we find that mutation of BRS motifs is inhibitory. Another possibility is that BRS-mediated association with negatively charged phospholipid influences the lateral distribution and/or compartmentalization of the TCR/CD3 complex within the plasma membrane. To investigate this possibility, we used fluorescence recovery after photobleaching to examine the effect of BRS mutation on TCR/CD3 mobility (Fig. 4A). The BRS mutant had a twofold faster recovery lifetime (Fig. 4B), indicating a modest increase in membrane mobility. It is possible that BRS-mediated association of TCRζcyt with the plasma membrane accounts for this reduced mobility.
Fig. 4.
Mutation of the BRS motifs in TCRζ alters TCR mobility and its distribution with respect to Lck. (A) Fluorescent recovery trace after photobleaching a 1.3 ×1.3 μm area on the surface of BW5δ cells expressing the indicated TCRζ* construct. An exponential recovery equation was fitted to the data to determine the recovery lifetime and mobile fraction (Materials and Methods). (B) Mean TCR recovery lifetimes from multiple (n = 30 cells) determinations. (C) Confocal imaging of BW5δ cells expressing Lck-DsRed and TCRζ* with or without (B−) the BRS motifs. The images obtained by confocal microscopy represent an optical section of 0.5 μm in z axis, focusing on the glass coverslip. (D) Pearson coefficients representing colocalization of Lck-DsRed with TCRζ* or TCRζ*B−. Data are means of multiple (n = 30 cells) determinations.
To investigate the possible role for the TCRζ BRS motifs on the compartmentalization of TCR/CD3, we examined the effect of BRS mutations on the relative spatial distribution of TCR/CD3 and Lck, which is associated with lipid rafts. TFP-labeled TCRζ* was cotransduced with DsRed-labeled Lck into BW5δ cells. Confocal imaging showed colocalization of Lck and TCRζ*, with a Pearson coefficient of 0.6 (Fig. 4 C and D). There was a twofold reduction in colocalization of Lck with the BRS mutant TCRζ* (Fig. 4 C and D). We next measured FRET between Lck-DsRed (acceptor) and TCRζ-TFP (donor) by examining dequenching following bleaching of DsRed. The TCRζ* showed fluorescent recovery after Lck-DsRed bleaching, but the BRS mutant consistently showed no recovery (Fig. S9), indicating that the very close proximity between TCRζ and Lck is lost in the BRS mutant. Taken together, these data suggest that mutation of BRS motifs alters the distribution of TCR, reducing its proximity to Lck-containing membrane domains
Discussion
The key findings reported here are (i) the C terminus of TCRζcyt is closely associated with the plasma membrane and this association is dependent on BRS motifs; (ii) TCR engagement induces dissociation of the TCRζcyt from the plasma membrane and this dissociation requires TCRζcyt phosphorylation; and (iii) mutation of TCRζcyt BRS motifs inhibits TCR signaling and alters its mobility and distribution within the plasma membrane.
Previous studies have shown that recombinant TCRζcyt peptide associates with artificial membranes and that this association involves electrostatic interactions (10, 11, 14). Using a FRET-based method, Xu et al. (6) showed that the cytoplasmic tail of a CD3ε chimera expressed in cells associates with the plasma membrane in a BRS-dependent manner. We have extended these studies by using the same FRET-based method to study TCRζcyt within the native TCR complex. We show that in the native TCR–CD3 complex the C-terminal end of the TCRζcyt is associated closely with the plasma membrane in at least 25% of TCRs and that this association is dependent on BRS motifs.
It has been proposed previously that the association of the TCRζ and CD3ε cytoplasmic domains with the membrane prevents TCR triggering by sequestering ITAM tyrosine residues from tyrosine kinases (6, 14). According to this safety model (16), ligand engagement stimulates phosphorylation of these cytoplasmic domains by inducing their dissociation from the plasma membrane. We therefore investigated the effect of TCR engagement in our system. We found that anti-TCR antibodies did indeed induce dissociation of TCRζcyt from the plasma membrane, but this dissociation was dependent upon the phosphorylation of ITAMs by Lck. Given that the interaction of TCRζcyt with the membrane is likely to be driven by electrostatic interactions, the most plausible explanation is that phosphorylation of the ITAMs alters the charge of the TCR sufficiently to prevent membrane association. In support of this notion, previous studies have showed that phosphorylation of the TCRζcyt peptide inhibits its association with acidic phospholipids (10, 11).
It is notable that, based on our results and the findings of Xu et al. (6), each TCR–CD3 complex could have up to four cytoplasmic domains physically associated with the plasma membrane in the resting state. It seems likely that the dissociation of the cytoplasmic domains from the plasma membrane following TCR triggering would decrease the effective radius and increase the mobility of each triggered TCR–CD3 complex. This change in radius and mobility could contribute to the aggregation of TCR–CD3 complexes that is observed following TCR triggering (18, 19).
Our third key finding was that mutation of the TCRζcyt BRS motifs reduced TCR signaling. This effect was fairly modest in the intact TCR but nevertheless was highly reproducible and was observed with both antibody and pMHC stimulation. The modest nature of the BRS mutations likely reflects the known redundancy between the multiple TCR–CD3 signaling subunits (17). Our results contrast with a recent report that mutation of BRS sites of TCRζcyt had little effect on signaling (11). A possible explanation for this discrepancy is that DeFord-Watts et al (11) mutated only a subset of the charged residues in the BRS motifs. In support of this notion, when DeFord-Watts et al mutated all three BRS motifs, similar to the BRS mutant used in this report, they observed a decrease in phosphorylation of TCRζ by Lck in a model system.
To facilitate further dissection of the BRS mutations, we used a simplified TCR construct comprising only a single ITAM from TCRζ. Ligand-induced TCRζ phosphorylation and downstream signaling events were virtually abolished when the BRS motifs in this construct were mutated. This effect was not simply the result of a disruption of TCRζ structure, because the BRS mutation had no effect on Lck phosphorylation in vitro. The fact that our BRS mutation altered lysine residues raises the possibility that our mutations affected TCRζ ubiquitination. However, the primary ubiquitination site on TCRζ is K54 (22), which was not mutated in this study. Furthermore, Huang et al. (22) found that disruption of ubiquitination by mutation of K54 increased TCR phosphorylation. Thus it is unlikely that our findings were the result of altered TCRζ ubiquitination.
The finding that mutation of TCRζ BRS motifs decreased signaling raises the question as to the mechanism. Given that these mutations also disrupt the association of the TCRζcyt with the plasma membrane, it is reasonable to speculate that these phenomena are related, but the question of how they are related remains. As noted previously, the safety model postulates that BRS motif-mediated association of TCRζ and CD3ε cytoplasmic domains inhibits their phosphorylation. However, because this model predicts that mutations that disrupt membrane association should enhance TCRζ phosphorylation, it cannot account for our results.
Another possible explanation for the effect of the BRS motif mutation is that BRS motif mutation perturbs the membrane environment of the TCR complex. Because the BRS motifs have been shown to mediate interactions with negatively charged phospholipids (6), it is likely that mutation of these motifs would affect the lipid environment of the TCR–CD3 complex. In support of this notion, we found that BRS mutation altered both TCR–CD3 mobility and colocalization of the TCR–CD3 complex with Lck, which itself localizes with lipid rafts. Because lipid rafts are believed to play an important role in TCR signaling it seems plausible that mutations that affect the interaction of the TCR with its lipid environment would effect TCR triggering.
Our finding that mutation of TCRζcyt BRS motifs disrupts signaling is analogous to studies on CD3ε (12, 23). However, although we were able to show mutation of TCRζcyt BRS motifs does not affect Lck phosphorylation of TCRζcyt in vitro, a recent study found that mutation of the CD3εcyt BRS motif does disrupt Lck phosphorylation in vitro even in the absence of membranes (21). Given that a major drawback of mutagenesis studies is that mutations can have indirect effects on overall protein structure, this discrepancy suggests that findings with CD3ε BRS mutants should be interpreted with caution until such indirect structural effects can be ruled out.
In conclusion our results, together with the findings of others (6, 11, 12, 14), suggest the following model for the role of BRS motifs and plasma membrane association of TCR cytoplasmic domains in triggering. In the unengaged TCR complex the TCRζ and CD3ε BRS motifs mediate association with negatively charged lipids in the plasma membrane. This association has two possible consequences: (i) it could affect the membrane environment or localization of the TCR–CD3 complex; (ii) it could affect the accessibility of the ITAMs to tyrosine phosphorylation. TCR engagement induces TCRζ and CD3ε phosphorylation by one of a number of possible mechanisms (reviewed in ref. 3), including binding-induced dissociation of TCRζ and CD3ε from the membrane, as proposed in the safety model. Dissociation of TCRζ and CD3ε from the plasma membrane, whether a cause or consequence of phosphorylation, may facilitate the TCR–CD3 clustering that follows TCR–CD3 triggering and which may be required for full T-cell activation.
Materials and Methods
Cell Lines, Transduction, Antibodies, and Stimulation.
BW5147-DC115.1 (referred to as “BW5δ") is a mouse T-cell hybridoma line that lacks endogenous TCRζ (17). A TCRζ-deficient 2B4 T-cell hybridoma (24) was obtained from Nicolai S. C. van Oers (University of Texas Southwestern Medical Center, Dallas, TX). The 2B4 TCR is specific for a moth cytochrome c (MCC) peptide (88-104, ANERADLIAYLKQATK) presented on I-Ek. Various TCRζ constructs were transduced using an ecotropic packaging system developed by the Nolan laboratory (Stanford University, Stanford, CA; http://www.stanford.edu/group/nolan/). Matching cell-surface expression levels were obtained via FACS. Jurkat E6.1, p116, and p116 cells reconstituted with Zap70 were a gift from O. Acuto (The Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom). Antibodies used for flow cytometry include staining anti-rat CD2-biotin (OX34; Serotec), streptavidin-phycoerythrin Serotec), and anti-pZap70 (PY319)/pSyk (PY352)-Alexa Fluor 647 (Phosflow; BD). Antibodies used for Western blotting include anti-pZap70 (PY319)/pSyk (PY352) (Cell Signaling), anti-Zap70 (Cell Signaling), anti-LAT (Upstate), anti-pLAT Y191 (Invitrogen), anti-phosphotyrosine 4G10 (Upstate), polyclonal rabbit anti-TCRζ (Acris Antibodies), anti-rabbit IgG-HRP (Cell Signaling), anti-mouse IgG-HRP (Cell Signaling), anti-GST (Abcam), goat anti-rabbit IR Dye 680 nm (Licor), and goat anti-mouse IR Dye 800 nm (Licor). Western blots were preformed as previously stated (25). Anti-rat CD2 (OX34; Serotec) was used for immunoprecipitation. Cells were stimulated with the anti-mouse CD3ε mAb 145–2C11 (Biolegend), the anti-rat CD2 mAb OX54, pervanadate (10 μM), or MCC-pulsed I-Ek–expressing CHO cells. Where indicated, cells were preincubated with PP2 (100 μM) for 10 min at 37 oC. Beads used for simulation or immunoprecipitation were coated with anti-mouse Ig (Invitrogen) or biotin (Bangs Laboratories Inc.). Lck kinase assay buffers and proteins were purchased commercially (Millipore).
Imaging by Laser-Scanning Microscope.
Maximum viability of the cells was achieved by frequent splitting. Usually 1 million cells were pelleted and resuspended in Tyrode solution. An optimal number of cells were transferred to a glass-bottomed imaging chamber (Attofluor cell chamber, A-7816; Invitrogen), which was mounted onto a laser-scanning microscope (LSM510; Zeiss). The stage containing the sample was kept at 37 °C during the entire imaging process. Fluorophores were excited and then collected with available optimal wavelengths and filters, respectively. Data were collected and quantified with bundled LSM510 software. Final images layouts were produced in Photoshop (Adobe) and Illustrator (Adobe). The color space of all images was Adobe 1998.
FRET Measurements.
The FRET pair analyzed were R18 (acceptor) (Invitrogen) and TFP (donor) (Allele Biotechnology). Maximum viability of the cells was maintained by splitting the cells frequently. FRET imaging was performed on a Zeiss LSM 510 confocal microscope. Microscope setup and procedure were as described previously (6). Briefly, FRET efficiency was measured by donor dequenching. BW5δ T cells expressing matched levels of TCRζ constructs were immobilized onto glass-bottomed open chambers at 37 °C. The plasma membranes for these cells were labeled with R18. A set of images with both TFP and R18 channels was recorded before (Q) and after (DQ) R18 was bleached by a 543-nm laser (full power) for 60–80 s. Mean dequenching efficiency (E) from multiple (n = 30) cells was calculated using the equation E = (DQ − Q)/DQ. In cells stimulated with anti-CD3–coated beads, the dequenching efficiency within the bead–cell interface and outside the interface are shown. FRET measurements were completed within 5 min anti-CD3–coated beads were added.
Statistics.
The error bars represent SD. P values for the significance of differences between means were calculated by student's t test.
Supplementary Material
Acknowledgments
We thank Marcus Bridge for technical assistance including DNA purification and the IL-2 assay and Kai Wucherpfennig for advice and communication of unpublished data. This work was supported by The E. P. Abraham Research Fund (H.Z, S-P.C., and P.A.v.d.M.), the Medical Research Council (S-P.C. and P.A.v.d.M), the Clarendon Fund (H.Z.), and a Newton Fellowship from the Royal Society (to O.D).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108052108/-/DCSupplemental.
References
- 1.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 2.Fooksman DR, et al. Functional anatomy of T cell activation and synapse formation. Annu Rev Immunol. 2010;28:79–105. doi: 10.1146/annurev-immunol-030409-101308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 4.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 5.Gil D, Schamel WW, Montoya M, Sánchez-Madrid F, Alarcón B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil D, Schrum AG, Alarcón B, Palmer E. T cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes. J Exp Med. 2005;201:517–522. doi: 10.1084/jem.20042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymczak AL, et al. The CD3epsilon proline-rich sequence, and its interaction with Nck, is not required for T cell development and function. J Immunol. 2005;175:270–275. doi: 10.4049/jimmunol.175.1.270. [DOI] [PubMed] [Google Scholar]
- 9.Mingueneau M, et al. The proline-rich sequence of CD3epsilon controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat Immunol. 2008;9:522–532. doi: 10.1038/ni.1608. [DOI] [PubMed] [Google Scholar]
- 10.Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry. 2006;45:15731–15739. doi: 10.1021/bi061108f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeFord-Watts LM, et al. The CD3 zeta subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR-CD3 complex at the immunological synapse. J Immunol. 2011;186:6839–6847. doi: 10.4049/jimmunol.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deford-Watts LM, et al. The cytoplasmic tail of the T cell receptor CD3 epsilon subunit contains a phospholipid-binding motif that regulates T cell functions. J Im munol. 2009;183:1055–1064. doi: 10.4049/jimmunol.0900404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risueno RM, Schamel WW, Alarcon B. T cell receptor engagement triggers its CD3epsilon and CD3zeta subunits to adopt a compact, locked conformation. PLoS ONE. 2008;3:e1747. doi: 10.1371/journal.pone.0001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 15.Sigalov AB, Hendricks GM. Membrane binding mode of intrinsically disordered cytoplasmic domains of T cell receptor signaling subunits depends on lipid composition. Biochem Biophys Res Commun. 2009;389:388–393. doi: 10.1016/j.bbrc.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhns MS, Davis MM. The safety on the TCR trigger. Cell. 2008;135:594–596. doi: 10.1016/j.cell.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wegener AM, et al. The T cell receptor/CD3 complex is composed of at least two autonomous transduction modules. Cell. 1992;68:83–95. doi: 10.1016/0092-8674(92)90208-t. [DOI] [PubMed] [Google Scholar]
- 18.Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 19.Hanke JH, et al. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 20.Lyons DS, et al. A TCR binds to antagonist ligands with lower affinities and faster dissociation rates than to agonists. Immunity. 1996;5:53–61. doi: 10.1016/s1074-7613(00)80309-x. [DOI] [PubMed] [Google Scholar]
- 21.Gagnon E, et al. Response multilayered control of T cell receptor phosphorylation. Cell. 2010;142:669–671. doi: 10.1016/j.cell.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, et al. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes RA, et al. What controls T cell receptor phosphorylation? Cell. 2010;142:668–669. doi: 10.1016/j.cell.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Sussman JJ, et al. Failure to synthesize the T cell CD3-zeta chain: Structure and function of a partial T cell receptor complex. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 25.Nika K, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32:766–777. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.