Abstract
Large-scale transcription profiling via direct cDNA sequencing provides important insights as to how foundation species cope with increasing climatic extremes predicted under global warming. Species distributed along a thermal cline, such as the ecologically important seagrass Zostera marina, provide an opportunity to assess temperature effects on gene expression as a function of their long-term adaptation to heat stress. We exposed a southern and northern European population of Zostera marina from contrasting thermal environments to a realistic heat wave in a common-stress garden. In a fully crossed experiment, eight cDNA libraries, each comprising ∼125 000 reads, were obtained during and after a simulated heat wave, along with nonstressed control treatments. Although gene-expression patterns during stress were similar in both populations and were dominated by classical heat-shock proteins, transcription profiles diverged after the heat wave. Gene-expression patterns in southern genotypes returned to control values immediately, but genotypes from the northern site failed to recover and revealed the induction of genes involved in protein degradation, indicating failed metabolic compensation to high sea-surface temperature. We conclude that the return of gene-expression patterns during recovery provides critical information on thermal adaptation in aquatic habitats under climatic stress. As a unifying concept for ecological genomics, we propose transcriptomic resilience, analogous to ecological resilience, as an important measure to predict the tolerance of individuals and hence the fate of local populations in the face of global warming.
Keywords: EST-library, extreme event, thermal tolerance
Global climate change is imposing increasing stress on many organisms. Thus, one central question in ecology and evolution is how species cope with such environmental challenges (1–4). Global climate change is characterized by both the change in mean variables and the increase in extreme events such as heat waves, droughts, and heavy precipitation (5). These extreme events, in particular, strongly impact ecosystems and associated species (5, 6). In habitat foundation species (sensu ref. 7), such as corals, trees, and seagrasses, entire ecosystems depend on the stability and performance of populations of single species (8). Therefore, their persistence in the light of climate change is of particular interest. Among these habitat foundation species are the 60 or so species of seagrasses (9), monocotyledonous plants which form the basis of productive marine ecosystems, providing habitat and nursery grounds for associated species (10). Seagrass-based ecosystems alter the physical environment by stabilizing the sediment, nutrient fixation, and current modification (11), sometimes exceeding the ecosystem services provided by coral reefs (12). Accelerating global declines of seagrasses therefore are of major concern (13). Currently, it is unclear how much ocean warming contributes to seagrass decline, potentially exacerbating the other known anthropogenic factors that negatively impact seagrass beds such as eutrophication, fishing, mechanical destruction, and aquaculture (13).
We focus here on the effects of extreme summer sea-surface temperature on eelgrass (Zostera marina), a widespread and often locally dominant seagrass species of the northern hemisphere (9). Field surveys and experimental studies suggest that high water temperatures (≥25 °C) during summer heat waves increasingly threaten seagrass performance and survival in northern European seagrass beds (14–17). In contrast, genetically divergent Z. marina populations (18, 19) persist in Mediterranean bays and lagoons where summer temperatures frequently exceed 26 °C (20), suggesting enhanced thermal tolerance of the high summer temperatures in these locations. Together these data emphasize the importance identifying the physiological and genetic basis of thermal tolerances across species’ distributions (1, 21) and addressing whether limits to distributions can evolve or represent fundamental constraints (22).
Global gene-expression profiling is one emerging approach toward understanding these differences in tolerance, because thermal tolerance often is reflected in the differential expression of particular genes under thermal exposure, and such variation between populations often is indicative of local adaptation (23–28). Studying populations along a thermal gradient, for example the northern and southern European seagrass populations, is particularly instructive, because we can test the basic hypothesis that differential expression among localities under heat-stress conditions reflects thermal adaptation (29). Few studies have applied large-scale gene expression in natural populations along an environmental gradient (but see refs, 24, 30–32), and we are unaware of studies using next-generation RNA sequencing technologies that do not require a priori genomic/transcriptomic information for digital gene-expression analysis (33, 34). Still fewer studies have covered the recovery phase after an extreme event explicitly, and those that have done so have considered only a short time scale of a few hours to days (e.g., refs. 35–37). We also are not aware of any study on ecologically important foundation species, which often are not genetic/genomic model species.
To bridge this gap, we used 454 direct cDNA sequencing (34, 38) for a global assessment of the transcription profiles of heat-stressed and nonstressed eelgrass (Z. marina) plants from two contrasting locations. The experimental heat stress mimicked an actual heat wave that struck Europe in 2003 (Fig. S1) (39) and led to considerable mortality in northern European seagrass populations (15, 17). Our goal was to identify putative genes and molecular functions involved in adaptation to the specific local conditions (24, 40), with special attention to a joint analysis of both the acute response to heat stress and the recovery phase. Although we expected classical heat-shock proteins (HSPs) and chaperones to play an essential role during the acute phase of the heat wave (41–43), longer-term effects on the cellular metabolism, including increasing expression of proteins involved in protein degradation needed for the turnover of irreversibly damaged proteins, may be as critical as changes during the stress event (44, 45). Such assessments are possible only with a more comprehensive gene-expression analysis that goes beyond available target-gene approaches (20).
Results
Assembly and Gene Annotation.
Our global gene-expression analysis using leaf tissue covered ∼one-third of all genes of a typical flowering plant. The number of unique genes was highest in the larger reference clone assembly with 11,135 gene identifications. The population-level assemblies identified 8,673/8,579 unique genes for the northern and southern populations, respectively (Table S1). We identified tentative unigenes by mapping de novo-assembled contigs against reference proteomes from Arabidopsis thaliana and Oryza sativa. Annotation success was high, with 76% and 78% of the Z. marina contigs yielding results with e-values <10−4 against the A. thaliana proteome for the northern and southern population, respectively (Table S1). Often, more than one of the obtained de novo-assembled Z. marina contigs mapped against the same Arabidopsis-annotated tentative unigene. The underlying causes were evaluated in a small case study mapping all contigs against a set of 14 nonredundant Z. marina coding sequences downloaded from the National Center for Biotechnology Information (NCBI). The observed contig redundancy was caused by a combination of polymorphisms in gene sequences obtained from multiple genotypes in the libraries, sequencing errors, and contigs that mapped to different, nonoverlapping sections of reference genes. Interestingly, the annotation success against the phylogenetically closer monocotyledonous plant species O. sativa (rice) was very similar (Table S1). We thus continued with A. thaliana as reference because it is the better-annotated plant species. When very lowly expressed genes (read count across libraries of ≤2) were eliminated, 5,908 genes remained, corresponding to 104,753–139,020 EST reads for each library (Table S2).
Validation of 454 Transcriptome Sequencing via Quantitative Real-Time PCR.
Expression profiles obtained by direct cDNA 454 sequencing were validated using a sample of candidate genes measured in both populations during the heat wave using an independent method. We quantified expression levels of 18 genes in replicated genotypes (n = 5) using quantitative real-time PCR [via the delta-delta cycle threshold (ΔΔCT) method (46)] and compared these levels with data obtained via direct cDNA sequencing (via log2 fold-change) (Fig. S2). The two methods corresponded well, with correlation coefficients r = 0.812 and 0.807 for A. thaliana and O. sativa as reference proteome, respectively (P < 0.001) (Fig. S2).
Multivariate Characterization of Gene-Expression Profiles.
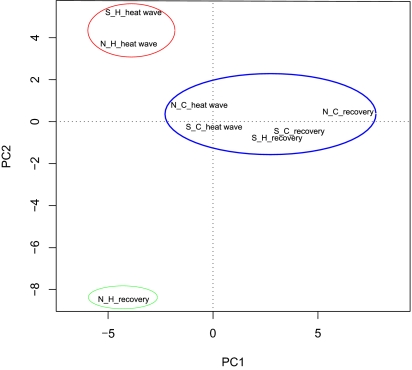
We identified genes that were tentatively differentially expressed (TDE) in response to the heat treatment by assembling sequence reads into contigs and then mapping those contigs against A. thaliana orthologs. We refer here to TDE genes because we do not wish to draw any conclusions based on particular genes; it is well known that genes identified in global transcription profiling need independent verification, for example using quantitative real-time PCR as described above. TDE genes were determined separately in all four library pairs (northern/southern population * during /after heat wave). Of 5,908 genes, 1,872 revealed evidence for significant changes in expression at least once and were kept for further analyses (Table S3). TDE genes with ∼40 mean read counts across libraries revealed fold-changes of ∼2, which we consider appropriate, although with higher transcript abundance, smaller fold-changes were detected as significant (Fig. S3). TDE genes were not interpreted at face value but only based on functional groupings that emerged from subsequent multivariate analyses. To identify patterns of similarity across all eight treatments (north/south * heat stress/control * during/after heat wave), a principal component analysis (PCA) of scaled expression profiles explained 41.84% of the variance and revealed three distinct clusters (Fig. 1). The largest cluster comprised libraries obtained from all four control conditions, along with the previously heat-stressed library of the southern population after the heat wave. A second well-defined cluster included the heat-stressed libraries of both populations during the heat wave. As a third cluster, genes in the northern population upon termination of the heat wave showed the most divergent expression of all libraries. The grouping of libraries was supported by analysis of similarity (ANOSIM) of the scaled expression profiles of TDE genes (r = 0.7968; P = 0.0057). We also performed a similar PCA on a smaller subset of TDE genes. When applying a more stringent filter of ≥10 reads in at least one of the libraries revealing significant expression change, 1,422 of the initial 1,872 TDE genes remained. When analyzed as above, full support of the three identified clusters in the PCA could be found (ANOSIM, r = 0.7112, P = 0.0059).
Fig. 1.
Multivariate grouping of experimental libraries based on the expression profiles of 1,872 TDE genes using PCA. TDE genes between heat and control treatment were assessed for four library combinations (southern vs. northern population; within vs. after heat wave). PCA was performed on the scaled expression matrix of genes (mean = 0; SD = 1). Populations: northern (N), southern (S); heat treatment (H), control treatment (C); time points: acute heat and recovery. Groupings are indicated by color: blue, expression during control conditions; red, expression during heat stress; green, divergent expression during early recovery. A list of all TDE genes is found in Table S3.
Identification and Functional Annotation of Indicator Genes.
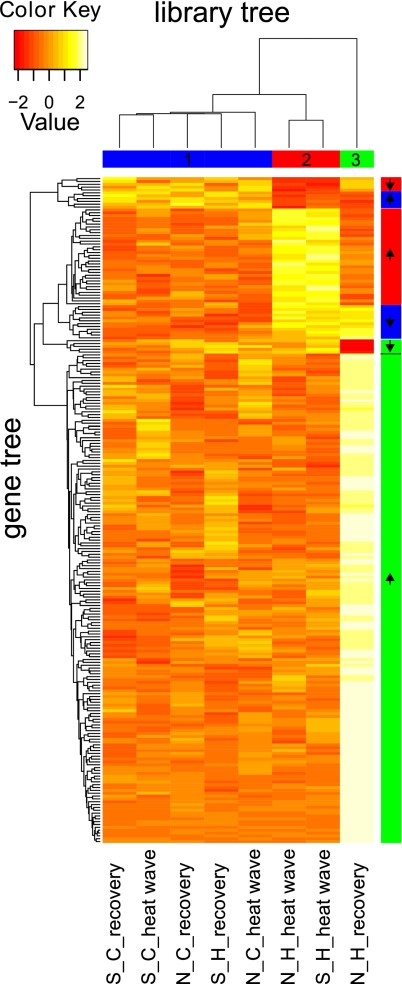
To identify TDE genes with the largest contribution to the identified grouping, an indicator gene analysis was conducted. Among the 234 TDE genes identified (all r ≥ 0.9 with above groupings; Table S4), 17 were specific for group 1 (the “control expression” group), with six indicating up-regulation and 11 indicating down-regulation of that group (Fig. 2). Group 2 (“during heat stress”) was supported by the expression of 39 genes, 34 showing group-specific up-regulation and five showing up-regulation. Group 3 (“divergent northern recovery”) contained 178 genes, 173 showing up-regulation and five showing down-regulation in comparison with the remaining groups.
Fig. 2.
Heatmap shows cross-correlation by treatment and similarity of gene-expression profiles of 234 heat-responsive genes that are strongly correlated (r ≥ 0.9) with the PCA-based clustering (Fig.1). x axis: columns display the cDNA libraries from the eight treatments, clustered by similarities among gene-expression profiles; y axis: each row displays the expression strength of a particular gene in the respective library, clustered by similarities across treatments. Expression strength was scaled for each gene across libraries (mean = 0; SD = 1). Values are color coded (white: highest expression strength; red: lowest expression strength). In combining library (treatment) and gene clustering, the following groups of genes are indicated: blue (1), control expression; red (2), during heat stress; green (3), divergent early recovery. Arrows along the gene order indicate up- and down-regulation of genes. Treatment codes are as in Fig. 1. Functional annotations of the six different gene sets that display characteristic up- or down-regulation of a group are shown in Fig. 3 and Fig. S4; a detailed list is given in Table S4.
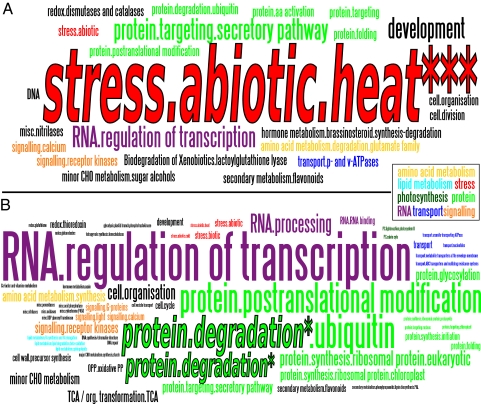
The six different subsets of indicator genes (respective PCA-based group * up-/down-regulation) were functionally annotated via MapMan (47). The dominant functional group of up-regulated genes during heat stress was associated with the category “stress.abiotic.heat” (25% of all annotations) and consisted of various HSPs (Fig. 3A), which were significantly overrepresented (Fisher's exact test; P < 0.001). Further categories were related to “protein” (22%) (belonging to the differing subcategories protein targeting, degradation, posttranslational modification, folding, and amino acid activation) and the categories “RNA.Regulation of transcription” (6%), “development” (6%), and “signaling” (6%). In contrast, HSPs played a only minor role during early recovery of the northern population (Fig. 3B). Here, genes of different putative functions were abundant. The category “protein.degradation” was most frequent (12%) and also was significantly overrepresented (Fisher's exact test; P = 0.034). Other frequently identified genes were associated with categories “RNA.regulation of transcription” (9%), “protein.postranslational modification” (6%), and “signaling” (7%). The functional annotation of the remaining four groups of indicator genes can be found in Fig. S4.
Fig. 3.
Functional annotation of gene sets showing up-regulation in a specific group of libraries in comparison with the remaining groups. Putative functions of (A) up-regulated genes in group 2, during heat stress (5.6% of genes not annotated) and (B) up-regulated genes in group 3, divergent early recovery (24.1% of genes not annotated). Genes were annotated with MapMan categories and are presented via term clouds; the annotation frequency is proportional to word size. Gene categories that are significantly enriched (Fisher's exact test) are marked (*P ≤ 0.05; ***P ≤ 0.001).
Expression of HSPs During Heat Stress.
We further scrutinized gene-expression patterns during the heat wave in both populations and searched for TDE genes that were annotated by MapMan category “stress.abiotic.heat” or directly with the key term “HSP”; this search yielded 27 genes. The expression of these 27 genes during heat stress was highly correlated in the two populations (r = 0.97; P < 0.001). Of these 27 genes, only two showed differential expression between populations during the heat treatment (Fig. S5).
Discussion
In this study we examined a nearly complete transcriptomic response to global warming by a foundation species that structures an entire ecosystem, including a recovery phase after a realistic heat-wave scenario. The experimental genotypes came from two contrasting regions in the European thermal cline, the Adriatic Sea (southern population, Italy), and the Limfjord (northern population, Denmark).
After systematic data reduction, two salient findings of the transcription profiling were the similarity of gene expression during the heat wave and the strong divergence between the two populations shortly thereafter. During heat stress, in both the northern and southern population, the transcriptomic syndrome was dominated by the up-regulation of genes associated with the MapMan annotation “stress.abiotic.heat” (Fig. 3A), comprised mainly of classical HSPs. This finding is in line with studies suggesting that the expression of HSPs and molecular chaperones is correlated with thermotolerance and thermal adaptation (41–43, 48). Most previous studies, however, came from terrestrial organisms, and the duration of the experiment and the period of gene induction were, on average, 10-fold shorter (35, 49–53). Gene-expression studies in aquatic plants that experience more gradual temperature changes remain largely unexplored (but see refs. 54 and 55). Aquatic organisms experience smaller extremes in temperature that are dampened because of the high specific heat of the aqueous medium (20, 41). Once critical temperatures are attained, however, these conditions typically last longer in aquatic habitats, with no possibility for evaporative cooling. This gradual onset and prolonged duration of critical temperatures may explain, at least in part, why differences in expression between the two populations during the heat-stress treatment were modest, with only two of 27 HSP genes revealing differences in expression between populations (Fig. S5).
In contrast, both populations showed drastic differences in gene expression 1 d after termination of the heat wave. The expression profile of the southern population revealed considerable resilience and rapidly returned to control expression levels, but the transcription profile of the northern population diverged even further from all control treatments. The functional annotation of up-regulated genes in the northern population suggests a nonadaptive syndrome of failed metabolic compensation in the northern plants. Accordingly, up-regulated functions after the heat stress were dominated by “protein.degradation” and “RNA.regulation of transcription,” suggesting that proteins were damaged irreversibly and needed to be degraded and removed from the cell (56). Similar up-regulation of protein degradation has been observed in other plant species under various stressors (44, 45, 57). That the southern population returned so rapidly to control levels of gene expression demonstrates that temperatures >26 °C do not represent a fundamental limit to the distribution of Z. marina (22), a finding that is significant for seagrass conservation and ecology (13).
The use of next-generation direct cDNA sequencing on the Roche 454 Titanium platform enabled us to perform global transcription profiling in a non-model foundation species (34, 38), at the cost of having to pool RNA over individuals within treatments. The digital expression measurement on cDNA libraries of pooled genotypes was verified by quantitative real-time PCR. The quantitative real-time PCR measurements in 18 genes used biological replicates (n = 5) sampled in the same experiment (Fig. S2). Because the level of concordance between the two methods was high (r ∼0.8; P < 0.001), we conclude that the pooling of genotypes within treatment had only a small effect on the biological signal.
Upon de novo transcriptome assembly, our actual gene identification was guided by mapping to orthologs in reference proteomes in well-characterized plant species. Although de novo transcriptome assemblies have been performed on a variety of higher plants (e.g., refs. 58–60), currently used assembly software and assembly strategies are still in need of improvement, particularly when reference genomes are unavailable (61). In these cases, annotations against reference proteomes of related plant species, as was used here, are the second-best alternative. A reassuring result was that gene identification success was largely congruent, whether we used a monocotyledonous (O. sativa) or dicotyledonous (A. thaliana) plant species as reference proteome; the former was phylogenetically closer than the latter to our target species (62).
One key assumption of our common-stress garden approach is that the observed differences in gene expression in the two populations have a heritable basis, at least in part (25, 28, 31). In allowing a relatively long acclimation period, we tried to minimize the likelihood that our common-stress garden design captures carry-over effects from the past environments of the experimental plants. However, environmental and epigenetic influences during the life history of the studied individuals cannot be ruled out completely (63, 64).
The transcriptional patterns observed here were in line with recent phenotypic measurements of photosynthetic performance using pulse amplitude-modulated fluorometry, in which southern Z. marina genotypes, in contrast to their northern counterparts, recovered to control values immediately after the heat wave (65). Because dramatic differences in gene responses were detectable only during recovery, we speculate that many previous studies investigating transcriptomic responses to acute stress missed critical gene-expression patterns. We posit that evolutionary ecology experiments addressing the physiological response among divergent populations should fulfill the following criteria: (i) inclusion of a reasonable acclimation phase; (ii) application of realistic stress scenarios including rate of increase, intensities, duration, and recovery; and (iii) inclusion of nonclassical stress genes to detect general deviation from normal cell homeostasis compared with a control treatment.
Because acute heat-stress responses were surprisingly similar among genotypes from two locations with widely diverging thermal conditions, we suggest that the transcriptomic patterns during recovery may be a better predictor as to how populations across latitudinal clines are adapted to thermal stress. As a unifying concept for ecological genomics, we propose transcriptomic resilience, describing the return to control levels of gene expression, analogous to ecological resilience, which describes the return of species abundance and performance to predisturbance conditions (7, 66).
Methods
Study Species and Experimental Setup.
Eelgrass (Z. marina) plants were collected from two different populations located in Doverodde, Denmark (Limfjord, 56°43.07′N 8°28.45′E, hereafter “north”) and Gabicce Mare, Italy (Adriatic Sea, 43°57.97′N 12° 45.86′E, hereafter “south”) in early spring 2008 and transported within 48 h to the laboratory. At each location ∼30 leaf shoots were sampled from each of ∼15 sampling spots to collect several different genotypes that were identified via microsatellite genotyping (20). Plants were planted in a mesocosm facility at the University of Münster (Münster, Germany) that has been described in detail elsewhere (20). In brief, the experimental set-up consisted of two temperature-controlled flumes, each with six large 1-m3 tanks filled with artificial seawater and silicate sediments to a height of 10 cm. Plants were grown at 31 practical salinity units and under light-saturated conditions (∼400 μmol photons s−1 m−2). Plants were allowed to acclimatize for ∼30 d while the temperature was raised slowly from 14 °C (collecting temperature) to 19 °C, the control temperature during the heat-wave treatment (Fig. S1).
Heat-Wave Simulation.
The heat-wave simulation followed a common-stress garden design. Half of the experimental units were kept at a water temperature of 19 °C after acclimation, and the other half was subjected to a simulated heat wave, where water temperature was increased by 1.5 °C/d until it reached 26 °C. This temperature was maintained for 3 wk, followed by a temperature decrease of 1.5 °C/d until the water temperature reached 19 °C again (Fig. S1). The temperature profile closely followed the summer heat wave of 2003 (17).
RNA Preparation, 454 Library Construction, and Sequencing.
RNA samples were taken from eight conditions: the northern vs. southern population, under heat stress vs. control conditions, with samples taken at two time points during the experiment, in the middle of the heat wave, and 1 d after return to control values (Fig. S1). For RNA extraction an ∼2-cm-long section from a young, growing leaf was cut from a randomly chosen plant from each condition and replicate, cleaned, frozen by dipping into liquid nitrogen, and immediately ground in a ball mill, followed by RNA extraction with the Invisorb RNA plant HTS 96 extraction kit (Invitek). For each condition RNA samples were pooled from six to eight genotypes. cDNA libraries were constructed using the Clontech SMART cDNA synthesis kit (Invitrogen). First-strand synthesis was performed with ∼0.5 μg total RNA for first-strand synthesis via anchored oligo(dT) priming followed by 15 amplification cycles. Sequencing libraries were constructed based on 3 μg of obtained cDNA. Every library was sequenced on a quarter of a slide using physical library separation with the 454 Genome Sequencer FLX using the Titanium chemistry (Roche and 454 Life Sciences).
EST Preprocessing and de Novo Transcriptome Assembly.
After removal of the adaptor sequence, EST reads were quality trimmed with standard settings of the 454 Genome Sequencer FLX proprietary software (Roche) software. These raw sequence data for the eight generated libraries were deposited at the Sequence Read Archive (accession no. SRP007220). cDNA primer contaminations introduced by the SMART cDNA synthesis kit (Invitrogen) were identified by Crossmatch (http://www.phrap.org/) using the parameters minmatch 10 and minscore 15.
Three different subsets of 454 Genome Sequencer ESTs originating from different Z. marina populations were assembled independently into contigs to minimize polymorphisms derived from multiple divergent genotypes for de novo assembly. Subsequently they were combined in a final mapping assembly of the contigs of the northern and the southern population assemblies against the assembled contigs with the sequence reads obtained from a single Z. marina clone (Falkenstein, Germany) used as a backbone. De novo assembly was performed using MIRA (version 3.0.3, http://sourceforge.net/projects/mira-assembler/files/) using standard parameter settings in the accurate assembly mode (67). Note that the ESTs for the third assembly came from a single clone (Falkenstein, Germany, Baltic Sea, 54°24′N 10°12′E), comprised of 866,838 Roche 454 Titanium EST reads (NCBI Sequence Read Archive, accession no. SRA002573) (Table S1). The final assembly was created by mapping the contigs obtained from the two population assemblies against those from the single-clone assembly. For the second-stage assembly, Mira was used in the mapping and accurate mode, also allowing the creation of new contigs.
Quantification and Verification of Library-Specific Gene-Expression Profiles.
Differential gene expression for each of the eight experimental conditions was assessed by mapping cleaned sequence reads first to the final transcriptome assembly with BLASTN (68) and then identifying tentative unigenes via annotation using the A. thaliana proteome, using Tair9 (69). We also mapped against rice O. sativa (Michigan State University Rice Annotation Project Release 6.0) using BLASTX. Our approach was verified via quantitative real-time PCR for a set of 18 genes. Log2 fold-changes between read counts of heat and control treatment and ΔΔCT values (20) for both populations in the middle of the heat wave were compared (Fig. S2).
Identification of TDE Genes.
Genes TDE in response to the heat treatment were identified via bootstrapping for all four pairwise comparisons of control vs. heat-stress treatment. A null model was created under the hypothesis that gene expression is not different in heat-stress and control treatment. Sequence reads were resampled with replacement from the expressed sequence read pool of the control library. For each of the 10,000 bootstrap replicates, reads were sampled until the number of reads in the respective heat-stress library was attained. For each gene, the read count of the heat-treated library then was compared with the read distribution of the created null model to calculate empirical P values. P values were calculated with subsequent one-sided tests of the area outside the null distribution when assuming higher as well as lower gene expression. To correct for multiple testing, the false discovery rate (FDR) of α = 0.01 was used (70); thus we expect only 1% of genes to be false positives per pairwise comparison and roughly 4% to be false positives on a per gene basis. At low absolute expression levels of a given gene (∼40 reads per library comparison), a log2 fold-change of ∼2 can be detected (Fig. S3). Most indicator genes (215/234) identified in the analysis described below, on which our data interpretation relies, have absolute transcript abundances of ≥10 reads in at least one of the libraries involved (Fig. S3).
Multivariate Analysis of Gene-Expression Profiles.
The profiles of all TDE genes were subjected to multivariate analysis to detect similarities and differences in the transcriptomic response to treatments. Expression profiles were normalized for differences in library sizes (Table S2) (71) and scaled across all eight libraries to a mean of zero and an SD of one. PCA was performed with the R package VEGAN [(http://cran.r-project.org/web/packages/vegan/index.html) version 1.17–6].The resulting groups then were tested with ANOSIM implemented into VEGAN. Euclidian distances were used to calculate similarities between libraries, with P values calculated based on 10,000 permutations (72). The identification of genes that were indicative for the identified grouping of libraries followed an indicator value analysis (73), calculating a correlation index for each gene between the given grouping and the expression values across the libraries. The method is implemented in the R package indicspecies (version 1.5.1). Subsets of indicator genes were functionally annotated via MapMan (47). Abundant functional groups within those subsets of genes were tested formally for overrepresentation against the entire set of functionally annotated indicator genes using one-sided Fisher's exact test.
Supplementary Material
Acknowledgments
We thank Ilka Dankert and Lothar Miersch for laboratory assistance and August Hämmerli for help with collecting plants. This project was supported by the Volkswagen Foundation (S.U.F.), by the Alexander-von-Humboldt foundation (J.G.), by the Helmut-Robert Foundation (U.C.K.), by the Kiel excellence cluster Future Ocean (P.R.) and by a grant from Deutsche Forschungsgemeinschaft-AQUASHIFT (to T.B.H.R.). G.W. received support from the MINERVA Foundation. The sequencing was cofunded by a grant by Marine Genomics Network of Excellence (to J. L. Olsen, University of Groningen).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequence data for the eight generated libraries have been deposited at the Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP007220, accession no. SRP007220). ESTs for the third assembly came from a single clone (National Center for Biotechnology Information Sequence Read Archive, http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP002573, accession no. SRA002573).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107680108/-/DCSupplemental.
References
- 1.Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- 2.Root TL, et al. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- 4.Lubchenco J. Entering the century of the environment: A new social contract for science. Science. 1998;279:491–497. [Google Scholar]
- 5.Easterling DR, et al. Climate extremes: Observations, modeling, and impacts. Science. 2000;289:2068–2074. doi: 10.1126/science.289.5487.2068. [DOI] [PubMed] [Google Scholar]
- 6.Walther G-R, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- 7.Dayton PK. Toward an understanding of community resilience and the potential effects of enrichments to the benthos at McMurdo Sound, Antarctica. In: Parker BC, editor. Proceedings of the Colloquium on Conservation Problems in Antarctica. Lawrence, KS: Allen; 1972. pp. 81–95. [Google Scholar]
- 8.Jones CG, Lawton JH, Shachak M. Organisms as ecosystem engineers. Oikos. 1994;69:373–386. [Google Scholar]
- 9.Den Hartog C. Seagrasses of the world. Verhandelingen der koninklijke Nederlandse Akademie van Wetenschappen, Afd. Natuurkunde. 1970;59:1–275. [Google Scholar]
- 10.Hughes AR, Williams SL, Duarte CM, Heck Jr KL, Waycott M. Associations of concern: Declining seagrasses and threatened dependent species. Front Ecol Environ. 2009;7:242–246. [Google Scholar]
- 11.Larkum AWD, Orth RJ, Duarte CM, editors. Seagrasses: Biology, Ecology and Conservation. Berlin: Springer; 2006. [Google Scholar]
- 12.Costanza R, et al. The value of the world's ecosystem services and natural capital. Nature. 1997;387:253–260. [Google Scholar]
- 13.Waycott M, et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc Natl Acad Sci USA. 2009;106:12377–12381. doi: 10.1073/pnas.0905620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nejrup LB, Pedersen MF. Effects of salinity and water temperature on the ecological performance of Zostera marina. Aquat Bot. 2008;88:239–246. [Google Scholar]
- 15.Ehlers A, Worm B, Reusch TBH. Importance of genetic diversity in eelgrass Zostera marina for its resilience to climate warming. Mar Ecol Prog Ser. 2008;355:1–7. [Google Scholar]
- 16.Greve TM, Borum J, Pedersen O. Meristematic oxygen variability in eelgrass. Limnol Oceanogr. 2003;48:210–216. [Google Scholar]
- 17.Reusch TBH, Ehlers A, Hämmerli A, Worm B. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc Natl Acad Sci USA. 2005;102:2826–2831. doi: 10.1073/pnas.0500008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen JL, et al. North Atlantic phylogeography and large-scale population differentiation of the seagrass Zostera marina L. Mol Ecol. 2004;13:1923–1941. doi: 10.1111/j.1365-294X.2004.02205.x. [DOI] [PubMed] [Google Scholar]
- 19.Reusch TBH, Stam WT, Olsen JL. A microsatellite-based estimation of clonal diversity and population subdivision in Zostera marina, a marine flowering plant. Mol Ecol. 2000;9:127–140. doi: 10.1046/j.1365-294x.2000.00839.x. [DOI] [PubMed] [Google Scholar]
- 20.Bergmann N, et al. Population-specificity of heat stress gene induction in northern and southern eelgrass Zostera marina populations under simulated global warming. Mol Ecol. 2010;19:2870–2883. doi: 10.1111/j.1365-294X.2010.04731.x. [DOI] [PubMed] [Google Scholar]
- 21.Davis MB, Shaw RG. Range shifts and adaptive responses to Quaternary climate change. Science. 2001;292:673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- 22.Kellermann V, van Heerwaarden B, Sgrò CM, Hoffmann AA. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science. 2009;325:1244–1246. doi: 10.1126/science.1175443. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann AA, Sørensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: Bringing together quantitative and molecular approaches. J Therm Biol. 2003;28:175–216. [Google Scholar]
- 24.Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci USA. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead A, Crawford DL. Variation within and among species in gene expression: Raw material for evolution. Mol Ecol. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- 26.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 27.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sørensen JG, Nielsen MM, Loeschcke V. Gene expression profile analysis of Drosophila melanogaster selected for resistance to environmental stressors. J Evol Biol. 2007;20:1624–1636. doi: 10.1111/j.1420-9101.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- 29.Reusch TBH, Wood TE. Molecular ecology of global change. Mol Ecol. 2007;16:3973–3992. doi: 10.1111/j.1365-294X.2007.03454.x. [DOI] [PubMed] [Google Scholar]
- 30.Oleksiak MF, Roach JL, Crawford DL. Natural variation in cardiac metabolism and gene expression in Fundulus heteroclitus. Nat Genet. 2005;37:67–72. doi: 10.1038/ng1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stillman JH, Tagmount A. Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol Ecol. 2009;18:4206–4226. doi: 10.1111/j.1365-294X.2009.04354.x. [DOI] [PubMed] [Google Scholar]
- 32.Polato NR, et al. Location-specific responses to thermal stress in larvae of the reef-building coral Montastraea faveolata. PLoS ONE. 2010;5:e11221. doi: 10.1371/journal.pone.0011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres TT, Metta M, Ottenwälder B, Schlötterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bräutigam A, Gowik U. What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol (Stuttg) 2010;12:831–841. doi: 10.1111/j.1438-8677.2010.00373.x. [DOI] [PubMed] [Google Scholar]
- 35.Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8:125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank G, et al. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot. 2009;60:3891–3908. doi: 10.1093/jxb/erp234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauhan H, Khurana N, Tyagi AK, Khurana JP, Khurana P. Identification and characterization of high temperature stress responsive genes in bread wheat (Triticum aestivum L.) and their regulation at various stages of development. Plant Mol Biol. 2011;75:35–51. doi: 10.1007/s11103-010-9702-8. [DOI] [PubMed] [Google Scholar]
- 38.Tautz D, Ellegren H, Weigel D. Next generation molecular ecology. Mol Ecol. 2010;19(Suppl 1):1–3. doi: 10.1111/j.1365-294X.2009.04489.x. [DOI] [PubMed] [Google Scholar]
- 39.Schär C, et al. The role of increasing temperature variability in European summer heatwaves. Nature. 2004;427:332–336. doi: 10.1038/nature02300. [DOI] [PubMed] [Google Scholar]
- 40.Cheviron ZA, Whitehead A, Brumfield RT. Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Mol Ecol. 2008;17:4556–4569. doi: 10.1111/j.1365-294X.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- 41.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 42.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: An overview. Environ Exp Bot. 2007;61:199–223. [Google Scholar]
- 43.Kotak S, et al. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Aro EM, Virgin I, Andersson B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- 45.Giardi MT, et al. Long-term drought stress induces structural and functional reorganization of photosystem II. Planta. 1996;199:118–125. [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Usadel B, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003;6:1025–1037. [Google Scholar]
- 49.Camejo D, et al. High temperature effects on photosynthetic activity of two tomato cultivars with different heat susceptibility. J Plant Physiol. 2005;162:281–289. doi: 10.1016/j.jplph.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 50.Tóth SZ, et al. Biophysical studies of photosystem II-related recovery processes after a heat pulse in barley seedlings (Hordeum vulgare L.) J Plant Physiol. 2005;162:181–194. doi: 10.1016/j.jplph.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem. 2009;47:785–795. doi: 10.1016/j.plaphy.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Ouyang Y, Chen J, Xie W, Wang L, Zhang Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol. 2009;70:341–357. doi: 10.1007/s11103-009-9477-y. [DOI] [PubMed] [Google Scholar]
- 53.Hüve K, Bichele I, Rasulov B, Niinemets U. When it is too hot for photosynthesis: Heat-induced instability of photosynthesis in relation to respiratory burst, cell permeability changes and H₂O₂ formation. Plant Cell Environ. 2011;34:113–126. doi: 10.1111/j.1365-3040.2010.02229.x. [DOI] [PubMed] [Google Scholar]
- 54.Henkel SK, Hofmann GE. Differing patterns of hsp70 gene expression in invasive and native kelp species: Evidence for acclimation-induced variation. J Appl Phycol. 2008;20:915–924. [Google Scholar]
- 55.Pearson GA, Lago-Leston A, Mota C. Frayed at the edges: Selective pressure and adaptive response to abiotic stressors are mismatched in low diversity edge populations. J Ecol. 2009;97:450–462. [Google Scholar]
- 56.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 57.Degenkolbe T, et al. Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Mol Biol. 2009;69:133–153. doi: 10.1007/s11103-008-9412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vega-Arreguín JC, et al. Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics. 2009;10:299. doi: 10.1186/1471-2164-10-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Wang Y, Zhang Q, Qi Y, Guo D. Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics. 2009;10:465. doi: 10.1186/1471-2164-10-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, Weber AP. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics. 2011;12:227. doi: 10.1186/1471-2164-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Blaxter ML. Comparing de novo assemblers for 454 transcriptome data. BMC Genomics. 2010;11:571. doi: 10.1186/1471-2164-11-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson CL, Janssen T. The Timetree of Life. New York, NY: Oxford Univ Press; 2009. pp. 203–212. [Google Scholar]
- 63.Wang Y, Lin X, Dong B, Wang Y, Liu B. DNA methylation polymorphism in a set of elite rice cultivars and its possible contribution to inter-cultivar differential gene expression. Cell Mol Biol Lett. 2004;9:543–556. [PubMed] [Google Scholar]
- 64.Rapp RA, Wendel JF. Epigenetics and plant evolution. New Phytol. 2005;168:81–91. doi: 10.1111/j.1469-8137.2005.01491.x. [DOI] [PubMed] [Google Scholar]
- 65.Winters G, Nelle P, Fricke B, Rauch G, Reusch TBH. The effects of a simulated heat wave on the photophysiology and gene expression of high and low-latitude populations of Zostera marina. Mar Ecol Prog Ser. 2011;435:83–95. [Google Scholar]
- 66.Pimm SL. The Balance of Nature? Chigago, IL: Univ of Chicago Press; 1991. [Google Scholar]
- 67.Chevreux B, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 69.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008;36(Suppl 1):D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 71.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D'haeseleer P. How does gene expression clustering work? Nat Biotechnol. 2005;23:1499–1501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- 73.De Cáceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.