Abstract
The tripartite motif (TRIM)-containing proteins are a family of proteins that have been known to be involved in divergent biological processes, including important roles in immune responses through regulating various signaling pathways. In this study, we identified a member of the TRIM family, TRIM8, as a positive regulator of tumor necrosis factor-α (TNFα) and interleukin-1β (IL-1β)–triggered NF-κB activation. Overexpression of TRIM8 activated NF-κB and potentiated TNFα- and IL-1β–induced activation of NF-κB, whereas knockdown of TRIM8 had opposite effects. Coimmunoprecipitations indicated that TRIM8 interacted with TGFβ activated kinase 1 (TAK1), a serine/threonine kinase essential for TNFα- and IL-β–induced NF-κB activation. Furthermore, we found that TRIM8 mediated K63-linked polyubiquitination of TAK1 triggered by TNFα and IL-1β. Our findings demonstrate that TRIM8 serves as a critical regulator of TNFα- and IL-1β–induced NF-κB activation by mediating K63-linked polyubiquitination of TAK1.
The transcription factor NF-κB plays a pivotal role in many cellular events such as cell proliferation, inhibition of apoptosis, and innate immunity. NF-κB activation requires the signal-induced phosphorylation and degradation of IκB proteins (1). The kinase that phosphorylates IκB, termed IκB kinase (IKK) complex, consists of the catalytic subunits IKKα, IKKβ, and the regulatory subunit NEMO/IKKγ. The IKK complex is activated by a large variety of stimuli, including the proinflammatory cytokines tumor necrosis factor α (TNFα) and interleukin-1β (IL-1β) (2).
Binding of TNFα to its receptor TNF-RI results in recruitment of the adaptor protein TRADD. TRADD further recruits TRAF2, TRAF5, and RIP1 to the receptor complex. TRAF2 mediates polyubiquitination of RIP1, mainly in a K63-linked manner. Ubiquitinated RIP further recruits TAB2 and TAK1 and mediates activation of the latter. The activated RIP1–TAK1–TAB2 complex subsequently activates the IKK complex, leading to phosphorylation of IκBα and activation of NF-κB (3). Similarly, binding of IL-1β to its receptor IL-1R leads to the recruitment of adaptor proteins and kinases, including MyD88, IRAK1, and IRAK4. IRAK4 phospholylates and activates IRAK1, which in turn recruits TRAF6. IRAK1 and TRAF6 form a complex that is released from the receptor. TRAF6 possesses an E3 ubiquitin ligase activity that mediates its K63-linked autopolyubiquitination. Ubiquitinated TRAF6 further recruits the TAK1–TAB2–TAB3 complex, resulting in the activation of TAK1. Activated TAK1 eventually phosphorylates and activates IKK, leading to activation of NF-κB (3, 4).
TAK1 is a member of the MAPK kinase kinase family that was originally found to function in the transforming growth factor β (TGFβ)-mediated MAPK activation (5). TAK1 has been demonstrated to be essential in TNFα- and IL-1β–mediated activation of NF-κB and JNK (6, 7). In all of these pathways, activation of TAK1 is an important event that transmits the upstream signal from the receptor complex to the downstream signaling molecules (3, 4, 6, 7).
So far, several reports have suggested that polyubiquitination of TAK1 is involved in the regulation of TAK1-mediated signaling pathways. For example, TRAF6-mediated K63-linked polyubiquitination of TAK1 at K34 residue has been shown to correlate with TAK1 activation in TGFβ signaling (8). It has also been demonstrated that Helicobacter pylori CagA activates NF-κB by targeting TAK1 for TRAF6-mediated polyubiquitination (9). However, the molecular regulation of TAK1 in TNFα- and IL-1β–induced signaling pathways remains unclear.
The tripartite motif (TRIM)-containing proteins play divergent roles in many biological processes, including important roles in immune responses through regulating various signaling pathways. It has been reported that several TRIM proteins are up-regulated during immune responses, such as TRIM5α and TRIM19/PML, which have been shown to restrict viral replication (10).
In this study, we identified a member of the TRIM family, TRIM8, as a positive regulator of TNFα- and IL-1β–triggered NF-κB activation. Furthermore, we found that TRIM8 targeted TAK1 for K63-linked polyubiquitination at K158, which was essential for TNFα- and IL-1β–triggered NF-κB activation. These findings reveal a regulatory mechanism of NF-κB activation by proinflammatory cytokines such as TNFα and IL-1β.
Results
Identification of TRIM8 as a Positive Regulator of TNFα- and IL-1β–Triggered Signaling Pathways.
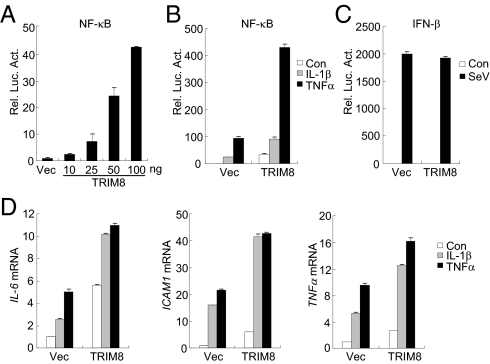
Ubiquitination is critically involved in regulation of several key components of the TNFα- or IL-1β–triggered NF-κB pathways (3). To identify additional enzymes that regulate NF-κB activation, we screened a cDNA array containing 352 expression clones of ubiquitin-related enzymes by NF-κB reporter assays. These experiments identified TRIM8. As shown in Fig. 1A, overexpression of TRIM8 activated NF-κB in a dose-dependent manner. Because NF-κB could be activated by various stimuli, we examined whether TRIM8 is involved in TNFα- or IL-1β–triggered NF-κB activation. The results indicated that TRIM8 potentiated TNFα- and IL-1β–induced activation of NF-κB in reporter assays (Fig. 1B). In reporter assays, TRIM8 did not potentiate Sendai virus (SeV)-triggered activation of the IFNβ promoter (Fig. 1C), suggesting that the regulation of TRIM8 in TNFα- or IL-1β–induced NF-κB activation is specific. Real-time PCR experiments also indicated that TRIM8 could potentiate TNFα- and IL-1β–induced expression of NF-κB downstream genes, such as IL-6, ICAM1, and TNFα (Fig. 1D). These data suggest that TRIM8 is involved in TNFα- and IL-1β–induced NF-κB activation.
Fig. 1.
Identification of TRIM8 as a positive regulator of TNFα- and IL-1β–triggered NF-κB and transcription induction of inflammatory genes. (A) Overexpression of TRIM8 activates NF-κB in a dose-dependent manner. A total of 293 cells (1 × 105) were transfected with NF-κB reporter (0.01 μg) and an increased amount of TRIM8 expression plasmid. Luciferase assays were performed 20 h after transfection. (B) Effects of TRIM8 on TNFα- and IL-1β–induced activation of NF-κB. A total of 293 cells (1 × 105) were transfected with NF-κB reporter (0.01 μg) and empty vector or TRIM9 expression plasmid. Twenty hours after transfection, cells were left untreated or stimulated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for 10 h before luciferase assays were performed. (C) TRIM8 is not involved in SeV-induced activation of the IFNβ promoter. Reporter assays were performed as described in B, except that the IFNβ promoter reporter plasmid was used. (D) TRIM8 potentiates TNFα- and IL-1β–induced transcription of endogenous IL-6, ICAM1, and TNFα genes. A total of 293 cells (2 × 105) were transfected with an empty vector or TRIM8 expression plasmid (0.1 μg). Twenty hours after transfection, cells were left untreated or stimulated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for 10 h before quantitative RT-PCR was performed.
Knockdown of TRIM8 Inhibits TNFα and IL-1β–Induced NF-κB Activation.
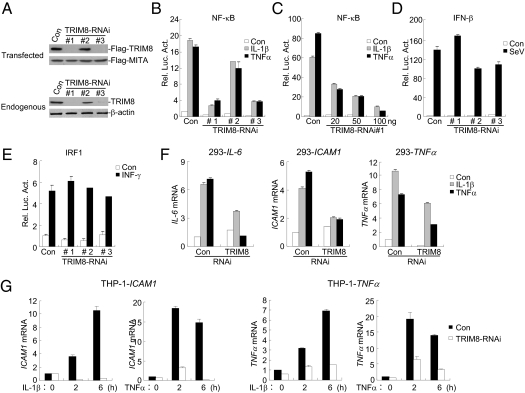
Because overexpression of TRIM8 potentiated TNFα- and IL-1β–induced NF-κB activation, we next determined whether endogenous TRIM8 regulates TNFα- and IL-1β–induced signaling. To do this, we constructed three human TRIM8 RNAi plasmids. Immunoblot analysis indicated that the #1 and #3 RNAi plasmids could markedly inhibit the expression of transfected and endogenous TRIM8 in 293 cells, whereas the #2 RNAi plasmid had little effect on TRIM8 expression (Fig. 2A). We transfected the TRIM8 RNAi plasmids into 293 cells and performed reporter assays. As shown in Fig. 2B, knockdown of TRIM8 expression inhibited TNFα- and IL-1β–induced NF-κB activation. The degrees of inhibition were correlated with the efficiencies of knockdown of TRIM8 by each RNAi plasmid (Fig. 2 A and B). We selected the #3 TRIM8-RNAi plasmid for additional experiments described below, and similar results were obtained with the #1 plasmid. Further experiments indicated that TRIM8-RNAi inhibited TNFα- and IL-1β–induced activation of the NF-κB promoter in a dose-dependent manner in 293 cells (Fig. 2C). However, knockdown of TRIM8 could not inhibit SeV-induced activation of the IFNβ promoter (Fig. 2D) and IFNγ-induced activation of the IRF1 promoter (Fig. 2E). Consistently, quantitative real-time PCR experiments indicated that knockdown of TRIM8 inhibited TNFα- and IL-1β–induced expression of endogenous IL-6, ICAM1, and TNFα genes in 293 cells (Fig. 2F). We further investigated whether TRIM8 plays a relevant role in inflammatory cells. The results indicated that knockdown of TRIM8 potently inhibited transcription of ICAM1 and TNFα genes in monocytic/macrophage THP1 (Fig. 2G) and U937 (Fig. S1) cells. Taken together, these data suggest that TRIM8 is required for TNFα- and IL-1β–triggered NF-κB activation and induction of inflammatory proteins.
Fig. 2.
Knockdown of TRIM8 inhibits TNFα- and IL-1β–induced NF-κB activation. (A) Effects of TRIM8 RNAi plasmids on expression of TRIM8. (Upper) A total of 293 cells (2 × 105) were transfected with Flag-TRIM8 (0.2 μg each), Flag-MITA (0.05 μg), and the indicated RNAi (1 μg) plasmids. Thirty-six hours later, immunoblot analysis was performed with anti-Flag. (Lower) A total of 293 cells (2 × 105) were transfected with the indicated RNAi plasmids (1 μg) for 48 h, and then immunoblot analysis was performed with the indicated antibodies. (B) Effects of TRIM8 RNAi plasmids on TNFα- and IL-1β–induced NF-κB activation. A total of 293 cells (1 × 105) were transfected with NF-κB reporter (0.01 μg) and the indicated RNAi plasmids. Thirty-six hours after transfection, cells were left untreated or stimulated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for 10 h before luciferase assays were performed. (C) TRIM8-RNAi inhibits TNFα- and IL-1β–induced activation of the NF-κB in a dose-dependent manner. A total of 293 cells (1 × 105) were transfected with NF-κB reporter (0.01 μg) and an increased amount of the #1 TRIM8-RNAi plasmid. Reporter assays were performed as described in B. (D and E) Effects of TRIM8 RNAi on SeV-triggered activation of the IFNβ promoter (D) or IFNγ-induced activation of the IRF1 promoter (E). A total of 293 cells (1 × 105) were transfected with the indicated reporter (0.05 μg) and TRIM-RNAi plasmids (0.5 μg) for 36 h. Cells were then subjected to the indicated stimulation for 8 h before luciferase assays were performed. (F) Effects of TRIM8 RNAi on TNFα- or IL-1β–induced transcription of endogenous IL-6, ICAM1, and TNFα genes in 293 cells. These 293 cells (2 × 105) were transfected with the indicated RNAi plasmids (2 μg). Thirty-six hours after transfection, cells were left untreated or stimulated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for 10 h before RT-PCR experiments were performed. (G) Effects of TRIM8 RNAi on TNFα- or IL-1β–induced transcription of endogenous ICAM1 and TNFα genes in THP-1 cells. THP-1 cells were transduced with GFP control or TRIM8 RNAi by retrovirus-mediated gene transfer. The cells were then either untreated or treated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for the indicated times before RT-PCR experiments were performed.
Mutant of TRIM8 Inhibits TAK1- but Not p65-Mediated NF-κB Activation.
We next examined the position of TRIM8 in TNFα- and IL1-β–induced signaling pathways. To do this, we constructed five TRIM8 truncation mutants and detected their abilities to activate NF-κB or regulate TNFα- and IL-1β–induced NF-κB activation. TRIM8 contains an N-terminal RING finger, followed by two B-boxes and a coil-coil domain. The C terminus of TRIM8 lacks a recognizable structural domain. The truncation mutants that we constructed are the following: TRIM8-RBC, which contains the RING finger, B-boxes, and the coil-coil domain; TRIM8-BC, which consists of the B-boxes and the coil-coil domain; TRIM8-ΔR, which lacks the RING finger; TRIM8-R, which contains only the RING finger; and TRIM-CT, which consists of the C terminus of TRIM8 (Fig. S2A). In reporter assays, full-length TRIM8 activated NF-κB and potentiated TNFα- and IL-1β–induced NF-κB activation. TRIM8-RBC, TRIM-BC, TRIM8-ΔR, and TRIM8-CT had minimal effect on NF-κB activation by themselves but retained their ability to potentiate TNFα- or IL-1β–induced NF-κB activation. However, the TRIM-R mutant completely lost the ability to activate NF-κB and potently inhibited TNFα- and IL-1β–induced NF-κB activation, suggesting that TRIM8-R acts as a dominant negative mutant to inhibit TNFα- or IL-1β–induced NF-κB activation (Fig. S2B).
To determine the position of TRIM8 in TNFα- and IL1-β–induced signaling pathways, we examined the effects of TRIM8-R on NF-κB activation mediated by various signaling molecules involved in these pathways. As shown in Fig. S2C, TRIM8-R inhibited NF-κB activation mediated by overexpression of TNF-R1, IL-1R, TRADD, FADD, TRAF2, TRAF6, RIP, TAK1, or IKKβ but not p65, suggesting that TRIM8 regulates TNFα- and IL1-β–induced signaling at a step between TAK1 and p65.
TRIM8 Interacts with TAK1.
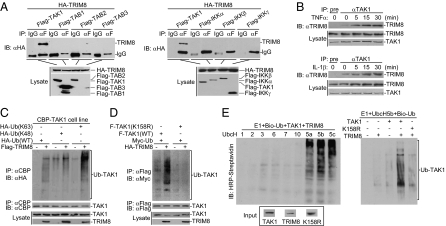
To further investigate the regulatory mechanism of TRIM8 in TNFα- and IL-1β–triggered signaling pathways, we examined the ability of TRIM8 to interact with different signaling components in these pathways. The results of transient transfection and coimmunoprecipitation experiments indicated that TRIM8 interacted with TAK1 but not with TAB1, TAB2, TAB3, IKKα, IKKβ, or IKKγ (Fig. 3A and Fig. S3A). We further performed endogenous coimmunoprecipitation experiments, and the results indicated that TRIM8 was weakly associated with TAK1 in unstimulated cells and that this association was increased after stimulation with TNFα or IL-1β (Fig. 3B and Fig. S3B). These results suggest that TNFα or IL-1β stimulation promotes endogenous association of TRIM8 with TAK1.
Fig. 3.
TRIM8 interacts with and ubiquitinates TAK1. (A) TRIM8 interacts with TAK1 in the mammalian overexpression system. A total of 293 cells (2 × 106) were transfected with the indicated plasmids (5 μg each), and coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies (Upper panels). Expression of the transfected proteins was analyzed by immunoblots with anti-HA and anti-Flag (Lower panels). (B) Endogenous TRIM8 is associated with TAK1 following TNFα or IL-1β stimulation. A total of 293 cells (2 × 107) were left untreated or stimulated with TNFα or IL-1β for the indicated times. Immunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (C) Overexpressed TRIM8 enhances wild-type and K63-linked polyubiquitination of TAK1. A total of 293 cells (2 × 106) stably expressing calmodulin-binding peptide (CBP)-tagged TAK1 were transfected with Flag-TRIM8 and the indicated ubiquitin plasmids. Twenty-four hours after transfection, cell lysates were immunoprecipitated with anti-CBP. The immunoprecipitates were analyzed by immunoblots with anti-HA, anti-TAK1, or anti-Flag as indicated. (D) TRIM8 enhances polyubiquitination of wild-type TAK1 but not of TAK1(K158R) in vivo. A total of 293 cells (2 × 106) were transfected with the indicated plasmids. Twenty-four hours after transfection, cell lysates were immunoprecipitated with anti-Myc. The immunoprecipitates were analyzed by immunoblots with anti-HA or anti-Flag as indicated. (E) TRIM8 enhances polyubiquitination of wild-type TAK1 but not of TAK1(K158R) in vitro. TAK1, TAK1(K158R), and TRIM8 were translated in vitro, and biotin–ubiquitin, E1, and the indicated E2s were added for ubiquitination assays. Ubiquitin-conjugated proteins were detected by immunoblot with HRP–streptavidin. Before ubiquitination analysis, the input levels of the translated proteins were detected by immunoblots.
TRIM8 Enhances Polyubiquitination of TAK1.
Because some members of the TRIM family are demonstrated to have E3 ubiquitin ligase activity, we determined whether TRIM8 has any effects on polyubiquitination of TAK1. As shown in Fig. 3C, overexpression of TRIM8 enhanced polyubiquitination of TAK1 in 293 cells. Furthermore, overexpression of TRIM8 potently promoted K63-linked polyubiquitination of TAK1 but had only a minimal effect on K48-linked polyubiquitination of TAK1 (Fig. 3C). Recently, it has been demonstrated that polyubiquitination of the K158 residue of TAK1 is required for TNFα- and IL-1β–induced NF-κB activation (11, 12). Interestingly, TRIM8 failed to mediate polyubiquitination of TAK1(K158R) in 293 cells (Fig. 3D). In vitro ubiquitination assays with UbcH5b as an E2 indicated TRIM8 could ubiquitinate TAK1 but not TAK1(K158R) (Fig. 3E). These results suggest that TRIM8 could mediate K63-linked polyubiquitination of TAK1 at residue K158. Interestingly, TAK1(K158R), in the presence of TAB1, significantly inhibited TNFα- and IL-1β–induced NF-κB activation (Fig. S4).
Knockdown of TRIM8 Reduces TNFα- and IL-1β–Induced K63 but Not K48-Linked Polyubiquitination of Endogenous TAK1.
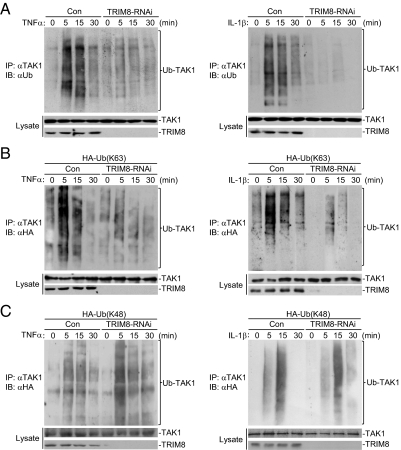
To determine whether endogenous TRIM8 is required for TNFα- and IL-1β–induced TAK1 polyubiquitination, we generated TRIM8 stable knockdown in 293 cell lines. Immunoprecipitation experiments indicated that knockdown of TRIM8 reduced both TNFα- and IL-1β–induced TAK1 polyubiquitination (Fig. 4A). We next determined whether endogenous TRIM8 mediates K48- or K63-linked polyubiquitination. Knockdown of TRIM8 reduced TNFα- or IL-1β–induced K63-linked (Fig. 4B), but not K48-linked (Fig. 4C), polyubiquitination of TAK1. These results suggest that TRIM8 mediates K63-linked polyubiquitination of endogenous TAK1 triggered by TNFα and IL-1β.
Fig. 4.
TRIM8-RNAi reduces TNFα- and IL-1β–induced K63-linked polyubiquitination of endogenous TAK1. (A) Effects of TRIM8 RNAi on TNFα- or IL-1β–induced polyubiquitination of endogenous TAK1. A total of 293 cells stably transduced with a control or TRIM8 RNAi were either untreated or stimulated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for the indicated times. Cell lysates were immunoprecipitated with anti-TAK1, and the immunoprecipitates were analyzed by immunoblots with anti-ubiquitin, anti-TRIM8, or anti-TAK1 as indicated. (B and C) Effects of TRIM8 RNAi on TNFα- or IL-1β–induced K63-linked (B) or K48-linked (C) polyubiquitination of endogenous TAK1. A total of 293 cells stably transduced with a control or TRIM8 RNAi were transfected with HA-tagged K63-only or K48-only ubiquitin plasmid. Twenty-four hours after transfection, cell lysates were immunoprecipitated with anti-TAK1. The immunoprecipitates were analyzed by immunoblots with anti-HA, anti-TAK1, and anti-TRIM8 as indicated.
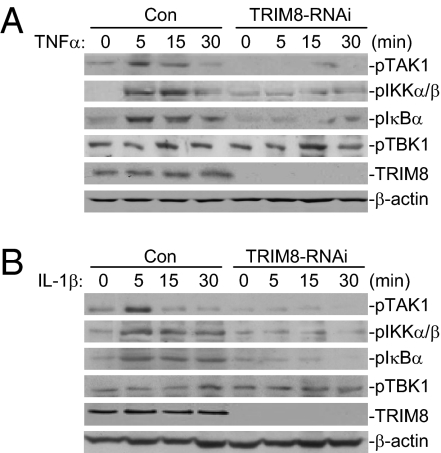
Knockdown of TRIM8 Reduces TNFα- and IL-1β–Induced Phosphorylation of TAK1, IKKα/β, and IκBα.
Previously, it has been demonstrated that phosphorylation of TAK1, IKKα, and IKKβ is required for their activation by TNFα or IL-1β (1–3). Therefore, we further analyzed the effects of TRIM8 knockdown on TNFα- and IL-1β–induced phosphorylation of TAK1 and IKK. As shown in Fig. 5, both TNFα and IL-1β–induced transient phosphorylation of TAK1 and IKK peaked at ∼5 min after cytokine stimulation, and the phosphorylation was inhibited and delayed in TRIM8 knockdown cells. Consistently, TNFα- or IL-1β–induced phosphorylation and degradation of IκBα, a downstream target of IKK, also was diminished in TRIM8 knockdown cells (Fig. 5 and Fig. S5). In these experiments, knockdown of TRIM8 had no inhibitory effect on basal phosphorylation of TBK1 (Fig. 5), indicating that TRIM8 did not affect global phosphorylation. These results suggest that TRIM8 is important for TNFα- and IL-1β–induced activation of TAK1 and IKK.
Fig. 5.
Knockdown of TRIM8 reduces TNFα- and IL-1β–induced phosphorylation of TAK1, IKKα/β, and IκBα. (A and B) TRIM8-RNAi inhibits TNFα-induced phosphorylation of TAK1, IKKα/β, IKBα but not of TBK1. A total of 293 cells stably transduced with a control or TRIM8 RNAi plasmid were either untreated or treated with TNFα (10 ng/mL) (A) or IL-1β (10 ng/mL) (B) for the indicated times and then subsequently analyzed by immunoblots with the indicated antibodies.
TRIM8 Is Involved in TNFα- and IL-1β–Induced AP1 but Not in ELK1 and CHOP Activation.
In addition to NF-κB, TNFα and IL-1β also activate the transcription factors AP1, ELK1, and CHOP via the serine/threonine kinases JNK, ERK1/2, and p38, respectively. In reporter assays, TRIM8 activated AP1 and potentiated TNFα- and IL-1β–induced AP1 activation (Fig. S6A). Knockdown of TRIM8 inhibited TNFα- and IL-1β–induced phosphorylation of JNK in THP1 and U937 cells. In contrast, overexpression or knockdown of TRIM8 had no marked effects on TNFα- and IL-1β–induced ELK1 and CHOP activation in reporter assays (Fig. S6B). These results suggest that TRIM8 is also involved in TNFα- and IL-1β–induced AP1 but not in ELK1 and CHOP activation.
Discussion
The transcription factor NF-κB plays critical roles in a broad range of physiological and pathological processes, such as inflammation, immune regulation, and cell survival. TNFα and IL-1β are two cytokines that induce inflammation through activation of NF-κB. In this study, we performed expression screens of ubiquitin-related enzymes for their abilities to regulate NF-κB activity. This effort identified TRIM8, a TRIM family protein, as an important regulatory component in the TNFα- and IL-1β–induced NF-κB activation pathways.
Overexpression of TRIM8 activated NF-κB in reporter assays. Knockdown of TRIM8 or a TRIM8 mutant inhibited TNFα- and IL-1β–induced NF-κB activation. These results suggest that TRIM8 is important for TNFα- and IL-1β–induced NF-κB activation. Our results also suggest that TRIM8 is important for TNFα- and IL-1β–induced AP1 but not for ELK1 and CHOP activation. These findings suggest that TRIM8 is differentially involved in TNFα- and IL-1β–induced signaling pathways.
TAK1 is a serine/threonine protein kinase involved in both TNFα- and IL-1β–induced NF-κB activation pathways. A TRIM8 mutant inhibited NF-κB activation mediated by overexpression of TAK1 as well as its upstream components but not downstream p65. Stimulation of cells with TNFα and IL-1β led to increased association of TRIM8 with TAK1. These results suggest that TRIM8 targets TAK1 in the TNFα- and IL-1β–induced pathways.
Previously, it has been demonstrated that TRAF6-mediated polyubiquitination of TAK1 at K158 is important for TNFα- and IL-1β–induced NF-κB activation (12, 13). In our experiments, we found that overexpression of TRIM8 caused increased K63-linked, but not K48-linked, polyubiquitination of TAK1, whereas knockdown of TRIM8 diminished TNFα- and IL-1β–induced K63-linked polyubiquitination of endogenous TAK1. In addition, knockdown of TRIM also inhibited TNFα- and IL-1β–induced phosphorylation of TAK1 as well as of the downstream components IKK and IκBα. These results suggest that TRIM8 is involved in TNFα- and IL-1β–induced NF-κB activation by mediating K63-linked TAK1 polyubiquitination and subsequent activation. Whether TRAF6 and TRIM8 collaborate or play redundant roles in this process is unknown at this point. It is also possible that TRAF6 and TRIM8 are differentially required for TAK1 activation in a temporal manner or different cell types.
The identification of TRIM8 as an important regulator of the TNFα- and IL-1β–induced NF-κB and AP1 activation pathways sheds light on the complicated regulatory processes of inflammation and immune regulation. Because TAK1 is also involved in signaling pathways mediated by certain pattern recognition receptors (14, 15), it is possible that TRIM8 has a broader role in innate immune responses. Nevertheless, our study provides evidence for a role of TRIM8 in signaling by the proinflammatory cytokines TNFα and IL-1β.
Materials and Methods
Reagents and Antibodies.
Recombinant TNFα, IL-1β, and IFNγ (R&D Systems); mouse monoclonal antibodies against FLAG (Sigma), HA (Origene), β-actin (Sigma), phosphorylated-TAK1, and IKKα/β; and mouse monoclonal antibodies against pIκBα, pTAK1, pTBK1, and pJNK (Cell Signaling) were purchased from the indicated companies. Rabbit anti-TAK1 was raised against recombinant human full-length TAK1 protein, and mouse anti-IKBα was raised against recombinant human full-length IKBα protein.
Constructs.
NF-κB, IRF1, AP1, and IFNβ promoter luciferase reporter plasmids and mammalian expression plasmids for IL-1R, IL1Racp, TRAF6, TAK1, TAB1, IKKβ, TNF-R1, RIP, TRAF2, TRADD, FADD, and p65 were previously described (16–18). pFB, pFA2-ELK1, and pFA2-CHOP plasmids in the Trans-Reporter Systems for ELK1 and CHOP were purchased from Stratagene. Mammalian expression plasmids for HA- or FLAG-tagged TRIM8 and its mutants were constructed by standard molecular biology techniques.
Expression Cloning.
The cDNA expression clones encoding 352 ubiquitin-related enzymes were obtained from the Ubiquitin-GFC Transfection Array purchased from Origene (catalog #UBGB19601). The clones were transfected together with an NF-κB luciferase reporter plasmid into 293 cells. Reporter assays were performed 16 h after transfection to identify clones that could markedly activate the NF-κB reporter.
Transfection and Reporter Assays.
A total of 293 cells (1 × 105) were seeded on 24-well dishes and transfected the following day by a standard calcium phosphate precipitation method. Empty control plasmid was added to ensure that each transfection receives the same amount of total DNA. To normalize for transfection efficiency, 0.05 μg of pRL-TK (Renilla luciferase) reporter plasmid was added to each transfection. Approximately 18 h after transfection, luciferase assays were performed using a dual-specific luciferase assay kit (Promega). Firefly luciferase activities were normalized on the basis of Renilla luciferase activities.
Real-Time PCR.
Total RNA was isolated from cells using TRIzol reagent (TAKARA), reverse transcribed, and subjected to real-time PCR analysis to measure mRNA expression levels of tested genes. Gene-specific primer sequences were as follows: IL-6—TTCTCCACAAGCGCCTTCGGTC (forward) and TCTGTGTGGGGCGGCTACATCT (reverse); ICAM1—TCAGTGTGACCGCAGAGGACGA (forward) and TTGGGCGCCGGAAAGCTGTAGAT (reverse); TNFα—GCCGCATCGCCGTCTCCTAC (forward) and CCTCAGCCCCCTCTGGGGTC (reverse); and GAPDH—GACAAGCTTCCCGTTCTCAG (forward) and GAGTCAACGGATTTGGTCGT (reverse).
RNAi Experiments.
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the pSuper.Retro RNAi plasmid (Oligoengine). The following sequences were targeted for human TRIM8 cDNA: #1 (5′-GAAGGAATGAAATCCGGAA-3′), #2 (5′-TTGATAAGACGGAGGATGT-3′), and #3 (5′-TGAACGAAGTGGDDAAGAA-3′).
Coimmunoprecipitation and Immunoblot Analysis.
For transient transfection and coimmunoprecipitation experiments, 293 cells (1 × 106) were transfected for 18–24 h. The transfected cells were lysed in 1 mL of lysis buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton, 1 mM EDTA, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 mM phenylmethylsulfonyl fluoride]. For each immunoprecipitation, a 0.4-mL aliquot of the lysate was incubated with 0.5 μg of the indicated antibody or control IgG and 25 μL of a 1:1 slurry of Protein G Sepharose (GE Healthcare) for 2 h. Sepharose beads were washed three times with 1 mL of lysis buffer containing 0.5 M NaCl. The precipitates were analyzed by standard immunoblot procedures.
For endogenous coimmunoprecipitation experiments, 293 cells (5 × 107) were stimulated with TNFα (10 ng/mL) or IL-1β (10 ng/mL) for the indicated times or left untreated. The coimmunoprecipitation and immunoblot experiments were performed as described above.
In Vitro Ubiquitination Assays.
The tested proteins were expressed with a TNT Quick Coupled Transcription/Translation Systems kit (Promega) following instructions of the manufacturer. Ubiquitination was analyzed with a ubiquitination kit (Enzo Life Sciences) following protocols recommended by the manufacturer.
RNAi-Transduced Stable U937 and THP-1 Cells.
The 293 cells were transfected with two packaging plasmids, pGag-Pol and pVSV-G, and the GFP control or TRIM8 RNAi retroviral plasmid by calcium phosphate precipitation. Cells were washed 12 h after transfection and new medium without antibiotics was added for 24 h. The recombinant virus-containing medium was filtered and used to infect U937 or THP-1 cells in the presence of polybrene (4 μg/mL). The infected U937 or THP-1 cells were selected using puromycin (0.5 μg/mL) for 2 wk before additional experiments were performed.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2012CB910200) and the National Natural Science Foundation of China (30921001, 91029302, and 31170792).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110946108/-/DCSupplemental.
References
- 1.Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol. 2002;22:7158–7167. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi K, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 6.Ninomiya-Tsuji J, et al. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 8.Sorrentino A, et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 9.Lamb A, et al. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10:1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNab FW, Rajsbaum R, Stoye JP, O'Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23(1):46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Fan YH, et al. USP4 targets TAK1 to downregulate TNFalpha-induced NF-kappaB activation. Cell Death Differ. 2011;18:1547–1560. doi: 10.1038/cdd.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y, et al. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1 beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J Biol Chem. 2010;285:5347–5360. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Y, Yu Y, Mao R, Zhang H, Yang J. TAK1 Lys-158 but not Lys-209 is required for IL-1β-induced Lys63-linked TAK1 polyubiquitination and IKK/NF-κB activation. Cell Signal. 2011;23:660–665. doi: 10.1016/j.cellsig.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen SS, et al. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: Dependence on TRAF2 and TAK1. J Biol Chem. 2009;284:10774–10782. doi: 10.1074/jbc.M807272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Xu LG, Li LY, Shu HB. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J Biol Chem. 2004;279:17278–17282. doi: 10.1074/jbc.C400063200. [DOI] [PubMed] [Google Scholar]
- 17.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Zhong B, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.