Abstract
Timing of organ development during embryogenesis is coordinated such that at birth, organ and fetal size and maturity are appropriately proportioned. The extent to which local developmental timers are integrated with each other and with the signaling interactions that regulate morphogenesis to achieve this end is not understood. Using the absolute requirement for a signaling pathway activity (bone morphogenetic protein, BMP) during a critical stage of tooth development, we show that suboptimal levels of BMP signaling do not lead to abnormal morphogenesis, as suggested by mutants affecting BMP signaling, but to a 24-h stalling of the intrinsic developmental clock of the tooth. During this time, BMP levels accumulate to reach critical levels whereupon tooth development restarts, accelerates to catch up with development of the rest of the embryo and completes normal morphogenesis. This suggests that individual organs can autonomously control their developmental timing to adjust their stage of development to that of other organs. We also find that although BMP signaling is critical for the bud-to-cap transition in all teeth, levels of BMP signaling are regulated differently in multicusped teeth. We identify an interaction between two homeodomain transcription factors, Barx1 and Msx1, which is responsible for setting critical levels of BMP activity in multicusped teeth and provides evidence that correlates the levels of Barx1 transcriptional activity with cuspal complexity. This study highlights the importance of absolute levels of signaling activity for development and illustrates remarkable self-regulation in organogenesis that ensures coordination of developmental processes such that timing is subordinate to developmental structure.
Keywords: heterochrony, odontogenesis, molar, shrew
Teeth are ectodermal organs that develop by an increasingly well-characterized series of reciprocal epithelial–mesenchymal interactions. Unlike most other organs that undergo a single program of morphogenesis to generate the shape of the organ, mammalian teeth have different crown shapes according to their positions in the jaw. Tooth morphogenesis programs are thus spatially regulated to generate the different crown shapes that make up the different tooth types: molar, incisor, etc.
Early tooth development proceeds through a series of events that are common to all tooth types. The oral epithelium thickens, forms a bud that invaginates into the underlying neural crest-derived mesenchyme, and eventually grows into a cap by inward curving of the tip of the bud. The subsequent stage of tooth development, in which is set up the number of cusps of the tooth crown, and therefore the tooth type, is different for multicusped (e.g., molar) and unicuspid (e.g., incisor) teeth. Morphogenesis to form the cusps of the tooth crown involves folding of the epithelium, regulated by signals from organizing centers, the enamel knots (1, 2). To form enamel knots and begin crown morphogenesis, tooth primordia (buds), have an absolute requirement for a mesenchymal to epithelial bone morphogenetic protein (BMP) signal. Mice lacking this BMP signal, such as mice mutant for type 1a BMP receptor in the epithelium (3) or the homeobox transcription factor Msx1 in the mesenchyme (4), exhibit a permanent arrest of tooth development at the bud stage.
Msx1 is expressed in the condensing mesenchyme cells of all tooth buds (5) and regulates the expression of BMP4 (6, 7). This BMP signal regulates the expression of epithelial genes such as Shh (8), in cells that form a transient signaling center, the primary enamel knot, required to coordinate cuspal morphogenesis. The importance of BMP activity in the formation of the correct cusp pattern is suggested from mathematical modeling of signaling changes in cusp abnormalities observed in mutants affecting BMP signaling (9).
Barx1 is a Bar-family homeobox gene, which has a unique expression pattern during tooth development that is different to all of the other genes expressed in the early jaw primordia mesenchyme (10, 11). In the early ectomesenchyme, Barx1 expression is highly restricted to a small patch of cells that corresponds to the position where molar teeth develop. At the bud stage of tooth development, in common with other homeobox genes such as Msx1, Barx1 is expressed in condensing mesenchyme cells around the epithelial tooth buds, but unlike the other genes, Barx1 is only expressed in molar tooth primordia and is not expressed in incisors at any time during their development.
We investigated the function of Barx1 during molar tooth development and found that Barx1 genetically and physically interacts with Msx1 to up-regulate the levels of BMP activity that are critical for the bud-to-cap transition. Interestingly, a lack of Barx1 results neither in an arrest of tooth development, as would be expected from the phenotype of Msx1 mutants, nor in the formation of abnormal teeth, as suggested by previous results (12). In the Barx1 mutants, BMP4 transcription and BMP4 activity drop, impairing the bud-to-cap transition. However, we show that this decrease in BMP signaling arrests molar development for only 24 h and that molar tooth development restarts after BMP levels have accumulated and reached a threshold allowing bud-to-cap transition. Strikingly, following this stalling, molar tooth development accelerates and catches up with other structures of the developing embryo, to eventually produce perfectly formed molars. Our findings therefore represent a unique example of an organ that self-regulates its developmental timing to adjust it to the one of the whole embryo. Finally, we also present evidence showing that Barx1 is expressed during development of both molars and premolars, with a transcription level that correlates with the degree of cusp complexity exhibited by tooth crowns, suggesting that Barx1 is necessary to fine-tune BMP signaling in all multicusped teeth.
Results
Temporal Arrest of Molar Tooth Development.
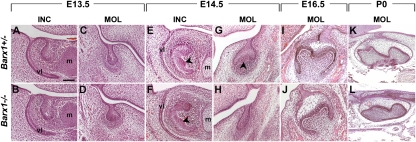
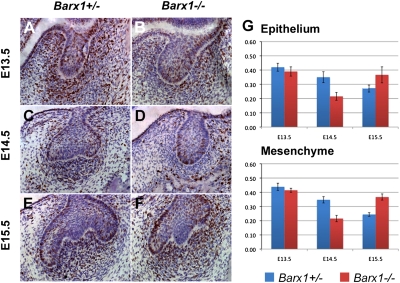
To analyze the role of Barx1 in molar tooth development, we generated mutant mice using gene targeting (Fig. S1) (13). Mutants die at birth most likely from cleft palate. Histological sections of jaws at birth in Barx1−/− animals showed all teeth to be present and at the appropriate stage (Fig. 1 K and L). At embryonic day 13.5 (E13.5), both molars and incisors had reached a bud stage in homozygous mutant animals (Fig. 1 B and D), similar to heterozygous animals (Fig. 1 A and C). At E14.5, when the molars had progressed to a cap stage (Fig. 1G), Barx1−/− molars were still at a bud stage, equivalent to E13.5 (Fig. 1H). This developmental arrest, which was fully penetrant (n > 10) and affected molars of all four quadrants, appeared to be molar specific, as Barx1−/− incisor tooth germs had reached a cap stage (Fig. 1 E and F). Between E13.5 and E14.5, tooth germs undergo the bud-to-cap transition that is regulated by signals such as BMP4 from the condensing mesenchyme to the epithelium. The main feature of this transition is the formation of the primary enamel knot signaling center that coordinates cusp formation. By E16.5, molar tooth development in Barx1−/− embryos looked almost identical to littermate controls (Fig. 1 I and J). First molar development in Barx1 mutant embryos thus undergoes a 24-h temporal arrest between E13.5 and E14.5. To begin to understand the basis of this temporal arrest, cell proliferation was assayed in mutants and heterozygous littermates at E13.5, E14.5, and E15.5. BrdU +ve and −ve cells were counted in serial sections through developing first molars. The changes in the numbers of BrdU +ve cells in epithelium and mesenchyme were similar, with a gradual decrease observed in controls between E13.5 and E15.5 (Fig. 2 A, C, E, and G). In Barx1−/− molar tooth germs, the number of BrdU +ve cells was less than controls at E13.5 and E14.5 (Fig. 2 B, D, and G) but at E15.5 it was considerably more (Fig. 2 F and G).
Fig. 1.
Temporal delay of molar tooth development in Barx1 homozygous mutants. Hematoxylin and eosin stained frontal (A–J) and sagittal (K and L) sections of lower E13.5 incisors (A and B) and first molars (C and D), E14.5 incisors (E and F), and first molars (G and H), E16.5 first molars (I and J), and postnatal day 0 (P0) first molars (K and L). At E13.5, all tooth germs have reached a bud stage both in the Barx1 homozygous mutant (B and D) and control littermate (A and C). Incisors develop normally in all Barx1 homozygous mutants, displaying a characteristic epithelial cap at E14.5 (E and F), whereas the molars of all four quadrants show a developmental delay between E13.5 and E14.5, exhibiting a bud shape instead of a cap (G and H) (n > 10). Arrowheads in E–G indicate the primary enamel knot, visualized as a bulge on the inside of the epithelial cap. At E16.5–P0, Barx1−/− molars are slightly smaller but otherwise normal (I–L). m, Meckel's cartilage; vl, vestibular lamina. (Scale bar, 100 μm in A–J and 200 μm in K and L.)
Fig. 2.
Barx1 mutant molar teeth exhibit changes in cell proliferation. (A–F) BrdU staining of frontal sections of developing first molar tooth germs at E13.5 (A and B), E14.5 (C and D), and E15.5 (E and F) in a Barx1+/− (A, C, and E) and Barx1−/− (B, D, and F) lower jaw. (G) Graphs comparing the numbers of BrdU-labeled cells in the epithelium and the condensed mesenchyme of developing lower first molars at E13.5, E14.5, and E15.5. Error bars show SD.
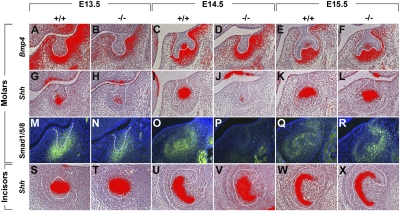
The bud-to-cap transition requires a mesenchyme-to-epithelium BMP4 signal that is responsible for directing epithelial cell differentiation and the formation of the primary enamel knot that is visible at the cap stage. We thus assayed expression of BMP4 as well as BMP signaling activity in the Barx1−/− molar tooth germs. In situ hybridization for BMP4 showed expression to be reduced in Barx1−/− E13.5 molar buds (Fig. 3 A and B) but by E14.5–E15.5, BMP4 expression was similar in mutant tooth mesenchyme (Fig. 3 D and F) and littermate controls (Fig. 3 C and E). Phospho-Smad1/5/8 immunohistochemistry was used to reveal the levels of BMP activity. At E13.5, phospho-Smad1/5/8 immunoreactivity was clearly reduced in Barx1−/− compared with controls (Fig. 3 M and N). By E14.5, immmunoreactivity could not be detected in the arrested mutant tooth buds but was visible in the control cap stage buds (Fig. 3 O and P). At E15.5, reactivity in the mutant tooth germs at the late cap stage was greater than in controls (Fig. 3 Q and R). To investigate the formation of primary enamel knots, expression of Shh was followed using in situ hybridization. At E13.5, expression of Shh at the tip of the tooth buds appeared considerably decreased in mutants compared with WT littermates (Fig. 3 G and H). By E14.5, strong Shh expression could be seen in the primary enamel knots of controls but very little expression was detected in Barx1−/− arrested tooth buds (Fig. 3 I and J). At E15.5, similar levels of Shh expression were observed in mutant and control tooth germs (Fig. 3 K and L). In contrast, Shh expression appeared unchanged in Barx1−/− incisors compared with Barx1+/+ incisors at similar stages of tooth development (Fig. 3 S–X). Changes in epithelial Shh expression in developing Barx1−/− molar tooth germs thus followed those of mesenchymal BMP4 activity. BMP4 expression and BMP signaling activity followed the same pattern that paralleled the changes in cell proliferation and epithelial cell differentiation, suggesting that a reduction in BMP4 expression at the molar bud stage results in a decrease in proliferation, a delay in primary enamel knot formation, and a consequent temporary arrest in development.
Fig. 3.
BMP4 expression and BMP activity changes in Barx1 mutant tooth development. (A–R) Expression of Bmp4 (A–F), Shh (G–L), and distribution of phospho-Smad1/5/8 (M–R) in lower first molar tooth germs at E13.5 (A, B, G, H, M, and N), E14.5 (C, D, I, J, O, and P), and E15.5 (E, F, K, L, Q, and R) in a WT (A, C, E, G, I, K, M, O, and Q) and Barx1 homozygous mutant (B, D, F, H, J, L, N, P, and R) littermates. (S–X) Expression of Shh in lower incisor tooth germs at E13.5 (S and T), E14.5 (U and V), and E16.5 (W and X). In situ hybridization was carried out on four separate samples for each genotype at each time point and immunostaining on two separate samples for each genotype at each time point. The epithelium of molar and incisor tooth germs is outlined in white.
Genetic Interaction Between Barx1 and Msx1.
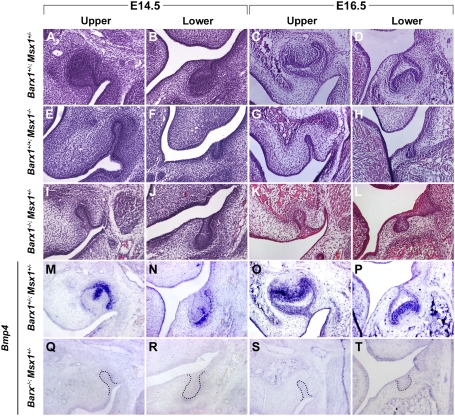
In Msx1 homozygous mutant embryos, tooth development is permanently arrested at the bud stage as a result of loss of BMP4. Because Barx1−/− molar tooth germs also showed a reduction in BMP signaling and arrest at the bud stage, we crossed Barx1 mutants with Msx1 mutants to identify any genetic interaction between these transcriptional regulators. Msx1+/− mice are normal but when combined with a Barx1−/− background, rather than molar tooth development showing the temporal arrest (Barx1−/− phenotype), molar tooth development was permanently arrested at the bud stage (Fig. 4 I–L), as observed in Msx1−/− (Fig. 4 E–H). As expected, development of Msx1+/−; Barx1−/− incisors proceeded normally because Barx1 expression is restricted to molars. Loss of a single allele of Barx1 and Msx1 had no effect on molar tooth development (Fig. 4 A–D). Thus, loss of a single allele of Msx1 on a Barx1 null background converts the temporal arrest of molar development into a permanent failure of development.
Fig. 4.
Arrest of molar tooth development associated with a lack of Bmp4 transcription in Barx1/Msx1 compound mutants. Frontal sections through upper (A, C, E, G, I, K, M, O, Q, and S) and lower (B, D, F, H, J, L, N, P, R, and T) developing first molars at E14.5 (A, B, E, F, I, J, M, N, Q, and R) and E16.5 (C, D, G, H, K, L, O, P, S, and T). Hematoxylin and eosin stained sections of Barx1+/−; Msx1+/− (A–D), Barx1+/+; Msx1−/− (E–H), and Barx1−/−; Msx1+/− (I–L). (M–T) Expression of BMP4 in the condensed mesenchyme of first molar tooth germs of Barx1+/−; Msx1+/− (M–P) and Barx1−/−; Msx1+/− (Q–T). Permanent arrest of molar tooth development was observed in three separate Barx1−/−; Msx1+/− animals and was highly penetrant. At E16.5, one molar tooth germ (n = 1/12) was occasionally observed at the cap stage (corresponding to E14.5).
In situ hybridization for BMP4 in the Barx1−/−; Msx1+/− molar tooth germs revealed a complete loss of expression at the bud stage (Fig. 4 Q–T). Loss of a single allele of Msx1 on a wild-type background does not affect expression of BMP4 similar to a loss of a single allele of Msx1 and Barx1 (Fig. 4 M–P). In Barx−/− molar tooth buds, BMP4 expression is reduced. A single allele of Msx1 on the Barx1−/− background reduces BMP4 expression to undetectable levels. Barx1 and Msx1 thus genetically interact to regulate the levels of BMP4 expression during molar tooth development.
Barx1–Msx1 Protein Interactions.
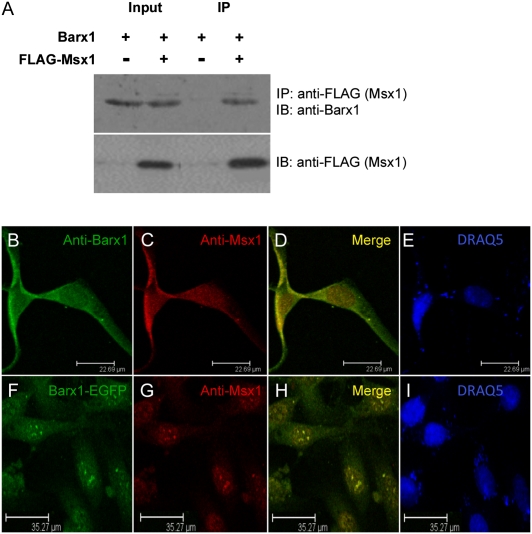
To investigate whether the genetic interaction between Msx1 and Barx1 can be reproduced in living cells as a physical protein–protein interaction, we performed coimmunoprecipitation assays in C3H10T1/2, a pluripotent embryonic mesenchymal cell line. We expressed exogenously constructs encoding Barx1 as a fusion protein with EGFP and Msx1 as a FLAG-tagged fusion protein. Barx1 protein was detected in the anti-FLAG immunoprecipitate from cells cotransfected with FLAG–Msx1, but not from cells cotransfected with empty vector (Fig. 5A). As control, an equal protein level of Barx1 was present in both input samples (20% input).
Fig. 5.
Msx1 interacts with molar tooth-specific transcription factor Barx1. (A) Msx1 interacts with Barx1 in living cells. C3H10T1/2 cells were cotransfected with pIRES2–Barx1–EGFP and either pCMV–FLAG–Msx1 or pCMV–FLAG–Tag2B empty control vector. Cell lysates were subjected to coimmunoprecipitations followed by Western blotting. Barx1 was detected only in the presence of FLAG–Msx1 in the IP sample. IP, immunoprecipitation; IB, immunoblotting. (B–I) Intracellular colocalization of Barx1 and Msx1 in C3H10T1/2 cells. (B–E) Intracellular colocalization of endogenously expressed Barx1 and Msx1. (B) Intracellular localization of Barx1 using anti-Barx1 (green); (C) intracellular localization of Msx1, using anti-Msx1 (red); (D) merged pictures showing intracellular colocalization of Barx1 and Msx1 (yellow); and (E) DNA staining using the fluorescence dye DRAQ5 (blue). (F–I) Intracellular colocalization of exogenously overexpressed Barx1 and Msx1 as EGFP and FLAG-tagged fusion proteins. (F) Intracellular localization of Barx1–EGFP (green); (G) intracellular localization of Msx1 using anti-Msx1 (red); (H) merged pictures show intracellular colocalization of Barx1–EGFP and FLAG–Msx1 (yellow); and (I) DNA staining using the fluorescence dye DRAQ5 (blue).
To determine whether these protein interactions occur endogenously, we performed protein colocalization analysis using confocal microscopy. C3H10T1/2 cells were used for the immunofluorescence staining to confirm the endogenous localization of Msx1 and its interacting protein Barx1. Using anti-Barx1 and anti-Msx1 antibodies, we show that C3H10T1/2 cells express Msx1 and Barx1 proteins sufficiently to detect their intracellular expression by immunofluorescence (Fig. 5 B and C). Merged pictures show endogenous Barx1 to be colocalized with Msx1 (Fig. 5D). The pattern of Msx1 immunofluorescence was identical with that of previous reports, whereas this is a unique report of Barx1 endogenous intracellular expression pattern. In addition, the transiently transfected cells with constructs expressing Barx1 and Msx1 as EGFP and FLAG-tagged fusion proteins used for our coimmunoprecipitation assays, were stained with anti-Msx1 antibodies, further confirming the intracellular colocalization of Barx1 with Msx1, using confocal microscopy (Fig. 5 F–I).
Barx1 Expression in Premolar Tooth Development.
In mouse tooth development, Barx1 expression is restricted to presumptive molar mesenchyme and throughout tooth development to molar mesenchyme cells (10). The role of the Barx1–Msx1 interaction in fine-tuning BMP activity supported the suggested importance of the level of BMP activity in regulating cusp formation (9). We argued that if the levels of BMP activity control cusp formation then the expression of Barx1 should correlate with tooth cusp pattern rather than being molar specific. To test this hypothesis we analyzed Barx1 expression in embryos of a mouse mutant that develops premolar teeth (Orpk) (14) and in a species that has natural premolars, the lesser shrew Cryptotis parva.
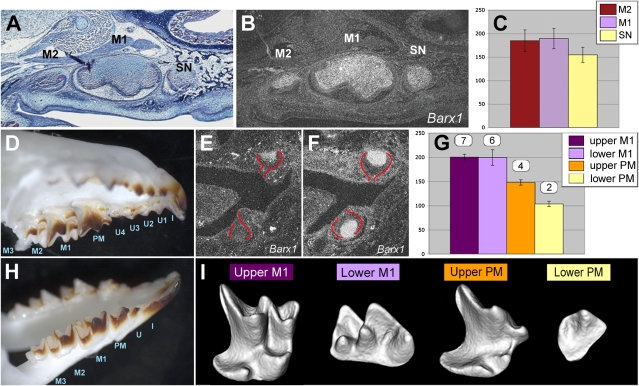
In Orpk embryos, Barx1 expression could be observed during development of the supernumerary teeth that develop mesial to the first molars and have a cusp pattern consistent with a premolar identity (Fig. 6 A and B). The lesser shrew has a more complete dentition than the mouse with premolar and unicuspid (canine-like) teeth (Fig. 6 D and H). Barx1 expression was observed in maxillary and mandibular molars (Fig. 6F), as well as in maxillary premolar tooth development but was barely detectable during mandibular premolar development (Fig. 6E) and absent during both unicuspid and incisor development. Grain counting of serial sections of premolar tooth primordia hybridized with Barx1 confirmed the impression from the in situ hybridization sections, namely that Barx1 expression was reduced in the Orpk premolar-like supernumerary tooth compared with Orpk first and second molars (Fig. 6C). Similarly, in shrew tooth primordia Barx1 expression was less in the upper premolar than the molars and less in the lower premolar than in the upper premolar (Fig. 6G). Comparison of the crown shape of adult shrew maxillary and mandibular premolars revealed that mandibular premolars only had two cusps, whereas maxillary premolars had a clear, molar-like pattern (Fig. 6I). Therefore, Barx1 expression is found only during development of multicuspid teeth and levels of expression correlate with cusp numbers, supporting a role in regulating signaling activity that controls cusp number.
Fig. 6.
Barx1 is expressed in all multicusped teeth with expression levels correlating with cusp numbers. (A–C) Supernumerary teeth forming in the diastema of mice homozygous for Tg737orpk display Barx1 expression levels lower than first and second molars. (A and B) Consecutive sagittal sections through the upper jaw of an E18.5 mouse homozygous for Tg737orpk showing from Left to Right a second molar (M2), first molar (M1), and supernumerary tooth (SN), the latter developing mesial to M1 in the normally toothless diastema. (A) Trichrome staining showing the premolar-like shape of the ectopic diastema tooth. (B) Radioactive in situ hybridization for Barx1. (C) Quantification of Barx1 expression level in the dental mesenchyme of Tg737orpk/Tg737orpk second molar (M2), first molar (M1), and supernumerary tooth (SN). (D–I) Level of cpBarx1 expression correlates with cusp number in shrew multicusped teeth. (D and H) Dentition of an adult shrew upper (D) and lower (H) jaw composed of molars (M), premolars (PM), unicusps (U), and incisors (I). (E and F) Radioactive in situ hybridization for cpBarx1 in shrew premolars (E) and molars (F). Developing molars and premolars (outlined in red) have reached a cap stage. (G) Quantification of Barx1 expression levels in the dental mesenchyme of shrew premolar and molar tooth primordia. Gene expression was quantified by analyzing consecutive sections spanning the whole dental papilla of each tooth using ImageJ 1.34s. The number of cusps and crests displayed by each tooth is indicated above their respective Barx1 expression level. (I) 3D reconstructions of micro-CT scans of the upper and lower first molars (M1) and premolars (PM) of a 24-d-old shrew. Teeth are viewed from a lingual side; distal is Right and proximal Left. The number of cusps of each tooth (indicated in G) was carefully assessed by rotating the 3D models.
Discussion
The regulation of crown morphology is a critical process in mammalian tooth development because it determines tooth shape (type) that begins with the transition from a tooth bud to a tooth cap. The formation of the primary enamel knot signaling center is regulated by BMP activity, with BMP4 protein being secreted by mesenchymal cells at the bud stage. This BMP4 expression is regulated by the transcription factor Msx1 in partnership with Pax9 and possibly other factors. Pax9 and Msx1 are coexpressed in the condensing dental mesenchyme and are critical for development of all tooth types, as in Msx1 and Pax9 homozygous null mutants tooth development is arrested at the bud stage (4, 15).
We identify here a developmental tooth type control of BMP signaling at the bud-to-cap transition whereby the optimal level of BMP activity required for developmental progression is fine-tuned by transcriptional activity of two interacting homeodomain transcription proteins, Barx1 and Msx1. Msx1 functions to regulate BMP4 expression in the development of all tooth types (incisors and molars), whereas Barx1 is only expressed in development of teeth with multicusped crowns (molariform teeth). In the complete absence of any Barx1–Msx1 interaction, (Barx1−/−; Msx1+/+), the resulting suboptimal level of BMP activity is insufficient to induce appropriate levels of primary knot signaling that controls cusp formation. Under these conditions, rather than the expected outcomes of abnormal morphogenesis or complete arrest, tooth development stalls until the optimal level is reached to form the correct cusp pattern. This identifies a developmental phenomenon where level of BMP activity is sensed by cells as a critical threshold (optimal) level for continued normal development. During this temporal arrest in molar development, development of other organs, including incisors, continues normally. Thus, molar teeth stall their development when BMP activity is below the threshold (suboptimal) and then restart when levels raise above the threshold, 24 h later. This autonomous self-regulation is thus a way for the embryo to cope with small inaccuracies in signaling that might otherwise lead to major abnormalities. The fact that following stalling of the intrinsic developmental clock, development then accelerates to be back in synchrony with the general timing of embryonic development, illustrates the importance of temporal coupling of developmental processes. Surprisingly, in vitro knockdown of Barx1 using lentiviruses expressing Barx1 siRNA led to a complete arrest of tooth development at the bud stage (16), suggesting that the ability to restart development is lost in this system. The subrenal culture of tooth rudiments is unlikely to be the cause of this definitive arrest of development, because Barx1−/− molar tooth rudiments grafted under a kidney capsule do form normal mineralized molars. Furthermore, in the Barx1 knockout, a transcriptional compensation through up-regulation of another Bar homeogene family member can be excluded, as Barx2, Barhl1, and Barhl2 are not expressed in E13.5 WT molar tooth buds (Fig. S2). Our data also suggest a role for different levels of BMP activity in the regulation of the cusp patterns that constitute different tooth types. This is consistent with theoretical modeling of cusp formation, on the basis of experimental data that indicate a key role for the level of BMP activity in cusp formation (9). Thus, development of different crown cusp patterns would be predicted to require particular individual thresholds of BMP activity for correct morphogenesis to be initiated.
Barx1 is specifically expressed only in teeth that develop multiple cusps (molars and premolars) and thus its role may be linked to cuspal morphogenesis. In the presence of Barx1, a single Msx1 allele is able to regulate normal BMP activity to ensure normal molar formation. However, in the absence of Barx1, a single allele of Msx1 is not sufficient and molar tooth development arrests at the bud stage. Barx1 and Pax9 are proteins that physically interact with Msx1 to regulate BMP activity (17, 18). Whereas both Msx1 and Pax9 are required for development of both molars and incisors to proceed through the bud-to-cap transition (4, 15), the Barx1–Msx1 interaction regulates or “fine-tunes” BMP activity only during molar development, and incisor development continues normally in the absence of Barx1 and one allele of Msx1. This may indicate that either another protein carries out a Barx1-like function in incisors or that the Msx1–Barx1 interaction is a molar-specific phenomenon. The latter would be consistent with the role of Barx1 in fine-tuning BMP levels to ensure correct cusp formation, a process that is not necessary in cuspless incisors. This was confirmed by the observation of Barx1 expression during development of premolars both in mutant mice and in the lesser shrew and lack of expression in canines (unicuspids). However, the reduced Barx1 expression in the development of the mandibular premolar of the shrew, whose crown has a reduced cusp number, shows that Barx1 expression correlates with cusp development rather than tooth type (position).
The transformation of incisor crown shape into a molariform shape following ectopic expression of Barx1 suggested that Barx1 would have an essential role in molar crown morphogenesis (10, 12). Clearly, molar teeth are formed in the absence of Barx1, albeit via an abnormal developmental route. This may be explained by the fact that the absolute requirement for Barx1 in molar tooth development is only fully manifested in the absence of an allele of Msx1 (Barx1−/−; Msx+/−). Thus, the dominant role of Msx1 in regulating BMP4 masks the more subtle, but nevertheless essential role of Barx1. This phenomenon has parallels with what has been observed in kidney development where the essential role of Gdnf/Ret signaling is masked by loss of Spry1 such that in Gdnf−/−; Spry1−/− and Ret−/−; Spry1−/− mice, kidney development shows only subtle alterations in branching (19). Thus, in kidney development, the balance of signaling pathway activities (Gdnf/Ret and FGF) is more important than the specific role of Gdnf.
Materials and Methods
Animals and Genotypes.
Barx1 mutant mice were made by homologous recombination targeting the Barx1 gene region from part of exon 2 to before the 3′-UTR of exon 4, including the DNA binding homeodomain (Fig. S1 and ref. 13). Barx1 mutant mice were bred into C57BL/6, 129SvEv, and CD1 breeding backgrounds for at least nine generations before analysis. A floxed-out allele of the Barx1 mutant was made by crossing the ubiquitous Cre line β-actin–Cre with the targeted allele to remove the NeoR cassette (Fig. S1). PCR primer 1: 5′-CGCAGTGTTCAAGTTCCCACT, primer 2: 5-CTATTCTGGAAAGAGTAACGCACA, and primer 3: 5′-GAGACTAGTGAGACGTGCTACTTCC were used for genotyping the Barx1 mutants with the NeoR, which amplify a 358-bp fragment for the wild type and 445-bp for the mutant, at an annealing temperature of 62 °C. Primer 4: 5′-CTTGGGCCAGTAGGTAACCA was used instead of primer 3 to amplify a 565-bp fragment for the NeoR floxed-out allele.
Tg737orpk and Msx1−/− mutant mice were produced as described previously (20, 21).
Time matings were set up such that noon of the day on which vaginal plugs were detected was considered as E0.5. All animal experiments were carried out in accordance with UK Home Office regulations.
In Situ Hybridization and Gene Expression Quantification.
In situ hybridization was carried out with riboprobes labeled with radioactive 35S-UTP or digoxygenin on 8-μm paraffin sections of paraformaldehyde-fixed tissue as previously described (22). Slides were counterstained with hematoxylin (Fluka) and examined in dark-field microscopy. Gene expression was quantified by using ImageJ 1.34s (23). For each multicusp tooth primordium, gene expression was analyzed in consecutive sections spanning the whole dental papilla. On each section, condensed mesenchyme of the dental papilla was outlined and white grains counted. A set of data were obtained for each multicusp tooth primordium. Mean of these values was plotted on a graph.
Generation of C. parva Shrew Barx1 Probe.
Total RNA was extracted from E16 C. parva shrew heads with TRIzol reagent (Invitrogen) and treated with DNA-free DNA removal kit (Ambion). C. parva shrew Barx1 probe was generated by RT-PCR with degenerate primers 5′-GCNGCNGTNTTYAARTTYCC-3′ and 5′-ACDATYTTYTTCCAYTTCAT-3′ using Access RT-PCR system (Promega). This was followed by one round of PCR with primers 5′-GCNGCNGTNTTYAARTTYCC-3′ and 5′-TTYTGRTACCANGTYTTNACYTG-3′. Degenerate primers were designed from conserved amino acid alignments generated using ClustalW (24).
Microcomputed Tomography (micro-CT) Analysis.
Specimens for micro-CT were scanned using a GE Locus SP micro-CT scanner. The specimens were immobilized using cotton gauze and scanned to produce 14-μm voxel size volumes. The specimens were characterized further by making 3D isosurfaces, generated and measured using Microview software (GE).
Immunostaining.
Immunofluorescence assays were performed in C3H10T1/2 cells to detect endogenous expression of Msx1 and Barx1. Cells were fixed using 4% PFA in PBS buffer for 5 min at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 8 min at room temperature, then blocked with 10% normal goat serum for 45 min, and incubated with appropriate primary and FITC- or TRITC-conjugated secondary antibodies for 1 h or overnight. C3H10T1/2 cells were sequentially immunostained with anti-Barx1 (H-55, rabbit polyclonal antibody; Santa Cruz Biotechnology) and anti-Msx1 antibody (rabbit polyclonal antibody; Santa Cruz Biotechnology). For exogenous expressions of Barx1–EGFP (green fluorescence) and FLAG–Msx1, cotransfected cells were singly immunostained with anti-Msx1 antibody for immunofluorescence. DRAQ5 (1:1,000 dilution in PBS) was used to stain nuclear DNA (blue). Immunofluorescence was visualized and images were collected in sequential scanning mode with a Leica TCS SP2 confocal microscope using different excitation wavelengths for green, red, and blue fluorescence.
Phospho-Smad1/5/8 (Cell Signaling Technology) and BrdU (Abcam) antibodies were used, respectively, with Tris buffer and citric acid antigen retrieval methods. Secondary antibodies conjugated with biotin or HRP (Vector) were used. Fluorescent signal was amplified with TSA Fluorescein system (PerkinElmer) and color reaction developed with ABC kit (Vector) using DAB.
BrdU Incorporation.
A total of 20 mg/kg BrdU (BD) was i.p. injected and mice were killed after 1 h. Tissues were fixed in modified Carnoy's (60% ethanol, 30% of 37% formaldehyde, and 10% of glacial acetic acid) and processed and embedded in paraffin wax for immunostaining. In molars, BrdU+ cells were counted in the whole epithelium and three mesenchymal areas randomly picked, all four quadrants were counted. BrdU+ cells were counted in two mice for each genotype, at each time point. Student's t test statistical analyses were used for regional estimation of proliferating cells and apoptotic cells.
Transfections and Coimmunoprecipitation Assays.
Murine mesenchymal cell line C3H10T1/2 (American Type Culture Collection; CCL-226) was cultured in high-glucose DMEM supplemented with 10% (vol/vol) FBS (Invitrogen). The cells in 60-mm dishes were cotransfected with plasmids of pIRES2–Barx1–EGFP and pCMV–FLAG–Msx1 or pIRES2–Barx1–EGFP and pCMV–Tag2B (Stratagene) using FuGENE 6 reagent (Roche) according to manufacturer protocol. Each transfection was repeated three times independently. The plasmid construct pCMV–FLAG–Msx1 expressed wild-type full-length Msx1 tagged with FLAG epitope at the N terminus. The plasmid construct pIRES2–Barx1–EGFP expressed a fusion protein Barx1–EGFP. After 36 h, C3H10T1/2 cells were lysed in RIPA lysis buffer [50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.5% Nonidet P-40, 0.1% sodium deoxycholate] with protease inhibitors (Roche), and proteins were immunoprecipitated by using EZview Red anti-FLAG M2 Affinity Gel beads (Sigma). The affinity gels were washed with the lysis buffer five times and eluted with 2× SDS sample buffer. For Western blotting of eluted protein, primary antibodies were used with 1:500 dilution of rabbit anti-Barx1 polyclonal antibody (Santa Cruz) or 1:1,000 dilution mouse anti-FLAG M2 monoclonal antibody (Sigma). The immunoprecipitated proteins were analyzed by immunoblotting, using ECL Western blotting detection reagent (Fisher).
Supplementary Material
Acknowledgments
We thank Benoit Robert for generously providing the Msx1 mutant mice and Jeremy Green for helpful discussion and comments. Work in the United Kingdom was supported by the Wellcome Trust (P.T.S.) and Research Councils UK (I.M.). S.C.A. had a scholarship from the Coordination for the Improvement of Higher Education Personnel (CAPES) Foundation BEX 5408/10-5.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112801108/-/DCSupplemental.
References
- 1.Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: Non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- 2.Vaahtokari A, Aberg T, Jernvall J, Keränen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- 3.Andl T, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 4.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 5.MacKenzie A, Ferguson MW, Sharpe PT. Hox-7 expression during murine craniofacial development. Development. 1991;113:601–611. doi: 10.1242/dev.113.2.601. [DOI] [PubMed] [Google Scholar]
- 6.Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. A new function of BMP4: Dual role for BMP4 in regulation of Sonic hedgehog expression in the mouse tooth germ. Development. 2000;127:1431–1443. doi: 10.1242/dev.127.7.1431. [DOI] [PubMed] [Google Scholar]
- 9.Salazar-Ciudad I, Jernvall J. A computational model of teeth and the developmental origins of morphological variation. Nature. 2010;464:583–586. doi: 10.1038/nature08838. [DOI] [PubMed] [Google Scholar]
- 10.Miletich I, Buchner G, Sharpe PT. Barx1 and evolutionary changes in feeding. J Anat. 2005;207:619–622. doi: 10.1111/j.1469-7580.2005.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tissier-Seta JP, et al. Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mech Dev. 1995;51:3–15. doi: 10.1016/0925-4773(94)00343-l. [DOI] [PubMed] [Google Scholar]
- 12.Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- 13.Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell. 2005;8:611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Ohazama A, et al. Primary cilia regulate Shh activity in the control of molar tooth number. Development. 2009;136:897–903. doi: 10.1242/dev.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters H, Neubüser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y, et al. Application of lentivirus-mediated RNAi in studying gene function in mammalian tooth development. Dev Dyn. 2006;235:1334–1344. doi: 10.1002/dvdy.20706. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa T, Kapadia H, Wang B, D'Souza RN. Studies on Pax9-Msx1 protein interactions. Arch Oral Biol. 2005;50:141–145. doi: 10.1016/j.archoralbio.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa T, et al. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J Biol Chem. 2006;281:18363–18369. doi: 10.1074/jbc.M601543200. [DOI] [PubMed] [Google Scholar]
- 19.Michos O, et al. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyer JH, et al. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. doi: 10.1126/science.8191288. [DOI] [PubMed] [Google Scholar]
- 21.Houzelstein D, Cohen A, Buckingham ME, Robert B. Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech Dev. 1997;65:123–133. doi: 10.1016/s0925-4773(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson DG. In Situ Hybridization: A Practical Approach. Oxford: IRL Press; 1992. [Google Scholar]
- 23.Rasband W. ImageJ (Natl Inst Health, Bethesda) 1997–2006 [Google Scholar]
- 24.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.