Abstract
In most carcinomas, invasion of malignant cells into surrounding tissues involves their molecular reprogramming as part of an epithelial-to-mesenchymal transition (EMT). Mutation of the APC gene in most colorectal carcinomas (CRCs) contributes to the nuclear translocation of the oncoprotein β-catenin that upon binding to T-cell and lymphoid enhancer (TCF-LEF) factors triggers an EMT and a proinvasive gene expression profile. A key inducer of EMT is the ZEB1 transcription factor whose expression promotes tumorigenesis and metastasis in carcinomas. As inhibitor of the epithelial phenotype, ZEB1 is never present in the epithelium of normal colon or the tumor center of CRCs where β-catenin remains membranous. We show here that ZEB1 is expressed by epithelial cells in intestinal tumors from human patients (familial adenomatous polyposis) and mouse models (APCMin/+) with germline mutations of APC that result in nuclear accumulation of β-catenin. However, ZEB1 is not expressed in the epithelium of hereditary forms of CRCs that carry wild-type APC and where β-catenin is excluded from the nucleus (Lynch syndrome). We found that β-catenin/TCF4 binds directly to the ZEB1 promoter and activates its transcription. Knockdown of β-catenin and TCF4 in APC-mutated CRC cells inhibited endogenous ZEB1, whereas forced translocation of β-catenin to the nucleus in APC-wild-type CRC cells induced de novo expression of ZEB1. Upregulation of MT1-MMP and LAMC2 by β-catenin/TCF4 has been linked to invasiveness in CRCs, and we show here that both proteins are activated by ZEB1 coexpressing with it in primary colorectal tumors with mutated APC. These results set ZEB1 as an effector of β-catenin/TCF4 signaling in EMT and tumor progression.
Keywords: transcriptional regulation, Wnt signaling
Loss of epithelial characteristics in favor of a mesenchymal phenotype—a process referred to as epithelial-to-mesenchymal transition (EMT)—bestows carcinoma cells at the tumor front with stem cell-like and more motile properties needed for their invasion into surrounding tissues (1). One of the signals initiating an EMT is the canonical Wnt pathway whose stimulation triggers the translocation of the oncoprotein β-catenin to the nucleus, where it acts as coactivator of T-cell and lymphoid enhancer (TCF–LEF) factors in the transcriptional activation of target genes (2). In the absence of nuclear β-catenin, TCF–LEF factors function instead as transcriptional repressors by binding to corepressors of the Groucho–TLE family. In normal epithelial cells, β-catenin associates at the cellular membrane with the adhesion molecule E-cadherin, while any free cytoplasmic β-catenin is phosphorylated and targeted for ubiquination-dependent degradation—thus preventing it from reaching the nucleus—by a protein complex formed by APC, GSK-3, CKIα, and Axin (2).
Aberrant activation of β-catenin/TCF signaling has been involved in a number of tumors, most notably colorectal carcinomas (CRCs) (2). Over 80% of sporadic CRCs and the vast majority of cases of familial adenomatous polyposis (FAP)—an autosomal dominant inherited syndrome displaying high predisposition to CRC—have loss-of-function mutations of the APC gene (3). A high predisposition to intestinal adenomas is also found in mice heterozygous for a stop codon mutation in APC (APCMin/+ mice) (4). In the normal intestinal epithelium of FAP patients and APCMin/+ mice, β-catenin remains membranous. However, in the tumors developed by FAP patients and APCMin/+ mice, a fraction of β-catenin—which levels vary among and within tumors—translocates to the cytoplasm and nucleus (5–7). Nuclear accumulation of β-catenin is higher and more homogenous in FAP carcinomas than in FAP adenomas and also increases as adenomas develop in APCMin/+ mice. In hereditary syndromes with predisposition to CRC but carrying wild-type APC, like the Lynch syndrome, β-catenin does not accumulate at the nucleus of CRC cells. Heterogeneity in the levels of nuclear β-catenin in the tumors of FAP patients and APCMin/+ mice is also observed in sporadic CRCs. Although in well-differentiated areas and the tumor center of CRCs β-catenin has a membranous/cytoplasmic distribution, it becomes mostly nuclear in scattered dedifferentiated cells undergoing an active EMT at the tumor invasive front (8). Genomic location analyses in CRC cells have identified a large number of genes that bind β-catenin at their regulatory regions (9). β-catenin/TCF activates a differential pattern of gene expression in the center and front of primary CRCs with the induction of proinvasive genes taking place only at the tumor front (10).
A key initial step in the EMT is the downregulation of E-cadherin, which at the transcriptional level is repressed by several factors; namely, ZEB1, ZEB2, SNAI1, SNAI2, Twist1, Twist2, and E12/E47 (1). Of all of them, ZEB1 (also known as δEF1) has the most consistent inverse correlation with E-cadherin across different types of carcinomas (11, 12). In inducing an EMT, ZEB1 not only transcriptionally represses epithelial markers like E-cadherin but also activates mesenchymal genes. ZEB1 expression determines resistance to chemotherapy drugs in carcinoma cell lines, promotes tumorigenesis and metastasis in mouse xenograft models, and associates to poorer clinical prognosis in human carcinomas (11–13). Control by ZEB1 of these cancer hallmarks involves not only direct transcriptional activation or repression of target genes but also indirect mechanisms. In most cell types, ZEB1 represses and is repressed by microRNAs of the miR-200 family that could also directly downregulate mesenchymal genes involved in cancer cell migration (11, 14, 15).
In normal and carcinomatous colonic mucosa, ZEB1 is mostly found in nonepithelial cells of diverse origin at the stromal compartment (16, 17). As expected from an inducer of EMT, ZEB1 is never expressed by epithelial cells of the normal mucosa or the tumor center of carcinomas. However, ZEB1 is present in the nucleus, and often also in the cytoplasm, of the above-mentioned dedifferentiated cancer cells at the tumor front of CRCs (13, 16, 18).
In this study, we show that ZEB1 is found in the epithelial cells of colorectal tumors from FAP patients and intestinal adenomas from APCMin/+ mice where it is coexpressed with nuclear β-catenin. By contrast, ZEB1 is not detected in the epithelium of Lynch syndrome CRCs with wild-type APC and lacking nuclear β-catenin. Using established cell lines of CRC with reportedly different levels of nuclear β-catenin (19), we found that endogenous β-catenin and TCF4 bind to the promoter region of ZEB1 and induces its expression. Finally, in CRC cells with mutated APC, ZEB1 activates MT1–MMP and LAMC2, two proinvasive genes jointly regulated by β-catenin/TCF4 at the tumor front of CRCs, which we also found coexpressed with ZEB1 in colorectal tumors from FAP patients.
Results
ZEB1 Coexpresses with Nuclear β-Catenin in the Epithelium of Tumors from FAP Patients and APCMin/+ Mice.
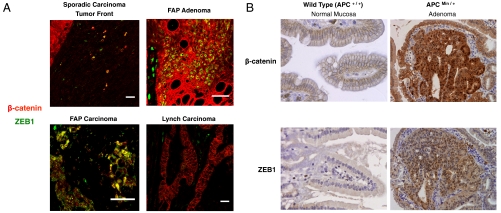
Expression of ZEB1 by dedifferentiated nuclear β-catenin-positive cancer cells at the tumor front of sporadic CRCs led us to question whether ZEB1 expression also correlates with nuclear β-catenin in colorectal tumors from FAP patients with mutated APC. In FAP adenomas and carcinomas, β-catenin is found in the cytoplasm and nucleus of epithelial tumor cells. Nuclear accumulation of β-catenin varies within and among FAP tumors but is higher and more homogeneously distributed in FAP carcinomas (5, 6) (red in Fig. S1 A and B). Interestingly, we found ZEB1 in the epithelial cells of FAP tumors (green in Fig. S1 A and B), where it was coexpressed with nuclear β-catenin (yellow in Fig. 1A and Fig. S1B). ZEB1 paralleled levels of nuclear β-catenin in tumor epithelial cells—highly expressed by the majority of FAP adenomas and virtually all FAP carcinomas (Fig. 1A and Fig. S1A). Obviously, no colocalization was observed in nonepithelial stromal cells, positive for ZEB1 but negative for β-catenin (green in Fig. 1A). ZEB1 was not detected in Lynch syndrome carcinomas (no epithelial cells labeled in green in Fig. 1A and Fig. S1 A and B) with wild-type APC and where β-catenin remains membranous/cytoplasmic (red in Fig. 1A and Fig. S1 A and B).
Fig. 1.
ZEB1 coexpresses with nuclear β-catenin. (A) Immunofluorescence staining for β-catenin (red, clone 14 or Ab6302) and ZEB1 (green, H-102 or E-20) in tumor front of sporadic CRCs, FAP colorectal adenomas and carcinomas, and Lynch CRCs. A representative merge picture is shown (yellow cells for colocalization). Single staining for β-catenin, ZEB1 and DAPI is shown in Fig. S1A and magnified in Fig. S1B. Scale bar, 25 μm. (B) Immunohistohemistry staining for β-catenin (clone 14) and ZEB1 (H-102) in normal intestinal mucosa and adenomas from 4-mo-old wild-type APC (magnification 400×) and APCMin/+ (200×) mice, respectively.
Expression of ZEB1 was then examined in APCMin/+ mice, considered as an animal model of human FAP. In the normal intestinal epithelium of wild-type and APCMin/+ mice, β-catenin stayed membranous and ZEB1 was absent (Fig. 1B). By contrast, in intestinal adenomas of APCMin/+ mice, nuclear accumulation of β-catenin was accompanied by expression of ZEB1 (Fig. 1B).
An important target of ZEB1 during EMT is the E-cadherin, expression of which is lost in those dedifferentiated cells at the invasive front of sporadic CRCs (Fig. S1C) (8). Here, we found that in epithelial cells of FAP colorectal tumors expression of ZEB1 also correlated with a loss/reduction of E-cadherin (Fig. S1C). Altogether, these results indicate that nuclear translocation of β-catenin in tumors with mutant APC correlates with ZEB1 expression and loss of E-cadherin.
β-Catenin/TCF4 Bind the Human ZEB1 Promoter and Activate Its Transcription.
The above results prompted us to investigate whether β-catenin signaling could induce ZEB1. To test this hypothesis, we examined the effect on ZEB1 transcription of exogenously overexpressing β-catenin and TCF4, the most prominent TCF–LEF factor in the intestine. In these experiments, we used two CRC cell lines with different levels of nuclear β-catenin (19). On the one hand, we used SW480 cells with mutant APC and where β-catenin is predominantly nuclear (Fig. S2A). As expected from the correlation we found in FAP tumors between nuclear β-catenin and ZEB1, SW480 cells contain high levels of ZEB1 and low levels of E-cadherin (Fig. S2B). On the other hand, we used HCT116 cells that have wild-type APC and low levels of β-catenin accumulated in the nucleus (Fig. S2A). In line with our results in Lynch samples, low nuclear β-catenin in HTC116 cells associated with low levels of ZEB1 and high levels of E-cadherin (Fig. S2B). A high pool of nuclear β-catenin in SW480 cells translates into strong β-catenin/TCF-mediated transcription as assessed by the activity of the pTOPFLASH luciferase reporter (20) (Fig. S2C), containing multimerized TCF binding sites. By contrast, in HCT116 cells Wnt signaling and pTOPFLASH transcription were low (20) (Fig. S2C).
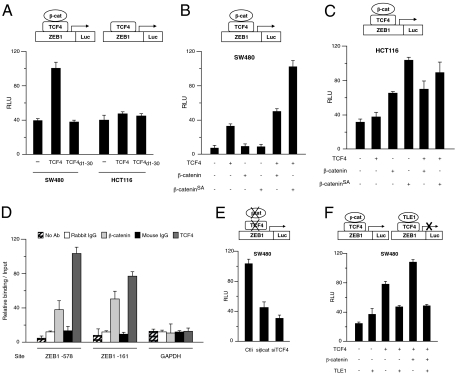
Overexpression of TCF4 in SW480 cells, but not its corresponding empty vector, increased transcription of the human ZEB1 promoter (Fig. 2A). This effect, however, was not found when we transfected a mutant form of TCF4 (TCF4d1-30) that lacks the β-catenin binding domain (Fig. 2A). These results suggest that the ability of TCF4 to activate ZEB1 transcription is dependent on the formation of a transcriptional activation complex with endogenous nuclear β-catenin.
Fig. 2.
β-catenin/TCF4 activates ZEB1 transcription. (A) SW480 and HCT116 cells were transfected with 0.5 μg of human ZEB1 promoter plus 1 μg of the empty expression vector (“−”) or equal molar amounts of expression vectors for TCF4 or TCF4d1-30 (“+”). Transcriptional assays were performed as described in SI Discussion. The relative luciferase activity (RLU) in the two cell lines with the empty vector was set to a common value for comparison. As for the TOPFLASH reporter (Fig. S2C), basal activity of the ZEB1 promoter is lower in HTC116 cells. (B) SW480 cells were transfected with ZEB1 promoter and combinations of 1 μg of empty expression vector or equal molar amounts of TCF4, β-catenin and/or β-cateninSA. (C) Same as in B, but in HCT116 cells. (D) TCF4 and β-catenin bind to the ZEB1 promoter. Real-time PCR quantification (qRT-PCR) of fragments of the ZEB1 and GAPDH promoters immunoprecipitated in ChIP assays from SW480 cells with Abs against TCF4, β-catenin, their respective control IgG, and with no Ab. Amplified ZEB1 promoter regions contain TCF binding sites at -578 and -161. Values represent relative binding in relation to input. (E) Endogenous β-catenin and TCF4 regulate ZEB1 transcription. As in A, SW480 cells were transfected with the ZEB1 promoter along with 100 nM siRNAs against either β-catenin (siβcat), TCF4 (siTCF4), or a control siRNA (siCtl). (F) As in B, SW480 cells were transfected with the ZEB1 promoter and different combinations of 1 μg of empty vector or equal molar amounts of TCF4, β-catenin, and TLE1.
Because SW480 cells have high endogenous levels of nuclear β-catenin, overexpression of β-catenin had no effect by itself on ZEB1 transcription (Fig. 2B)—presumably, most endogenous TCF4 is already bound to endogenous β-catenin. Exogenous β-catenin did synergize with TCF4 when the latter was also overexpressed, although only modestly (Fig. 2B). However, their synergistic activation of ZEB1 transcription was strong when TCF4 was coexpressed with a mutant and more stable form of β-catenin (β-cateninSA) that is not degradable by the APC-GSK3-Axin complex (Fig. 2B). It is of note that the effect on ZEB1 transcription of overexpressing in SW480 cells TCF4 and/or β-catenin mirrored that obtained with the pTOPFLASH reporter (Fig. S2D).
The same set of experiments was then performed in HCT116 cells. Because these cells have low levels of nuclear β-catenin, overexpression of TCF4—let alone TCF4d1-30—had by itself limited impact on the ZEB1 promoter (Fig. 2A). By contrast, exogenous expression of wild-type β-catenin and β-cateninSA activated ZEB1 transcription (Fig. 2C). Again, the results obtained with the ZEB1 promoter in HTC116 cells paralleled those observed with the pTOPFLASH reporter (Fig. S2E).
We next wondered whether the ability of TCF4 and/or β-catenin to activate ZEB1 transcription was mediated by their direct binding to the ZEB1 promoter. Examination of the human ZEB1 promoter sequence revealed the existence of several potential TCF binding sites, two of them—located at -578 and -161 and similar to those reported in the human laminin 5 γ2 chain/LAMC2 promoter (21)—within the ZEB1 promoter reporter used above. We therefore tested through ChIP assays if endogenous β-catenin/TCF4 complexes assemble on the ZEB1 promoter. Indeed, it was found that antibodies against TCF4 and β-catenin, but not their respective control IgG, immunoprecipitated regions of the ZEB1 promoter containing the TCF binding sites (Fig. 2D and Fig. S2F). Lower binding by β-catenin vis-à-vis TCF4 is consistent with the fact that β-catenin interacts with DNA only indirectly via TCF4. TCF4 and β-catenin antibodies failed, however, to immunoprecipate a fragment of the GAPDH promoter, which lacks TCF binding sites (Fig 2D). These results indicate that endogenous β-catenin/TCF4 complexes bind directly to the ZEB1 promoter.
To test whether these endogenous β-catenin/TCF4 complexes activate ZEB1 transcription, we examined the effect of eliminating both proteins. In SW480 cells, where high levels of nuclear β-catenin correlate with strong endogenous ZEB1 expression, knockdown of either endogenous β-catenin or TCF4 with specific siRNAs—but not a siRNA control—downregulated the activity of the ZEB1 promoter (Fig. 2E) just as it occurred with the TOPFLASH reporter (Fig. S2G).
β-catenin and members of the Groucho–TLE family of transcriptional corepressors competitively displace one another from their binding to TCF4. We therefore questioned whether TLE proteins could regulate ZEB1 transcription in the opposite direction. Overexpression of TLE1 in SW480 cells efficiently blocked β-catenin/TCF4-mediated transcription of the ZEB1 promoter (Fig. 2F) and the pTOPFLASH reporter (Fig. S2H).
Regulation of Endogenous ZEB1 by β-Catenin/TCF4.
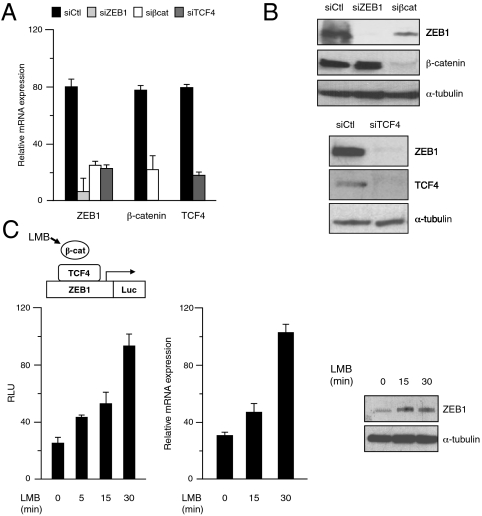
Next, we examined if β-catenin/TCF4 could regulate endogenous ZEB1. Knockdown of endogenous β-catenin or TCF4 in SW480 cells drastically downregulated both endogenous ZEB1 mRNA (Fig. 3A) and protein (Fig. 3B).
Fig. 3.
Endogenous regulation of ZEB1 by β-catenin/TCF4. (A) Endogenous β-catenin and TCF4 regulate endogenous ZEB1 mRNA. SW480 cells were transfected with 50–100 nM of a control siRNA or specific siRNAs for ZEB1, β-catenin, and TCF4. Relative mRNA levels were determined by qRT-PCR respect to GAPDH. (B) Endogenous β-catenin and TCF4 regulate endogenous ZEB1 protein. SW480 cells were transiently transfected with control and specific siRNAs and cell lysates were immunoblotted for ZEB1 (H-102), β-catenin (clone 14), TCF4 (H-125), as well as α-tubulin (B5-1-2) to control for loading. (C) Nuclear accumulation of β-catenin induces endogenous ZEB1. (Left) HTC116 cells were transfected with 0.5 μg of ZEB1 promoter and, after 48 h, treated with 10 ng/mL of LMB for different periods before cell lysates were processed for luciferase activity quantification (see Supporting Information). (Center) HCT116 cells, treated with 10 ng/mL of LMB for different periods, were assessed for ZEB1 mRNA levels by qRT-PCR. (Right) HCT116 cells, treated with 10 ng/mL of LMB for different periods, were lysed, and protein levels were determined by Western blot with ZEB1 (H-102) and α-tubulin (B5-1-2) Abs.
We also wondered whether forced nuclear translocation of endogenous β-catenin in HTC116 cells induces endogenous ZEB1. Leptomycin-B (LMB)—a drug that specifically blocks CRM1/exportin-dependent nuclear export and triggers a shift of β-catenin from the cell membrane/cytoplasm to the nucleus in APC–wild-type cells (19, 20)—caused a rapid accumulation of nuclear β-catenin in HCT116 cells (Fig. S3A). Nuclear translocation of β-catenin in these cells stimulated ZEB1 transcription (Fig. 3C) with a temporal pattern that resembled the activation of the TOPFLASH reporter (Fig. S3B). Accumulation of nuclear β-catenin in HCT116 cells also induced endogenous ZEB1 mRNA and protein (Fig. 3C), and, consequently, reduced E-cadherin mRNA levels (Fig. S3C).
β-Catenin/TCF4 Signaling Regulates ZEB1 Activation of Proinvasive Markers.
Activation of the β-catenin/TCF pathway upregulates the expression of proinvasive genes (10). Likewise, repression of epithelial genes and induction of mesenchymal markers by ZEB1 is required for the acquisition of an invasive phenotype by carcinoma cells at the tumor front (11, 12). E-cadherin is a direct target for ZEB1 repression, and knockdown of ZEB1 in SW480 cells induced an upregulation of endogenous E-cadherin mRNA that, in line with our results above, was also observed upon the elimination of β-catenin and TCF4 (Fig. S4A). These results are consistent with the loss of E-cadherin in cells positive for nuclear β-catenin and ZEB1 at the tumor front of sporadic CRCs and FAP adenomas and carcinomas (Fig. S1C; also see Discussion). On the other hand, cyclin D1 is a known target of β-catenin/TCF4 in CRCs (10) but not of ZEB1 and, accordingly, cyclin D1 mRNA levels were downregulated upon knockdown of β-catenin and TCF4 but not by a siRNA against ZEB1 (Fig. S4A).
The literature reveals some overlap in the gene expression pattern activated by β-catenin/TCF and ZEB1. For instance, vimentin is activated by both β-catenin/TCF and ZEB1 (22, 23), while knockdown of ZEB1 in CRC cells downregulates LAMC2 mRNA levels, a well-characterized target of β-catenin/TCF4 (16, 21).
A key mechanism by which β-catenin/TCF4 promotes invasiveness in sporadic CRCs is through the coordinated induction of MT1–MMP and LAMC2 at the tumor front (10, 16, 21, 24, 25). In addition, cleavage of LAMC2 by MT1–MMP promotes CRC cell migration (24). Given that nuclear β-catenin and ZEB1 are coexpressed at the tumor front of CRCs, we examined the ability of ZEB1 also to regulate MT1–MMP and LAMC2 in APC mutant CRC cells as well as their possible coexpression in FAP tumors.
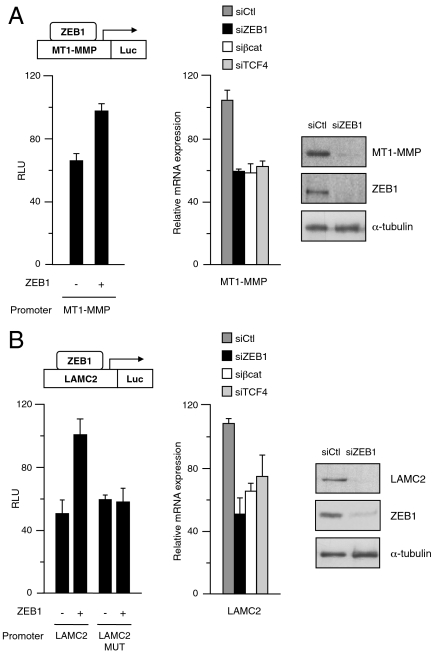
Because SW480 cells exhibit strong β-catenin/TCF-mediated transcription and ZEB1 expression, the basal activity of the MT1–MMP promoter in these cells was high (Fig. 4A). Still, overexpression of ZEB1 was able to further activate MT1–MMP transcription. It was also found that knockdown not only of β-catenin and TCF4 but also of ZEB1 reduced mRNA levels of MT1–MMP (Fig. 4A). Likewise, transient and stable elimination of ZEB1 in SW480 cells downregulated endogenous MT1–MMP protein expression (Fig. 4A and Fig. S4B).
Fig. 4.
Regulation of MT1–MMP and LAMC2 by ZEB1 in CRC cells with mutant APC. (A) ZEB1 activates MT1–MMP expression in CRC cells. (Left) ZEB1 activates MT1–MMP transcription. SW480 cells were transfected with 0.8 μg of the human MT1–MMP promoter along with 1.0 μg of empty expression vector or equal molar amounts of ZEB1 expression vector. (Center) ZEB1 activates endogenous MT1–MMP mRNA. SW480 cells transfected with a siRNA control or specific siRNAs for ZEB1, β-catenin, and TCF4 were assessed for relative levels of MT1–MMP mRNA by qRT-PCR. (Right) ZEB1 regulates endogenous MT1–MMP protein. SW480 cells were transiently transfected with 50 nM of a siRNA for ZEB1 or a control siRNA. Protein expression was assessed by Western blot with Abs for MT1–MMP (LEM2/15.8), ZEB1 (H-102), and α-tubulin (B5-1-2). (B) ZEB1 activates LAMC2 expression in CRC cells. (Left) ZEB1 activates LAMC2 transcription. Same as in A, but using both wild-type LAMC2 promoter (LAMC2) or a mutant version where two ZEB1 binding sites had been mutated (LAMC2-MUT) (Center) ZEB1 activates endogenous LAMC2 mRNA. Same as in A, but for LAMC2 mRNA. (Right) ZEB1 regulates endogenous LAMC2 protein. Same as in A, but a LAMC2 (D4B5) Ab was used.
We next explored the regulation of LAMC2 by ZEB1 in CRC cells. As for MT1–MMP, despite a high basal activity of the LAMC2 promoter in SW480 cells its transcription was stimulated by overexpression of ZEB1 (Fig. 4B). The LAMC2 promoter contains several consensus ZEB1 binding sites, two of them—at -213 and -96—within our promoter reporter and whose mutation abrogated the ability of ZEB1 to activate LAMC2 transcription in SW480 cells (Fig. 4B). ChIP assays showed that endogenous ZEB1 in SW480 cells binds to the LAMC2 promoter but not to the GAPDH promoter, expression of which is not regulated by ZEB1 (Fig. S4 C and D). In the same line, elimination of endogenous ZEB1 in these cells downregulated endogenous LAMC2 mRNA and protein (Fig. 4B and Fig. S4B). These results indicate that in CRC cells ZEB1 directly activates LAMC2 expression, although these results do not exclude the possibility of concurrent indirect regulation via miR-200.
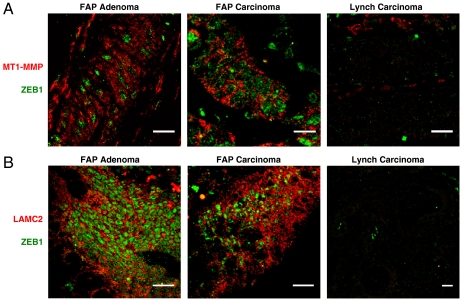
The expression of ZEB1 was then examined in tandem with that of MT1–MMP and LAMC2 in FAP adenomas and carcinomas. High levels of MT1–MMP (in red) were detected at the cell membrane and cytoplasm of tumor cells in FAP adenomas and carcinomas that also expressed ZEB1 (in green) (Fig. 5A and Fig. S5A). Despite lack of nuclear β-catenin, some expression of MT1–MMP was observed among stromal cells in Lynch CRCs where, obviously, there was no correlation with nonexistent ZEB1. Atypical nuclear localization of MT1–MMP has been associated with increased tumor aggressiveness (26). Remarkably, we found that MT1–MMP (in red) colocalized at the nucleus with ZEB1 (in green) precisely on dedifferentiated nuclear β-catenin-positive invading cancer cells at the tumor front of sporadic CRCs (yellow cells in Fig. S5B).
Fig. 5.
ZEB1 correlates with MT1–MMP and LAMC2 in FAP adenomas and carcinomas. (A) FAP adenomas and carcinomas and Lynch carcinomas were immunostained for MT1–MMP (red) and ZEB1 (green). Representative merge pictures are shown. Single staining for MT1–MMP, ZEB1 and DAPI is shown in Fig. S5A. (B) Same as in A, but for LAMC2 (red) and ZEB1 (green). Single staining captures are shown in Fig. S5C. Scale bars, 25 μm.
Similarly, when we examined ZEB1 and LAMC2 in FAP colorectal adenomas and carcinomas, we evidenced the coexpression of both proteins (green and red, respectively, in Fig. 5B and Fig. S5C). Again, neither expression nor correlation of ZEB1 and LAMC2 was found in CRCs from Lynch syndrome patients. Altogether these data show that ZEB1 and β-catenin/TCF4 regulate common proinvasive target genes in colorectal cancer.
Discussion
Aberrant activation of the β-catenin/TCF pathway—mostly by inactivating mutations of APC—is a necessary step in the genesis of many human cancers, including most CRCs. In this article, we show that ZEB1, a key inducer of EMT, is a direct target of β-catenin/TCF4. As an inhibitor of epithelial phenotype, ZEB1 is not present in the epithelium of normal colonic mucosa or in the center of sporadic CRCs but is expressed by dedifferentiated cells with nuclear β-catenin at their invasive front. In this study, ZEB1 was found in epithelial cells of intestinal tumors from FAP patients and APCMin/+ mice, both carrying germline mutations of APC and displaying accumulation of nuclear β-catenin. By contrast, ZEB1 is absent in Lynch syndrome CRCs with wild-type APC and lacking nuclear β-catenin. TCF4 and β-catenin bind to the ZEB1 promoter to activate its transcription that is, in turn, repressed by TLE1. β-catenin/TCF4 complexes regulate ZEB1 at the endogenous level in APC mutant CRC cells as knockdown of endogenous β-catenin and TCF4 resulted in the downregulation of endogenous ZEB mRNA and protein. On the other hand, forced nuclear translocation of β-catenin in wild-type APC CRC cells activated ZEB1 transcription and induced endogenous ZEB1 mRNA and protein. We also showed that MT1–MMP and LAMC2—two genes whose coordinated induction by β-catenin/TCF4 contributes to the invasiveness of sporadic CRCs—are also activated by ZEB1 in CRC cells and coexpress with ZEB1 in colorectal FAP adenomas and carcinomas.
The upstream signals regulating most other EMT-inducing transcription factors (e.g., SNAI1, SNAI2, Twist1, Twist2) are well characterized (1), but it is only recently that the regulation of ZEB1 has begun to be understood. ZEB1 is induced by several pathways triggering an EMT (11, 12), and here we report that ZEB1 is directly activated by β-catenin/TCF4. Redundant regulation of ZEB1 through several EMT-inducing signals suggests that its expression is key in the initiation and/or maintenance of the EMT process.
Although in established CRC cell lines mutation of APC is accompanied by nuclear translocation of β-catenin and activation of β-catenin/TCF4 transcription, most cells in primary CRCs with mutated APC have membranous β-catenin and do not exhibit mesenchymal characteristics (8). The fact that nuclear β-catenin is only found in dedifferentiated cells at the invasive front indicates that APC mutation is a necessary but not sufficient step for nuclear translocation of β-catenin and induction of an EMT (8). Recent evidence has shown that the microenvironment at the invasive front of CRCs—and, more specifically, secretion of factors like hepatocyte growth factor by stromal myofibroblasts in the vicinity of dedifferentiated cancer cells—play an important role in the nuclear translocation of β-catenin and activation of β-catenin/TCF signaling (27). Although not as strong and homogeneous as in FAP carcinomas, β-catenin also accumulates in the nucleus—and starts inducing ZEB1 expression—in relatively well-differentiated adenomas from FAP patients and APCMin/+ mice (5–7). This observation suggests that additional mechanisms beyond the microenvironment at the carcinoma front may regulate β-catenin translocation in FAP and APCMin/+ adenomas. This study found that ZEB1 expression in these samples correlates with levels of nuclear β-catenin across and within tumors (see SI Discussion).
Lynch syndrome patients have germline mutations in mistmach repair genes resulting in microsatellite instability and increased risk of developing CRCs (3). Compared to sporadic CRCs, Lynch CRCs are less invasive and metastatic and have a more favorable prognosis. Over 90% of Lynch CRCs also present mutations of the TGFβ receptor type II compromising their ability to undergo an EMT in response to TGFβ, thus contributing to explain their lower invasiveness (28). Here, we have shown that, contrary to FAP tumors, Lynch CRCs with wild-type APC do not express ZEB1. In this regard, it is worth noting that ZEB1 is not only induced by TGFβ but ZEB1 synergizes with R-Smads to activate TGFβ-dependent genes and is required for TGFβ-mediated induction of EMT (29, 30). It is therefore possible that in the same way redundancy of upstream EMT-inducing signals would ensure ZEB1 expression during tumor progression, lower invasive and metastatic behavior in Lynch CRCs requires the loss of ZEB1 through inactivation of several of these pathways (e.g. wild-type APC plus mutation of TGFβ receptor type II).
Proteolytic cleavage of LAMC2 by MT1–MMP stimulates migration of CRC cells (24), and the coordination induction of both proteins by β-catenin/TCF4 at the front of sporadic CRCs associates to increased invasiveness (25). We found that both genes are upregulated by ZEB1 in APC mutant CRC cells and coexpress with ZEB1 in FAP tumors. (see SI Discussion).
Altogether, our results support a model where APC mutation, factors in the microenvironment, and, potentially, additional signals bring about the nuclear translocation of β-catenin, which upon binding to TCF4 induces ZEB1. ZEB1 would then initiate the EMT process—repressing E-cadherin and other epithelial markers and activating mesenchymal and proinvasive genes, some jointly with β-catenin/TCF4—to promote tumor progression and metastasis. An alternative model would be that induction first of ZEB1 through some unrelated mechanism would downregulate E-cadherin, thus increasing the availability of β-catenin to translocate the nucleus. However, this second possibility could be rebutted because ZEB1 is known to be unable to activate the TOPFLASH reporter (16).
In sum, this work has shown that in intestinal primary tumors and CRC cell lines with mutant APC ZEB1 is a direct target of β-catenin/TCF4 and acts also as an effector of this signaling pathway in regulating genes associated to tumor invasiveness. Given the salience of the β-catenin/TCF4 pathway in CRCs and recent evidence about the role of ZEB1 in tumorigenesis, metastasis, and chemotherapy resistance, the establishment of such link sets ZEB1 not only as an important diagnostic predictor but also as a potential therapeutic target in CRCs.
Materials and Methods
Details of the materials and experimental procedures used are given in SI Materials and Methods. SI Discussion, Figs. S1–S5, and additional references are also included in Supporting Information.
Supplementary Material
Acknowledgments.
We are indebted to researchers who generously provided us with reagents (see Supporting Information). We are also grateful to A. Corbi and R. Gasa for critical reading of the paper. We apologize to researchers whose work was cited indirectly through reviews because of space limitations. This project was funded by grants from Olga Torres Foundation, Spanish Ministry of Science and Innovation (BFU2007-60302, BFU2010-15163), Spanish Association Against Cancer, La Caixa Foundation, and the European Commission to A.P. The salary of E.S.T was partly funded by La Caixa Foundation and the European Commission.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108977108/-/DCSupplemental.
References
- 1.Polyak K, Weinberg R. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 5.Inomata M, et al. Alteration of β-catenin expression in colonic epithelial cells of familial adenomatous polyposis patients. Cancer Res. 1996;56:2213–2217. [PubMed] [Google Scholar]
- 6.Kobayashi M, et al. Nuclear translocation of β-catenin in colorectal cancer. Br J Cancer. 2000;82:1689–1693. doi: 10.1054/bjoc.1999.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng H, et al. Nuclear translocation of β-catenin in hereditary and carcinogen-induced intestinal adenomas. Carcinogenesis. 1998;19:543–549. doi: 10.1093/carcin/19.4.543. [DOI] [PubMed] [Google Scholar]
- 8.Brabletz T, et al. Variable β-catenin expression in colorectal cancer indicates a tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38:5735–5745. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hlubek F, et al. Heterogeneous expression of Wnt/β-catenin target genes within colorectal cancer. Int J Cancer. 2007;121:1941–1948. doi: 10.1002/ijc.22916. [DOI] [PubMed] [Google Scholar]
- 11.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop: A motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Tillo E, et al. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am J Cancer Res. 2011;1:897–912. [PMC free article] [PubMed] [Google Scholar]
- 13.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 14.Cochrane DR, et al. Loss of miR-200c: A marker of aggressiveness and chemoresistance in female reproductive cancers. J Oncol. 2010 doi: 10.1155/2010/821717. 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13:R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spaderna S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Aigner K, et al. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Tillo E, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 19.Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 20.Rosin-Arbesfeld R, Cliffe A, Brabletz T, Bienz M. Nuclear export of the APC tumour suppressor controls β-catenin function in transcription. EMBO J. 2003;22:1101–1113. doi: 10.1093/emboj/cdg105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hlubek F, et al. Expression of the invasion factor laminin γ2 in colorectal carcinomas is regulated by β-catenin. Cancer Res. 2001;61:8089–8093. [PubMed] [Google Scholar]
- 22.Gilles C, et al. Transactivation of vimentin by β-catenin in human breast cancer cells. Cancer Res. 2003;63:2658–2664. [PubMed] [Google Scholar]
- 23.Eger A, et al. δEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 24.Koshikawa N, Giannelli G, Cirulli V, Miyazaki K, Quaranta V. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J Cell Biol. 2000;148:615–624. doi: 10.1083/jcb.148.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hlubek F, Spaderna S, Jung A, Kirchner T, Brabletz T. β-catenin activates a coordinated expression of the proinvasive factors laminin-5 γ2 chain and MT1-MMP in colorectal carcinomas. Int J Cancer. 2004;108:321–326. doi: 10.1002/ijc.11522. [DOI] [PubMed] [Google Scholar]
- 26.Ip YC, Cheung ST, Fan ST. Atypical localization of membrane type 1-matrix metalloproteinase in the nucleus is associated with aggressive features of hepatocellular carcinoma. Mol Carcinog. 2007;46:225–230. doi: 10.1002/mc.20270. [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 28.Pino MS, et al. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology. 2010;138:1406–1417. doi: 10.1053/j.gastro.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by δEF1 proteins in epithelial mesenchymal transition induced by TGF-β. Mol Biol Cell. 2007;18:3533–3544. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFβ/BMP signaling pathway. EMBO J. 2003;22:2443–5242. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.