Abstract
Plastids are the semiautonomous organelles that possess their own genome inherited from the cyanobacterial ancestor. The primary function of plastids is photosynthesis so the structure and evolution of plastid genomes are extensively studied in photosynthetic plants. In contrast, little is known about the plastomes of nonphotosynthetic species. In higher plants, plastid genome sequences are available for only three strictly nonphotosynthetic species, the liverwort Aneura mirabilis and two flowering plants, Epifagus virginiana and Rhizanthella gardneri. We report here the complete sequence of a plastid genome of nonphotosynthetic mycoheterotrophic orchid Neottia nidus-avis, determined using 454 pyrosequencing technology. It was found to be reduced in both genome size and gene content; this reduction is however not as drastic as in the other nonphotosynthetic orchid, R. gardneri. Neottia plastome lacks all genes encoding photosynthetic proteins, RNA polymerase subunits but retains most genes of translational apparatus. Those genes that are retained have an increased rate of both synonymous and nonsynonymous substitutions but do not exhibit relaxation of purifying selection either in Neottia or in Rhizanthella.
Keywords: Neottia nidus-avis, Orchidaceae, plastid genome, mycoheterotrophy, gene loss, substitution rate
Introduction
The information on chloroplast genome sequences is essential in different fields of plant biology: plant physiology, population genetics, phylogenetics, and evolution. As a consequence, the number of published chloroplast genome sequences greatly increased in past three years allowing an assessment of general trends of their evolution (reviewed in Wicke et al. 2011). In contrast, little is known about the structure of the plastid genome in nonphotosynthetic species. In higher plants, complete plastid genome sequences are available now for only three completely nonphotosynthetic species: two angiosperms—the parasitic plant Epifagus virginiana from Orobanchaceae (Wolfe et al. 1992) and mycoheterotrophic orchid, Rhizanthella gardneri (Delannoy et al. 2011), and a mycoheterotrophic liverwort Aneura mirabilis (Wickett, Zhang, et al. 2008). While there is about 3,000 nonphotosynthetic plants representing more than ten families, this is obviously insufficient to infer general patterns of the evolution of plastome in the absence of photosynthetic activity. We have increased this set by sequencing the entire plastome of a mycoheterotrophic orchid, Neottia nidus-avis. Neottia belongs to the same family as Rhizanthella whose plastid genome was recently characterized (Delannoy et al. 2011) but the shift to heterotrophy occurred independently in these two lineages (Molvray et al. 2000). Thus, this information is useful for revealing both general features of nonphotosynthetic plant plastomes and for comparing the plastid genome structure under the parallel loss of photosynthetic activity in a specific plant group, the family Orchidaceae.
The sequence of Neottia plastome was assembled from the partial genomic DNA sequence produced using high-throughput pyrosequencing technology (454 sequencing) complemented with Sanger sequencing. As expected, the plastid genome of Neottia is highly reduced in length (92 Kb compared with 146–149 in photosynthetic orchids) and in gene content (fig. 1). All genes encoding photosystem I and II components are lost or pseudogenized. The same is true for the genes of the cytochrome b6f components and photosystem I assembly proteins; ccsA (involved in c-type cytochrome synthesis) and cemA (chloroplast envelope membrane protein) genes are also lost. All ndh genes are completely lost or turned to pseudogenes. The latter does not however seem to be related to heterotrophic way of life since it is characteristic for several photosynthetic angiosperms, including orchids (Chang et al. 2006; Wu et al. 2010; Blazier et al. 2011) and for gymnosperms (Braukmann et al. 2009). Also, Neottia plastome apparently lacks functional matK gene. It contains a region with high similarity to matK but its comparison with orthologous sequences from photosynthetic orchids reveals strong divergence of its 5′end (including the substitution in the start codon); this suggests that in Neottia, matK is a pseudogene. Despite this, the genes that are supposed to require matK protein activity for the splicing of their mRNAs (rpl2, rps12, clpP, trnA-UGC, trnG-UCC) have retained their introns. This is in contrast with the genus Cuscuta where matK loss observed in the species of subgenus Grammica is correlated with the loss of group IIa introns (McNeal et al. 2009). There are two possible explanations. First, we can imagine that matK-like region retained in Neottia is still functional—this is possible under the condition that alternative start codon is adopted or that multiple RNA editing events occur in the 5′end of this region. Second, the involvement of matK in the splicing of group IIa introns could be not as essential in orchids as it is Cuscuta (and, apparently, in most plants). This hypothesis is supported by the fact that Rhizanthella, another nonphotosynthetic orchid, totally lacks any matK-like regions but also retains group IIa introns and that matK is pseudogenized in Corallorhiza trifida (Freudenstein and Senyo 2008), a mycoheterotrophic species that partially retains photosynthetic activity.
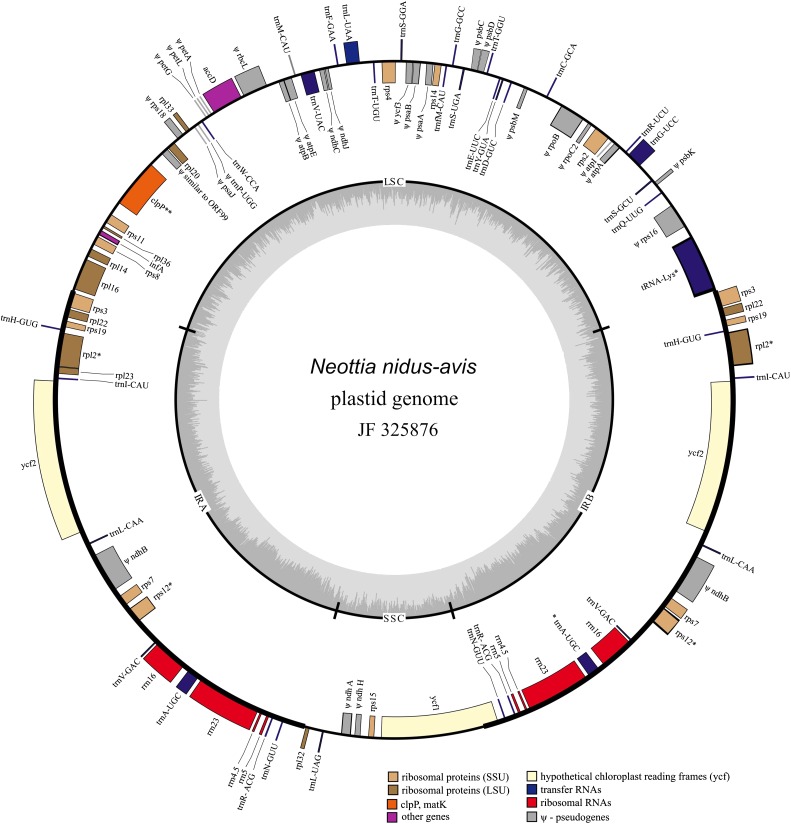
FIG. 1.—
Circular map of the plastid genome of Neottia nidus-avis. Genes shown inside the circle are transcribed clockwise, those outside the circle are transcribed counterclockwise. Asterisks indicate intron-containing genes, dark gray bars inside the inner circle indicate guanine-cytosine content.
The only class of genes that is unaffected by the reduction is the ribosomal RNA genes. All four rRNA genes characteristic for typical plant plastomes are present in Neottia and share very high similarity (96–99%) with their orthologs from other orchids. Genes encoding another essential component of the ribosome, the ribosomal proteins, are also mostly retained: Neottia plastome encodes a complete set of large subunit proteins genes (including rpl23 and rpl22 that are turned to pseudogenes in many photosynthetic species) and most small subunit proteins genes. The functionality of two small subunit proteins genes—rps16 and rps18—is questionable. Though they are highly similar to the orthologous genes from photosynthetic orchids in silico translation reveals internal stop codons in both sequences. These stop codons are however in frame and thus are potential targets of RNA editing so additional experiments are required to confirm the nonfunctionality of rps16 and rps18. In Anthoceros formosae, conversion of nonsense codons into sense is found in 52 genes, including rps18 (Kugita et al. 2003). RNA editing system was shown to be active in Rhizanthella so it is presumably active in Neottia plastids too. Another possible target of RNA editing is the rpl2 gene that has atypical start codon ACG. As for transfer RNA, we were able to find the sequences with high similarity for all tRNAs characteristic for plant plastomes with the exception of trnI-GAU. Most of them seem to be functional genes because they share high similarity with their orthologs from photosynthetic orchids (overall similarity of tRNA sequences between Neottia and Phalaenopsis is 0.95) and have conserved secondary structure typical for tRNAs. trnV-UAC and trnP-UGG are putative pseudogenes since they differ from their orthologs by multiple substitutions and indels. infA gene, which encodes translation initiation factor 1 is also present. In contrast to translation apparatus that seems to be almost unaffected, genes of the transcription machinery—those encoding plastid RNA polymerase subunits—are lost (rpoA, rpoC1) or pseudogenized (rpoB, rpoC2). Two genes involved in plastid metabolism—accD and clpP—are retained, as well as two large ORF encoding proteins of the unknown function—ycf1 and ycf2. The retention of ycf1 is the most enigmatic since it is presumably pseudogenized in photosynthetic orchids Oncidium and Phalaenopsis. The regions with strong similarity to ycf1 are present in their plastomes but their in silico translation reveals multiple stop codons due to nontriplet indels. Pseudogenization of ycf1, as well as another large plastid ORF, ycf2 occurs in grasses where a region similar to ycf1 is present but highly reduced due to multiple deletions (Hiratsuka et al. 1989). The structure of ycf1 in Phalaenopsis and Oncidium may represent a first stage of gene degradation. Another explanation of the frameshifts might be related to sequencing errors because ycf1 is rich in homopolymer regions, which are prone to errors. Broader survey of ycf1 integrity in orchids is required to see if this gene is indeed nonfunctional in either lineage of orchids.
Despite about one-third reduction in length, the overall structure of Neottia plastid genome is conserved and collinear with that of photosynthetic orchids. The only alteration of the gene order is the position of inverted repeat-large single copy border that resides in the rps3-rpl16/rps3-trnK spacer in Neottia. The expansions of the IR are well documented in different lineages of angiosperms; in particular in orchids IR was found to include complete sequence of rps19, trnH, and a part of rpl22 (Wang et al. 2008). In Neottia, further expansion of the IR is observed—it includes rpl22 and rps3. Such structure of JIR-LSC is by now unique however it is possible that increased taxon sampling will reveal similar structures in other orchids, either photosynthetic or not.
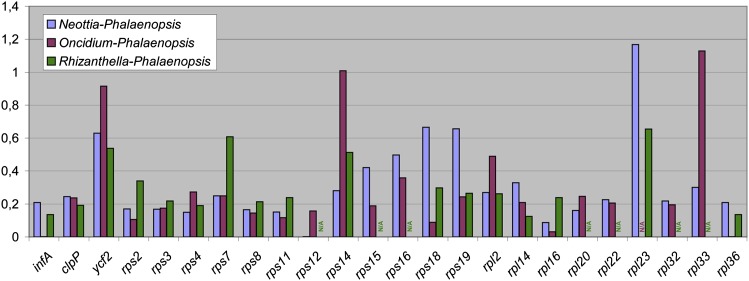
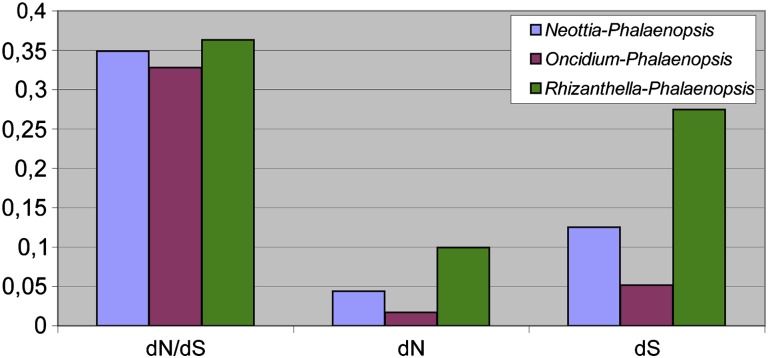
To assess the selection constraint on plastid genes in Neottia, we performed the analysis of synonymous and nonsynonymous substitutions in protein-coding genes. Rhizanthella, another nonphotosynthetic orchid was also included in the analysis. The examination of dN/dS ratio for individual genes that are shared between Neottia, Rhizanthella, and photosynthetic orchids demonstrates that most of them are evolving under purifying selection in both nonphotosynthetic species (fig. 2). The only Neottia gene where dN/dS is greater than 1 is rpl23. The sequence of this gene is highly conserved between Phalaenopsis and Oncidium, differing by single substitution (nonsynonymous). rpl23 is a pseudogene in several photosynthetic plants, for example, Caryophylalles (Logacheva et al. 2008). High dN/dS in this gene may indicate on the relaxation of selection constraint as initial stage of pseudogenization. However, in rps14 and rpl33, dN/dS is found to be increased in photosynthetic species. Such increase may be caused by the unreliability of dN/dS estimates for short genes (Haddrill et al. 2007) and thus not be biologically relevant. To get a generalized view on substitution rate in Neottia and Rhizanthella, we calculated substitution values for all genes combined in one sequence. This demonstrated that dN/dS ratios are very similar between two photosynthetic species and between photosynthetic and nonphotosynthetic ones. In contrast, the number of both synonymous and nonsynonymous substitutions differs greatly being about 2.5 times much in Neottia and 5 times in Rhizanthella (fig. 3). The apparent effect of higher substitution rate is often observed in annual plants when compared with perennials (e.g., Yue et al. 2010); however, both Neottia and Rhizanthella are perennials as well as Phalaenopsis and Oncidium. This suggests that this effect is related to their heterotrophic way of life and that plastid genes in nonphotosynthetic orchids have higher mutation rate but retain their functionality.
FIG. 2.—
Pairwise dN/dS (the ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site) for all genes shared between photosynthetic orchids and Neottia and/or Rhizanthella. N/A indicates the cases where it was not possible to estimate the ratio due to the absence of synonymous substitutions or the absence of gene in the plastid genome.
FIG. 3.—
dN/dS ratio and dN and dS values for all genes shared between Neottia, Rhizanthella, Oncidium, and Phalaenopsis, combined in one sequence.
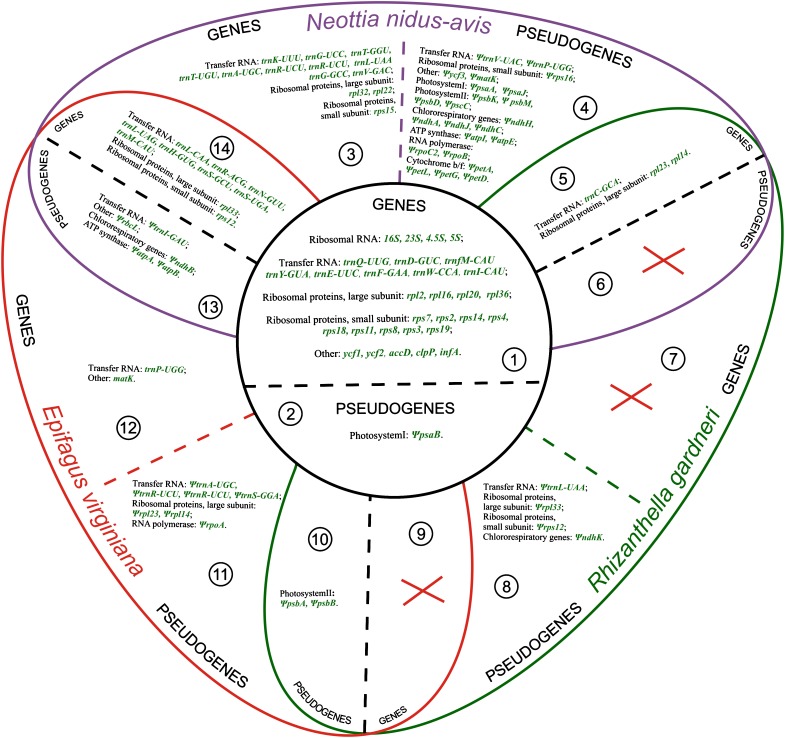
The broader comparison that includes all nonphotosynthetic higher plant plastomes—two orchids, parasitic dicot Epifagus and liverwort Aneura—reveals both parallelisms and dissimilarities in their structure. The plastome sizes differ almost twice—from about 108 Kb in Aneura to 59,190 bp in Rhizanthella. The degree of gene loss and pseudogenization also differs—in Aneura, the plastome is much less affected than in angiosperms. It retains all transfer RNA, RNA polymerase, and ribosomal protein genes and also many photosynthesis-related genes (Wickett, Zhang, et al. 2008) being similar in this respect to the species of Cuscuta that retain photosynthetic activity (Funk et al. 2007; McNeal et al. 2007). In strictly nonphotosynthetic angiosperms not a single photosynthesis-related gene is retained as intact reading frame, even in Neottia that is characterized by the least reduction in both size and gene content and is only 16 Kb less than in Aneura. In general, the structure and gene content of plastome are more similar within nonphotosynthetic angiosperms than in any of them compared with Aneura. It is well known that the activity of the plastid is not provided by plastid genome only but also is highly dependent on and coordinated with that of nuclear genome (reviewed in Taylor 1989 and Woodson and Chory 2008). The results from an ongoing genomic project of a liverwort, Marchantia polymorpha, indicate that the organization of the nuclear genomes in angiosperms and in liverworts differs greatly; in particular, the diversity of gene families is lower in Marchantia (Yamato et al. 2011). This and other differences in nuclear genome organization can account for the difference in the structure of plastid genomes in nonphotosynthetic angiosperms and nonphotosynthetic liverworts. Other possible explanation is the time past from the moment of switch to heterotrophy. Though there are no direct (paleontological) observations on this subject, we suggest that heterotrophy is the most recent in A. mirabilis—since another species of the genus Aneura are photosynthetic (Wickett, Fan, et al. 2008) and the most ancient in Epifagus which shares the holoparasitic way of life with related genera Conopholis and Orobanche (dePamphilis et al. 1997). Neottia and Rhizanthella represent genera that include only nonphotosynthetic species but their most closely related genera are completely or partially photosynthetic. Moreover, in some systematic treatments, Neottia is merged with photosynthetic genus Listera (Chase et al. 2003) that suggests recent switch to heterotrophy, similar to Aneura. In terms of gene content, Neottia is more similar to Epifagus than to Rhizanthella (fig. 4) or Aneura. Phylogenetically Neottia is close to Rhizanthella thus the dissimilarity of their plastid genomes is unlikely to be related to major dissimilarities in the organization of their nuclear genomes but rather to lineage-specific variations in rate and pattern of plastid genome evolution. This phenomenon is well known with regard to single nucleotide substitutions (reviewed in Muse 2000), and it is likely to be applicable to length mutations as well.
FIG. 4.—
Gene and pseudogene content in the plastomes of nonphotosynthetic angiosperms, numbers indicate different groups of genes/pseudogenes according to their presence in the plastomes: (1) genes that are present in all plastid genomes of nonphotosynthetic angiosperms, (2) pseudogenes that are present in all plastid genomes of nonphotosynthetic angiosperms, (3) genes that are unique for Neottia, (4) pseudogenes that are unique to Neottia, (5) genes present only in Neottia and Rhizanthella, (6) pseudogenes present only in Neottia and Rhizanthella (no), (7) genes unique to Rhizanthella (no), (8) pseudogenes unique to Rhizanthella, (9) genes present only in Rhizanthella and Epifagus (no), (10) pseudogenes present only in Rhizanthella and Epifagus, (11) pseudogenes unique to Epifagus, (12) genes unique to Epifagus, (13) pseudogenes present only in Epifagus and Neottia, and (14) genes present only in Epifagus and Neottia.
Among about 100 of genes encoded in typical plastid genome, only 29 are shared in all the plastid genomes of nonphotosynthetic plants. This core set of genes constitutes of ribosomal and transfer RNA and ribosomal protein genes, translation initiation factor (infA), protease subunit (clpP), acetyl-CoA carboxylase subunit (accD) genes, and two large ORF of unknown function—ycf1 and ycf2 (fig. 4). This is in contrast with the situation observed in experimental systems of transition to heterotrophy. Calli cultured for a long time on sucrose-rich media also undergo multiple deletions in plastid DNA and extensive gene losses. However, their size and location drastically differ in different calli samples and do not seem to converge to a gene set shared by all samples. Moreover, the loss of circular structure typical for plastid DNAs, is reported (Harada et al. 1992; Abe et al. 2002). The stability of plastid genomes of nonphotosynthetic plants—the conservation of typical quadripartite structure, the existence of shared gene set, and the evidence of purifying selection acting on these genes—suggests that the evolution of plastid genomes in nonphotosynthetic plants is dictated by the constraints that are obviously different from those acting on photosynthetic plant plastomes but are not less strong.
Orchidaceae is a perfect model system for deeper study of the evolution of nonphotosynthetic plants’ plastomes because it contains many cases of independent transition to nonphotosynthetic lifestyle (Molvray et al. 2000), from rather ancient occurring at the level of tribe to extremely recent occurring in certain individuals within the population (Tranchida-Lombardo et al. 2010). Moreover, many photosynthetic orchids are able to use fungi as secondary carbon source being mixotrophic rather than fully autotrophic (Bidartondo et al. 2004; Abadie et al. 2006). Mixotrophy is thought to be the preadaptation that mediates the transition to completely heterotrophic lifestyle (Selosse and Roy 2009). It is interesting to infer if any of the changes in the plastome characteristic for strictly nonphotosynthetic plants (e.g., gene loss, increase of substitution rate) are observed in mixotrophic species. We expect that further sampling of the plastid genome sequences from the species of Orchidaceae representing different stages and different times of transition to mycoheterotrophy will provide valuable information about the evolution of plastomes in nonphotosynthetic plants.
Materials and Methods
Total DNA was extracted from the above-ground part of three Neottia plants using NucleoSpin Plant II kit (Macherey-Nagel, Germany). DNA (10 μg) was used for sequencing with Roche Genome Sequencer FLX system using the Titanium kit (454 Life Sciences). Sequencing was performed at the University of Illinois at Urbana-Champaign, W.M. Keck Center for Comparative and Functional Genomics. The sample was run on a half of a picotiter plate. The sequencing resulted in 590640 reads with an average read length of 528 bp. The output from the sequencing system in Standard Flowgram Format was converted to FASTA. Then a BLAST database was made from the resulting FASTA file and a plastome of Oncidium, the photosynthetic orchid was queried against it. All reads with e-value lower then 10−10 were used for de novo assembly using MIRA assembler ver. 3.2.0 (Chevreux et al. 1999). The assembly resulted in 23 contigs with length more than 1,000 nucleotides. Two largest contigs (11,141 and 49,963 nucleotides) were used as a basis for the generation of draft version of the Neottia plastome. They were aligned with Oncidium plastome using the whole VISTA genome alignment tool (Frazer et al. 2004). Then Sanger sequencing was used to fill the gaps. To check the accuracy of the assembly and to correct possible 454 sequencing errors associated with homopolymer runs, several regions were sequenced by Sanger sequencing using the primers designed on the base of the assembly (supplementary table S1, Supplementary Material online). Average coverage for regions derived from 454 assembly is assessed as 29.8×; for regions sequenced by Sanger sequencing it is 2×. Polymerase chain reaction (PCR) amplification was performed on Biometra T300 thermal cycler using Encyclo PCR kit (Evrogen, Russia). PCR conditions were as follows: initial denaturation 3 min at 94 °C, then 35 cycles of 15 s at 94 °C, 25 s at 59 °C, and 1–5 min (depending on the expected length of the product) at 72 °C. Sanger sequencing was performed in the interinstitutional sequencing center at Engelhardt Institute of Molecular Biology (Moscow, Russia) using ABI PRISM BigDye Terminator kit v. 3.1 with following analysis on ABI PRISM 3730 genetic analyzer (Applied Biosystems). Initial annotation was produced using DOGMA (Wyman et al. 2004). Then manual correction and adjustment, that included alignment of every Oncidium and Phalaenopsis gene with Neottia plastome sequence was performed. The regions with similarity to known protein-coding genes but lacking intact ORF were classified as pseudogenes. To detect tRNAs pseudogenes, each tRNA-like sequence was analyzed using tRNAscan-SE (Lowe and Eddy 1997) and by comparison with its putative orthologs from Oncidium and Phalaenopsis. Those sequences that lack typical tRNA folding and/or differ from their orthologs by multiple indels or substitutions were considered to be pseudogenes. The map of Neottia plastome was visualized using OGDRAW online tool (Lohse et al. 2007) with further manual correction. Assembled and corrected sequence of Neottia plastome was deposited in the GenBank under accession number JF325876.
For dN/dS calculation, all protein-coding plastid genes that are shared between Neottia, Rhizanthella, Oncidium, and Phalaenopsis were included in the analysis. Alignment was performed using ClustalW (Thompson et al. 1994); dN/dS ratios were calculated using codeml program from the package PAML 4.3 (Yang 2007). In the alignments of several genes, nontriplet indels or in-frame stop codons were found near 3′-end. In these cases, only the part of sequence before presumable indel or stop codon was used for dN/dS calculation.
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Dmitry Sokoloff and Margarita Remizowa (Moscow State University, Russia) for providing plant material for the study; Alvaro Hernandez (University of Illinois at Urbana-Champaign) for 454 sequencing; Viktoria Shtratnikova (Center of Innovation and Technology of Biologically Active Compounds, Russia); Georgiy Bazykin (Institute for Information Transmission Problems, Russia) for helpful discussion; and three anonymous reviewers for the suggestions and comments on the manuscript. This work was supported by the Russian Foundation for Basic Research (project no. 11-04-02031) and Ministry of Education and Science of Russian Federation (state contract no.1054 and project 11.G34.31.0008).
References
- Abadie J-C, et al. Cephalanthera longifolia (Neottieae, Orchidaceae) is mixotrophic: a comparative study between green and nonphotosynthetic individuals. Can J Bot. 2006;84(9):1462–1477. [Google Scholar]
- Abe T, Ii N, Togashi A, Sasahara T. Large deletions in chloroplast DNA of rice calli after long-term culture. J Plant Physiol. 2002;159:917–923. [Google Scholar]
- Bidartondo MI, Burghardt B, Gebauer G, Bruns TD, Read DJ. Changing partners in the dark: isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proc Biol Sci. 2004;271(1550):1799–1806. doi: 10.1098/rspb.2004.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier CJ, Guisinger MM, Jansen RK. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae) Plant Mol Biol. 2011;76(3–5):263–272. doi: 10.1007/s11103-011-9753-5. [DOI] [PubMed] [Google Scholar]
- Braukmann TW, Kuzmina M, Stefanović S. Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr Genet. 2009;55(3):323–337. doi: 10.1007/s00294-009-0249-7. [DOI] [PubMed] [Google Scholar]
- Chang CC, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Chase MW, Freudenstein JF, Cameron KM. DNA data and Orchidaceae systematics: a new phylogenetic classification. In: Dixon KW, Kell SP, Barrett RL, Cribb PJ, editors. Orchid conservation. Kota Kinabalu (Malaysia): Natural History Publications; 2003. pp. 69–89. [Google Scholar]
- Chevreux B, Wetter T, Suhai S. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics. 1999. Genome sequence assembly using trace signals and additional sequence information. Vol. 99; 1999 Oct 4--6; Hanover (Germany) [Google Scholar]
- Delannoy E, Fujii S, des Francs CC, Brundrett M, Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28(7):2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis CW, Young ND, Wolfe AD. Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rate variation. Proc Natl Acad Sci U S A. 1997;94(14):7367–7372. doi: 10.1073/pnas.94.14.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenstein JV, Senyo DM. Relationships and evolution of matK in a group of leafless orchids (Corallorhiza and Corallorhizinae; Orchidaceae: Epidendroideae) Am J Bot. 2008;95:498–505. doi: 10.3732/ajb.95.4.498. [DOI] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8(2):R18. doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Ishikawa R, Niizeki M, Saito K. Pollen-derived rice calli that have large deletions in plastid DNA do not require protein synthesis in plastids for growth. Mol Gen Genet. 1992;233(1–2):145–150. doi: 10.1007/BF00587572. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217(2–3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003;31(9):2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Samigullin TH, Dhingra A, Penin AA. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale—a wild ancestor of cultivated buckwheat. BMC Plant Biol. 2008;8:59. doi: 10.1186/1471-2229-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS One. 2009;4(6):e5982. doi: 10.1371/journal.pone.0005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molvray M, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influences of floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Collingwood (Australia): CSIRO Publishing; 2000. pp. 441–448. [Google Scholar]
- Muse SV. Examining rates and patterns of nucleotide substitution in plants. Plant Mol Biol. 2000 42 (1): 25–43. [PubMed] [Google Scholar]
- Selosse MA, Roy M. Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci. 2009;14(2):64–70. doi: 10.1016/j.tplants.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Taylor WC. Regulatory interactions between nuclear and plastid genomes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:211–233. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranchida-Lombardo V, et al. Spatial repartition and genetic relationship of green and albino individuals in mixed populations of Cephalanthera orchids. Plant Biol (Stuttg). 2010;12(4):659–667. doi: 10.1111/j.1438-8677.2009.00252.x. [DOI] [PubMed] [Google Scholar]
- Wang RJ, et al. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. Evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76(3–5):273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Fan Y, Lewis PO, Goffinet B. Distribution and evolution of pseudogenes, gene losses, and a gene rearrangement in the plastid genome of the nonphotosynthetic liverwort, Aneura mirabilis (Metzgeriales, Jungermanniopsida) J Mol Evol. 2008;67(1):111–122. doi: 10.1007/s00239-008-9133-1. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, et al. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 2008;25(2):393–401. doi: 10.1093/molbev/msm267. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 1992;89(22):10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9(5):383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yamato KT, Ishizaki K, Kohchi T, Floyd S, Bowman J. Abstracts of the XVIII International Botanical Congress; 2011 Jul 23–30; Melbourne (Australia) 2011. The genome of Marchantia polymorpha L; p. 107. Available from: http://www.ibc2011.com/downloads/IBC2011_Abstract_Book.pdf. [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yue JX, et al. Genome-wide investigation reveals high evolutionary rates in annual model plants. BMC Plant Biol. 2010;10:242. doi: 10.1186/1471-2229-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]