Abstract
The formation of new transcription factor–binding sites (TFBSs) has a major impact on the evolution of gene regulatory networks. Clearly, single nucleotide mutations arising within genomic DNA can lead to the creation of TFBSs. Are molecular processes inducing single nucleotide mutations contributing equally to the creation of TFBSs? In the human genome, a spontaneous deamination of methylated cytosine in the context of CpG dinucleotides results in the creation of thymine (C → T), and this mutation has the highest rate among all base substitutions. CpG deamination has been ascribed a role in silencing of transposons and induction of variation in regional methylation. We have previously shown that CpG deamination created thousands of p53-binding sites within genomic sequences of Alu transposons. Interestingly, we have defined a ∼30 bp region in Alu sequence, which, depending on a pattern of CpG deamination, can be converted to functional p53-, PAX-6-, and Myc-binding sites. Here, we have studied single nucleotide mutational events leading to creation of TFBSs in promoters of human genes and in genomic regions bound by such key transcription factors as Oct4, NANOG, and c-Myc. We document that CpG deamination events can create TFBSs with much higher efficiency than other types of mutational events. Our findings add a new role to CpG methylation: We propose that deamination of methylated CpGs constitutes one of the evolutionary forces acting on mutational trajectories of TFBSs formation contributing to variability in gene regulation.
Keywords: CpG methylation, CpG deamination, evolution of transcription factor–binding sites, evolution of gene regulatory elements, Alu transposon
Introduction
CpG Deamination and Evolution of Transcription Factor–Binding Sites
The creation of transcription factor–binding sites (TFBSs) is a fundamental evolutionary process shaping gene regulatory networks. Two mechanisms have been recognized to facilitate generation of new TFBSs: insertional activity of transposable elements resulting in spreading of TFBSs (Bourque 2009) and mutational events. Some insight into the robustness of the latter mechanism has been gained in the context of evolutionary studies reporting widespread turnover of TFBSs in mammalian genomes (Dermitzakis and Clark 2002; Horvath et al. 2007; Odom et al. 2007; Kasowski et al. 2010).
However, up to this point, the unequal contribution of molecular processes inducing single nucleotide mutational events in DNA has not been taken into account when deciphering evolutionary trajectories leading to creation of TFBSs. In vertebrates, the mutation of cytosine to thymine (C → T) in the context of CpG dinucleotides has the highest rate among all base substitutions and is reflected in the ongoing genomic depletion of CpG dinucleotides. On the molecular level, this is triggered by methylation of cytosine followed by a spontaneous deamination event creating TpG or CpA dinucleotides (Razin and Riggs 1980).
We have previously shown that methylation and deamination of CpGs embedded within Alu transposons in the human genome resulted in generation of thousands of p53-binding sites with the preferred core motif composed of CpA and TpG dinucleotides (Zemojtel et al. 2009). We reasoned that this phenomenon should not be limited to Alu sequences and thus were expecting to gather evidence that CpG deamination has created multiple binding sites in multiple regions of the human genome. Second, as CpG deamination–driven mutagenesis is characterized by the highest rate among all substitutions, we wanted to learn if it is more efficient in creating binding sites than the nondeamination-driven mutagenesis. How can we define a proper creation efficiency measure in this context? Here, we address these issues and provide evidence that CpG deamination is indeed the important evolutionary process impacting on TFBS formation.
Materials and Methods
Ancestral Sequence Reconstruction
We reconstructed human and chimp ancestral sequences extending a model developed by Arndt and Hwa (2005). Initially, we downloaded multiple alignments of vertebrate genomes to hg18 human genome from the University of California–Santa Cruz (UCSC) Genome Bioinformatics Site. In the analysis, we considered only alignments which contained aligned sequences of all three species: human, chimp, and rhesus. We extracted alignment fragments overlapping the putative promoter regions and the in -vivo bound regions.
We inferred the nucleotide substitution frequencies along the two terminal branches from the human–chimp ancestor to human and chimp as well as the trinucleotide distribution for the human–chimp ancestor using the maximum likelihood-based method developed by Duret and Arndt (2008).
The nucleotide substitution frequencies were then used to compute the transition probabilities of any trinucleotide at the human–chimp ancestor into a trinucleotide in human and likewise in chimp. Given the states of 5′ and 3′ adjacent nucleotides in human and their orthologous partners in chimp and rhesus, we could then infer the ancestral nucleotide distribution of the nucleotide in the middle position. We reconstructed ancestral nucleotides according to this distribution.
Prediction of TFBS
Position frequency matrices (PFMs) available in the Transfac 9.4 database as well as de novo profiles constructed from ChIP-Seq data (Celniker et al. 2009; Kunarso et al. 2010) were used to model binding of transcription factors.
Predictions with PFMs
We predicted binding sites using the method described by Rahmann et al. (2003). First, based on a positional frequency matrix, we calculated a corresponding score matrix and a cutoff threshold. Next, we scanned both strands of a DNA sequence and checked whether it contained a motif of a score at least equal to the threshold. We normalized the thresholds for each matrix expecting the same number of false positives within studied sequence regions.
Predictions with IUPAC Consensus Sequence
In order to identify binding sites of c-Myc transcription factor in genome-wide ChIP-Seq regions (Zeller et al. 2006), we used a consensus-match strategy. For a site prediction, we demanded a perfect match to one of the two c-Myc consensuses: CACGTG (canonical motif) and CACATG or CATGTG (noncanonical).
Identification of Potential Creation Events
We scanned the ancestral sequences in order to identify “potential creation events,” that is, all sequence locations where a single nucleotide mutation could lead to creation of a new TFBS. Specifically, the latter is achieved by enumerating all possible single nucleotide mutations in the ancestral sequence and then checking whether a new binding site is predicted for the mutated sequence.
Estimation of Number of TF Recognizing Products of Deamination
Based on PFMs, we estimated the fraction of TFs recognizing products of deamination. For each of 217 Transfac PFMs, we calculated how many CA or TG dinucleotides will be on average present in the predicted motifs, using the formula: ∑i {f[ i,C] * f[ i + 1, A] + f[ i,T] * f[i + 1, G]}, where f(i, N) denotes the frequency of observation of nucleotide N at position i within the PFM. We observed that on average at least 85% of PFMs recognize a motif with CA or TG dinucleotides.
Classification of Putative Promoter Regions
We defined putative promoter regions based on the gene coordinates provided by EnsEMBL database (version 47). After Weber et al. (2007), we extracted regions from 700 bp upstream to 200 bp downstream around transcription start sites of the genes.
We classified the promoters into two groups: high CpG set (HCP) and low CpG set (LCP) depending on their sequence composition. We adopted a classification introduced by Weber et al. (2007). That is, a promoter is added to the HCP if it contains a fragment of 500 bp with GC content larger than 0.55 and ratio of observed to expected CpG dinucleotides higher than 0.75. Moreover, if the ratio is never larger than 0.48 for any 500 bp fragment of a promoter, it is classified in the LCP.
Annotation of Known TF-Binding Motifs in Putative Promoter Regions
Due to redundancies observed in the Transfac database, we used a subset of available vertebrate PFMs. For each transcription factor which has more than a single PFM assigned, we chose only the PFM with the highest information content. This selection process resulted in a set of 217 PFMs used in our study. We annotated this set of TF-binding motifs in the putative promoter regions.
Predicted Creation Events in the LCP and HCP Promoters
We provide two text files listing predicted TFBS creation events in the LCP and HCP promoters (supplementary text file S1 and S2, Supplementary Material online). The data columns contain the following information: name of a transcription factor, genomic location of the new site in the human genome (hg18), the reconstructed ancestral sequence at the location, the corresponding sequence in human hg18 genome, a list of differences/mutations between the ancestral sequence and the corresponding human sequence, and the list of the differences that are interpreted as deamination events. Finally, the last column describes potential single mutation events, which when occurred in the ancestral sequence could result in a binding site. Events marked with a star are interpreted as deamination events.
Analyses of In Vivo Bound Sequences of Oct-4, Nanog, Ctcf, and c-Myc
Genome-wide regions bound in human embryonic stem cells by Oct-4, Nanog, and Ctcf TFs have been identified in the ChIP-Seq experiments described in Kunarso et al. (2010). In total, respectively, 2.1 Mbase, 7.6 Mbase, and 7.5 Mbase of sequence alignable to chimp and rhesus genomes have been studied (supplementary table 2, Supplementary Material online).
Genome-wide regions bound by the c-Myc TF in a model human B cell line, P493 (Zeller et al. 2006) have been downloaded from the UCSC database (table wgEncodeGisChipPetMycP493 for the human genome version hg18). We studied only regions identified by at least three independent ChIP-PET tags. It has been demonstrated in 29 ChIP-qPCR assays that clusters associated with at least three ChIP-PET tags (PET-3+) always corresponded to Myc binding (Zeller et al. 2006). In total, we analyzed 1168 regions, which represented 1.26 Mbp of sequence alignable to chimp and rhesus genomes.
Analyses of In Vivo Bound Sequences of TFs from ENCODE Consortium Data Set
We downloaded from the UCSC genome browser database the genome wide–binding locations of TFs identified in conducted by the ENCODE consortium ChIP-Seq experiments (in human cell lines) (Celniker et al. 2009). We used our pipeline to calculate binding site creation efficiencies for these TFs. In order to reduce estimation error of creation efficiencies, we selected only those experiments for which the number of identified deamination-driven creation events or nondeamination-driven creation events was at least 5. This resulted in evolutionary analysis of nine TF-binding data sets (wgEncodeHudsonalphaChipSeqPeaks tracks: Rep2Gm12878Irf4, Rep1Gm12878Nrsf, Rep2Gm12878Pax5n19, Rep2Gm12878Pbx3, Rep2Hepg 2RxraPcr1x, Rep1K562Usf1, and wgEncodeYaleChIPseqPeaks tracks: K562Nfya, K562Nfyb, and K562bYy1). The characteristics of the ENCODE ChIP-Seq regions are reported in (supplementary table 2, Supplementary Material online). We used MEME algorithm to identify motifs recognized by the TFs. The identified motifs were matched by these previously published in the Transfac database (Transfac ID numbers: Irf4: M00972, Nfy: M00209, NRSF: M01028, Pax5: M00143, Pbx3: M00998, Rxra: M00518, Usf1: M00121, and YY1: M00069).
Results/Discussion
Identification of TFBS Creation Events
In order to reveal the role of CpG deamination in TFBS formation, we were interested in tracing the mutational events that led to the creation of TFBSs in the human promoters after the split of human and chimp. Thus, we reconstructed the sequences of the human–chimp ancestor promoters employing multiple sequence alignments of human, chimp, and rhesus (see Material and Methods). In this approach, rhesus constitutes an outgroup to infer the directionality of mutations. Using 217 TF matrices obtained from the TRANSFAC database (constituting a diverse repertoire of motifs recognized by transcription factors, see Materials and Methods), we annotated binding sites in the human promoters and in the reconstructed human–chimp ancestor promoters (see Material and Methods). Of all annotated TFBSs in the human promoters, ∼96.5% also get annotated at the orthologous positions in the ancestral sequence. The remaining ∼3.5% (48861) of sites comprises a group of candidates for TFBSs created in the human promoters since the split of human and chimp lineages. Out of those putative creation events, the vast majority (∼96%) was due to a single nucleotide mutation event and thus we focused on such creation events in our further analysis. We further subdivided the single nucleotide creation events into two groups. These are, likely “CpG deamination–driven creation events” encompassing CpG → CpA or TpG mutational events, accounting for ∼22% of all single nucleotide creation events and “nondeamination-driven creation events” harboring all other mutational events.
High Efficiency of CpG Deamination–Driven Creation Events in Human Promoters
On the first look, the CpG deamination–driven creation events might seem to constitute a rather modest fraction of all creation events specific to the human lineage. However, in order to interpret this result, the following has to be taken into account. The number of TFBSs created by CpG deamination events after the split of human and chimp lineages depends primarily on the following two factors:
The mutational rate of CpGs.
The number and the composition of the CpG-containing motifs residing in the human–chimp ancestral sequence.
In connection with point 2, it has to be emphasized that CpG dinucleotides are underrepresented in the mammalian genomes, including promoter regions (Saxonov et al. 2006).
We have thus set out to derive a new measure of a propensity of CpG deamination events to create binding sites that account for above factors. First, building on our observation that the vast majority (96%) of creation events in the human lineage were single nucleotide mutation events and point 2 above, we annotated all single nucleotide mutation events that could potentially lead to creation of a TFBS within the reconstructed human–chimp ancestral sequence. Any such mutation event, we call a “potential TFBS creation event.” Depending on their origin, we distinguish two classes of potential TFBS creation events: potential CpG deamination–driven creation events (defined as mutations of Cs within CpGs) and potential nonCpG deamination–driven creation events (encompassing all the remaining mutations). Clearly, only a fraction of all annotated potential creation events in the ancestral sequence occurred after the split of human and chimp lineages. We compute this fraction by introducing the measure of creation efficiency (CE). We define CEdeam for CpG deamination–driven creation events as the ratio of the number of CpG deamination–driven creation events that occurred after the split of human and chimp to the number of all potential CpG deamination–driven creation events in the ancestral sequence.
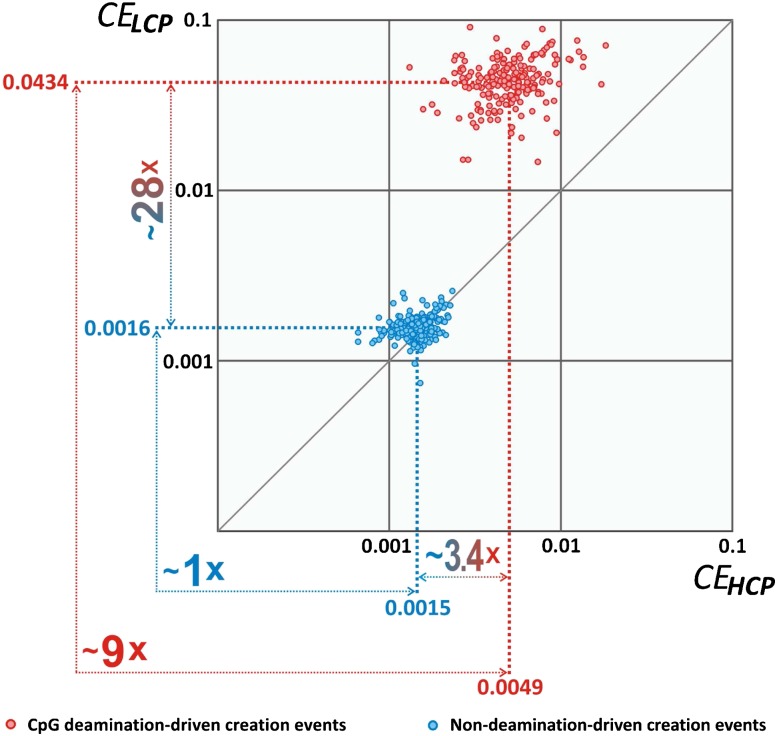
Analogously, we define CEnondeam for nonCpG deamination–driven creation events. How efficient are CpG deamination–driven creation events in comparison with nondeamination–driven creation events? In order to answer this question, we compute for each of 217 TFs (represented by a set of 217 TF matrices) two types of CE: CEdeam and CEnondeam. In addition, we determine average CEdeam and CEnondeam values over all 217 TFs. We performed the calculation separately on two distinct classes of human promoters (Saxonov et al. 2006) characterized by low (LCPs) and high contents of CpG dinucleotide (HCPs). Importantly, LCPs were shown to be mostly methylated, whereas the vast majority of HCPs are hypomethylated in somatic cells (Weber et al. 2007). As depicted in figure 1, the average CEdeam value in LCPs was ∼0.04 meaning that on average 4 of 100 potential CpG deamination–driven creation events took place in LCPs. This was ∼9-fold higher than in HCPs where the average CEdeam value was ∼0.005. In contrast, the average CEnondeam value was approximately equal in HCPs and LCPs (fig. 1). In LCPs, CpG deamination–driven creation events occurred ∼28-fold more frequently than nondeamination-driven creation events, and in HCPs, we obtained a value of ∼3.4-fold enrichment. These data are compatible with a ∼20.4-fold higher rate of CpG mutation (into products of deamination CpA or TpG) in LCPs when compared with HCPs (not shown). In line with these observations, it has been established that in germ line cells (the cells contributing genetic material to the next generation), CpGs are constitutively methylated in LCPs and, in contrast, CpGs are largely unmethylated in HCPs (Weber et al. 2007).
FIG. 1.—
CpG deamination creates TFBSs with high efficiency in human promoters. Each point represents a CE value calculated for each of 217 TFs. The horizontal axis represents CE values recorded in HCPs, and the vertical axis represents CE values obtained in LCPs. Average CE values for CpG deamination–driven creation events (CEdeam) and nondeamination–driven creation events (CEnondeam) are highlighted in bold red and blue digits, respectively.
These results strongly indicate that CpG deamination–driven depletion of CpG dinucleotide, which has been recognized to contribute to variation in regional methylation (Feinberg and Irizarry; Kerkel et al. 2008), serves as an efficient mechanism for generation of new TFBSs.
CpG Deamination Creates In Vivo Binding Sites
Since the evolutionary analysis of binding sites annotated in the human promoter regions revealed a remarkable efficiency of CpG deamination–driven creation events, we were interested if the same could be observed for TFBSs detected in “genome-wide” experiments. Interestingly, we found that >85% of 217 TF-binding matrices recognize on average at least one product of deamination, CpA or TpG (Materials and Methods) and reasoned that a large number of TFs is capable of interacting with these dineuclotides in vivo.
We identified 13 TF-binding data sets generated in genome-wide ChIP-Seq experiments in human cells (in large part produced by the ENCODE consortium; Celniker et al. 2009, table 1) for which we could reliably estimate creation efficiencies (see Materials and Methods). The 13 TFs belong to the following major classes: helix-loop-helix/leucine zipper, helix-turn-helix/homeodomain/Paired box/Tryptophan clusters, Cys2His2 zinc finger domain, Cys4 zinc finger of nuclear type, and histone fold class. The values of CEdeam for the 13 TFs were ∼4-fold to ∼25-fold higher (and on average ∼11-fold higher) than the corresponding values of CEnondeam (table 1). The latter values fall in the range observed for the annotated binding sites in the promoter regions (fig. 1). On average, per TF, CpG deamination–driven creation events comprised ∼25% of all binding site creation events (table 1). This all indicated that CpG deamination events strongly contributed to creation of in vivo TFBSs.
Table 1.
Numbers of Created In Vivo Binding Sites and Corresponding Creation Efficiencies
| Factor | d+ | d− | Nond+ | Nond− | CEdeam | CEnondeam | CEdeam/CEnondeam | P value | % of d+ |
| c-Myc_canonical | 5 | 187 | 13 | 4050 | 0.026042 | 0.003200 | 8.0 | 9.4E−4 | 27.8 |
| c-Myc_noncanonical | 26 | 976 | 21 | 11706 | 0.025948 | 0.001791 | 14.5 | 3.4E−17 | 55.3 |
| Ctcf | 90 | 3356 | 832 | 199548 | 0.026117 | 0.004152 | 6.3 | 5.2E−40 | 9.8 |
| Irf4 | 24 | 769 | 327 | 136133 | 0.030265 | 0.002396 | 12.6 | 1.9E−18 | 6.8 |
| Nanog | 70 | 972 | 246 | 91794 | 0.067178 | 0.002672 | 25.1 | 2.2E−111 | 22.2 |
| Nfya | 21 | 1207 | 50 | 27906 | 0.017101 | 0.001788 | 9.6 | 7.1E−13 | 29.6 |
| Nfyb | 22 | 1417 | 70 | 34235 | 0.015288 | 0.002041 | 7.5 | 9.4E−12 | 23.9 |
| Nrsf | 5 | 195 | 10 | 4207 | 0.025000 | 0.002371 | 10.5 | 3.5E−4 | 33.3 |
| OCT4 | 33 | 382 | 99 | 24123 | 0.079518 | 0.004039 | 19.7 | 2.4E−29 | 25.0 |
| Pax5 | 83 | 9321 | 244 | 146697 | 0.008826 | 0.001661 | 5.3 | 1.4E−29 | 25.4 |
| Pbx3 | 10 | 609 | 35 | 15217 | 0.016155 | 0.002294 | 7.0 | 6.1E−6 | 22.2 |
| Rxra | 26 | 1299 | 83 | 55298 | 0.019622 | 0.001498 | 13.1 | 4.0E−19 | 23.8 |
| Usf1 | 52 | 506 | 272 | 17701 | 0.093190 | 0.015134 | 6.2 | 2.1E−23 | 16.0 |
| YY1 | 8 | 1354 | 27 | 19203 | 0.005874 | 0.001404 | 4.2 | 1.3E−3 | 22.9 |
NOTE.—The following values are listed: the numbers of observed new site creations due to CpG deamination events (d+) and nondeamination events (nond+); the numbers of potential deamination and nondeamination-driven creation events which could lead to site creation but have not been observed (d− and nond−, respectively); creation efficiencies for deamination-driven creation events (CEdeam), nondeamination-driven creation events (CEnondeam), and percentages of deamination-driven creation events (% of d+). The P value was calculated with Fisher’s exact test.
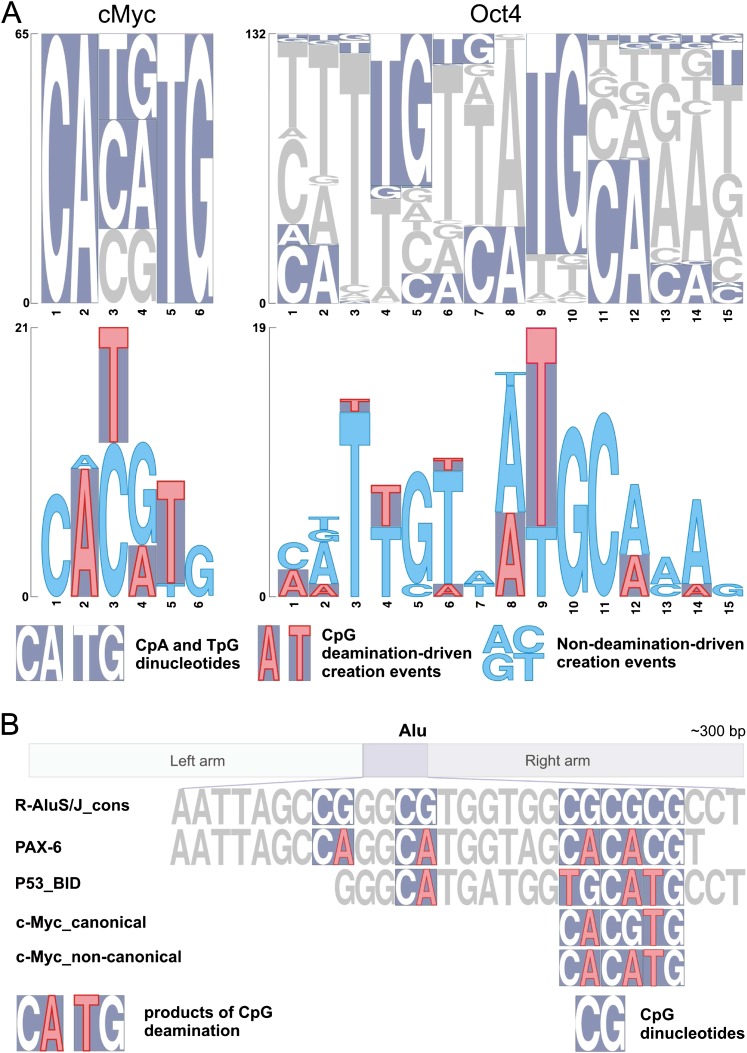
In the following, we illustrate the significance of CpG deamination–driven creation events with an evolutionary analysis of in vivo TFBSs of such key transcription factors as c-Myc, Nanog, Oct4, and Ctcf. It is important to note that these factors contain CpA and TpG dinucleotides as part of their binding motifs (fig. 2A; supplementary figs. 1 and 2, Supplementary Material online). c-Myc is known to bind to two different E-box motifs composed of CpA, TpG, and CpG dinucleotides: the so called canonical Myc E-box 5′-CACGTG-3′ and noncanonical Myc E-box 5′-CACATG-3′/5′-CATGTG-3′ (Zeller et al. 2006). Our analysis of in vivo c-Myc sites detected in a chromatin immunoprecipitation with the paired-end ditag experiment (ChIP-PET) (Zeller et al. 2006) revealed that CpG deamination–driven creation events make up as much as 56% of noncanonical and 28% of canonical c-Myc site creation events, and as expected, they are localized at positions 2, 3, 4, and 5 within c-Myc motifs (fig. 2A and table 1). The values of CEdeam for canonical and noncanonical c-Myc sites were 8- and 14-fold higher, respectively, than the values of CEnondeam. Likewise, the strong contribution of CpG deamination events was also observed for c-Myc sites annotated in the two classes of human promoters (supplementary table 1, Supplementary Material online). In LCPs, CpG deamination–driven creation events constituted as much as 78% of all noncanonical c-Myc site creation events, and the ratio of the CEdeam and CEnondeam values was ∼51.
FIG. 2.—
CpG deamination drives creation of in vivo TFBSs. (A) Upper panel: Histogram of c-Myc- and Oct4-binding sites created via single nucleotide mutation events. The presence of the CpA and TpG dinucleotides within individual binding sites is highlighted by a violet background. Lower panel: Histogram of single nucleotide mutation events leading to creation of TFBSs. CpG deamination–driven creation events and non-CpG deamination–driven creation events are depicted in red and blue, respectively. (B) PAX-6-, c-Myc-, and p53-binding sites are created via deamination of methylated CpGs in Alu transposons. R-AluS/J_cons: consensus sequence of the CpG-containing region in the right arm of AluS and AluJ subfamilies.
Moreover, we found further evidence suggesting that CpG deamination created Myc E-box sites. Interestingly, a recent study reported the identification of a canonical Myc E-box site in AluS sequences (Wang et al. 2009). Likewise, we identified noncanonical Myc E-box sites to reside in sequences of AluS transposons (fig. 2B). Our literature searches identified experimental studies in which two canonical Myc E-box sites within AluJ and AluS elements inserted in the second intron of the CDC25A gene (Galaktionov et al. 1996) and in the distal promoter region of the KIR gene (Cichocki et al. 2009), respectively, were shown to function as Myc-responsive elements. Provocatively, we found that all these canonical and noncanonical c-Myc sites are located in one particular location in AluS/J, which overlaps with the p53-binding site previously described by us (Zemojtel et al. 2009) (fig. 2B). The reconstructed consensus sequences of AluS and AluJ subfamilies contain the CGCGCG sequence at the location corresponding to the Myc-binding site. Thus, we conclude that like p53 sites, they were also created via CpG deamination after the insertion of Alu transposons into the genome. Specifically, two and three CpG deaminations are required to create from a CGCGCG template the canonical and the noncanonical c-Myc sites, respectively. Interestingly, it has been reported that methylation of the CpG dinucleotide present in the canonical E-box inhibits Myc binding both in vitro (Prendergast et al. 1991) and in vivo (Perini et al. 2005). In light of this, spontaneous deamination of the methylated CpG in a canonical Myc-binding site would result in the creation of a noncanonical-binding site, which is desensitized to methylation.
Likewise, evolutionary analysis of TFBSs of Oct4, Nanog, and Ctcf detected in human ES cells (Kunarso et al. 2010) also pointed to a strong contribution of CpG deamination–driven creation events. For Oct4, Nanog, and Ctcf, we obtained, respectively, a 24-, 22-, and 6-fold enrichment of CpG deamination–driven creation events when compared with nondeamination–driven creation events (table 1). CpG deamination events constituted 25%, 22%, and 10% of all creation events for Oct4, Nanog, and Ctcf, respectively (table 1 and fig. 2A; supplementary fig. 3, Supplementary Material online). For example, it can be seen that CpG deamination–driven creation events were abundant at positions corresponding to nucleotides 4, 8, 9, and 12, where CA and TG dinucleotides are present within the Oct4-binding motif (fig. 2A, supplementary fig. 1, Supplementary Material online). We provide here experimental evidence supporting the notion that single nucleotide mutations from CpG to TpG (resulting from CpG deamination) such as seen at position 9 in figure 2A (bottom panel) can create functional Oct4-binding motifs. Recently, an SNP at position 9 in a putative Oct4 motif (as referenced in fig. 2A) was discovered in a patient with Beckwith–Wiedemann syndrome (Demars et al. 2010). The mutation from T to C occurred in the context of a TpG dinucleotide and resulted in the creation of a CpG dinucleotide in the patient sequence; WT: GTTTGAGATGCTAAT → P: GTTTGAGACGCTAAT. The study used in vitro GEMSA (Gel Electrophoretic Mobility Shift Assay) with nuclear extract from Oct4-overexpressing cells to provide evidence that Oct4 did not bind to the sequence variant containing CpG (P) but only to the one having a TpG instead of CpG.
Together, these results highlight the efficiency of CpG deamination events in the creation of TFBS.
Originally, it has been postulated that a key role of CpG methylation and deamination is in the inactivation of transposons and thus in protecting mammalian genomes from their insertional activity (Yoder et al. 1997; Zemach and Zilberman 2010). One-third of all CpGs in the human genome are located within Alu transposons. Over time, CpG deamination permanently inactivates initially CpG-rich Alus. As a side effect of this process, decaying Alu sequences give birth to new regulatory elements. In particular, the CpG-rich ∼20 nt long template sequence residing in Alu elements (fig. 2B), can be converted via means of deamination into p53 (Zemojtel et al. 2009), PAX-6 (Zhou et al. 2002), and c-Myc TFBSs as documented here (fig. 2B).
This phenomenon is not limited to these three TFs and Alu transposons. By analyzing mutational events leading to creation of TFBSs in human promoters (217 TFBSs) and in genome-wide regions bound in vivo (14 TFBSs), we document that CpG deamination events create TFBSs with much higher efficiency than other single nucleotide mutational events. In a recent study reporting genome-wide locations of Ctcf sites in human genome, it has been noticed that orthologous Ctcf-binding sequences in vertebrate genomes accumulated at a very high rate C → T mutations at the position where the CpG dineuclotide is located in the Ctcf-binding motif (position 15 as referenced in the supplementary fig. 2, Supplementary Material online) (Kim et al. 2007). This observation is compatible with CpG deamination as a driving process. In light of this, it is tempting to speculate that CpG deamination might in fact constitute a double-edged sword involved not only in creating but also in inactivating binding sites. In this context, we propose that CpG deamination, which is known to induce variable regional methylation in human populations, constitutes an evolutionary benefit facilitating innovation in gene regulation.
Supplementary Material
Supplementary figures 1–3, Supplementary tables 1 and 2, and Supplementary files 1 and 2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors gratefully acknowledge the funding received from the Max Planck Society.
References
- Arndt PF, Hwa T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics. 2005;21:2322–2328. doi: 10.1093/bioinformatics/bti376. [DOI] [PubMed] [Google Scholar]
- Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki F, et al. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demars J, et al. Analysis of the IGF2/H19 imprinting control region uncovers new genetic defects, including mutations of OCT-binding sequences, in patients with 11p15 fetal growth disorders. Hum Mol Genet. 2010;19:803–814. doi: 10.1093/hmg/ddp549. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kim TH, et al. Analysis of the vertebrate insulator protein Ctcf-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perini G, Diolaiti D, Porro A, Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc Natl Acad Sci U S A. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Rahmann S, Muller T, Vingron M. On the power of profiles for transcription factor binding site detection. Stat Appl Genet Mol Biol. 2003;2 doi: 10.2202/1544-6115.1032. Article7. [DOI] [PubMed] [Google Scholar]
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bowen NJ, Mariño-Ramírez L, Jordan IK. A c-Myc regulatory subnetwork from human transposable element sequences. Mol Biosyst. 2009;5:1831–1839. doi: 10.1039/B908494k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Zilberman D. Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol. 2010;20:R780–R785. doi: 10.1016/j.cub.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Zemojtel T, Kielbasa SM, Arndt PF, Chung HR, Vingron M. Methylation and deamination of CpGs generate p53-binding sites on a genomic scale. Trends Genet. 2009;25:63–66. doi: 10.1016/j.tig.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Zheng JB, Gu X, Saunders GF, Yung WK. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 2002;12:1716–1722. doi: 10.1101/gr.188302. [DOI] [PMC free article] [PubMed] [Google Scholar]