Abstract
Purpose
The development and implementation of a pharmacist-managed Clinical Pharmacogenetics service is described.
Summary
Therapeutic drug monitoring (TDM) is a well-accepted role of the pharmacist. Pharmacogenetics, the study of genetic factors that influence the variability in drug response among patients, is a rapidly evolving discipline that integrates knowledge of pharmacokinetics and pharmacodynamics with modern advances in genetic testing. There is growing evidence for the clinical utility of pharmacogenetics, and pharmacists can play an essential role in the thoughtful application of pharmacogenetics to patient care.
A pharmacist-managed Clinical Pharmacogenetics service was designed and implemented. The goal of the service is to provide clinical pharmacogenetic testing for gene products important to the pharmacodynamics of medications used in our patients. The service is modeled after and integrated with an already established Clinical Pharmacokinetics service. All clinical pharmacogenetic test results are first reported to one of the pharmacists, who reviews the result and provides a written consult. The consult includes an interpretation of the result and recommendations for any indicated changes to therapy. In 2009, 136 clinical pharmacogenetic tests were performed, consisting of 66 TPMT tests, 65 CYP2D6 tests, and 5 UGT1A1 tests. Our service has been met with positive clinician feedback.
Conclusion
Our experience demonstrates the feasibility of the design and function of a pharmacist-managed Clinical Pharmacogenetics service at an academic specialty hospital. The successful implementation of this service highlights the leadership role that pharmacists can take in moving pharmacogenetics from research to patient care, thereby potentially improving patient outcomes.
Therapeutic drug monitoring (TDM) involves the measurement of drug concentrations in biological fluid and the interpretation of those concentrations in order to individualize therapy. TDM applies the principles of both pharmacokinetics (the time course of drug concentrations) and pharmacodynamics (the concentration-effect relationship). Traditional TDM requires close collaboration between the clinical laboratory and the pharmacist to ensure appropriate sample collection and timely interpretation of results.1 The benefits of pharmacist-directed TDM through a Clinical Pharmacokinetics service have been well-documented and include decreased adverse effects which translate into decreased length of treatment, length of hospital stay, and reduced costs.2–4
Pharmacist-managed Clinical Pharmacokinetics services are now common in most hospitals, and ASHP states that clinical pharmacokinetic monitoring is a “fundamental responsibility of all pharmacists.”5 Results of a 2008 ASHP survey showed that pharmacists at 83.7% of hospitals surveyed provide clinical pharmacokinetics consultations, and 97.2% of respondents agreed that pharmacokinetic dose adjustment is an essential pharmacy service.6 Reports in the literature have demonstrated clear clinical and economic benefits of these pharmacist-managed services.7–9
Pharmacogenetics is the study of genetic factors that influence the variability in drug response among patients. As a discipline, pharmacogenetics integrates knowledge of pharmacokinetics and pharmacodynamics with modern advances in genetic testing. Of particular relevance to this science are polymorphisms in genes encoding drug-metabolizing enzymes, drug transporters, and drug targets, and the effect of these polymorphisms on drug efficacy and toxicity in individual patients.10 Over the past decade, pharmacogenetics has been widely incorporated into pharmacological research and drug development initiatives.11 The potential role for pharmacogenetics in reducing adverse drug reactions (ADRs) was highlighted in a systematic review which found that the disposition of the majority of 27 drugs frequently involved in ADRs is influenced by genetic factors, suggesting that many ADRs may be prevented by individualizing drug therapy based on patients’ genetic profiles.12
In 2001, Ensom et al. predicted that the “TDM of the future” would include pharmacogenetics-oriented TDM and that pharmacists would play a major role in interpreting the results of these new clinical laboratory-based tests.13 In recent years, the Food and Drug Administration (FDA) has approved incorporation of genetic information to guide dosing in product labeling for specific drugs, such as mercaptopurine, irinotecan, cetuximab, trastuzumab, abacavir, clopidogrel, and warfarin. 14,15 The incorporation of pharmacogenetics into product labeling, combined with the increasing availability of genotyping tests from Clinical Laboratory Improvement Amendments of 1988 (CLIA)-certified reference laboratories, has facilitated the transition of pharmacogenetics from a research endeavor into clinical practice.16,17
Pharmacogenetics-based testing can help determine which individuals may benefit from a specific drug and the most appropriate dose of a drug.18 Unlike traditional TDM, which is not performed until after a drug is administered to the patient, clinical pharmacogenetics-oriented TDM can be conducted even before treatment begins, decreasing the “trial and error” period or even eliminating certain drugs from consideration.13 Optimally, clinical pharmacogenetics and TDM can play complementary roles in managing patients’ drug therapy to achieve positive outcomes. The following variables may be considered in assessing the importance of clinical pharmacogenetic testing, as suggested by Phillips et al.: 1) widely used drugs with a high incidence of toxicity (i.e. narrow therapeutic index); 2) prevalence of variant alleles of relevant gene is high enough to warrant use of genetic information; 3) severe toxicity associated with use; 4) current methods for monitoring response or evaluating toxicity are inefficient or inadequate; 5) sufficient evidence exists for a relationship between genetic factors and response or patient outcomes; 6) an assay is available that can rapidly, reliably and inexpensively detect the variant alleles; and 7) clinicians are able to interpret the results and use the information.19
Several studies have suggested clinical pharmacogenetic testing can result in cost reduction.20 Screening of patients treated with thiopurines for thiopurine methyltransferase (TPMT) polymorphisms has been shown to be cost-effective in the treatment of several disease states.21–23 Other research has shown CYP2C19 genotyping to be cost-effective when incorporated into treatment decisions for Helicobacter pylori infection, 24 HLA-B*5701 genotyping to be cost-effective in preventing abacavir hypersensitivity in patients with HIV,25 and HER2 testing to be cost-effective when incorporated into treatment decisions for metastatic breast cancer.26
Research studies conducted over 2 decades have shown that TPMT-deficient patients treated with conventional doses of thiopurines are predisposed to drug-induced complications due to accumulation of excessive intracellular concentrations of thioguanine nucleotide metabolites.27 Clinical interest in TPMT pharmacogenetics is based on studies showing that TPMT genotype or phenotype can identify patients at high risk of hematopoietic toxicity after thiopurine therapy.28,29 Moreover, studies have shown that ALL patients with at least one TPMT-variant allele may respond better to thiopurine therapy when compared to those with two wild-type TPMT alleles, with no difference in long-term treatment efficacy.30–32
With these findings firmly established in the research setting, we sought to move the use of genotype testing for relevant genes, such as TPMT, into the clinical care of our patients.
The purpose of this report is to describe the development and implementation of a pharmacist-managed clinical pharmacogenetics service at an academic pediatric hospital. The review of patient charts was approved by the Institutional Review Board.
Description of the Program
St. Jude Children’s Research Hospital is a 60-bed inpatient facility that also has nearly 60,000 outpatient visits per year. Since the early 1980’s, the Pharmaceutical Department at St. Jude Children’s Research Hospital has operated both the clinical and laboratory aspects of a Clinical Pharmacokinetics service, which provides TDM for a variety of drugs administered to patients with catastrophic illnesses (eg. cancer, HIV) in a clinical research setting. The Pharmaceutical Department employs 10 advanced practitioner pharmacists with post-PharmD residency training and substantial professional experience, who spend the majority of their time focused on clinical activities. These pharmacists practice as part of multidisciplinary inpatient and outpatient care teams and also are involved in clinical research.
TDM samples are usually ordered by pharmacists through collaborative practice agreements, and samples are subsequently assayed by one of 5 medical technologists in the Clinical Pharmacokinetics laboratory. All TDM results are first reported to one of our pharmacists who reviews and evaluates the results to provide a consultation documented within the computerized medical record regarding any indicated changes to drug therapy. A pharmacist provides a consult for every concentration or series of concentrations for a course of therapy. In 2009, our Clinical Pharmacokinetics laboratory processed 6,405 clinical specimens, and our pharmacists conducted 4,205 clinical pharmacokinetics consults for aminoglycosides/glycopeptides, chemotherapy agents, antiepileptic agents, azole antifungals, calcineurin/mTOR inhibitors, 99mTc-DTPA, and others.
The Clinical Pharmacogenetics service, which we describe herein, was designed to integrate with and mirror the functions of our Clinical Pharmacokinetics service. Any member of the hospital’s clinical staff may order these tests when clinically indicated. As with TDM samples, pharmacists may order pharmacogenetic tests under a collaborative practice agreement. The genotyping tests offered through the Clinical Pharmacogenetics service are clinical tests and do not require special consent to be obtained. All clinical pharmacogenetic test results are first reported to a pharmacist who reviews the result and provides a written consult. The consult includes an interpretation of the test result and recommendations for any changes to therapy.
Development and Implementation
In 2005, the initial steps in implementing a Clinical Pharmacogenetics service were undertaken. We began by communicating the usefulness of clinical pharmacogenetic testing to pharmacists and other clinicians involved in direct patient care. The possibility of offering this service was discussed at weekly multi-disciplinary clinical research program meetings and clinical pharmacy conferences. Enthusiastic support for the service was provided by clinicians throughout the institution.
By reviewing the hospital’s formulary, drugs were identified that are metabolized by polymorphic enzymes with commercially available genotyping tests. We identified drugs meeting the criteria previously described by Phillips et al. that are used in our patient population.33 Based on these criteria, initial efforts were focused on two clinical pharmacogenetic tests, thiopurine methyltransferase (TPMT) and uridine glucuronosyltransferase 1A1 (UGT1A1). TPMT catalyses the S-methylation of azathioprine, mercaptopurine and thioguanine,34 which are all inactive prodrugs that require metabolic conversion to thioguanine nucleotides or inactivation via TPMT. UGT1A1 is a polymorphic gene involved in the inactivation pathway of irinotecan, a camptothecin analogue approved for treatment of colorectal cancer. This polymorphism is also is associated with Gilbert syndrome, a mild form of indirect hyperbilirubinemia. In 2007, we expanded the service to include a third test for cytochrome P450 2D6 (CYP2D6). CYP2D6 is a polymorphic gene involved in the transformation of up to 25% of clinically useful drugs, including codeine, tamoxifen, antiarrythmics, neuroleptics, and tricyclic antidepressants (Table 1).
Table 1.
Pharmacogenetic tests selected for inclusion in Clinical Pharmacogenetics Service and key affected drugs.
| Gene | Key Affected Drugs on Institution’s Formulary |
Effect of variant allele | FDA-approved genetic information in product labeling? |
|---|---|---|---|
| Thiopurine methyltransferase (TPMT) 30–32 | 6-mercaptopurine Thioguanine Azathioprine |
Severe, sometimes fatal, hematological toxicity24,33–35 |
Yes |
| Cytochrome P450 2D6 (CYP2D6)36,37 | Codeine | CYP2D6 poor metabolizers and receive no analgesic effect from codeine;38 ultrarapid metabolizers of CYP2D6 are at very high risk of toxicity to codeine39 |
Yes |
| Uridine glucuronosyltransferase 1A1 (UGT1A1) |
Irinotecan | Increased potential for hematological or gastrointestinal toxicity40 |
Yes |
Over 3 months in Fall 2005, our faculty conducted a series of ACPE-accredited educational seminars for our pharmacists to establish competencies in providing pharmacogenetic consults for the genes to be tested by the Clinical Pharmacogenetics service. Understanding of the material was assessed by use of an exam. The pharmacists most likely to use pharmacogenetics in their practice took advantage of additional self-study and collaboration with other clinicians. Laboratory staff members were trained on proper pharmacogenetic sample handling and reporting of results. Test names and written consult templates were created in our institution’s electronic medical record system for each of the genes to be measured. The templates facilitate patient-specific written consultations by pharmacists for each pharmacogenetic test result. Each consult contains relevant test data, patient specific information, and the pharmacist’s clinical assessment. As part of the electronic medical record, the completed consult is available electronically to all clinicians.
A cost analysis was performed to determine the feasibility of conducting the selected genotyping tests in-house. Based on our institution’s anticipated usage (anticipated < 100 tests per year for each test) and associated costs, we chose to send our samples for genotyping to an outside laboratory selected from several reference laboratories that offer pharmacogenetic testing services. Insurance reimbursement was established according to the American Medical Association’s Current Procedural Terminology (CPT) codes for each test; however no insurance reimbursement was established for the pharmacist’s consults.
The mission statement of the Clinical Pharmacokinetics Laboratory was broadened to include pharmacogenetic testing. The mission statement now reads, “Provide state of the art therapeutic drug monitoring and pharmacogenetic testing that will be interpreted by pharmacists to assure optimal drug dosing.” A new departmental policy was created to provide direction to pharmacists and laboratory staff involved in the ordering and reporting of clinical pharmacogenetic test results. The policy lists the genes available for testing, the relevant drugs metabolized by each polymorphic enzyme, and mandates that a pharmacy consultation accompany all clinical pharmacogenetic test results in the electronic medical record. Each test requires between 2 and 5 mL of whole blood which is sent overnight to the testing laboratory, where DNA is extracted for the genotyping tests and the genotyping test is performed using a polymerase chain reaction-based method. Results are then transmitted to the Clinical Pharmacokinetics laboratory.
The availability of this new service was communicated to medical staff. Because many of our patients are treated on clinical trials incorporating pharmacogenetics research objectives, our medical staff is familiar with the concept of pharmacogenetics, and overall has been accepting of the services provided by the Clinical Pharmacogenetics service. Information about the available tests and pharmacist services was placed on the Clinical Pharmacokinetics Laboratory’s intranet site, which is accessible by all hospital employees.
To assess use and relevance of the pharmacogenetic tests and services offered, a steering committee was formed, consisting of department leaders and the Clinical Pharmacokinetics laboratory director. The committee meets to evaluate use of available tests, new evidence for implementation of additional tests, and other service quality metrics (eg. test turn-around times).
Experience with the Program
Implementation of the Clinical Pharmacogenetics service occurred in Fall 2005, with tests for TPMT and UGT1A1 offered. In 2007, a genotyping test for CYP2D6 was added. Table 2 summarizes the number of each type of pharmacogenetic test ordered by year. In 2009, 136 clinical pharmacogenetic tests were performed, consisting of 66 TPMT tests, 65 CYP2D6 tests, and 5 UGT1A1 tests. It is estimated that each clinical pharmacogenetics pharmacy consult requires between 20 minutes and 2 hours of a pharmacist’s time, depending on the complexity of the result and of the consult. The median turn-around time for these send-out tests has been 11 days (range 4–25 days). The relatively long turn-around time associated with sending tests to a reference lab could mean that the results might not be available swiftly enough to be useful to avoid toxicity with early doses of the monitored drug and to maximize efficacy. For this reason, as much as possible, we have implemented pre-emptive pharmacogenetic testing of TPMT and CYP2D6 genotyping beginning with patients on our frontline treatment protocol for acute lymphoblastic leukemia, a population that will receive thiopurines and who is at risk for pain control issues. With pre-emptive testing, a blood sample is obtained from a patient who is likely to need a medication. The genotyping results can be generated “up front,” (i.e. soon after a patient begins treatment and before the patient is expected to require the pharmacogenetically-monitored drug) and could then be available to the clinician at the time of the point-of-care decision on whether the drug should be prescribed and what dosage should be given. The proactive use of pharmacogenetic tests is one of the ultimate goals for integration of pharmacogenetics into clinical care.
Table 2.
Number of pharmacogenetic tests ordered by calendar year
| Gene | Number of Tests Performed in 2005 |
Number of Tests Performed in 2006 |
Number of Tests Performed in 2007 |
Number of Tests Performed in 2008 |
Number of Tests Performed in 2009 |
|---|---|---|---|---|---|
| Thiopurine methyltransferase (TPMT) |
64 | 54 | 62 | 62 | 66 |
| Cytochrome P450 2D6 (CYP2D6) |
0 | 0 | 8 | 60 | 65 |
| Uridine glucuronosyltransferase 1A1 (UGT1A1) |
0 | 8 | 3 | 9 | 5 |
One challenge in implementing a Clinical Pharmacogenetics service is the speed with which the science and information advance. The Clinical Pharmacogenetics Service steering committee has proved valuable to review the tests offered on a regular basis to be certain the tests remain relevant and useful. Requests from clinicians to implement new pharmacogenetic tests are evaluated by this committee. In one such use, the steering committee evaluated the evidence for UGT1A1 clinical genotyping. In the intervening time since UGT1A1 testing was made available by our service, clinical findings emerged to indicate that the UGT1A1 genotype does not correlate closely with irinotecan toxicity in pediatric cancer patients receiving the schedule of irinotecan used at our institution.35 After evaluating the clinical data, the steering committee elected to continue offering UGT1A1 testing, but not to advocate its use for all patients receiving irinotecan.
A second potential obstacle to incorporating pharmacogenetics into clinical practice is the issue that many practicing health care professionals have not had formal training in the field of pharmacogenetics.36 For pharmacogenetics to be a useful clinical tool, clinicians must be able to interpret the results and appropriately use the information to make decisions about drug therapy. We addressed this concern by ensuring that our pharmacists were trained and educated in the pharmacogenetic considerations for the drugs covered by our service. The written policy of our service guarantees that a pharmacogenetic test result will always appear in the electronic medical record along with a pharmacy consultation, which provides interpretation and suggestions for changes to drug therapy, when indicated. The clinical laboratory staff alerts the pharmacist on service of each new pharmacogenetics test that is ordered, so that the order can be reviewed for appropriateness, and discussed with the patient’s care team if necessary.
Of the 66 patients whom we genotyped for TPMT through the Clinical Pharmacogenetics service in 2009, 5 (7.6%) patients were determined to have one variant allele, which results in intermediately low TPMT enzyme activity, placing these patients at increased risk for toxicity to thiopurines.37 For each patient determined to be heterozygous for a variant TPMT allele, the pharmacists' recommendation was to presecribe a decreased dose of mercaptopurine at a maximum of 80% of a normal daily dose for patients with ALL (e.g., maximum dose of 60 mg/m2/day for heterozygous patients instead of a full dose of 75 mg/m2/day). By genotyping for TPMT pre-emptively, the information of each patient’s heterozygous status was placed in the chart before the first dose of mercaptopurine was prescribed.
We conducted CYP2D6 genotyping for 65 patients in 2009. Of those, 49 (75.4%) patients were determined to be extensive metabolizers, defined as individuals carrying 1 or 2 (normal) functional alleles; 4 (6.2%) patients were determined to be intermediate metabolizers, defined as individuals carrying either 2 reduced-function alleles or one non-functional and one reduced-function allele; 4 (6.2%) patients were determined to be poor metabolizers, defined as individuals carrying 2 non-functional alleles; and 3 (4.6%) patients were determined or presumed to be ultrarapid metabolizers, defined as individuals carrying 3 or more (normal) functional alleles. Our service takes action on patients determined to be poor metabolizers by alerting the clinical team that these patients are at high risk for a lack of response to codeine, as the ability to form the active metabolite from codeine requires a functional CYP2D6 gene. Likewise, we take action on patients determined to have a duplication of the CYP2D6 gene by alerting the clinical team that this patient may be an "ultrarapid metabolizer" of drugs metabolized by CYP2D6, such as codeine, and that these patients are at high risk for toxicity to "normal" doses of codeine. A choice of a nother analgesic is recommended for pain control in these patients. We have had a 100% acceptance rate by physicians of the therapeutic recommendations made by pharmacists in the pharmacogenetics consults.
While new clinical findings have brought into question the usefulness of the UGT1A1 genotype in choosing an irinotecan dose in our patient population,38 our experience is that the UGT1A1 genotype test remains a valuable clinical tool. This test is beneficial for diagnosing Gilbert syndrome, which aids in ruling out drug-related toxicity in patients presenting with persistent hyperbilirubinemia. We determined UGT1A1 genotype in 5 patients being treated for cancer who had hyperbilirubinemia in 2009; the results revealed that all 5 patients had two variant *28 alleles. This clinical information has been added to each patient’s permanent medical record, allowing the patient care team to make informed decisions about whether to hold further doses of chemotherapy and to individualize future therapy for drugs which are metabolized by UGT1A1.
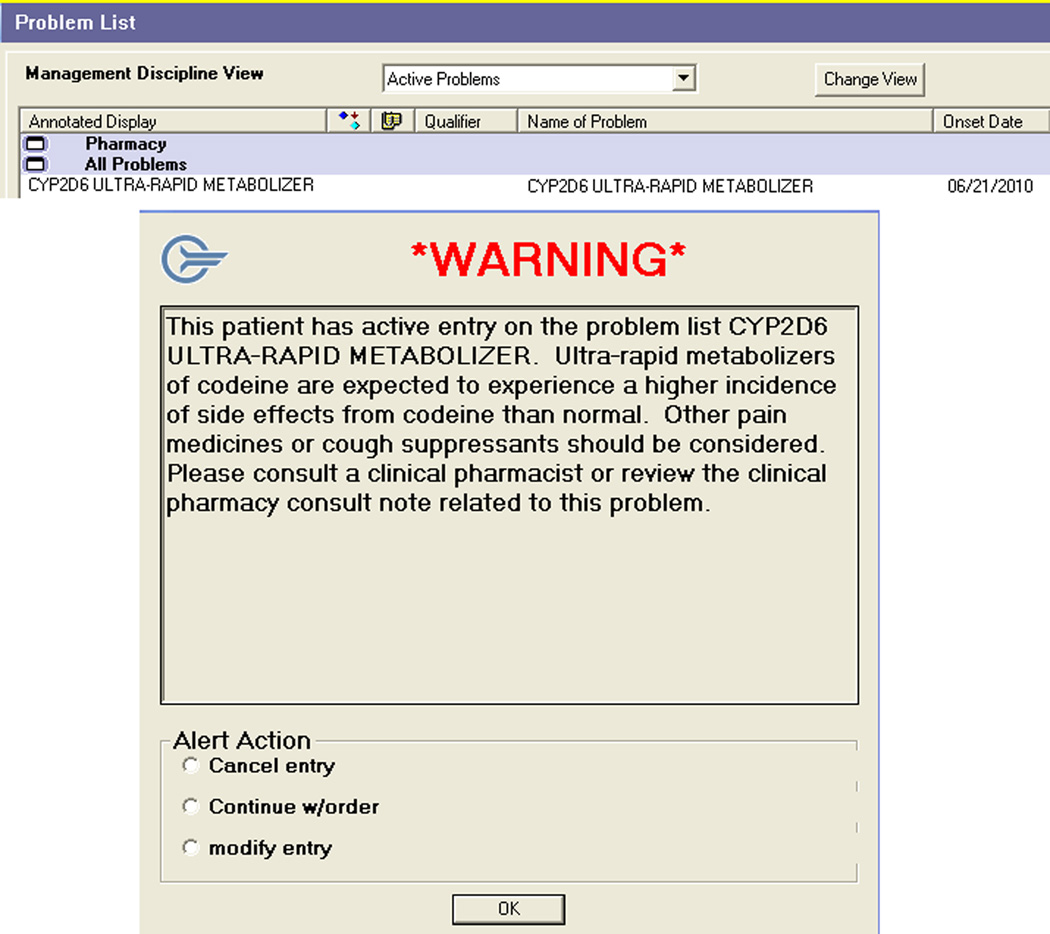
To make certain that genetic information is used for drug therapy decision-making throughout a patient’s care at our hospital, automated clinical decision support rules have been created to facilitate the consistent application of pharmacogenetic information in specific instances. For example, in the case of CYP2D6 and codeine, when the pharmacist interprets the genetic test result and determines a patient to be a poor or ultra-rapid metabolizer, this information is recorded in our hospital’s electronic medical record in 2 ways: through a clinical pharmacy consult and as a codified problem list entry. When any codeine-containing medication is ordered electronically for a patient with a problem list entry of CYP2D6 ultra-rapid metabolizer, a warning message reminds the clinician that this patient may be expected to have a higher incidence of side effects than normal and that alternative drug therapy should be considered. (Figure 1).
Figure 1.
Discussion
Our goal was to establish a Clinical Pharmacogenetics service to improve the dosing of drugs which are metabolized by polymorphic enzymes and to help catalyze the appreciation of pharmacogenetics as a key clinical tool by other healthcare providers in our system. The involvement of the clinical laboratory was essential to the establishment of the clinical pharmacogenetics service, as the laboratory staff set up the tests in the electronic medical record and established a method for posting results to the medical record. In our situation, laboratory aspects of the service were expedited since the clinical laboratory for these services is part of the Pharmaceutical Department, but other pharmacy departments can achieve similar results through close collaboration with the clinical laboratory at their hospital. Because not all clinicians are familiar with the interpretation of pharmacogenetic test results, we trained pharmacists to provide test result interpretation.
To date, the Clinical Pharmacogenetics service at our institution focuses on pharmacogenetics in patients with cancer, but clinical pharmacogenetics also plays a role in an increasing number of disease states and drug therapies. A recent study reviewed 1,200 FDA approved drug labels, and found that 121 labels contained pharmacogenomics information.39 The authors then queried the medication use of 36.1 million patients who filled prescriptions through a major pharmacy benefit manager and showed that approximately one fourth of patients received a prescription for a medication that included pharmacogenomic information in the label.
In the new era of personalized drug therapy, Clinical Pharmacogenetics services will assist in identifying the safest and most effective drug and dose from the outset of therapy, and pharmacists are well-positioned to lead these services.
Conclusion
Our experiences demonstrate the feasibility of the design and function of a pharmacist-managed Clinical Pharmacogenetics service at an academic specialty hospital. This Clinical Pharmacogenetics service, one of the first of its kind, can serve as a model for other health systems. Pharmacogenetic testing has been made readily available to our patients receiving relevant drugs through collaboration between the clinical laboratory and pharmacists.
The development and implementation of this service illustrates the maturation of pharmacogenetics as a discipline and the growing awareness of the value of incorporating pharmacogenetic testing into clinical best practices and quality initiatives. As the science of pharmacogenetics advances, some clinical application of pharmacogenetic testing will likely exist in every patient population and health-system. Existing pharmacokinetic services can serve as a platform for the introduction of clinical pharmacogenetic testing. As drug therapy experts, pharmacists should lead the effort to incorporate pharmacogenetic information into patient care.
Acknowledgements
Supported in part by grant GM-61393 from the National Institutes of Health and by the American Lebanese Syrian Associated Charities. The authors acknowledge the pharmacists who operate the Clinical Pharmacogenetics service, as well as all the other clinicians, patients and families involved in this effort.
Footnotes
The authors have declared no potential conflicts of interest.
Contributor Information
Kristine R. Crews, Translational Research Laboratory Director, Pharmaceutical Department, St. Jude Children's Research Hospital, and Assistant Professor of Clinical Pharmacy, University of Tennessee Health Science Center, College of Pharmacy, Memphis, TN.
Shane J. Cross, Clinical Pharmacist, Pharmaceutical Department, St. Jude Children's Research Hospital and Assistant Professor of Clinical Pharmacy, University of Tennessee Health Science Center, College of Pharmacy, Memphis, TN.
John N. McCormick, Clinical Pharmacy Specialist, Pharmaceutical Department, St. Jude Children's Research Hospital, and Assistant Professor of Clinical Pharmacy, University of Tennessee Health Science Center, College of Pharmacy, Memphis, TN.
Donald K. Baker, Clinical Decision Support Officer, Department of Information Sciences, St. Jude Children's Research Hospital, and Assistant Professor of Clinical Pharmacy, University of Tennessee Health Science Center, College of Pharmacy, Memphis, TN.
Alejandro R. Molinelli, Director, Clinical Pharmacokinetics Laboratory, Pharmaceutical Department, St. Jude Children's Research Hospital, Memphis, TN.
Richard Mullins, Director, Clinical Pharmacokinetics Laboratory, Pharmaceutical Department, St. Jude Children's Research Hospital, Memphis, TN at the time of manuscript preparation; currently, Director, Core Laboratories, Children’s Healthcare of Atlanta, Atlanta, GA.
Mary V. Relling, Member and Chair, Pharmaceutical Department, St. Jude Children's Research Hospital, Professor of Clinical Pharmacy and Pediatrics, University of Tennessee Health Science Center, Colleges of Pharmacy and Medicine, Memphis, TN.
James M. Hoffman, Medication Outcomes & Safety Officer, Pharmaceutical Department, St. Jude Children's Research Hospital, and Assistant Professor of Clinical Pharmacy, University of Tennessee Health Science Center, College of Pharmacy, Memphis, TN.
References
- 1.Radomski KM, Bush BA, Ensom MH. Collaboration between pharmacy and laboratory: defining total allowable error limits for therapeutically monitored drugs. Annals of Pharmacotherapy. 1998;32:170–175. doi: 10.1345/aph.17187. [DOI] [PubMed] [Google Scholar]
- 2.Streetman DS, Nafziger AN, Destache CJ, Bertino AS., Jr Individualized pharmacokinetic monitoring results in less aminoglycoside-associated nephrotoxicity and fewer associated costs. Pharmacotherapy. 2001;21:443–451. doi: 10.1592/phco.21.5.443.34490. [DOI] [PubMed] [Google Scholar]
- 3.Bond CA, Raehl CL. Clinical and economic outcomes of pharmacist-managed aminoglycoside or vancomycin therapy. Am J Health Syst Pharm. 2005;62:1596–1605. doi: 10.2146/ajhp040555. [DOI] [PubMed] [Google Scholar]
- 4.Schumock GT, Butler MG, Meek PD, Vermeulen LC, Arondekar BV, Bauman JL. Evidence of the economic benefit of clinical pharmacy services: 1996–2000. Pharmacotherapy. 2003;23:113–132. doi: 10.1592/phco.23.1.113.31910. [DOI] [PubMed] [Google Scholar]
- 5.American Society of Health-System Pharmacists. ASHP statement on the pharmacist's role in clinical pharmacokinetic monitoring. Am J Health-Syst Pharm. 1998;55:1726–1727. doi: 10.1093/ajhp/55.16.1726. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen CA, Schneider PJ, Scheckelhoff DJ. ASHP national survey of pharmacy practice in hospital settings: dispensing and administration--2008. Am J Health Syst Pharm. 2009;66:926–946. doi: 10.2146/ajhp080715. [DOI] [PubMed] [Google Scholar]
- 7.Streetman DS, Nafziger AN, Destache CJ, Bertino AS., Jr Individualized pharmacokinetic monitoring results in less aminoglycoside-associated nephrotoxicity and fewer associated costs. Pharmacotherapy. 2001;21:443–451. doi: 10.1592/phco.21.5.443.34490. [DOI] [PubMed] [Google Scholar]
- 8.Bond CA, Raehl CL. Clinical and economic outcomes of pharmacist-managed aminoglycoside or vancomycin therapy. Am J Health Syst Pharm. 2005;62:1596–1605. doi: 10.2146/ajhp040555. [DOI] [PubMed] [Google Scholar]
- 9.Schumock GT, Butler MG, Meek PD, Vermeulen LC, Arondekar BV, Bauman JL. Evidence of the economic benefit of clinical pharmacy services: 1996–2000. Pharmacotherapy. 2003;23:113–132. doi: 10.1592/phco.23.1.113.31910. [DOI] [PubMed] [Google Scholar]
- 10.Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orr MS, Goodsaid F, Amur S, Rudman A, Frueh FW. The experience with voluntary genomic data submissions at the FDA and a vision for the future of the voluntary data submission program. Clin Pharmacol Ther. 2007;81:294–297. doi: 10.1038/sj.clpt.6100053. [DOI] [PubMed] [Google Scholar]
- 12.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 13.Ensom MH, Chang TK, Patel P. Pharmacogenetics: the therapeutic drug monitoring of the future? Clin Pharmacokinet. 2001;40:783–802. doi: 10.2165/00003088-200140110-00001. [DOI] [PubMed] [Google Scholar]
- 14.Frueh FW, Amur S, Mummaneni P, et al. Pharmacogenomic biomarker information in drug labels approved by the United States food and drug administration: prevalence of related drug use. Pharmacotherapy. 2008;28:992–998. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]
- 15.U.S. Food and Drug Administration. Drugs@FDA: FDA-approved drug products. Silver Spring, MD: U.S. Food and Drug Administration; 6-9-2010. 6-9-2010 Ref Type: Electronic Citation. [Google Scholar]
- 16.AmpliChip CYP450 test. Med Lett Drugs Ther. 2005;47:71–72. [PubMed] [Google Scholar]
- 17.Invader UGT1A1 molecular assay for irinotecan toxicity. A genetic test for an increased risk of toxicity from the cancer chemotherapy drug irinotecan (Camptosar) Med Lett Drugs Ther. 2006;48:39–40. [PubMed] [Google Scholar]
- 18.Dervieux T, Meshkin B, Neri B. Pharmacogenetic testing: proofs of principle and pharmacoeconomic implications. Mutat Res. 2005;573:180–194. doi: 10.1016/j.mrfmmm.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 19.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 20.Dervieux T, Meshkin B, Neri B. Pharmacogenetic testing: proofs of principle and pharmacoeconomic implications. Mutat Res. 2005;573:180–194. doi: 10.1016/j.mrfmmm.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Marra CA, Esdaile JM, Anis AH. Practical pharmacogenetics: the cost effectiveness of screening for thiopurine s-methyltransferase polymorphisms in patients with rheumatological conditions treated with azathioprine. J Rheumatol. 2002;29:2507–2512. [PubMed] [Google Scholar]
- 22.Oh KT, Anis AH, Bae SC. Pharmacoeconomic analysis of thiopurine methyltransferase polymorphism screening by polymerase chain reaction for treatment with azathioprine in Korea. Rheumatology (Oxford) 2004;43:156–163. doi: 10.1093/rheumatology/keh001. [DOI] [PubMed] [Google Scholar]
- 23.Dubinsky MC, Reyes E, Ofman J, Chiou CF, Wade S, Sandborn WJ. A cost-effectiveness analysis of alternative disease management strategies in patients with Crohn's disease treated with azathioprine or 6-mercaptopurine. Am J Gastroenterol. 2005;100:2239–2247. doi: 10.1111/j.1572-0241.2005.41900.x. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann DF, Medicis JJ, Franklin PD. Polymorphisms and the pocketbook: the cost-effectiveness of cytochrome P450 2C19 genotyping in the eradication of Helicobacter pylori infection associated with duodenal ulcer. J Clin Pharmacol. 2003;43:1316–1323. doi: 10.1177/0091270003259389. [DOI] [PubMed] [Google Scholar]
- 25.Hughes DA, Vilar FJ, Ward CC, Alfirevic A, Park BK, Pirmohamed M. Cost-effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics. 2004;14:335–342. doi: 10.1097/00008571-200406000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Elkin EB, Weinstein MC, Winer EP, Kuntz KM, Schnitt SJ, Weeks JC. HER-2 testing and trastuzumab therapy for metastatic breast cancer: a cost-effectiveness analysis. J Clin Oncol. 2004;22:854–863. doi: 10.1200/JCO.2004.04.158. [DOI] [PubMed] [Google Scholar]
- 27.Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J Pediatr. 1991;119:985–989. doi: 10.1016/s0022-3476(05)83063-x. [DOI] [PubMed] [Google Scholar]
- 28.Lennard L, Lilleyman JS, Van LJ, Weinshilboum RM. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 29.Relling MV, Hancock ML, Rivera GK, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 30.Stanulla M, Schaeffeler E, Flohr T, et al. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. JAMA. 2005;293:1485–1489. doi: 10.1001/jama.293.12.1485. [DOI] [PubMed] [Google Scholar]
- 31.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–844. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 32.Stocco G, Cheok MH, Crews KR, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;85:164–172. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 34.Krynetski EY, Evans WE. Pharmacogenetics of cancer therapy: getting personal. American Journal of Human Genetics. 1998;63:11–16. doi: 10.1086/301941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart CF, Panetta JC, O'Shaughnessy MA, et al. UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J Clin Oncol. 2007;25:2594–2600. doi: 10.1200/JCO.2006.10.2301. [DOI] [PubMed] [Google Scholar]
- 36.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 37.Relling MV, Hancock ML, Rivera GK, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 38.Stewart CF, Panetta JC, O'Shaughnessy MA, et al. UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J Clin Oncol. 2007;25:2594–2600. doi: 10.1200/JCO.2006.10.2301. [DOI] [PubMed] [Google Scholar]
- 39.Frueh FW, Amur S, Mummaneni P, et al. Pharmacogenomic biomarker information in drug labels approved by the United States food and drug administration: prevalence of related drug use. Pharmacotherapy. 2008;28:992–998. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]