Abstract
Ethanol (ETOH) can cause apoptotic death of neurons by depleting GSH with an associated increase in oxidative stress. The current study illustrates a means to overcome this ETOH-induced neurotoxicity by enhancing GSH through boosting Nrf2, a transcription factor that controls GSH homeostasis. ETOH treatment caused a significant increase in Nrf2 protein, transcript expression, Nrf2-DNA binding activity, and expression of its transcriptional target, NQO1, in primary cortical neuron (PCNs). However, this increase in Nrf2 did not maintain GSH levels in response to ETOH, and apoptotic death still occurred. To elucidate this phenomenon, we silenced Nrf2 in neurons and found that ETOH-induced GSH depletion and the increase in superoxide levels were exacerbated. Furthermore, Nrf2 knockdown resulted in significantly increased (P < 0.05) caspase 3 activity and apoptosis. Adenovirus-mediated overexpression of Nrf2 prevented ETOH-induced depletion of GSH from the medium and high GSH subpopulations and prevented ETOH-related apoptotic death. These studies illustrate the importance of Nrf2-dependent maintenance of GSH homeostasis in cerebral cortical neurons in the defense against oxidative stress and apoptotic death elicited by ETOH exposure.
Introduction
Exposure to ethanol during pregnancy, a debilitating public health problem, is associated with adverse outcomes ranging from fetal alcohol syndrome to alcohol-related neurodevelopmental disorder, which are collectively known as fetal alcohol spectrum disorder (FASD) (Sokol et al., 2003). A variety of animal- and cell-based experimental FASD models used in identifying the underlying mechanisms behind ethanol-induced oxidative stress-dependent developmental deficits have been comprehensively reviewed (Brocardo et al., 2011), yet such mechanisms and interventions remain elusive.
Previous studies from our laboratory and others in both in vivo and in vitro models have documented that the ethanol-elicited developmental deficits affecting multiple regions of the central nervous system are linked to reduced progenitor cell proliferation, disordered neuronal migration, disrupted neurotransmitter function, and enhanced neuron death (Gressens et al., 1992; Henderson et al., 1995; Jacobs and Miller, 2001; Ramachandran et al., 2001, 2003; He et al., 2005). Although programmed cell death of neurons is an obligatory process in the developing brain for its optimal development (Sastry and Rao, 2000), exposure to the pro-oxidant, ethanol, elicits an apoptotic response concomitant with a loss of redox homeostasis in neurons (Ramachandran et al., 2003).
In general, NF-E2-related factor 2 (NFE2L2/Nrf2), a basic leucine-zipper transcription factor, tightly controls redox homeostasis and facilitates the adaptation of neurons to a hostile oxidative environment (Lee et al., 2005). Under the basal condition, Nrf2 is sequestered in the cytoplasm by a chaperone molecule, Keap-1, and, upon oxidant stimulation, escapes from Keap-1, binding to ARE elements in the nucleus and up-regulating the transcription of NQO1, phase II genes including those involved in GSH biosynthesis (Itoh et al., 1999; Shih et al., 2003; Kraft et al., 2004). Although converging evidence from several fields demonstrated that Nrf2 deficiency in cells or animals is linked to increased susceptibility to oxidant-mediated injury, the intact Nrf2 pathway is indispensable for mounting defense mechanisms against those toxic stressors (Shih et al., 2005a,b; Satoh et al., 2006; Kensler et al., 2007). In a recent study, Dong et al. (2008) demonstrated a significant role for Nrf2 signaling in ETOH-induced teratogenesis in intact embryos. In addition, there are many studies elucidating a range of mechanisms involved in ethanol-induced apoptosis in numerous cell types, viz., cerebellar granule neurons (Bhave et al., 2000), neocortical neurons (Jacobs and Miller, 2001), and midbrain neurons (Crews et al., 1999). Evidence generated from our laboratory has illustrated that ethanol can accelerate apoptotic cell death of neurons in developing brain (Ramachandran et al., 2001, 2003), and such an increase in neuron loss can be correlated to a reduction in progressive brain tissue volume shrinkage and in cortical gray matter volume and ultimately to altered cortical functions (Pfefferbaum et al., 1998). Thus, connections between ethanol-mediated damage to cells within the developing cerebral cortex and functional consequences of ethanol exposure are in place. Our current study is the first of its kind to focus specifically on the cerebral cortical region response of fetuses and neurons isolated from fetal cortices to ETOH with respect to a vital signaling event(s) involving Nrf2. It is further shown that adenovirus-mediated overexpression of Nrf2 can protect fetal primary cortical neurons against ETOH-induced oxidative stress and apoptosis.
Materials and Methods
Materials.
Minimum essential medium (MEM), Dulbecco's minimum essential medium, Hanks' balanced salt solution, fetal bovine serum, and TRIzol were purchased from Invitrogen (Carlsbad, CA). Horse serum (HS), trypsin, DNase, antibiotics, poly-d-lysine, uridine, monochlorobimane, hydroethidine, actinomycin D, and tubulin monoclonal antibody were from Sigma-Aldrich (St. Louis, MO). Fisher-Costar cell culture inserts for coculture were obtained from Thermo Fisher Scientific (Waltham, MA). 4,6-Diamidino-2-phenylindole (DAPI) and annexin V FITC apoptosis detection kits were obtained from BD Biosciences (San Jose, CA). Nrf2 (C-20), GAPDH antibodies, and NFE2/Nrf2/ARE consensus and mutant oligonucleotides were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The anti-microtubule-associated protein 2 (MAP2), GSH detection kit was purchased from Millipore Bioscience Research Reagents (Temecula, CA). Anti-glial fibrillary acidic protein was obtained from Millipore Corporation (Billerica, MA). SMARTPool small interfering RNA (siRNA) against Nrf2 and nontargeting siRNA pool was purchased from Dharmacon RNA Technologies (Lafayette, CO). The QuantiTect reverse transcription kit for first-strand synthesis was purchased from QIAGEN (Valencia, CA). The cytosolic and nuclear protein extraction kit and ECL detection kit were purchased from Thermo Fisher Scientific. The Caspase-Glo 3/7 assay kit was obtained from Promega (Madison, WI). siPORT amine was from Ambion (Austin, TX). The TransAM ELISA kit for Nrf2 was purchased from Active Motif Inc. (Carlsbad, CA).
Primary Cortical Neuron Cultures.
Primary cortical neurons (PCNs) were prepared from embryonic day 16 to 17 timed pregnant Sprague-Dawley rats as described earlier (Dutton, 1990; Ramachandran et al., 2003). In brief, embryos from the amniotic sac were taken out, and the cerebral cortex was carefully dissected out. Fetal cortex was mechanically dissociated in Hanks' balanced salt solution, and the cells were suspended in MEM containing 10% fetal bovine serum and 10% HS. The cells were seeded at a density of 1.75 × 106 cells/well in a six-well plate previously coated with poly-d-lysine, and the neurons were maintained in a humidified atmosphere of 95% air and 5% CO2. After 1DIV, the cells were given an “inhibitory” feeding with uridine (10 mg/ml) containing MEM supplemented with 10% HS to suppress the growth of astrocytes. After 48 h, old medium was replaced by fresh MEM containing 10% HS and, subsequently, PCNs were subjected to treatment either on the 4th day of culture (for siRNA transfection and adenovirus infection experiments) or on the 5th day [for ETOH and actinomycin D (Act D) treatment]. This is a well established and documented primary neuronal culture system, which is essentially free of glia. Dual immunostaining with MAP2 (for neurons) and glial fibrillary acidic protein (for astrocytes) were performed and the isolation procedure adopted yielded ∼95% enriched neuronal culture (Supplemental Fig. 1).
In Vivo Model.
A 2-day exposure regimen in an animal model of acute ethanol exposure was used to mimic an alcohol binge in humans (Henderson et al., 1995). Sprague-Dawley rats were intubated with ethanol [4 g/kg b.wt., 25% (v/v)] at 12-h intervals on days 10 and 17 of gestation. Pair-fed control rats were weight-matched to the ethanol-fed dams and were intubated with isocaloric dextrose. On day 18, 1 h before sacrifice, a final dose was administered. Both control and ethanol-fed dams had full access to standard laboratory chow and water ad libitum, whereas pair-fed controls had full access to water but received the weight of chow consumed by the corresponding ethanol dam during the previous 24-h period. The gestational age of the pair-fed control and ethanol rats were staggered by a day to ensure that animals from the pair-fed controls received chow at the same stage of gestation as did the corresponding ethanol-treated dams. At the end of treatment, animals were sacrificed; Blood alcohol levels were determined using an Analox AM1 analyzer. Fetal brain cortices were carefully isolated and stored in −80°C until use. All animal protocols were approved by the institutional animal care and use committee. Handling and treatment of animals were carried out according to guidelines for the use and care of laboratory animals promulgated by the National Institutes of Health (Institute of Laboratory Animal Resources, 1996).
Ethanol Treatment of PCNs.
On 5DIV, PCNs were treated with ETOH (4 mg/ml) for different time points (30 min, 1 h, 2 h, 4 h, 8 h, 12 h, and 24 h) in an incubator saturated with ETOH to maintain medium ETOH (monitored using the Analox AM1 alcohol analyzer) (Rathinam et al., 2006). The in vitro experiments involving ETOH in the current study are short-term (24 h), whereas alcohol abuse is usually prolonged. This is a clinically relevant, albeit high, concentration, which is at or below that reported in mouse and rat models that expressed prominent brain apoptotic responses (Olney et al., 2002; Rathinam et al., 2006).
Actinomycin D Treatment.
PCNs on 5DIV were coincubated with ETOH (4 mg/ml) and Act D (1 μg/ml). No adverse effects of Act D on PCNs were seen at this concentration and duration. After treatment, cells were incubated for the above time periods, after which they were harvested, total RNA was isolated, and real-time qRT-PCR for Nrf2 and GAPDH determinations was performed.
Adenovirus Infection.
PCNs seeded at a density of 1.75 × 106 cells/well in six-well plates were infected at 4DIV with either Ad GFP, Ad WT Nrf2, or Ad DN Nrf2. The viruses were generously provided as a gift by Dr. Jeffrey Johnson (University of Wisconsin, Madison, WI). Virus infection was performed in complete medium at a multiplicity of infection of 200 for 1 h at room temperature. The plates were returned to the incubator for additional 1 h, and the medium was replenished with fresh 10% MEM. The infected plates were then returned to a 37°C humidified 5% CO2 incubator, and the cells were harvested after 24 and 48 h for Western analysis of Nrf2 expression. All experiments involving 24-h ETOH exposure were performed at 24 h after infection.
siRNA Transfection.
siRNA transfection experiments were performed in 4DIV PCNs in six-well plates. Then 5 μl of siPORT amine diluted in Opti-MEM was gently mixed either with 100 nM SMARtPool siRNA against Nrf2 or nontargeting siRNA pool. After complex formation according to the manufacturer's instructions (Ambion), the mixture was gently added to PCNs. Silencing was observed with Western analysis for Nrf2 protein expression after 24 and 48 h. For the experiments involving ETOH, 24 h after transfection of siRNA, the cells were exposed to ETOH for an additional 24 h and processed for various downstream applications such as Western and FACS analysis for detection of MCB, HET, and annexin V FITC/PI and the Caspase-Glo assay.
RNA Extraction and Real-Time qRT-PCR Analysis.
Total RNA was isolated from PCNs or cerebral cortex using the TRIzol reagent according to the manufacturer's recommendations (Invitrogen). Then 1.5 μg of total RNA was effectively removed of genomic DNA contamination and reverse-transcribed with a QuantiTect reverse transcription kit (QIAGEN). One-tenth of the cDNA was used in real-time RT-PCR analysis of mRNA expression for Nrf2, γ-glutamylcysteine ligase (GCLC), and GAPDH. TaqMan gene expression assays consisting of the primers and probes specific for rat Nrf2 and rat GAPDH were from Applied Biosystems (Foster City, CA). Real-time PCR amplification was performed in 96-well optical plates in a final volume of 20 μl containing 10 μl of TaqMan Universal Master Mix (Applied Biosystems), 20 pmol of the respective primers, and 1/10 of the reverse-transcribed RNA. For the experiment involving GCLC mRNA quantitation, an SYBR Green-based real-time PCR assay was performed using real-time primers for rat GCLC and rat GAPDH according to the manufacturer's instructions (SABiosciences, Frederick, MD). Real-time PCR was conducted in an ABI Prism 7300 system (Applied Biosystems). The expression of the target genes (Nrf2 and GCLC) was determined relative to GAPDH as an internal control, and the relative fold change in the mRNA expression was calculated using the 2−ΔΔCT, where ΔCT = CTtarget gene − CTGAPDH and ΔΔCT = ΔCTtreated condition − ΔCTuntreated condition.
Reverse Transcription-PCR Analysis.
Total RNA was isolated from untreated and ETOH-treated PCNs and in utero brain cortices, and cDNA was prepared as described above. One-tenth of the cDNA was used in the PCR using primers specific for rat NQO1 [forward, 5′-ACTCGGAGAACTTTCAGTACC-3′; reverse, 5′-TGGAGCAAAGTAGAGTGGT-3′ (Yan et al., 2008)] and rat GAPDH (forward, 5′-AGACAGCCGCATCTTCTTGT-3′; reverse, 5′-TACTCAGCACCAG CATCACC-3′). After an initial denaturation at 95°C for 3 min, PCR specific for NQO1 was performed in a 25-μl reaction volume for 35 cycles under the following conditions: 95°C for 30 s, 53°C for 30 s, 72°C for 60 s, and finally an extension at 72°C for 5 min. GAPDH cycling parameters were the same except for the annealing temperature, which was 55°C for 30 s. Aliquots of the PCR product were run on a 1% agarose gel, and the products for NQO1 and GAPDH were visualized at 492 and 323 base pairs, respectively, by ethidium bromide staining using a gel documentation system (UVP, Inc., Upland, CA). The images were photographed and quantified using ImageJ software (http://rsbweb.nih.gov/ij/).
Western Blot Analysis.
PCNs or cerebral cortices were lysed in radioimmunoprecipitation assay buffer at 4°C for 30 min, and after sonication and centrifugation at 15,000g for 15 min, the supernatant was used for immunoblot assay. In brief, 25 μg of equal amounts of lysates from various treated groups were resolved by electrophoresis on 8% polyacrylamide gels, electrotransferred to a polyvinylidene difluoride filter, and blocked with 5% nonfat dry milk. After incubation with primary antibodies against rat Nrf2, GAPDH, tubulin, and lamin, the membranes were then washed three times in PBS-Tween 20 and incubated with horseradish peroxidase-conjugated anti-rabbit or mouse IgG or goat IgG for 1 h. The antigen-antibody complex was detected using an ECL chemiluminescence kit. Nrf2 bands were quantified by scanning densitometry using ImageJ software and were normalized to the signal intensity of GAPDH.
Double Immunofluorescence.
PCNs cultured on poly-d-lysine-coated coverslips were treated with or without ETOH (4 mg/ml, 30 min), washed with PBS, fixed in 2% paraformaldehyde, and blocked as needed, followed by Nrf2 and MAP2 dual immunostaining. Nrf2 and MAP2 expression was detected using Cy3-conjugated donkey anti-rabbit antibodies and FITC-tagged donkey anti-mouse secondary antibodies, respectively. DAPI was used to stain the nuclei to determine nuclear localization of Nrf2, if any. The cells were visualized and imaged using Olympus FluoView 500.
Electrophoretic Mobility Shift Assay.
Nuclear proteins were extracted from PCNs or cerebral cortex using a nuclear and cytoplasmic extraction kit (Thermo Fisher Scientific). EMSA binding reactions were performed at room temperature in a final volume of 20 μl containing 10 μg of the nuclear extract from both control and various time points of ETOH-treated samples, 5 × 104 cpm of 32P-labeled Nrf2 oligonucleotide probe, and 1 μg of poly(dI-dC). Likewise, in a separate reaction, the corresponding mutated Nrf2 oligonucleotide was also incubated with the nuclear extracts. To determine the specificity of protein binding to the Nrf2/ARE sequence, competition assays were performed in the presence of a 100-fold excess of the unlabeled competitor oligo (Nrf2 consensus) along with nuclear extract for 10 min before the addition of radiolabeled probe. The supershift assays were performed by preincubating nuclear extracts with 2.5 μg of Nrf2 antibody in ice for 15 min, and the reaction was performed at room temperature for 30 min after addition of the radiolabeled probe. The DNA-protein complexes were resolved in a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate, and EDTA buffer and gels were dried and autoradiographed with intensifying screens at −80°C.
ELISA-Based Measurement of Nrf2 Activity.
The TransAM Nrf2 Kit was used to assay the DNA-binding activity of Nrf2 in the nuclear extracts that were obtained from both the untreated and ETOH-treated cells/brain cortices. In brief, 5 μg of nuclear extracts from each sample in triplicate were incubated in a 96-well plate that was coated with oligonucleotide containing a consensus binding site for Nrf2. For competitive binding experiments, which measure the specificity of the assay, 5 μg of nuclear extract from the ETOH-treated cells were assayed in the presence of wild-type or mutated competitor oligonucleotides. After 1 h of incubation, the wells were washed and incubated with 100 μl of a 1:1000 dilution of rabbit polyclonal antibody against Nrf2. Incubation with normal rabbit polyclonal IgG was also performed separately to determine the specificity of the Nrf2 antibody. The wells were then washed, followed by incubation with 100 μl of a 1:1000 dilution of horseradish peroxidase-conjugated, anti-rabbit secondary antibody at room temperature. The wells were developed using 100 μl of TMB substrate for 6 and 3 min for cell lysates and tissue lysates, respectively, before addition of 100 μl of stop solution. Optical density was read at 450 nm with a reference wavelength of 650 nm using a plate reader from Bio-Rad Laboratories (Hercules, CA).
GSH Measurement by Flow Cytometry.
Monochlorobimane, a nonfluorescent reagent that reacts with GSH to form a highly fluorescent derivative, was used to detect free GSH in individual viable cells by flow cytometry (Maffi et al., 2008). At the end of treatment, neurons were incubated with 10 μM MCB for 30 min in a cell culture incubator, and cells were scraped, washed, and resuspended in ice-cold PBS. Acquisition and analysis were performed on a FACS flow cytometer with excitation and emission settings of 360 and 460 nm, respectively.
GSH Measurement by Fluorometric Method.
Intracellular levels of reduced GSH were assessed by a GSH detection kit (Millipore Bioscience Research Reagents). Nrf2 knockdown cells treated with or without ETOH for 24 h were washed with 1.5 ml of wash buffer followed by lysis and clarification using centrifugation. Then 90 μl of lysate were mixed with 10 μl of MCB and incubated in a black 96-well plate at room temperature in a light-protected environment. Fluorescence was read in a fluorometer using a 380/460 filter set, and the results are expressed as relative fluorescence units.
Measurement of Cellular Superoxide by Flow Cytometry.
Generation of superoxide radicals as a measure of oxidative stress was estimated by flow cytometry. The relative levels of intracellular superoxide were analyzed using the cell-permeable redox-sensitive fluorochrome HET. Cells were incubated with 1 μM HET for 15 min at 37°C and subsequently washed twice in ice-cold PBS. The cells were harvested in PBS containing 1% bovine serum albumin and filtered through a 70-μm filter, and superoxide accumulation was analyzed in terms of the increase in ethidium fluorescence using flow cytometry. A total of 104 events was collected, and the level of superoxide in cells was expressed as the percentage of the control (untreated) samples.
Annexin-V Staining and FACS Analysis.
Annexin V binding was used as one measure of apoptotic neurons. After treatment, both detached and attached cells were harvested and centrifuged. The cell pellet was washed with ice-cold PBS and resuspended in binding buffer containing annexin V-FITC and PI. The cells were gently vortexed and incubated in the dark. Untreated cells that were either unstained or stained with PI or stained with annexin V-FITC were included along with the experimental samples to correct the background signals. For overexpression experiments with adenovirus, annexin V PE/ 7-amino-actinomycin D was used to avoid false-positives that could arise as a result of green fluorescent protein expressed by the bicistronic adenoviral construct. Data were collected on a flow cytometer and analyzed using Cell Quest (BD Biosciences) software.
Caspase-Glo 3/7 Assay.
Caspase 3 and 7 activities were estimated using Caspase-Glo 3/7. After treatment, neurons were washed with PBS, and 300 μl of Caspase-Glo 3/7 reagent was added to each well. Cells were scraped and collected in a microfuge tube. The collected lysate was incubated in the dark, and the resultant luminescence was read in a GloMax luminometer (Promega). Relative luminescence units were noted, and the results are expressed as fold change in caspase 3/7 activity from control.
Statistical Analysis.
Data are presented as means ± S.E.M. Statistical differences were determined using one-way ANOVA when experiments involved more than two groups. Significant differences among the different groups were analyzed using the Student-Newman-Keuls test. For the experiments involving only two groups (untreated control and ETOH), a paired Student's t test was used. P < 0.05 was considered as statistically significant.
Results
Ethanol Induces Endogenous Nrf2 Protein by Transcriptional Regulation.
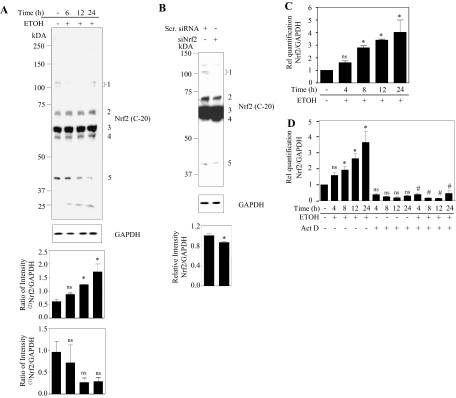
Prior studies in our laboratory, and others have illustrated that ethanol, both in brain and in cultured neurons, elicits oxidative stress (Jacobs and Miller, 2001; Ramachandran et al., 2001, 2003; Olney et al., 2002), a setting that can activate the Nrf2 cytoprotective system (Dong et al., 2008). To determine whether ETOH activates Nrf2 in cerebral cortical neurons (PCNs), we initially measured Nrf2 protein levels in PCNs treated with ETOH for different periods (Fig. 1A). Immunological detection of Nrf2 using an antibody specific against Nrf2 (C-20) resulted in the detection of multiple bands and they are marked as 1, 2, 3, 4, and 5 in the blot (Fig. 1A) corresponding to molecular masses, 100 and 110, 68, 57, 53, and 41 kDa, respectively. Upon Nrf2 silencing using the siPORT amine, only the faint 100- and 110-kDa bands and the strong 68-kDa band were found to be down-regulated whereas the 57-, 53-, and 41-kDa bands (Fig. 1B) did not show any correspondence to Nrf2 silencing and were not regulated. The transfection agent used herein, siPORT amine, did not have any toxic effect as measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium assay (Supplemental Fig. 2). Treatment of PCNs with a proteosomal inhibitor, MG-132, was found to increase only the 100- and 110- and 68-kDa bands in accordance with the previous studies (Furukawa and Xiong, 2005; Kapeta et al., 2010) with the other bands showing no change (Supplemental Fig. 3). This finding suggests that the products detected at 57, 53 and 41 kDa could be nonspecific. At this point, it is not clear whether the ∼100 kDa observed is native Nrf2 or modified Nrf2. However, the high-molecular-mass form could be an anomalous migration of Nrf2 in SDS-polyacrylamide gel electrophoresis owing to its inherent high acidic charges (Moi et al., 1994). Previous reports by others have also provided evidence that the ∼100-kDa form is a Nrf2-actin dimer (Kang et al., 2002), ubiquitinated Nrf2 (Li et al., 2005). Thus, in our experiments, a molecular mass of 68 kDa, which is the actual predicted size for Nrf2, is considered. Studies are underway in our laboratory to further identify the nature of both high- and low-molecular-mass Nrf2 forms.
Fig. 1.
Ethanol induces Nrf2 expression in a time-dependent manner in primary cortical neurons. PCNs were treated with ETOH (4 mg/ml) for the times indicated. A, Nrf2 protein expression was determined by immunoblot analyses and equal loading demonstrated by anti-GAPDH. Bottom, densitometric scanning analysis ratio of Nrf2 to GAPDH. Superscripts (1) and (2) in the densitometric plot represent the ratio intensity of 100- to 110- and 68-kDa products corrected to GAPDH, respectively. B, PCNs were transfected with either nontargeting scramble siRNA (Scr. siRNA) or a SMARTpool mix of four siNrf2 using siPORT amine. Cells were lysed 48 h after transfection, and immunoblots were analyzed with anti-Nrf2 and GAPDH. Bottom, densitometric scanning analysis ratio of 68-kDa Nrf2 to GAPDH. C, PCNs were treated with ETOH (4 mg/ml) for the times indicated and real-time qRT-PCR analysis for Nrf2 transcript expression normalized to GAPDH. D, cells were incubated with Act D (1 μg/ml) along with ETOH (4 mg/ml) for a given time in hours. Total RNA was then extracted and subjected to qRT-PCR for Nrf2 and GAPDH. A to C, results are the mean ± S.E.M. of experiments performed in triplicate. One-way ANOVA for A, C, and D and a t test for B was performed to establish statistical significance. *, P < 0.05 compared with untreated control; ns, not significant compared with untreated control; #, P < 0.05 compared with respective ETOH time treatment. Rel, relative.
As shown in Fig. 1A, exposure of PCNs to 4 mg/ml ETOH resulted in a time-dependent increase in Nrf2 protein levels. This increase, although clearly evident at the 6-h time points, ultimately became statistically significant at 12 h of treatment and remained elevated for 24 h (a 2-fold increase). To determine whether this induction of Nrf2 protein expression by ETOH is due to an increase in the mRNA levels, total RNA was isolated from PCNs treated with or without ETOH for the same ETOH exposure periods. Figure 1C illustrates that transcript levels are expressed in a pattern similar to that of protein, although the increase reached statistical significance within 8 h of treatment. To determine whether this reflects an ETOH effect on transcriptional activation of Nrf2, we coincubated PCNs with and without ETOH and Act D, which prevents transcription by binding to single-stranded DNA. Shown in Fig. 1D is the effect of Act D on the ETOH-related induction of Nrf2 mRNA. Act D treatment abolished the ETOH-induced Nrf2 mRNA levels at each time point measured, and a significant (P < 0.05) inhibitory effect was observed as early as 4 h, indicating that ETOH regulates Nrf2 by transcriptional mechanisms.
Ethanol Activates Nrf2/ARE Binding and Nuclear Localization in PCNs.
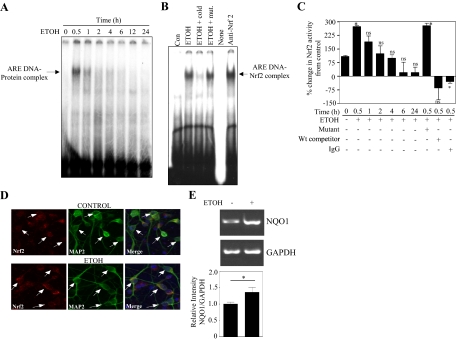
To examine whether the above induction of Nrf2 expression in PCNs in response to ETOH resulted in functional Nrf2 activity, we assessed Nrf2/ARE DNA-binding ability by a band shift assay (Fig. 2). In the mobility shift assay with the radiolabeled Nrf2/ARE probe, nuclear extracts obtained from 30-min ETOH treatments showed enhanced ability to form the ARE-DNA protein complex (Fig. 2A, lane 1 versus lane 2). A progressive decline was noted at subsequent time intervals. To confirm the specificity of the consensus Nrf2/ARE oligonucleotide, we performed gel shift assays in the presence of mutant and unlabeled competitor along with 30-min ETOH nuclear extracts (Fig. 2B). As expected, binding competition by the Nrf2/ARE unlabeled probe significantly decreased the labeled ARE DNA-protein complex (Fig. 2B, lane 3 versus lane 2), whereas the binding of the latter was unaltered when it was incubated with an ARE core mutated oligo (Fig. 2B, lane 4 versus lane 2), confirming the specificity of complex formation. To detect the presence of Nrf2 in this complex formation, we performed supershift analysis using Nrf2 antibody. We observed an enhanced complex signal instead of an expected supershifted band (Fig. 2B, lane 6 versus lane 2). Repeated attempts failed to generate any shift, and similar limitations with the use of this Nrf2 antibody have been reported by Cho et al. (2002). Subsequent supershift assays were performed using biotin-based EMSA with multiple newly available Nrf2 antibodies, out of which anti-Nrf2 (OriGene, Rockville, MD) produced a supershift (Supplemental Fig. 4). In addition, a TransAm ELISA for Nrf2 was performed to validate the Nrf2 specific binding. As shown in Fig. 2C, a statistically significant 175% increase in Nrf2 binding activity was found in the nuclear extracts within 30 min of ETOH treatment. Prolonged ETOH treatment resulted in a similar decline in Nrf2 activity as seen in Fig. 2A. ELISA studies were tightly controlled for antibody and Nrf2/ARE oligo specificity.
Fig. 2.
Ethanol induces Nrf2/ARE binding activity in PCN nuclear extracts. A, nuclear protein obtained from controls and PCNs exposed to ETOH (4 mg/ml) for different time points was used to detect ARE-specific oligonucleotide-protein complexes by EMSA. B, supershift analysis was performed using Nrf2 antibody and the presence of Nrf2 in the ARE DNA-protein complex was assayed. Excess unlabeled and mutant oligonucleotide was used to determine specificity of the Nrf2/ARE binding. C, TransAM ELISA for Nrf2 was performed to confirm Nrf2 DNA binding activity in control and ETOH-treated PCN nuclear extracts (mean ± S.E.M., n = 3). Statistical significance was established by one-way ANOVA. D, a representative immunofluorescence photomicrograph (original magnification, 20×) from control and 30-min ETOH (4 mg/ml)-treated PCNs for Nrf2 nuclear localization. Anti-MAP2 was used as a neuronal marker, and the nucleus was identified by DAPI staining. Data are representative of three independent experiments. E, RT-PCR image and statistical analysis for NQO1 and GAPDH mRNA from PCNs treated with and without ETOH (4 mg/ml) (n = 4). *, P < 0.05 compared with untreated control; ns, not significant compared with untreated control. Con, control; Wt, wild-type.
Because oxidants and xenobiotics activate Nrf2-related systems by promoting nuclear transport (Itoh et al., 1999), we determined whether ETOH-induced Nrf2 activity, as shown by EMSA and ELISA, reflected nuclear Nrf2 localization. A dual immunofluorescence study showed Nrf2 (stained red) distribution throughout the MAP2-positive neurons (stained green) including cytoplasm and nucleus in untreated controls (Fig. 2D). Treatment with ETOH for 30 min resulted in accumulation of Nrf2 in the nuclear compartment (Fig. 2D; compare control versus ETOH merge image). Further 24-h ETOH treatment significantly up-regulated the mRNA level of one of the Nrf2-regulated gene products, NQO1 (Fig. 2E). Taken together, these results demonstrate a role for ETOH in Nrf2 activation and promoting nuclear localization in primary cortical neurons.
In Utero Binge Ethanol Exposure Activates Nrf2 in Fetal Brain Cortices.
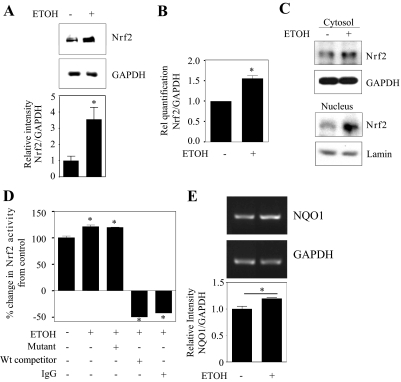
Having shown that ETOH up-regulates Nrf2 expression in vitro in PCNs, we next extended the in vitro findings to the in vivo setting by testing for Nrf2 expression and activation in an animal model of acute ethanol exposure that mimics binge drinking in humans. First, Nrf2 protein expression was determined by Western blot analysis in total cortical protein extracts from day 18 fetal brains whose dams were intubated (gastric) with either isocaloric dextrose or ETOH. As Fig. 3A shows, cortical Nrf2 protein expression was increased by almost 3-fold in the ETOH-exposed group compared with the isocaloric dextrose-administered control (P < 0.05). Real-time PCR analysis for Nrf2 mRNA illustrated an ETOH-mediated increase in the Nrf2 transcript, suggesting that the up-regulation of Nrf2 protein is due to increased mRNA levels as occur in primary cultures of cerebral cortical neurons. In utero exposure of ETOH elicited a significant increase (P < 0.05) in Nrf2 transcript levels by approximately 1.5-fold (Fig. 3B). Altered Nrf2 localization associated with its defective signaling may play a critical role in oxidative stress-related experimental and clinical neurological diseases such as Alzheimer's and Parkinson's disease (Ramsey et al., 2007). Hence, Western blot analyses for Nrf2 in both cytoplasmic and nuclear enriched fractions from fetal brain cortices were performed (Fig. 3C). GAPDH and lamin B1 were included as markers for cytoplasmic and nuclear fractions, respectively, and Nrf2 levels in each fraction were normalized to these. ETOH administration strongly increased endogenous Nrf2 expression compared with that in controls (P < 0.05). This occurred primarily in the nuclear compartment, but cytoplasmic Nrf2 expression was also increased. Figure 3D is a measure of Nrf2 binding activity by TransAM ELISA for Nrf2 in the nuclear fractions. Activated Nrf2 in nuclear extracts from ETOH-treated animals was significantly increased (25%; P < 0.05) compared with that in dextrose-administered animals (Fig. 3D, lane 1 versus lane 2). To determine specificity of binding, ETOH-treated nuclear extracts were incubated with either wild-type Nrf2 oligonucleotide or normal rabbit IgG or mutated Nrf2 oligonucleotide. This illustrated that Nrf2 DNA binding activity was reduced in wild-type and IgG-incubated samples (Fig. 3D, lanes 4 and 5 versus lane 2), whereas the mutated Nrf2 oligonucleotide did not change the Nrf2-DNA binding activity (lane 3 versus lane 2). RT-PCR analysis using primers that specifically amplified the NQO1 gene, a well known Nrf2-transcriptional target, revealed that the NQO1 transcript was significantly increased by ∼20% in ETOH-treated fetal brain cortices compared with controls (Fig. 3E). These results suggest that in utero ETOH administration resulted in up-regulation of Nrf2 and its activation in brain cortex of fetuses similar to that observed in in vitro cortical neuron cultures.
Fig. 3.
Prenatal ethanol exposure increases Nrf2 expression and activation in fetal brain cortices. Pregnant rats (Sprague-Dawley) at embryonic day 16 were administered ETOH (4 g/kg b.wt.) or isocaloric dextrose by gastric intubation at 12-h intervals for 2 days. At embryonic day 18 brain cortex from embryos were dissected and processed for Nrf2 protein expression by immunoblotting (A, n = 6), Nrf2 mRNA expression by qRT-PCR (B, n = 6). A, bottom, densitometric scanning ratio of Nrf2/GAPDH intensities. A and B, Student's t test (paired) was performed to determine the significance of treatment. Nuclear and cytosolic extracts were used for immunoblot analyses of Nrf2 to determine localization (C, mean ± S.E.M., n = 5), and nuclear extracts were used in TransAM ELISA for Nrf2 activity determination (D, mean ± S.E.M., n = 3). Statistical significance was established by one-way ANOVA. E, RT-PCR image and statistical analysis for NQO1 and GAPDH mRNA from control and ETOH-treated fetal brain cortices (n = 4). *, P < 0.05 compared with isocaloric dextrose-treated control; ns, not significant compared with isocaloric dextrose-treated animals. Rel, relative; Wt, wild-type.
Nrf2 Silencing Increases Sensitivity to ETOH-Related Oxidative Stress and Apoptosis in PCNs.
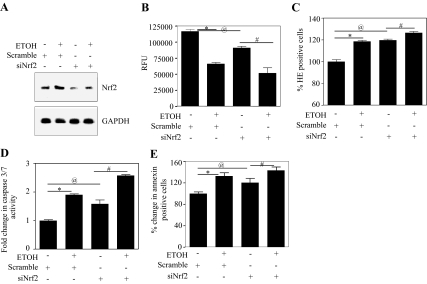
The significance of perturbations of Nrf2 expression on ethanol-related damage to neurons was addressed using siRNA-based Nrf2 knockdown experiments. The Nrf2 content of PCNs was reduced by treatment with 100 nM Nrf2 SMARTpool siRNA to target endogenous Nrf2 and a nontargeting scrambled siRNA pool to show that silencing of Nrf2 is specific and not due to nonspecific effects. siRNA targeting of Nrf2 remarkably reduces the endogenous Nrf2 protein expression in PCNs (Fig. 4A, lane 3 versus lane 1), and this knockdown further abolished the ETOH-induced Nrf2 expression (Fig. 4A, lane 4 versus lane 2). Nrf2/ARE activation regulates cellular GSH homeostasis machinery (Shih et al., 2005a), and ethanol-mediated apoptotic death is dependent on this antioxidant system (Dong et al., 2008). Figure 4B illustrates that reduced Nrf2 expression decreases baseline GSH content (22%; column 1 versus column 3), and this is further amplified by 44% (column 3 versus column 4) upon ETOH treatment as assessed by a reduction in MCB fluorescence. Concomitant with this response is a significant increase in superoxide radical levels by 18% in siNrf2 to 27% with ETOH-treated Nrf2-depleted neurons (Fig. 4C). Lipid hydroperoxide levels, a measure of lipid peroxidation, were also significantly increased in Nrf2-compromised neurons (Supplemental Fig. 6A). The effect of this response on ETOH-induced apoptosis is reflected by two apoptotic markers, caspase 3/7 activation and annexin V binding. The reduction in Nrf2 expression further increased (P < 0.05) caspase 3/7 activity by 1.5-fold (Fig. 4D) and annexin V binding by 1.2-fold (Fig. 4E) over that occurring in “normal” PCNs.
Fig. 4.
siRNA-mediated Nrf2 knockdown exacerbates ethanol-induced oxidative stress and neuronal death. A, PCNs were transfected with either nontargeting scramble siRNA or a SMARTpool mix of four siNrf2s using siPORT amine. Twenty-four hours after transfection of scrambled siRNA or siNrf2, the cells were treated with or without ETOH (4 mg/ml) for an additional 24 h. Protein extracts were then immunoblot-analyzed with anti-Nrf2 and GAPDH. B, live cells were loaded with MCB and cells were fluorometrically analyzed for GSH content, and data are presented as relative fluorescence units (RFU) (mean ± S.E.M., n = 6). C, live cells were loaded with HET followed by FACS for ethidium positivity (measure of superoxide) to indicate level of oxidative stress (mean ± S.E.M., n = 6). D, cell extracts were used in the Caspase-Glo 3/7 assay and caspase 3/7 activity was determined as luminescence units (mean ± S.E.M., n = 6). E, live cells were stained using annexin V-FITC conjugate/PI, and FACS was used to measure apoptotic cell death (mean ± S.E.M., n = 6). One-way ANOVA was performed to establish statistical significance. *, @, P < 0.05 compared with scramble untreated control; #, P < 0.05 compared with siNrf2.
Overexpression of Nrf2 Rendered Neuroprotection against ETOH Toxicity.
The above findings (Fig. 4) support an important role for Nrf2 in protection of neurons in the developing brain from ETOH-induced oxidative stress and subsequent apoptotic death. Thus, a key issue is that although ETOH exposure increases Nrf2 protein expression and subsequent early DNA/ARE binding (Figs. 1–3), a portion of these neurons will develop oxidative stress that ultimately elicits apoptotic death (Maffi et al., 2008) (Fig. 4). One explanation for this is that the basal Nrf2/ARE cytoprotective machinery in some neurons is incapable of a response that is sufficient to provide protection from ETOH-mediated oxidative stress. It is to be noted that several mechanisms have been reported to be the cause for FASD phenotypes in vivo and elicit toxic responses in cultured neuron models, which include modulation of retinoic acid signaling (Deltour et al., 1996), induction of oxidative stress (Ramachandran et al., 2001, 2003), calcium-based signaling events (Fischer et al., 2003), and others. Herein experiments were performed to address the role of Nrf2/ARE pathway in regulating ETOH-induced oxidative stress-mediated neuronal injury.
Ad Nrf2 Increased Nrf2 Protein and γ-Glutamylcysteine Ligase Content of PCNs.
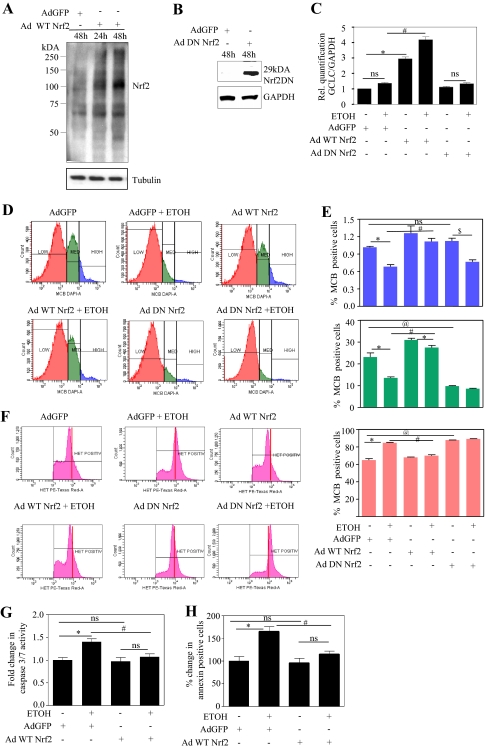
PCNs were infected either with Ad Nrf2, Ad DN Nrf2, or a control virus (Ad GFP) on 4DIV with a titer of 200 MOI based on the previous study (Shih et al., 2003, 2005a,b). This titer did not cause significant neuronal cell death compared with virus-free conditions (uninfected control PCNs) as measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (Supplemental Fig. 5) and FACS analysis for annexin V/PE (data not shown). Nrf2 overexpression was confirmed at 24 and 48 h postinfection by determinations of Nrf2 protein levels. Figure 5A illustrates that in PCNs infected with Ad GFP, there was only a background low basal level of Nrf2 protein, whereas expression of Nrf2 in neurons infected with Ad Nrf2 the protein expression increased in a time-dependent manner (Fig. 5A, lanes 2 and 3 versus lane 1). Overexpression of DN Nrf2, which lacks an N-terminal transactivating domain was detected as a 29-kDa immunoreactive band in Western blot detection (Fig. 5B) using the same C-terminal Nrf2 antibody that was used to detect full-length Nrf2 (Shih et al., 2003). To address the ultimate effectiveness of the Nrf2 overexpression, its impact on neuron content of a major determinant of GSH synthesis capacity, GCLC mRNA (Yang et al., 2005) was determined. Ad Nrf2 overexpression resulted in a 3-fold increase in GCLC mRNA expression compared with the Ad GFP-infected control (Fig. 5C, column 3 versus column 1). Ad Nrf2-infected PCNs, when treated with ETOH, showed further enhancement in GCLC mRNA by approximately 50% (P < 0.05) compared with of Ad Nrf2 alone-infected PCNs (Fig. 5C, column 4 versus column 3). Neither the DN Nrf2 alone-infected PCNs nor the ETOH-exposed DN Nrf2-infected PCNs showed an increase in GCLC transcript levels (Fig. 5C).
Fig. 5.
Adenovirus-mediated overexpression of Nrf2 contained ethanol-induced oxidative stress and neuronal death. A and B, PCNs (4DIV) in serum containing medium were infected with adenovirus encoding Nrf2 cDNA (A) or DN Nrf2 (B) at 200 MOI. Twenty-four and 48 h after infection, cells were processed for protein and immunoblot-analyzed for Nrf2 overexpression and normalized with anti-tubulin/GAPDH expression. Adenovirus encoding GFP cDNA was used as the control virus in this study. C to J, PCNs were infected with either Ad GFP/Ad Nrf2/Ad DN Nr2 for 24 h at 4DIV and followed by 24 h of ETOH (4 mg/ml) treatment. C, total RNA was extracted and subjected to qRT-PCR for GCLC, an Nrf2 transcriptional target and a constitutively expressed gene, GAPDH. Relative (Rel.) quantification of GCLC/GAPDH transcripts is illustrated (mean ± S.E.M., n = 6). D, neurons infected and treated as in C were subjected to flow cytometric determination of MCB staining to measure cellular GSH. A representative experiment is shown (D) wherein neuronal population were divided into low (orange), medium (green), and high GSH (blue) containing cells based on differential MCB staining. E, percentage of cells positive for MCB derived from high, medium, and low GSH populations are represented in blue, green, and orange graphs, respectively (mean ± S.E.M., n = 6). F, a representative FACS diagram, portraying the effect of Nrf2 on ethanol-induced superoxide generation measured in terms of oxidation of HET into the fluorescent product ethidium. Red vertical bars were introduced to visualize shift in peaks. G, estimation of effector caspase 3/7 activity in neuronal extracts obtained from the aforementioned treatment by the Caspase-Glo assay (mean ± S.E.M., n = 4). H, neurons were stained with annexin V-PE conjugate/7-amino-actinomycin and read immediately by flow cytometry to measure the extent of apoptosis (mean ± S.E.M., n = 6). One-way ANOVA was performed to establish statistical significance. *, @ P < 0.05 versus Ad GFP control; #, P < 0.05 versus Ad GFP + ETOH; ns, not significant.
Nrf2 Overexpression Prevented the ETOH-Mediated Decrease in PCN GSH Content.
Previous findings have illustrated that prevention of the ETOH-related reduction of GSH in PCNs mitigates subsequent cell death and that GSH heterogeneity in a single cell type can dictate PCN sensitivity to ETOH (Ramachandran et al., 2003; Watts et al., 2005; Maffi et al., 2008). Cultured PCNs express a striking heterogeneity of GSH content and overexpression of Nrf2 clearly influences ETOH-induced changes in this GSH heterogeneity (Fig. 5D). Flow cytometric segregation of PCNs based on MCB positivity illustrates three populations of cells that are classified as high, medium, and low GSH.
The mean distribution of high, medium, and low GSH in Ad GFP-infected control cells is 1.1, 28.7, and 70.2%, respectively (Fig. 5E, column 1). Treatment with ETOH resulted in a clear (P < 0.05) shift in GSH distribution, which presents as a decrease in both medium GSH (Fig. 5, D and E, area under green peak; Ad GFP versus Ad GFP + ETOH) and high GSH population (Fig. 5, D and E, area under blue peak; Ad GFP versus Ad GFP + ETOH) and a shift to the low GSH population (Fig. 5, D and E, area under orange peak; Ad GFP versus Ad GFP + ETOH). By considering the high GSH population (which is 1.1% of the total) as 100%, Nrf2-augmented neurons showed a 20% increase compared with the Ad GFP control. This significantly prevented ETOH-induced depletion of high and medium GSH. In contrast to overexpression of wild-type Nrf2, use of DN Nrf2, which would be expected to mimic Nrf2 knockdown, likewise showed a dramatic increase in the low GSH population with a concomitant decline in the medium and high GSH populations (Fig. 5D). Although glutathione transferase (GST) activity is required for the measurement of intracellular GSH using MCB detection, it has been shown that MCB fluorescence intensity versus time peak labeling in different cell types, viz., meninges, astrocytes, and differentiated cortical layer II neurons occurs within 20 min. In other words, the reaction between MCB and GSH proceeds to completion within 20 min (Sun et al., 2006). Thus, the protocol used in our study measures the plateau level of GSH-MCB (30-min MCB incubation; refer to Materials and Methods), ensuring completion of the initial GST-mediated conjugation to detect the GSH-MCB fluorescence resulting from the cellular GSH concentrations, thereby not allowing GST activity as a limiting factor.
Nrf2 Overexpression Prevented ETOH-Mediated Oxidative Stress and Apoptosis of PCNs.
Prior studies by our laboratory and others have connected ETOH-mediated apoptotic death of neurons to oxidative stress (Jacobs and Miller, 2001; Ramachandran et al., 2001; Olney et al., 2002; Watts et al., 2005). FACS analysis was used with HET as an estimate of superoxide radical content. ETOH increased the HET signal (Fig. 5, F and Ad GFP + ETOH versus Ad GFP), which was normalized in Nrf2-overexpressing neurons (Fig. 5, F and Ad Nrf2 + ETOH versus Ad GFP + ETOH). Conversely, in Nrf2-compromised neurons, a robust increase in superoxide generation occurred (Fig. 5F, shift in peak). Associated with normalization of superoxide levels, the lipid hydroperoxide levels were also attenuated in Nrf2-overexpressing neurons (Supplemental Fig. 6B). Both caspase 3/7 activation and annexin V binding were used as evidence of ETOH-mediated apoptotic death of PCNs. Augmentation of Nrf2 significantly prevented the ETOH-induced caspase 3/7 activity (Fig. 5G, column 4 versus column 2) and annexin V binding to neurons (Fig. 5H, column 4 versus column 2). The above findings illustrate that Nrf2 overexpression can mitigate the ETOH-induced oxidative stress and prevent the associated neuronal injury.
Discussion
A major determinant of the neurotoxicity of ETOH is depletion of neuronal GSH content (Watts et al., 2005; Maffi et al., 2008), and modulation of the Nrf2/ARE/GSH pathways (Yang et al., 2005) has been shown to prevent ethanol-induced liver injury in mice (Gong and Cederbaum, 2006). In our current study, we assessed the involvement of Nrf2 and the effect of heterologous Nrf2 overexpression in regulating GSH-mediated protection against ETOH in fetal neurons.
Up-Regulation of Nrf2 by Ethanol in Cultured Fetal Cerebral Cortical Neurons.
In vitro exposure of PCNs to ETOH (4 mg/ml, 24 h) resulted in a significant increase in both Nrf2 message and protein, which is in agreement with previous findings that oxidative stress increases Nrf2 levels and the resultant endogenous antioxidant system (Zhang et al., 2006). In the absence of alcohol, Act D, a potent inhibitor of RNA polymerase II-dependent transcription did not alter Nrf2 mRNA levels, whereas Act D coincubation with ETOH resulted in a strong decrease in Nrf2 mRNA levels. This result illustrates that the observed ETOH-related increased Nrf2 mRNA is newly synthesized (i.e., a transcriptionally favored regulation). It is noteworthy that the current Nrf2 regulation paradigm is mainly attributed to post-translational mechanisms (Purdom-Dickinson et al., 2007; Kang et al., 2010). If regulation at the level of stability is playing a role, then coincubation of ETOH + Act D could have resulted in the maintenance of high levels of Nrf2 mRNA transcripts compared with those in Act D alone-treated groups. In line with our findings, Nrf2 mRNA was reported to be up-regulated in livers and hepatocytes of chronic alcohol-fed rats (Gong and Cederbaum, 2006).
The early increase in nuclear accumulation (Fig. 2D) and DNA binding of Nrf2 (Fig. 2, A and C) could be attributed largely to the cytoplasmic release of Nrf2 from Keap-1, which could result from redox-related modifications of Keap-1 cysteines and/or phosphorylation of Nrf2 by kinases such as protein kinase C, phosphatidylinositol 3-kinase, and mitogen-activated protein kinase (Yu et al., 1999; Huang et al., 2002; Kang et al., 2002).
Ethanol Up-Regulates Nrf2 in the Intact Fetal Cerebral Cortex.
The in utero binge model demonstrated that the increased Nrf2 expression combined with functional activation in brain cortices of fetuses supported its congruity to in vitro cortical neuronal cultures. Our findings are consistent with a previous study showing that maternal ETOH exposure increases Nrf2 expression and ARE binding/signaling pathways in mouse embryos (Dong et al., 2008). It is noteworthy that the transcriptional activation of Nrf2 observed in response to ETOH in vitro at 30 min (increase) and at 24 h (little to none) is not mirrored by in utero observations (which were determined at the end of the treatment period). In general, the temporal effects on transcriptional activity seen in vitro in cellular systems cannot be directly correlated to in vivo observations. In our studies, these deviations may be accounted for by 1) the difference in dosing patterns between the two experimental settings, in vitro with a single high dose of ETOH and in utero with a well documented binge exposure, which uses multiple doses administered at 12-h intervals and 2) the possible influences of developmental stage, maternal dependence, and physiological environment in in utero settings, which might differentially affect binding affinities of the respective transcription factors to relevant consensus sites. Although Nrf2 transcriptional activity did not temporally correlate between the in utero and in vitro settings, our results illustrate that an event occurring in vitro in response to ethanol is also occurring in utero with a pattern of maternal ethanol consumption that elicits damage to the developing brain. In response to oxidative stress, Nrf2 accumulates in the nucleus with an associated decrease in cytoplasm. However, prenatal ETOH exposure increased Nrf2 in both cytoplasmic and nuclear fractions (Fig. 3C). Although the mechanism underlying this increase is not established, it could be attributed to the newly synthesized Nrf2 mRNA (Fig. 3B). In response to tert-butylhydroquinone, a similar increase in Nrf2 expression in both cytoplasm and nucleus was observed (He et al., 2007), and ETOH might activate a similar mechanism to regulate the localization and function of Nrf2.
Nrf2 Targeting Exacerbates ETOH-Induced Oxidative Stress.
A critical insight from gene-specific RNA interference-based knockdown experiments showed that ETOH-induced neurotoxicity is dependent on Nrf2 signaling in cortical neurons. This is reflected by a significant decrease in intracellular GSH content, concomitant with increased levels of superoxide and hydroperoxide (Fig. 4, B and C; Supplemental Fig. 6A). In support of our findings, data obtained in Nrf2 knockout mice and mammalian cells showed that Nrf2 modulates GSH levels by regulating the expression of GCLC, an enzyme involved in GSH biosynthesis (Yang et al., 2005). Cells deficient in Nrf2 display a disturbed redox homeostasis and are exquisitely sensitive to various oxidants as reviewed previously (Kensler et al., 2007). This observation shows that Nrf2 could serve as a key control point in both basal and ETOH-induced oxidative defenses.
Ethanol Elicits Neuron Death in the Presence of Its Nrf2 Up-Regulation.
Although ETOH enhanced Nrf2 expression both in vitro and in utero, neuron survival was still impaired (Ramachandran et al., 2001, 2003). A reasonable explanation for this impairment is that the ETOH-related Nrf2 up-regulation is simply insufficient for protection. Other plausible reasons for circumvention of Nrf2/antioxidant protection during prolonged ETOH treatment (24 h) include nuclear exclusion of Nrf2, direct caspase activation, and persistent reactive oxygen species generation (Ramachandran et al., 2003; current study). Nevertheless, the increases in Nrf2 may reflect a critical adaptive response to counteract the ETOH-induced oxidative stress and an effort to maintain redox homeostasis. Consistent with our findings, Hirota et al. (2005) demonstrated that UVA irradiation triggered Nrf2 expression and activation but failed to limit apoptosis in fibroblasts. The exaggerated caspase 3/7 activity and apoptosis (Fig. 4, D and E) seen in ETOH-treated Nrf2-deficient neurons could be ascribed to the enhanced accumulation of superoxide radical (Fig. 4C) and a subsequent loss in GSH levels (Fig. 4B). Thus, our results reveal that there is a definitive role for Nrf2 in ETOH-induced apoptosis and the deficiency of Nrf2 could sensitize fetal cortical neurons to the proapoptotic effects of ETOH. Although a definitive role for Nrf2 is evident, a tight quantitative correlation between manipulations of Nrf2 expression (siNrf2 knockdown) and subsequent neuroprotection (apoptosis measures) is not achieved in our experimental conditions. The contribution from residual Nrf2 toward survival signals could explain this result. It has been demonstrated that overexpression of Nrf2 was not able to prevent death of neurons treated with staurosporine, a potent inhibitor of phospholipid/Ca2+-dependent protein kinase (Shih et al., 2003), suggesting that modulation of Nrf2 may not necessarily protect neurons from apoptosis induced by certain pathways that do not involve reactive oxygen species. Overall, our results suggest that there is an explicit involvement of Nrf2 in ETOH-mediated apoptosis; however, the influence of Nrf2-independent events cannot be ruled out.
Overexpression of Nrf2 Thwarts ETOH-Induced Apoptosis in Fetal Cortical Neurons.
Under the basal condition, two molecules of Keap-1 sequester one molecule of Nrf2 in the cytoplasm and target Nrf2 for proteasomal degradation (Wakabayashi et al., 2004), leaving fewer Nrf2 molecules compared with Keap-1 molecules. Although ETOH induces endogenous Nrf2 and activates defense, this increase may not be sufficient to outweigh Keap-1 levels, which in turn can keep Nrf2 under control. In a recent study, treatment with tert-butylhydroquinone, a known inducer of Nrf2, has been shown to override ETOH-induced oxidative stress and prevent neural crest cell apoptosis (Yan et al., 2010). Herein, we adopted a direct up-regulation strategy by heterologous overexpression of Nrf2, with the intent of overwhelming the binding capacity of Keap-1 and freeing Nrf2 molecules to activate ARE-dependent genes before ETOH insult. A previous study demonstrated that direct Nrf2 overexpression resulted in robust ARE-dependent transcription and cytoprotection (Shih et al., 2005b). Furthermore, because DNA-based transfection of primary neurons in culture can be experimentally challenging, we adopted a well suited and efficient adenoviral strategy to overexpress Nrf2. In the Nrf2-overexpressed condition, the GSH-depleting effects of ETOH (Fig. 5, D and E) appeared to be compensated for by an increase in the rate-limiting enzyme in GSH synthesis, namely GCLC (Fig. 5C). However, the ethanol-related increase in Nrf2 expression did not generate a significant up-regulation of GCLC. One potential explanation is as follows. ETOH, a complex toxin is known to increase transforming growth factor-β1 in neurons (Kuhn and Sarkar, 2008), and the latter is known to decrease GCLC mRNA (Franklin et al., 2003). Thus, ETOH could stimulate both GCLC mRNA synthesis (by increasing Nrf2) as well as GCLC down-regulation by a transforming growth factor-β1-mediated mechanism, thereby mitigating a significant increase in the net copies of GCLC mRNA. This would not happen with adenovirus-mediated Nrf2 overexpression.
Supporting the importance of Nrf2 in neuroprotection from ETOH is the increased sensitivity to this compound associated with dysregulation of GSH (Ramachandran et al., 2003). A functional consequence of this is a selective vulnerability of cerebral cortical neurons to ETOH, which is dependent on heterogeneity of GSH homeostasis (Maffi et al., 2008). The present study supports our earlier findings that a 24-h ETOH exposure dramatically reduced the high and medium GSH populations (Fig. 5, D and E), along with increasing superoxide (Fig. 5F), hydroperoxide (Supplemental Fig. 6A), caspase 3/7 activity (Fig. 5G), and apoptosis (Fig. 5H). All of the aforementioned events were corrected by Nrf2 overexpression, thus helping in maintenance of intracellular redox balance. In accordance with our results, up-regulation of Nrf2 was correlated to increased intracellular GSH and decreased caspase 3 activity rendering neuroprotection against various oxidative stressors in several models including neuron-glia (Cho et al., 2002; Shih et al., 2003; Kraft et al., 2004; Li et al., 2005; Dong et al., 2008). It remains to be determined whether differential Nrf2 levels likewise exist in neurons, such a scenario being responsible for phenotypic divergences in GSH homeostasis and sensitivity to toxins.
In conclusion, our current studies demonstrate that although ETOH transcriptionally induces Nrf2 activation as a potentially protective stress response, this response is insufficient to prevent damage to select populations of these cells. Yet, the importance of the neuron Nrf2-mediated cytoprotection systems is clearly illustrated by multiple loss- and gain-of-function experiments. The functional relevance to ETOH-related damage to the developing brain is that the periods of highest sensitivity to ethanol are the second and early third trimester equivalents during which there is little to no astrocyte-mediated maintenance of GSH homeostasis. Thus, interventions to provide protection of neurons at these sensitive developmental stages must be at the neuron level.
Supplementary Material
Acknowledgments
We thank the flow cytometry and optical imaging core facility of University of Texas Health Science Center at San Antonio. We thank Dr. Ina Urbatsch, Texas Tech University Health Sciences Center, for granting us access to the fluorometry facility and also Dhyanesh Patel, Texas Tech University Health Sciences Center, for assisting with biotin EMSAs.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grant R01-AA010114] (to G.I.H.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/mol.111.073262.
- FASD
- fetal alcohol spectrum disorder
- ARE
- antioxidant response element
- ETOH
- ethanol
- MEM
- minimal essential medium
- HS
- horse serum
- DAPI
- 4,6-diamidino-2-phenylindole
- FITC
- fluorescein isothiocyanate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MAP2
- microtubule-associated protein 2
- siRNA
- small interfering RNA
- ECL
- enhanced chemiluminescence
- ELISA
- enzyme-linked immunosorbent assay
- PCN
- primary cortical neuron
- DIV
- days in vitro
- Act D
- actinomycin D
- q
- quantitative
- RT
- reverse transcriptase
- PCR
- polymerase chain reaction
- Ad GFP
- adenovirus for green fluorescent protein
- Ad WT Nrf2
- adenovirus for wild-type Nrf2
- Ad DN Nrf2
- adenovirus for dominant-negative Nrf2
- FACS
- fluorescence-activated cell sorting
- MCB
- monochlorobimane
- HET
- hydroethidine/dihydroethidium
- PI
- propidium iodide
- GCLC
- γ-glutamylcysteine ligase
- PBS
- phosphate-buffered saline
- EMSA
- electrophoretic mobility shift assay
- PE
- phycoerythrin
- ANOVA
- analysis of variance
- siNrf2
- small interfering RNA against Nrf2
- DN
- dominant negative
- MG-132
- N-[(phenylmethoxy)carbonyl]-l-leucyl-N-[(1S)-1-formyl-3-methylbutyl]-l-leucinamide
- GST
- glutathione transferase.
Authorship Contributions
Participated in research design: Narasimhan, Mahimainathan, and Henderson.
Conducted experiments: Narasimhan, Mahimainathan, Rathinam, and Riar.
Performed data analysis: Narasimhan, Mahimainathan, and Rathinam.
Wrote or contributed to the writing of the manuscript: Narasimhan, Mahimainathan, and Henderson.
References
- Bhave SV, Snell LD, Tabakoff B, Hoffman PL. (2000) Chronic ethanol exposure attenuates the anti-apoptotic effect of NMDA in cerebellar granule neurons. J Neurochem 75:1035–1044 [DOI] [PubMed] [Google Scholar]
- Brocardo PS, Gil-Mohapel J, Christie BR. (2011) The role of oxidative stress in fetal alcohol spectrum disorders. Brain Res Rev 67:209–225 [DOI] [PubMed] [Google Scholar]
- Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. (2002) Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol 26:175–182 [DOI] [PubMed] [Google Scholar]
- Crews FT, Waage HG, Wilkie MB, Lauder JM. (1999) Ethanol pretreatment enhances NMDA excitotoxicity in biogenic amine neurons: protection by brain derived neurotrophic factor. Alcohol Clin Exp Res 23:1834–1842 [PubMed] [Google Scholar]
- Deltour L, Ang HL, Duester G. (1996) Ethanol inhibition of retinoic acid synthesis as a potential mechanism for fetal alcohol syndrome. FASEB J 10:1050–1057 [PubMed] [Google Scholar]
- Dong J, Sulik KK, Chen SY. (2008) Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: implications for the prevention of fetal alcohol spectrum disorders. Antioxid Redox Signal 10:2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton GR. (1990) Isolation, culture, and use of viable central nervous system perikarya, in Methods in Neuroscience (Conn PM. ed) pp 87–102, Academic Press, New York [Google Scholar]
- Fischer W, Franke H, Illes P. (2003) Effects of acute ethanol on the Ca2+ response to AMPA in cultured rat cortical GABAergic nonpyramidal neurons. Alcohol Alcohol 38:394–399 [DOI] [PubMed] [Google Scholar]
- Franklin CC, Rosenfeld-Franklin ME, White C, Kavanagh TJ, Fausto N. (2003) TGFβ1-induced suppression of glutathione antioxidant defenses in hepatocytes: caspase-dependent post-translational and caspase-independent transcriptional regulatory mechanisms. FASEB J 17:1535–1537 [DOI] [PubMed] [Google Scholar]
- Furukawa M, Xiong Y. (2005) BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol 25:162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Cederbaum AI. (2006) Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology 43:144–153 [DOI] [PubMed] [Google Scholar]
- Gressens P, Lammens M, Picard JJ, Evrard P. (1992) Ethanol-induced disturbances of gliogenesis and neuronogenesis in the developing murine brain: an in vitro and in vivo immunohistochemical and ultrastructural study. Alcohol Alcohol 27:219–226 [PubMed] [Google Scholar]
- He J, Nixon K, Shetty AK, Crews FT. (2005) Chronic alcohol exposure reduces hippocampal neurogenesis and dendritic growth of newborn neurons. Eur J Neurosci 21:2711–2720 [DOI] [PubMed] [Google Scholar]
- He X, Lin GX, Chen MG, Zhang JX, Ma Q. (2007) Protection against chromium (VI)-induced oxidative stress and apoptosis by Nrf2. Recruiting Nrf2 into the nucleus and disrupting the nuclear Nrf2/Keap1 association. Toxicol Sci 98:298–309 [DOI] [PubMed] [Google Scholar]
- Henderson GI, Devi BG, Perez A, Schenker S. (1995) In utero ethanol exposure elicits oxidative stress in the rat fetus. Alcohol Clin Exp Res 19:714–720 [DOI] [PubMed] [Google Scholar]
- Hirota A, Kawachi Y, Itoh K, Nakamura Y, Xu X, Banno T, Takahashi T, Yamamoto M, Otsuka F. (2005) Ultraviolet A irradiation induces NF-E2-related factor 2 activation in dermal fibroblasts: protective role in UVA-induced apoptosis. J Invest Dermatol 124:825–832 [DOI] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, Pickett CB. (2002) Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem 277:42769–42774 [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources (1996) Guide for the Care and Use of Laboratory Animals, 7th ed, Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council, Washington DC [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JS, Miller MW. (2001) Proliferation and death of cultured fetal neocortical neurons: effects of ethanol on the dynamics of cell growth. J Neurocytol 30:391–401 [DOI] [PubMed] [Google Scholar]
- Kang HJ, Hong YB, Kim HJ, Bae I. (2010) CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. J Biol Chem 285:21258–21268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang KW, Lee SJ, Park JW, Kim SG. (2002) Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol Pharmacol 62:1001–1010 [DOI] [PubMed] [Google Scholar]
- Kapeta S, Chondrogianni N, Gonos ES. (2010) Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem 285:8171–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116 [DOI] [PubMed] [Google Scholar]
- Kraft AD, Johnson DA, Johnson JA. (2004) Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci 24:1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P, Sarkar DK. (2008) Ethanol induces apoptotic death of beta-endorphin neurons in the rat hypothalamus by a TGF-β1-dependent mechanism. Alcohol Clin Exp Res 32:706–714 [DOI] [PubMed] [Google Scholar]
- Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. (2005) Nrf2, a multi-organ protector? FASEB J 19:1061–1066 [DOI] [PubMed] [Google Scholar]
- Li J, Johnson D, Calkins M, Wright L, Svendsen C, Johnson J. (2005) Stabilization of Nrf2 by tBHQ confers protection against oxidative stress-induced cell death in human neural stem cells. Toxicol Sci 83:313–328 [DOI] [PubMed] [Google Scholar]
- Maffi SK, Rathinam ML, Cherian PP, Pate W, Hamby-Mason R, Schenker S, Henderson GI. (2008) Glutathione content as a potential mediator of the vulnerability of cultured fetal cortical neurons to ethanol-induced apoptosis. J Neurosci Res 86:1064–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. (1994) Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc Natl Acad Sci USA 91:9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney JW, Tenkova T, Dikranian K, Qin YQ, Labruyere J, Ikonomidou C. (2002) Ethanol-induced apoptotic neurodegeneration in the developing C57BL/6 mouse brain. Brain Res Dev Brain Res 133:115–126 [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Rosenbloom MJ, Mathalon DH, Lim KO. (1998) A controlled study of cortical gray matter and ventricular changes in alcoholic men over a 5-year interval. Arch Gen Psychiatry 55:905–912 [DOI] [PubMed] [Google Scholar]
- Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. (2007) Translational control of nrf2 protein in activation of antioxidant response by oxidants. Mol Pharmacol 72:1074–1081 [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Perez A, Chen J, Senthil D, Schenker S, Henderson GI. (2001) In utero ethanol exposure causes mitochondrial dysfunction, which can result in apoptotic cell death in fetal brain: a potential role for 4-hydroxynonenal. Alcohol Clin Exp Res 25:862–871 [PubMed] [Google Scholar]
- Ramachandran V, Watts LT, Maffi SK, Chen J, Schenker S, Henderson G. (2003) Ethanol-induced oxidative stress precedes mitochondrially mediated apoptotic death of cultured fetal cortical neurons. J Neurosci Res 74:577–588 [DOI] [PubMed] [Google Scholar]
- Ramsey CP, Glass CA, Montgomery MB, Lindl KA, Ritson GP, Chia LA, Hamilton RL, Chu CT, Jordan-Sciutto KL. (2007) Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol 66:75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam ML, Watts LT, Stark AA, Mahimainathan L, Stewart J, Schenker S, Henderson GI. (2006) Astrocyte control of fetal cortical neuron glutathione homeostasis: up-regulation by ethanol. J Neurochem 96:1289–1300 [DOI] [PubMed] [Google Scholar]
- Sastry PS, Rao KS. (2000) Apoptosis and the nervous system. J Neurochem 74:1–20 [DOI] [PubMed] [Google Scholar]
- Satoh T, Okamoto SI, Cui J, Watanabe Y, Furuta K, Suzuki M, Tohyama K, Lipton SA. (2006) Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophilic [correction of electrophillic] phase II inducers. Proc Natl Acad Sci USA 103:768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AY, Imbeault S, Barakauskas V, Erb H, Jiang L, Li P, Murphy TH. (2005a) Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J Biol Chem 280:22925–22936 [DOI] [PubMed] [Google Scholar]
- Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. (2003) Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci 23:3394–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AY, Li P, Murphy TH. (2005b) A small-molecule-inducible Nrf2-mediated antioxidant response provides effective prophylaxis against cerebral ischemia in vivo. J Neurosci 25:10321–10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol RJ, Delaney-Black V, Nordstrom B. (2003) Fetal alcohol spectrum disorder. JAMA 290:2996–2999 [DOI] [PubMed] [Google Scholar]
- Sun X, Shih AY, Johannssen HC, Erb H, Li P, Murphy TH. (2006) Two-photon imaging of glutathione levels in intact brain indicates enhanced redox buffering in developing neurons and cells at the cerebrospinal fluid and blood-brain interface. J Biol Chem 281:17420–17431 [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA 101:2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts LT, Rathinam ML, Schenker S, Henderson GI. (2005) Astrocytes protect neurons from ethanol-induced oxidative stress and apoptotic death. J Neurosci Res 80:655–666 [DOI] [PubMed] [Google Scholar]
- Yan D, Dong J, Sulik KK, Chen SY. (2010) Induction of the Nrf2-driven antioxidant response by tert-butylhydroquinone prevents ethanol-induced apoptosis in cranial neural crest cells. Biochem Pharmacol 80:144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Wang HD, Hu ZG, Wang QF, Yin HX. (2008) Activation of Nrf2-ARE pathway in brain after traumatic brain injury. Neurosci Lett 431:150–154 [DOI] [PubMed] [Google Scholar]
- Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, Lu SC. (2005) Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-κB and AP-1. Mol Cell Biol 25:5933–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, Wu J, Kong AN. (1999) Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J Biol Chem 274:27545–27552 [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu H, Iles KE, Liu RM, Postlethwait EM, Laperche Y, Forman HJ. (2006) 4-Hydroxynonenal induces rat γ-glutamyl transpeptidase through mitogen-activated protein kinase-mediated electrophile response element/nuclear factor erythroid 2-related factor 2 signaling. Am J Respir Cell Mol Biol 34:174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.