Abstract
Previous experiments suggested that trafficking of the a-factor transporter Ste6 of Saccharomyces cerevisiae to the yeast vacuole is regulated by ubiquitination. To define the ubiquitination-dependent step in the trafficking pathway, we examined the intracellular localization of Ste6 in the ubiquitination-deficient doa4 mutant by immunofluorescence experiments, with a Ste6-green fluorescent protein fusion protein and by sucrose density gradient fractionation. We found that Ste6 accumulated at the vacuolar membrane in the doa4 mutant and not at the cell surface. Experiments with a doa4 pep4 double mutant showed that Ste6 uptake into the lumen of the vacuole is inhibited in the doa4 mutant. The uptake defect could be suppressed by expression of additional ubiquitin, indicating that it is primarily the result of a lowered ubiquitin level (and thus of reduced ubiquitination) and not the result of a deubiquitination defect. Based on our findings, we propose that ubiquitination of Ste6 or of a trafficking factor is required for Ste6 sorting into the multivesicular bodies pathway. In addition, we obtained evidence suggesting that Ste6 recycles between an internal compartment and the plasma membrane.
INTRODUCTION
Many cellular proteins are modified by the attachment of ubiquitin (Hochstrasser, 1996; Hershko and Ciechanover, 1998). One well established function of ubiquitination is to mark proteins for degradation by the 26S proteasome. Recent studies of the internalization of yeast plasma membrane proteins, however, suggested a nonproteasomal role for ubiquitination of cell surface proteins (Hicke, 1999). Experiments with the yeast a-factor transporter Ste6, a member of the ATP-binding cassette (ABC) transporter family (Higgins, 1992), provided the first evidence that ubiquitination might be linked to endocytosis (Kölling and Hollenberg, 1994). Although Ste6 is required for secretion of the mating pheromone a-factor, it is mainly associated with internal membranes. In endocytosis mutants, however, it accumulates at the plasma membrane in a ubiquitinated form, indicating that it travels to the plasma membrane but resides there only transiently because of efficient endocytosis. The Ste6 ubiquitination observed in endocytosis mutants does not appear to be connected with proteasomal degradation because the half-life of Ste6 is not altered in mutants with severely affected proteasome function (Kölling and Losko, 1997). Instead, Ste6 is strongly stabilized in a pep4 mutant, which inhibits vacuolar proteolysis. These experiments suggested that ubiquitination could play a role in targeting Ste6 to the vacuole for degradation.
Subsequently, a number of other yeast cell surface proteins have been shown to be ubiquitinated. The most suggestive evidence for a role of ubiquitination as a trigger of endocytosis has been gained from experiments on the α-pheromone receptor Ste2 (Hicke and Riezman, 1996). Additional evidence for a role of ubiquitination in endocytosis has been obtained for several other yeast plasma membrane proteins, including the a-factor receptor Ste3 (Roth and Davis, 1996), uracil permease (Fur4) (Galan et al., 1996), the general amino acid permease (Gap1; Hein et al., 1995), and the multidrug transporter Pdr5 (Egner and Kuchler, 1996). Ubiquitination has also been directly implicated in endocytosis and lysosomal degradation of the mammalian growth hormone receptor (Strous et al., 1996). Although this study established a correlation between the ubiquitin system and growth hormone receptor endocytosis, ubiquitination of the receptor itself may not be absolutely required for endocytosis (Govers et al., 1999).
Information about the substrate features that specify recognition by the ubiquitination machinery is still limited. For Ste6, we could show that the linker region, which connects the two homologous halves of Ste6, contains a signal that mediates its ubiquitination and fast turnover (Kölling and Losko, 1997). This signal was also functional in the context of another plasma membrane protein. Ste6 deletion mutants with reduced ubiquitination accumulate at the cell surface, supporting the view that Ste6 ubiquitination is required for efficient endocytosis from the plasma membrane. However, this finding does not yet establish a causal relationship between ubiquitination and endocytosis. The deletions could as well remove separate signals for endocytosis and ubiquitination.
Another way to obtain information about the role of ubiquitination in Ste6 trafficking is to interfere with the ubiquitination machinery. In this study, we used the doa4 mutant as a tool to examine the effects of a general reduction in ubiquitination on Ste6 trafficking. Doa4 (or UBP4) is a member of a large family of ubiquitin-specific processing proteases that remove ubiquitin from ubiquitin-protein conjugates (Papa and Hochstrasser, 1993). Doa4 plays an important role in recycling ubiquitin from proteolytic substrates destined for degradation by the 26S proteasome or the vacuole. Loss of Doa4 function results in a depletion of ubiquitin, particularly as the cells enter stationary phase, presumably because ubiquitin gets degraded along with the proteolytic substrate proteins (Swaminathan et al., 1999). The doa4 mutation should, therefore, interfere with all ubiquitin-dependent processes. The doa4 mutant has been used before to demonstrate effects of ubiquitination on the internalization of yeast plasma membrane proteins (Galan and Haguenauer-Tsapis, 1997; Terrell et al., 1998). Here, we show that Ste6 accumulates at the vacuolar membrane in the doa4 mutant and not as expected at the plasma membrane. Our data suggest that ubiquitination is required for the uptake of Ste6 into the lumen of the vacuole. Additional data indicate that Ste6 recycles between internal compartments and the plasma membrane.
MATERIALS AND METHODS
Plasmids
The plasmid pRK182 is based on the single-copy vector YCp50 (Rose et al., 1987) and contains a 6.2-kb BglII/SalI STE6 fragment. To place STE6 under the control of the CUP1 promoter, pRK158 was constructed by inserting a 436-bp BamHI/EcoRI CUP1 promoter fragment and a 5.5-kb BspHI/SalI STE6 fragment between the ClaI and SalI sites of YCp50. To construct the STE6-green fluorescent protein (GFP) plasmid pRK599, a new SalI site was introduced into the STE6 gene by polymerase chain reaction (PCR) just upstream of the stop codon. A 4.5-kb BglII/SalI fragment of this modified STE6 gene and a 740-bp XhoI/SalI PCR fragment, encoding the S65G/S72A GFP variant, were cloned into the vector YEplac195 (Gietz and Sugino, 1988) upstream of a 350-bp CYC1 terminator fragment. To construct pRK628, the 1.2-kb PstI STE6 fragment of pRK599 was replaced by the 1.05-kb PstI fragment of pRK256 encoding the STE6 ΔA-box variant (Kölling and Losko, 1997). Plasmid pRK567 was obtained by insertion of a 1.8-kb chromosomal HindIII VPS4 fragment into the vector YEplac181 (Gietz and Sugino, 1988).
Yeast Strains
The yeast strains used are listed in Table 1. The Δdoa4, Δpep4, and Δste6 deletion strains are directly derived either from JD52 or from strains obtained by backcrossing to the JD52 background. The Δdoa4 strains JD116 and RKY1587 were generated by one-step gene replacement with a disruption cassette described previously (Papa and Hochstrasser, 1993). The strains were verified by PCR with a specific set of primers. In addition, JD116 was tested for complementation of the doa4 temperature sensitive (ts) phenotype by the wild-type DOA4 gene. RKY959 was obtained by transformation of JD52 with a disruption cassette consisting of the LEU2 gene and flanking STE6 sequences. This disruption removes the whole STE6-coding sequence. The STE6 disruption in strain RKY1001 was constructed with a disruption cassette described previously (Kuchler et al., 1989). Both STE6 deletions were confirmed by mating tests and western blotting. The Δpep4 strains RKY975, RKY1449, and RKY1582 were generated with PEP4 disruption cassettes containing the HIS3 and URA3 markers, respectively. The PEP4 deletions were verified by a Fast Garnet test for protease activity. The end4 and sec1-1 alleles from RH268-1C and HMSF1 were backcrossed to our JD52 background at least two times. The sec1-1 mutation was confirmed by complementation of the ts phenotype with a SEC1 plasmid.
Table 1.
Yeast strains
| Yeast | ||
| Genetic | ||
| Stock | ||
| HMSF1 | MATa sec1-1 | Center |
| JD52 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 | J. Dohmen |
| JD53 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 | J. Dohmen |
| JD116 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 Δdoa4∷LEU2 | J. Dohmen |
| RH268-1C | MATa bar1-1 his4 leu2 ura3 end4 | H. Riezman |
| RKY959 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 Δste6∷LEU2 | This work |
| RKY975 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 Δpep4∷HIS3 | This work |
| RKY1001 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 Δste6∷HIS3 | This work |
| RKY1190 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 sec1-1 Δste6∷HIS3 | This work |
| RKY1177 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 end4 sec1-1 Δste6∷HIS3 | This work |
| RKY1203 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-521 end4 Δste6∷HIS3 | This work |
| RKY1392 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 GAL1p-STE6-c-myc suc2∷(kanR-CUP1p-STE6-HIS3) | This work |
| RKY1449 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 Δdoa4∷LEU2 Δpep4∷HIS3 | This work |
| RKY1582 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 GAL1p-STE6-c-myc suc2∷(kanR-CUP1p-STE6-HIS3) Δpep4∷URA3 | This work |
| RKY1587 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 end4 Δdoa4∷LEU2 | This work |
| RKY1651 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 suc2∷(kanR-CUP1p-STE6- HIS3) Δdoa4∷LEU2 | This work |
Immunofluorescence and GFP Staining
The immunofluorescence experiments were performed essentially as described by Pringle et al. (1989). Cells were grown to exponential phase (OD600 = 0.5–0.8, 3–4 × 107/ml) and fixed directly for 4 h with formaldehyde (final concentration 5%). The fixed cells were spheroplasted and extracted with 0.1% Triton X-100 for 5 min and then attached to a multiwell slide treated with 0.1% poly-lysine (Sigma, St. Louis, MO). The cells were first incubated with the anti–c-myc mouse monoclonal primary antibody (9E10, Berkeley Antibody Co, Berkeley, CA; 1:200 dilution in phosphate-buffered saline plus 1 mg/ml bovine serum albumin) for 90 min and then another 90 min with fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibodies (Dianova, Hamburg, Germany; 1:300 dilution in phosphate-buffered saline/bovine serum albumin). Finally, the cells were incubated for 5 min with 4,6-diamidino-2-phenylindole (1 mg/ml). Fluorescence was visualized with a confocal laser scanning microscope (Leica, Deerfield, IL). For GFP fluorescence, cells were grown overnight to exponential phase (OD600 = 0.5–0.8, 3–4 × 107/ml) in minimal medium (YNB, Difco, Detroit, MI) supplemented with required nutrients. Cells were fixed on microscope slides by mixing with low-melting agarose and examined with a Axioskop fluorescence microscope (Zeiss, Oberkochen, Germany) using the FITC filter set. Images were acquired with a couple-charged device camera (Sony, Tokyo, Japan).
Cell Fractionation
Exponentially growing cells (OD600 < 0.8, 5 × 107 cells/ml) from a 100-ml culture were harvested and washed in STED10 (10% [wt/wt] sucrose, 10 mM Tris-Cl [pH 7.6], 10 mM EDTA, 1 mM dithiothreitol). The cells were lysed by agitation with glass beads in 0.25 ml of STED10 (plus protease inhibitors: 0.5 μg/ml each of aprotinin, antipain, chymostatin, leupeptin, and pepstatin A and 1.6 μg/ml benzamidine, 1 μg/ml phenanthroline, and 170 μg/ml phenylmethylsulfonyl fluoride) for 3 min. After the addition of another 1 ml of STED10, the extract was transferred to a fresh tube and spun at 500 × g for 5 min to remove cell debris. The cleared cell extract was loaded on top of a sucrose gradient. The gradient was prepared as follows. In an SW40 centrifuge tube, 4 ml of STED50 (50% sucrose), STED35 (35% sucrose), and STED20 (20%) sucrose were layered on top of each other. The centrifuge tubes were slowly turned into a horizontal position. After 3–5 h of horizontal diffusion, the gradients were again turned into an upright position, loaded with extract, and spun at 4°C for 15 h at 30,000 rpm in an SW40 rotor; 700-μl fractions were collected from the top of the gradients and examined for marker enzymes by Western blotting.
Pulse-Chase Experiments and Immunoprecipitation
The experiments were basically performed as described previously (Franzusoff et al., 1991). Cells were grown overnight to exponential phase in synthetic minimal medium (YNB) with required nutrients; 5 OD600 units (3 × 108 cells) were harvested and resuspended in fresh medium to a density of OD600 = 2. After a 20-min preincubation, the cells were labeled for 15 min with 70 μCi of Tran 35S-label (ICN, Costa Mesa, CA). Chase was initiated by the addition of 1/50 volume of concentrated chase solution (0.3% cysteine, 0.4% methionine) to the culture. Aliquots of 1 OD600 unit of cells were removed at 20-min intervals, washed once in 1 ml of cold 10 mM NaN3, and lysed by agitation with 400 mg of glass beads in 110 μl of lysis buffer (0.3 M sorbitol, 50 mM 3-(N-morpholino)propanesulfonic acid, pH 7.5, 10 mM NaN3 plus protease inhibitors) for 3 min. Proteins were solubilized with 1 volume of 2× sample buffer (4% SDS, 125 mM Tris-Cl [pH 6.8], 20% glycerol, 40 mM dithiothreitol, 0.02% bromphenol blue) for 30 min at 50°C. The 4 volumes of immunoprecipitation (IP)-dilution buffer (1.25% Triton X-100, 6 mM EDTA, 60 mM Tris-Cl [pH 7.4]) were added, and insoluble material was removed by centrifugation for 5 min in a table top centrifuge. The samples were incubated overnight at 4°C with 10 μl of Ste6 antiserum and another 3 h at 4°C with 100 μl of a 20% suspension of protein A-Sepharose beads (Pharmacia, Piscataway, NJ). The beads were washed three times with IP buffer (1% Triton X-100, 5 mM EDTA, 50 mM Tris-Cl [pH 7.4]) and finally resuspended in 50 μl of IP buffer. Proteins were removed from the beads by incubation with sample buffer for 30 min at 50°C. About half of the immunoprecipitated material was loaded onto a 7.5% SDS-PAGE gel. The gels were fixed in a mixture of acetic acid (7%) and methanol (20%), soaked in Amplify (Amersham, Arlington Heights, IL) to enhance the signal, dried, and exposed to X-Omat AR film (Kodak, Rochester, NY).
RESULTS
Ubiquitinated Ste6 Is Associated with Internal Membranes
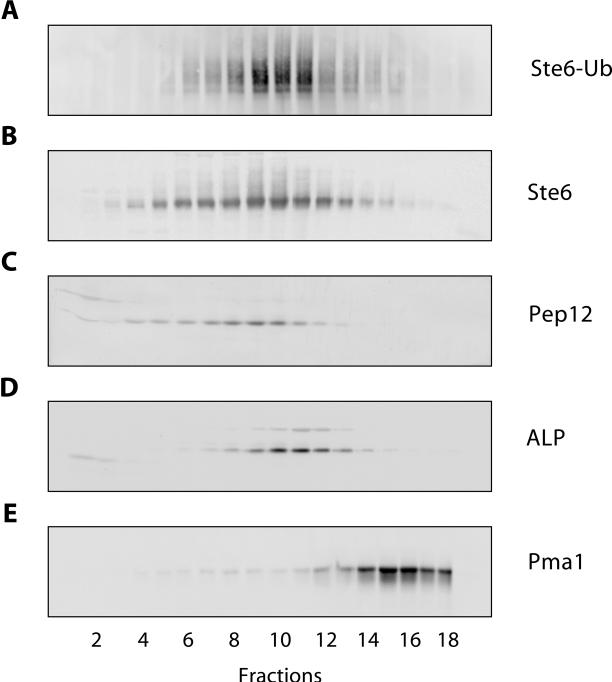
Information about the site and function of Ste6 ubiquitination may be gained from the analysis of the intracellular distribution of ubiquitinated Ste6 forms. To determine the intracellular distribution of ubiquitinated Ste6, cell fractionation experiments were performed. Ubiquitinated forms of Ste6 are not easily detected by Western blotting with cell extracts prepared from wild-type cells, because only a minor fraction of Ste6 appears to be ubiquitinated at any given time. It is possible that this minor fraction has a different intracellular distribution than the bulk of Ste6. To be able to detect ubiquitinated Ste6, the plasmid YEp112 (Hochstrasser et al., 1991) carrying a hemeagglutin (HA)-tagged ubiquitin variant, under the control of the copper-inducible CUP1 promoter, was cotransformed into the Δste6 strain RKY959 together with a multicopy STE6 plasmid. Ste6 overproduced from a multicopy plasmid showed the same half-life and distribution on sucrose gradients as Ste6 expressed form the chromosomal copy of the STE6 gene. Whole cell extracts prepared from this strain were fractionated on sucrose density gradients. Fractions were collected from the gradients and assayed for the presence of marker enzymes by Western blotting (Figure 1). In addition, Ste6 was immunoprecipitated from the gradient fractions with anti-Ste6 antibodies and the immunoprecipitates where analyzed by Western blotting with anti-HA antibodies for the presence of HA-tagged ubiquitin, covalently linked to Ste6. As reported previously, Ste6 (Figure 1B) did not cofractionate with the plasma membrane marker Pma1 (Figure 1E), which indicates that most of Ste6 is associated with internal membranes and not with the cell surface. The Ste6 distribution (peak in fractions 9 and 10), however, closely matched the distribution of the endosomal t-SNARE Pep12 (peak in fraction 9, Figure 1C), which suggests that a fraction of Ste6 is associated with endosomal membranes. The distribution of the vacuolar marker alkaline phosphatase (ALP; peak in fractions 10 and 11, Figure 1D) was slightly shifted toward the denser fractions of the gradient compared with Ste6. The distribution of ubiquitinated Ste6 (Figure 1A) closely matched the distribution of unmodified Ste6. This result indicates that ubiquitinated Ste6 accumulates at the bottleneck of the Ste6 degradative pathway. Interestingly, no ubiquitinated Ste6 was detected in the plasma membrane fraction. If ubiquitination triggers Ste6 endocytosis, internalization must be a very rapid process. Alternatively, these findings are also compatible with an internal role of ubiquitination in Ste6 trafficking.
Figure 1.
Intracellular distribution of ubiquitinated Ste6. Whole cell extracts of the Δste6 strain RKY959 transformed with YEp112 (2μ-CUP1-UBI-HA) and pYKS2 (2μ-STE6-c-myc) were fractionated on a sucrose density gradient (20–50% [wt/wt] sucrose, fraction 1: low sucrose density). The gradient fractions were analyzed by Western blotting for the presence of marker proteins with specific antibodies against HA-tag (BAbCo) (A) , Ste6 (B), Pep12 (C), ALP (D; Molecular Probes, Eugene, OR), and Pma1 (E). In A, immunoprecipitates were analyzed for the presence of HA-tagged ubiquitin covalently linked to Ste6 after immunoprecipitation with anti-Ste6 antibodies.
Effects of the doa4 Mutation on Ste6 Transport and Turnover
The evidence obtained so far strongly implicates ubiquitination in the regulation of Ste6 trafficking to the vacuole (Kölling and Hollenberg, 1994; Kölling and Losko, 1997). However, the exact step in the transport pathway to the vacuole affected by ubiquitination is still unknown. If we assume that ubiquitination is required for a certain transport step (e.g., internalization from the plasma membrane), Ste6 should accumulate at that point in the transport pathway when ubiquitination is blocked. As a tool to interfere with Ste6 ubiquitination, we chose a mutant defective in the deubiquitinating enzyme Doa4 (or Ubp4). All ubiquitin-depending processes, e.g., Ste6 ubiquitination, should be affected in this mutant because of its reduced ubiquitin level (Swaminathan et al., 1999).
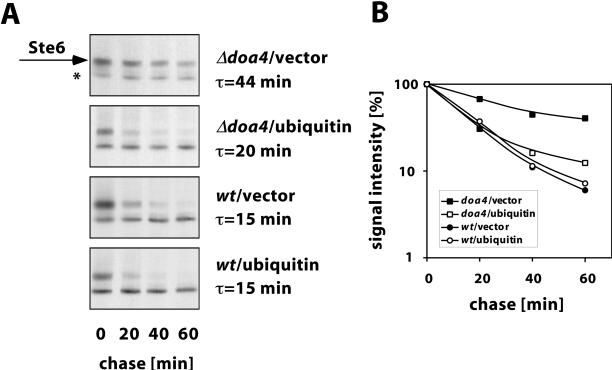
To test whether doa4 deficiency has an effect on Ste6 transport to the vacuole, the Ste6 half-life was determined in a wild-type strain and in a congenic doa4 mutant by pulse-chase experiments. After a 15-min pulse with [35S]methionine, the chase was initiated and samples were taken at time intervals and analyzed for the presence of Ste6 by immunoprecipitation, SDS-PAGE, and autoradiography (Figure 2A). In the wild-type strain JD52, Ste6 was quickly degraded with a half-life of 15 min. In the doa4 mutant JD116, however, Ste6 was strongly stabilized (half-life: 44 min). A quantification of the data is shown in Figure 2B. This experiment shows that Ste6 is indeed stabilized in a doa4 mutant. A similar finding was published previously (Loayza and Michaelis, 1998).
Figure 2.
Effect of ubiquitin overexpression on Ste6 turnover in the doa4 mutant. Ste6 half-life was determined by pulse-chase experiments in the doa4 strain JD116 and the wild-type strain JD52 transformed with the ubiquitin-overexpressing plasmid YEp96 or the control vector YEplac112. Cells grown at 30°C in the presence of 100 μM CuSO4 were labeled for 15 min with Tran35S-label (ICN) and were then chased with an excess of cold methionine and cysteine. Ste6 was immunoprecipitated from cell extracts prepared at time intervals as indicated. The precipitated proteins were analyzed by SDS-PAGE and autoradiography. (A) From top to bottom: JD116 (doa4)/YEplac112 (vector), JD116 (doa4)/YEp96 (CUP1-UBI), JD52 (wt)/YEplac112 (vector), JD52 (wt)/YEp96 (CUP1-UBI). The Ste6 band is marked with an arrow; a background band is marked with an asterix. (B) Signal intensities (logarithmic scale) plotted against the chase time. ▪, JD116/YEplac112; □, JD116/YEp96; ●, JD52/YEplac112; ○, JD52/YEp96.
Many of the doa4 phenotypes appear to be the result of a reduction in free ubiquitin level (Swaminathan et al., 1999). If the observed Ste6 stabilization is caused by reduced ubiquitination, it should be possible to rescue the defect in Ste6 degradation by ubiquitin overexpression. To test this prediction, Ste6 half-life was determined in the doa4 strain JD116 transformed with the multicopy plasmid YEp96 carrying the ubiquitin gene under the control of the copper-inducible CUP1 promoter (Hochstrasser et al., 1991). Ubiquitin expression was induced by the addition of 100 μM CuSO4 to the growth medium. As can be seen in Figure 2, almost wild-type turnover was observed upon ubiquitin overexpression in the doa4 strain. The half-life was only slightly longer (20 versus 15 min) compared with the wild-type strain under the same conditions. From this experiment, we conclude that the defect in Ste6 degradation is primarily the result of a lowered ubiquitin level and thus of reduced ubiquitination. In addition, Doa4 may also have a direct effect on Ste6 turnover, because the turnover could not be completely restored to wild-type levels by ubiquitin overexpression. This effect, however, appears to be small compared with the effect of reduced ubiquitination.
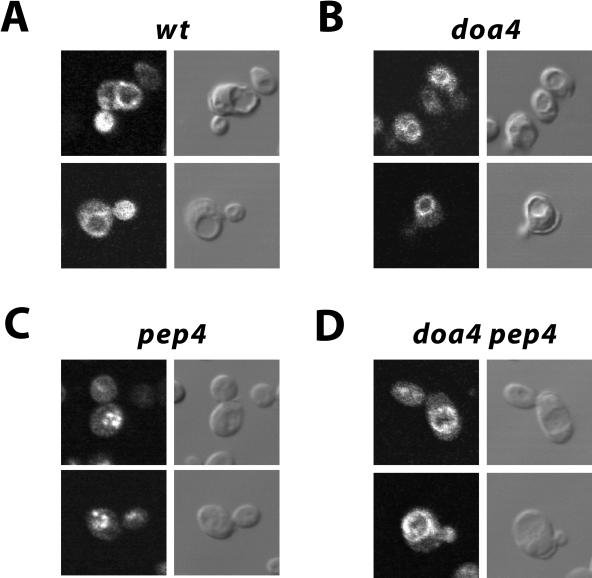
To identify the transport step blocked in the doa4 mutant, the intracellular localization of c-myc–tagged Ste6, expressed from the multicopy plasmid pYKS2 (Kuchler et al., 1993), was examined by immunofluorescence microscopy. As reported previously (Kölling and Losko, 1997), a ring-like Ste6 staining surrounding the vacuole was observed in the wild-type strain JD52/pYKS2 (Figure 3A). The vacuoles can be seen as indentations in the Nomarsky image. In addition, some of the cells showed a few stained patches close to the vacuole. This staining pattern is in line with an endosomal/vacuolar localization of Ste6. No signal was observed with JD52 transformed with a vector control. The congenic doa4 strain JD116/pYKS2 gave a staining very similar to wild type (Figure 3B), except that the staining was much brighter than with wild type because of the higher steady-state level of Ste6 in this strain (the signals in Figure 3 were adjusted to the same intensities). Most conspicuously, despite a much brighter signal, there was no surface staining visible in the doa4 strain. Surface staining would have been expected, if the doa4 mutation blocked internalization, as suggested for other yeast plasma membrane proteins (Hicke, 1999). It is unlikely that Ste6 present at the cell surface could have escaped detection because in endocytosis mutants it is readily detected at the cell surface (Kölling and Hollenberg, 1994). We therefore conclude that the doa4 mutation does not affect internalization of Ste6. Instead, it appears to block a trafficking step late in the degradation pathway of Ste6. This step could be uptake into the vacuole.
Figure 3.
Localization of Ste6 by immunofluorescence. The c-myc-tagged Ste6 variant, encoded by pYKS2, was detected with anti-myc primary antibodies and FITC-conjugated anti-mouse secondary antibodies in different strains. (A) JD52 (wild type). (B) JD116 (doa4). (C) RKY975 (pep4). (D) RKY1449 (doa4 pep4). Left, FITC-fluorescence; right, Nomarsky image. The cells were grown at 30°C in minimal medium with 1% casamino acids.
There are two possible destinations for membrane proteins targeted to the vacuole. The proteins may end up in the vacuolar membrane surrounding the vacuole or they may be targeted to the vacuolar lumen by the recently described multivesicular bodies (MVB) pathway (Odorizzi et al., 1998). If Ste6 is indeed targeted to the lumen of the vacuole, it should accumulate inside the vacuole when vacuolar proteolysis is blocked by a pep4 mutation. We therefore examined the Ste6 immunofluorescence staining in the pep4 mutant RKY975/pYKS2 and in the pep4 doa4 double mutant RKY1449/pYKS2. Both strains are derived from the wild-type strain JD52. With the pep4 strain, we saw brightly staining patches in the area where the vacuoles were located in the Nomarsky image (Figure 3C). In contrast to the pep4 mutant, the doa4 pep4 double mutant again showed a ring-like staining around the vacuole (Figure 3D), just like wild type and the doa4 single mutant. Thus, genetically, the doa4 mutation is epistatic to the pep4 mutation. From these results, we conclude that the doa4 mutation blocks uptake of Ste6 into the vacuole.
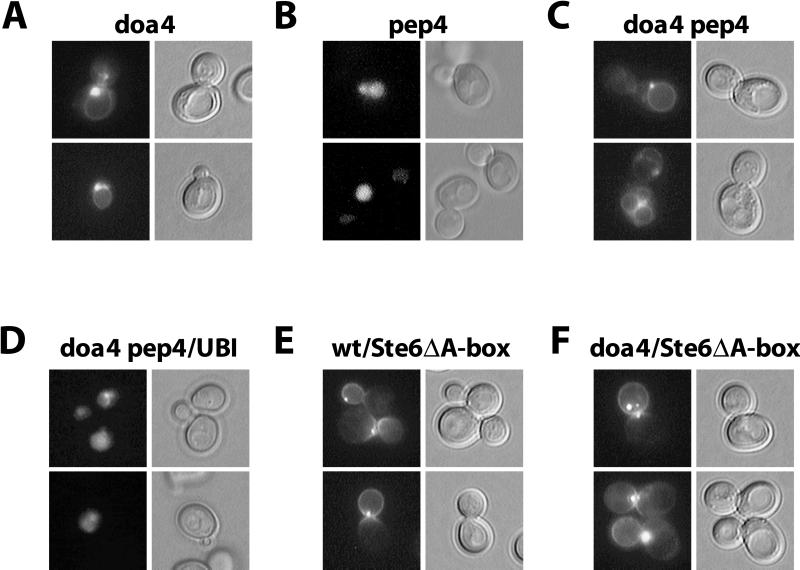
To exclude fixation artifacts, we also examined the intracellular Ste6 distribution in the living cell using an Ste6-GFP fusion. The Ste6-GFP fusion was tested in a mating assay and proved fully functional. There is one problem, however, with using GFP for the detection of Ste6. Because Ste6 is a very short-lived protein with a half-life of approximately 15 min and because the GFP chromophore is formed with a much longer half-life of approximately 80 min (Tsien, 1998), no meaningful information can be obtained with the Ste6-GFP fusion in a wild-type background. The Ste6-GFP fusion proved very useful, however, for the analysis of the Ste6 distribution in mutants in which Ste6 is strongly stabilized. The STE6-GFP fusion on a multicopy plasmid (pRK599) was transformed into different mutant strains. In the doa4 mutant JD116/pRK599, we observed a ring-like staining around the vacuole, very similar to the one observed in the immunofluorescence experiment (Figure 4A). In addition, most of the cells showed a brightly staining dot close to the vacuole. In the pep4 mutant RKY975/pRK599, the interior of the vacuole was diffusely and uniformly stained (Figure 4B). The doa4 pep4 double mutant RKY1449/pRK599 showed a staining indistinguishable from the staining of the doa4 single mutant (Figure 4C). Again, this demonstrates that doa4 is epistatic to pep4, i.e., that Ste6 uptake into the vacuole is blocked in the doa4 mutant.
Figure 4.
Intracellular localization of Ste6-GFP fusions. GFP fluorescence was observed in exponentially growing cells transformed with pRK599 (wild-type STE6 fused to GFP) or pRK628 (STE6 ΔA-box fused to GFP). (A) JD116/pRK599 (doa4). (B) RKY975/pRK599 (pep4). (C) RKY1449/pRK599 (doa4 pep4). (D) RKY1449/pRK599/YEp96 (doa4 pep4, 2μ-ubiquitin). (E) JD52/pRK628 (wild type). (F) JD116/pRK628 (doa4). Left, GFP-fluorescence; right, differential interference contrast (DIC) image. The cells were grown at 30°C in minimal medium with 1% casamino acids.
Like in the immunofluorescence experiment, no cell surface staining was observed in the doa4 mutant. One possible explanation for the lacking endocytosis defect is that ubiquitination functions as a plasma membrane-targeting signal for Ste6. In that case, Ste6 would never reach the plasma membrane. To test whether Ste6 is able to reach the plasma membrane in the doa4 mutant, we constructed a GFP fusion with an endocytosis-defective Ste6 variant (pRK628). We had shown previously that this Ste6 ΔA-box variant accumulates at the cell surface (Kölling and Losko, 1997). In both wild-type (Figure 4E) and doa4 strain (Figure 4F), cell surface staining was observed with this GFP fusion. The staining was mainly concentrated in the bud. In addition, a brightly staining dot was seen at the cell periphery in most cells. This experiment shows that Ste6 can travel to the plasma membrane in the doa4 mutant. It therefore appears unlikely that ubiquitination is required for Ste6 sorting to the plasma membrane. Surface staining was also observed with the wild-type Ste6-GFP fusion in an doa4 end4 strain (not shown). This experiment, however, may be uninformative because it has been shown that endocytosis mutants can suppress the ubiquitin deficiency of the doa4 mutant (Swaminathan et al., 1999).
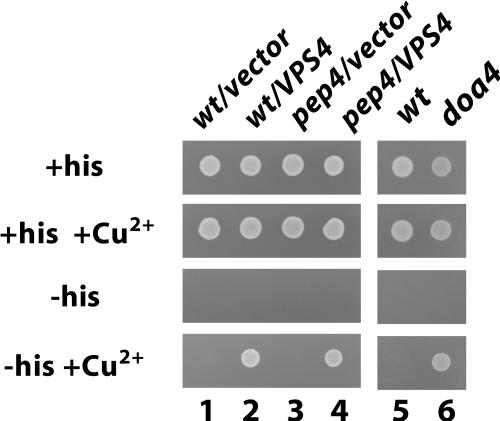
Our experiments clearly show that Ste6 is transported into the lumen of the vacuole. In a previous report, however, it had been claimed, that Ste6 accumulates in the vacuolar membrane in the pep4 mutant (Loayza and Michaelis, 1998). Observations made with an Ste6-His3 fusion protein further substantiate our conclusions. The His3 portion of this hybrid protein, which is fused to the C terminus of Ste6, should be oriented toward the cytoplasm, according to the topological model of Ste6. If His3 is accessible from the cytoplasm, the fusion should be able to complement the histidine auxotrophy of a his3 deletion strain. Under normal conditions, the level of Ste6-His3, expressed from the fully induced CUP1 promoter is not sufficient to support growth of a his3 strain. But, when the level of Ste6-His3 is increased, e.g., because of an Ste6-stabilizing mutation, the strains are able to grow on media lacking histidine (Losko, Kopp, Kranz, and Kölling, unpublished observations). We have shown before that Ste6 is strongly stabilized in a pep4 mutant (∼10-fold; Kölling and Hollenberg, 1994). If Ste6-His3 accumulated in the vacuolar membrane in a pep4 mutant, the cells should be able to grow on plates lacking histidine. As can be seen in Figure 5, the Δpep4 cells were not able to grow without histidine in the presence of 0.5 mM CuSO4 (to induce the CUP1 promoter), indicating that Ste6-His3 is sequestered from the cytoplasm. As a positive control, the strains were transformed with a multicopy plasmid carrying the VPS4 gene. Overproduction of VPS4 stabilizes Ste6 approximately threefold (Kranz, Kinner, Kölling, unpublished data). Although the stabilization caused by overproduction of VPS4 was far less than the stabilization through Δpep4, the transformants could grow without histidine. This experiment, therefore, supports our conclusion that Ste6 accumulates in the lumen of the vacuole in a pep4 mutant. We also tested whether Ste6-His3 results in histidine prototrophy in doa4 cells. As can be seen in Figure 5, the Ste6-His3 cassette, integrated into the doa4 strain JD116, supports growth on plates lacking histidine. This result shows that Ste6-His3 is accessible from the cytoplasm under these conditions and supports our conclusion that Ste6 accumulates in the vacuolar membrane in doa4 cells.
Figure 5.
Ste6 is sequestered from the cytoplasm in a pep4 mutant. The wild-type strain RKY1392 (lanes 1, 2, and5) and the congenic Δpep4 strain RKY1582 (lanes 3 and4), which both contain a CUP1p-STE6-HIS3 cassette integrated into their genome, were transformed with the vector plasmid YEplac181 (lanes 1 and 3) or the 2μ-VPS4 plasmid pRK567 (lanes 2 and 4). Lane 6, doa4 strain RKY1651 containing the CUP1-STE6-HIS3 cassette. An equal amount of cells was spotted onto minimal plates containing different supplements (required nutrients, +/−histidine, +/−0.5 mM CuSO4), as indicated. Plates were incubated for 3 d at 30°C.
The defect in Ste6 degradation in the doa4 strain can largely be suppressed by ubiquitin overexpression (Figure 2). We were therefore interested to see whether this is also true for the observed uptake defect into the vacuole. To this end, the doa4 pep4 strain RKY1449/pRK599 was transformed with the multicopy plasmid YEp96 carrying the ubiquitin gene under the control of the copper-inducible CUP1 promoter (Hochstrasser et al., 1991). The transformants were examined for their Ste6-GFP fluoresence pattern. In contrast to the cells without additional ubiquitin (Figure 4C), which showed exclusively vacuolar membrane staining, ∼70% of the cells now showed a pep4-staining pattern with a diffuse labeling of the vacuolar lumen (Figure 4D). This degree of suppression was already observed without the addition of copper (which induces the CUP1 promoter). The fraction of suppression, however, could not be further increased by the addition of copper. A similar situation was observed in the pulse-chase experiments, in which the Ste6 half-life in the doa4 strain was still slightly longer in the presence of additional ubiquitin than in the wild-type strain under the same conditions. But still, the finding that Ste6 uptake into the vacuole could be restored to a large extent by additional ubiquitin strongly suggests that the doa4 phenotypes on Ste6 turnover are mainly the result of a ubiquitination defect. In addition, it is possible that Doa4 also has a small direct effect on Ste6 uptake into the vacuole.
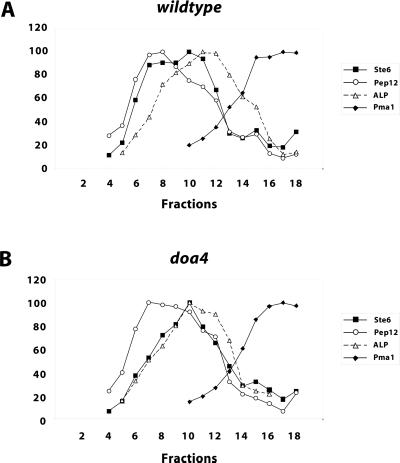
To determine the intracellular distribution of Ste6 expressed from the chromosomal copy of the STE6 gene, cell fractionation experiments were performed. Whole cell extracts were fractionated on sucrose density gradients. Fractions were collected and assayed for the presence of marker enzymes by Western blotting. A quantification of the Western blot signals is shown in Figure 6. Ste6 prepared from the wild-type strain JD52 (Figure 6A) was found in two overlapping peaks (fractions 8 and 10) in the middle of the gradient. Again, Ste6 did not cofractionate with the plasma membrane marker Pma1. The first of the two peaks (fraction 8) closely matched the peak of the endosomal marker Pep12, which suggests that this peak could correspond to endosome-localized Ste6. The second peak (fraction 10) overlapped but did not coincide with the peak of ALP, the vacuolar marker protein (fractions 11 and 12). The marker proteins from the doa4 strain JD116 showed a very similar distribution on the sucrose gradient compared with wild type. The Ste6 distribution, however, was different. With the doa4 strain (Figure 6B), only a single Ste6 peak was observed (fraction 10), which closely matched the peak of the vacuolar marker ALP. The “endosomal” peak observed with wild type was missing. In line with the immunofluorescence and GFP experiments, no accumulation of Ste6 in the plasma membrane fraction was observed. The cell fractionation experiments, therefore, support the conclusion that Ste6 accumulates at the vacuole in the doa4 mutant.
Figure 6.
Distribution of marker proteins from wild-type and doa4 strains on sucrose gradients. Whole cell extracts of the wild-type strain JD52 (A) and the doa4 strain JD116 (B) grown at 30°C in minimal medium with 1% casamino acids were fractionated on a sucrose density gradients (20–50% [wt/wt] sucrose, fraction 1: low sucrose density). The gradient fractions were analyzed by Western blotting for the presence of marker proteins with specific antibodies. The Western blots were scanned and the signal intensities were quantified with the program NIH Image 1.61. The strongest signals were set to 100%. ▪, Ste6; ○, Pep12; ▵, ALP; ♦, Pma1.
Sec1-independent Ste6 Transport to the Plasma Membrane
The results presented so far point to an internal role of ubiquitination in Ste6 trafficking. How can this be reconciled with our earlier findings that Ste6 accumulates in a ubiquitinated form at the plasma membrane upon block of endocytosis? One possible explanation is that ubiquitinated Ste6 continuously cycles between an internal compartment (e.g., endosomes) and the plasma membrane and becomes trapped at the plasma membrane after block of endocytosis. Transport to the plasma membrane via an endosomal recycling pathway should be independent of the late secretory pathway. We therefore tested whether ubiquitinated Ste6 still accumulates at the plasma membrane in an endocytosis mutant when the late secretory pathway is blocked. To be able to simultaneously block endocytosis and secretion, an end4 sec1-1 double mutant was constructed. Both mutations are conditional, i.e., support growth at low temperature (25°C) and confer a ts phenotype at nonpermissive temperature (37°C). At nonpermissive temperature, the end4 mutation inhibits internalization of yeast plasma membrane proteins (Raths et al., 1993), whereas sec1-1 blocks the fusion of secretory vesicles with the plasma membrane (Novick and Schekman, 1979).
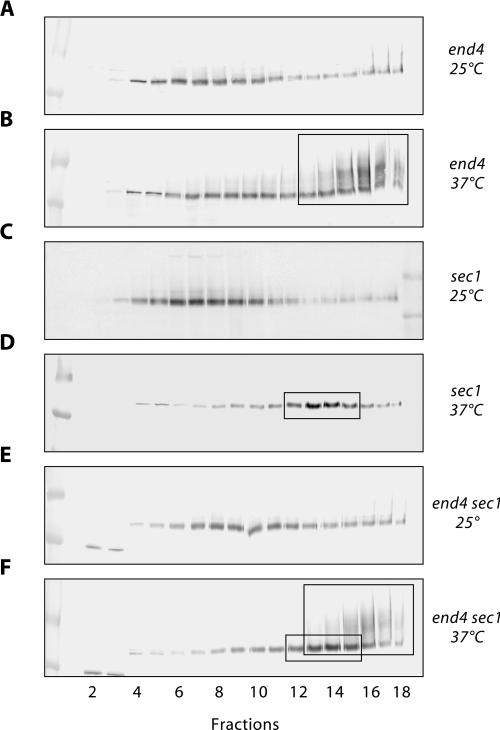
The intracellular Ste6 distribution in the end4 and sec1-1 single mutants and the in the end4 sec1-1 double mutant was examined by cell fractionation on sucrose density gradients. All strains used in this experiment carry a STE6 deletion and were, therefore, transformed with a single-copy STE6 plasmid (pRK182). The gradient fractions were analyzed for the presence of Ste6 and the plasma membrane marker protein Pma1 by Western blotting. As in the gradients described above, Pma1 was found in the gradient fractions with the highest density (fractions 13–18, not shown). In the end4 mutant RKY1203/pRK182 grown at 25°C, Ste6 was mainly found in the middle of the gradient (Figure 7A), just like in the wild-type strain JD52 (Figure 6A). A small amount of Ste6 was also detected in the plasma membrane fraction (fractions 13–18) because of a partial endocytosis defect of the mutant at permissive temperature. After growth for 1 h at 37°C, a massive accumulation of ubiquitinated Ste6 was observed in the plasma membrane fraction (boxed in Figure 7B). Ubiquitinated Ste6 can be seen as slower migrating diffusely staining material on top of the normal Ste6 band (“ubiquitin-smear”). At 25°C, the sec1-1 mutant RKY1190/pRK182 showed a wild-type Ste6 distribution (Figure 7C). However, when it was shifted to 37°C for 1 h, Ste6 accumulated in a new peak (fraction 13 and 14), which did not coincide with any of the Ste6 peaks described so far (boxed in Figure 7D). This peak most likely corresponds to secretory vesicles that accumulate in the sec1-1 mutant at nonpermissive temperature. Does the sec1-1 mutation prevent accumulation of ubiquitinated Ste6 at the plasma membrane in the end4 sec1-1 double mutant at nonpermissive temperature? As can be seen in Figure 7F, ubiquitinated Ste6 still accumulated at the plasma membrane in the double mutant RKY1177/pRK182, after a 1-h shift to 37°C. In fact, the Ste6 distribution in the double mutant looked like a superimposition of the Ste6 patterns seen with the single mutants at 37°C (boxes in Figure 7F). This result shows that Ste6 can travel to the plasma membrane by a Sec1-independent pathway.
Figure 7.
Sec1-independent Ste6 transport to the plasma membrane. The intracellular distribution of Ste6 in different strains, transformed with the single-copy STE6 plasmid pRK182, was examined by sucrose density gradient fractionation (20–50% [wt/wt] sucrose, fraction 1: low sucrose density) and Western blotting with anti-Ste6 antibodies. The strains were either continuously grown at 25°C (A, C, and E) or were first grown at 25°C and then shifted to 37°C for 1 h (B, D, and F) before extract preparation. (A and B) RKY1203/pRK182 (end4). (C and D) RKY1190/pRK182 (sec1-1). (E and F) RKY1177/pRK182 (end4 sec1-1). The area where the plasma membrane marker Pma1 is found in the gradients is marked by large boxes in B and F. The putative secretory vesicle peak is highlighted by small boxes in D and F.
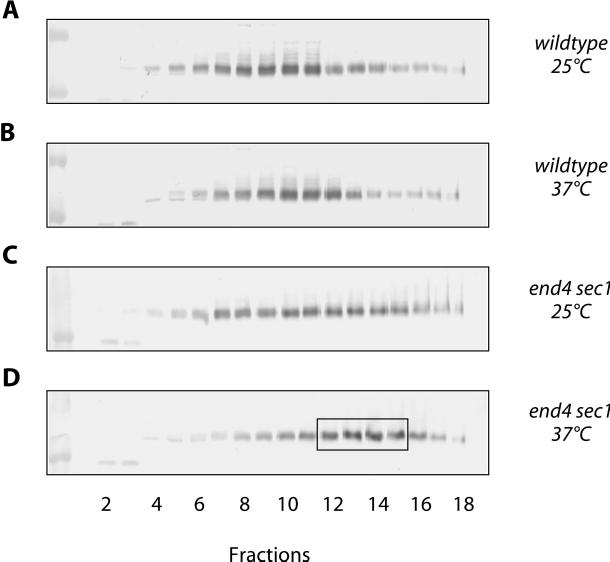
These findings, however, are not yet proof of an endosome to plasma membrane-recycling pathway for Ste6. An alternative interpretation of the data is that Ste6 transport to the plasma membrane occurs exclusively via the secretory pathway but that it is only partially (or not at all) inhibited by the sec1-1 mutation. This would imply that Ste6 behaves very differently from other exocytic proteins, e.g., invertase, whose secretion is completely blocked by sec1-1. The results of the following experiment argue against this second, alternative interpretation. In this experiment, we examined the intracellular distribution of Ste6 protein synthesized in the end4 sec1-1 double mutant after the shift to nonpermissive temperature. To be able to switch on STE6 expression at a defined time point, the end4 sec1-1 strain RKY1177 was transformed with a single-copy plasmid (pRK158), which carries STE6 under the control of the copper-inducible CUP1 promoter. With this construct, only a low level of expression was observed without the addition of copper. STE6 expression was induced by the addition of Cu2+ ions 15 min after the shift to nonpermissive temperature. After further incubation at 37°C for 45 min, whole cell extracts were prepared and fractionated on sucrose density gradients. The fractions were analyzed for the presence of Ste6 and Pma1 by Western blotting. Again, Pma1 was found in the dense fractions of the gradient. The Ste6 distribution (Figure 8D), however, was markedly different compared with the previous experiment with steady-state STE6 expression (Figure 7F). This time no ubiquitinated Ste6 could be detected in the plasma membrane fraction (fractions 13–18). Only the secretory vesicle peak (fractions 12–15, boxed in Figure 8D) was observed. When the temperature shift to 37°C was omitted (Figure 8C), the Ste6 distribution was very similar to the one observed with steady-state STE6 expression (Figure 7E). Under these conditions, a small amount of ubiquitinated Ste6 could be seen in the plasma membrane fraction. With the wild-type strain RKY1001/pRK158 the Ste6 peak was found in the middle of the gradient at both temperatures (Figure 8, A and B). From these experiments, we conclude that only Ste6 protein pre-existing at the time of the temperature shift can be translocated to the plasma membrane in the end4 sec1-1 double mutant. This translocation event is Sec1 independent. On the contrary, the Ste6 protein synthesized after the temperature shift accumulates in secretory vesicles and cannot reach the plasma membrane. This demonstrates that transport of newly synthesized Ste6 to the plasma membrane is indeed absolutely dependent on Sec1. Our data further suggest that Ste6 gains access to the “recycling compartment” via a Sec1-dependent step (i.e., via the plasma membrane).
Figure 8.
Newly synthesized Ste6 is not transported to the cell surface in a sec1-1 mutant at nonpermissive temperature. The intracellular Ste6 distribution in STE6 deletion strains transformed with the single-copy CUP1p-STE6 plasmid pRK158 was examined by sucrose density gradient fractionation (20–50% [wt/wt] sucrose, fraction 1: low sucrose density) and by Western blotting with anti-Ste6 antibodies. The cells were either continuously grown at 25°C (A and C) or were first grown at 25°C and then shifted to 37°C 1 h (B and D) before extract preparation. STE6 expression from the CUP1 promoter was induced 45 min before extract preparation by the addition of 0.5 mM CuSO4 to the cultures, i.e., copper was added 15 min after the 37°C cultures had been shifted to nonpermissive temperature. (A and B) RKY1001/pRK158 (wild type). (C and D) RKY1177/pRK158 (end4 sec1-1). The putative secretory vesicle peak is boxed (D).
DISCUSSION
We have presented evidence pointing to an internal role of ubiquitination in Ste6 trafficking. Our data suggest that ubiquitination is required for the uptake of Ste6 into the lumen of the vacuole
Phenotypes of the doa4 Mutant
How does the doa4 mutant interfere with Ste6 trafficking? One obvious possibility is that doa4 interferes with a step that requires deubiquitination of Ste6. Several lines of evidence, however, suggest that doa4 primarily affects ubiquitination by lowering the free ubiquitin level in the cell (Swaminathan et al., 1999). In line with this notion, we found that the defect in Ste6 turnover in the doa4 mutant could be suppressed by ubiquitin overexpression. Likewise, uptake into the vacuole in the doa4 pep4 mutant could be restored by expression of extra ubiquitin. This strongly suggests that the Ste6-trafficking defects observed in the doa4 mutant are primarily the result of a lowered ubiquitin level and thus of reduced ubiquitination. Suppression of the doa4 defects, however, was not complete. Therefore, it is possible that Doa4 may also have a direct effect on Ste6 trafficking and turnover. This effect, however, appears to be small compared with the effect of reduced ubiquitination.
Is there evidence for reduced Ste6 ubiquitination in the doa4 mutant? This question is difficult to answer, because detection of ubiquitinated species, as it is now widely performed by most researchers in the field, entails overexpression of epitope-tagged ubiquitin variants, which in turn suppresses most doa4 phenotypes. From the appearance of the Ste6 band on Western blots, there are no indications for a Ste6 hyperubiquitination in the doa4 mutant. As in wild type, only a distinct Ste6 band and no “ubiquitin smear” is observed. Because the ubiquitination level of Ste6 in the doa4 mutant cannot be determined, we cannot decide at present whether doa4 acts directly by reducing Ste6 ubiquitination or by affecting the function or turnover of another factor required for Ste6 uptake into the vacuole.
The effect of doa4 on Ste6 turnover has also been investigated by another group (Loayza and Michaelis, 1998). However, those researchers arrived at completely different conclusions. They concluded that Ste6 is degraded, while situated in the vacuolar membrane, by a concerted action of vacuolar hydrolases and the proteasome. Our combined immunofluorescence and GFP experiments and the behavior of the Ste6-His3 fusion, however, clearly show that Ste6 is localized to the vacuolar lumen in pep4 mutants, whereas it is located in or at the vacuolar membrane in doa4 mutants. Our findings are in line with results obtained for the α-factor receptor Ste2, which has been shown to be sorted to the lumen of the vacuole for degradation via the MVB pathway (Odorizzi et al., 1998). Loayza and Michaelis (1998) themselves note that their suggested degradation mechanism would be quite unique. We think that it is more likely that all cell surface proteins in yeast follow the same degradation pathway.
Ubiquitination and Internalization from the Cell Surface
If ubiquitination were required for Ste6 internalization from the cell surface, as suggested previously (Kölling and Hollenberg, 1994; Kölling and Losko, 1997), surface accumulation should be observed in the doa4 mutant. There are several possible explanations for the lack of cell surface accumulation in the doa4 strain. For instance, ubiquitination could be required for Ste6 sorting to the plasma membrane. However, this does not appear to be the case, because an endocytosis-defective Ste6 mutant can still be detected at the cell surface in the doa4 mutant. Alternatively, ubiquitination could have a dual function in trafficking of Ste6 to the vacuole. Although the free ubiquitin level is reduced at least threefold under exponential growth conditions in the doa4 mutant (Swaminathan et al., 1999), the residual ubiquitin concentration may still be sufficient to allow for some Ste6 ubiquitination. Internalization of underubiquitinated Ste6 may still proceed normally, whereas other steps in the trafficking pathway, such as uptake into the vacuole, may require the full extent of ubiquitination. The finding that monoubiquitination may be sufficient for endocytosis of the α-factor receptor Ste2 (Terrell et al., 1998) is in line with this interpretation. But it is also possible that Ste6, in contrast to other yeast cell surface proteins, does not at all require ubiquitination for internalization.
What is the site of Ste6 ubiquitination? The massive accumulation of ubiquitinated Ste6 at the cell surface in the end4 mutant suggested that Ste6 ubiquitination occurs at the plasma membrane. In this report, however, we have presented evidence that Ste6 cycles between endosomes and the plasma membrane. It is therefore well possible that Ste6 ubiquitination occurs at an intracellular compartment (e.g., the endosome) and that ubiquitinated Ste6 is transported from there to the cell surface via the recycling pathway and becomes trapped at the plasma membrane by the end4 block.
Possible Role of Ubiquitination in Ste6 Trafficking
Membrane proteins destined for the vacuole can be either sorted to the delimiting vacuolar membrane or to the lumen of the vacuole. Sorting to the vacuolar lumen occurs by the recently described MVB pathway (Odorizzi et al., 1998). Because we have shown that Ste6 ends up in the lumen of the vacuole, it is reasonable to assume that it also enters the MVB pathway. The same appears to be the case for the Ste2 protein (Odorizzi et al., 1998). When ubiquitination is compromised in a doa4 mutant, Ste6 accumulates in the delimiting membrane of the vacuole. Based on these findings, we propose that ubiquitination is required for Ste6 sorting into the MVB pathway. In the doa4 mutant transfer to the MVB pathway is prevented, and as a consequence, Ste6 gets stuck in the vacuolar membrane.
While this manuscript was under revision, a report was published that also suggests a role for Doa4 at the late endosomal/vacuolar compartment (Amerik et al., 2000). Hochstrasser and coworkers isolated extragenic suppressors of doa4. All of the cloned suppressor functions code for class E vacuolar protein sorting proteins thought to be required for MVB formation at the late endosome. In addition, they presented evidence that a large fraction of Doa4 is associated with the late endosome in cells lacking Vps4 function. Further evidence from yeast and animal cells that ubiquitination may regulate sorting at the endosome and transGolgi network is summarized in a recent review (Lemmon and Traub, 2000). Our findings present another strong argument that ubiquitination/deubiquitination plays an important role at the endosomal/vacuolar compartment.
ACKNOWLEDGMENTS
Plasmids and strains were generously provided by Jürgen Dohmen, Mark Hochstrasser, and Howard Riezman. Furthermore, we thank Hugh Pelham for the gift of Pep12 antibodies. We are also grateful to Daniel Fein for his assistance with some of the experiments, to Jürgen Dohmen for stimulating discussions, and to Cor Hollenberg for his support. This work was supported by Deutsche Forschungsgemeinschaft grant Ko 963/2-3 to R.K.
REFERENCES
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Rothblatt J, Schekman R. Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 1991;194:662–674. doi: 10.1016/0076-6879(91)94048-h. [DOI] [PubMed] [Google Scholar]
- Galan J, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Moreau V, André B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Govers R, ten Broeke T, van Kerkhof P, Schwartz AL, Strous GJ. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 1999;18:28–36. doi: 10.1093/emboj/18.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein C, Springael JY, Volland C, Haguenauer TR, André B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L. Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Ellison MJ, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R, Hollenberg CP. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R, Losko S. The linker region of the ABC-transporter Ste6 mediates ubiquitination and fast turnover of the protein. EMBO J. 1997;16:2251–2261. doi: 10.1093/emboj/16.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Dohlman HG, Thorner J. The a-factor transporter (STE6 gene product) and cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1993;120:1203–1215. doi: 10.1083/jcb.120.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Sterne RE, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol, 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Papa FR, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–434. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol, 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Roth AF, Davis NG. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous GJ, van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]