Abstract
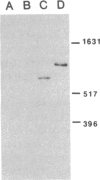
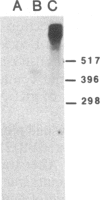
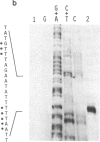
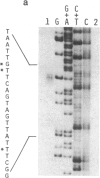
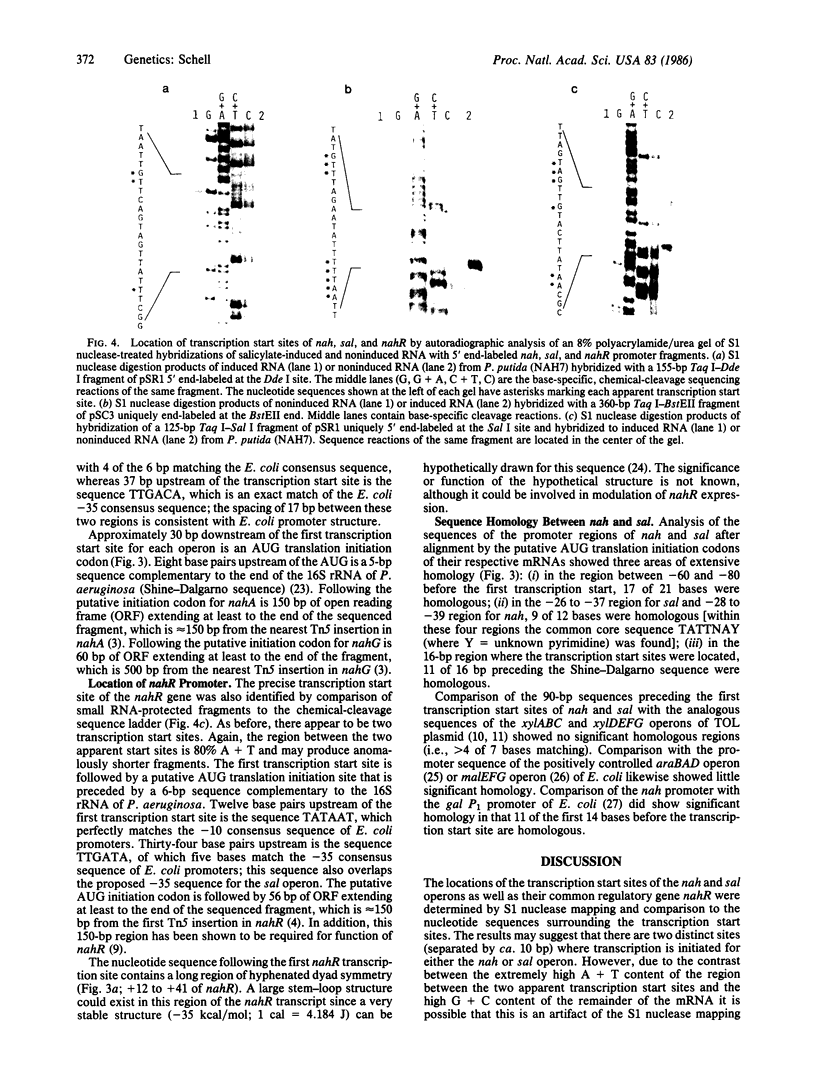
The in vivo transcription start sites of the nah and sal operons of the NAH7 plasmid were determined by S1 nuclease mapping and the nucleotide sequence surrounding these transcription start sites was determined. Since expression of both of these operons is coordinately controlled by the product of the transcriptional activator gene nahR, the sequences were compared to locate potential sites involved in common regulation. In the 100-base-pair region preceding transcription start sites of both operons, three regions of extensive homology were found and may be involved in nahR-mediated transcriptional control: between -80 and -60 with 81% homology; between -40 and -28 with 75% homology; between -1 and +15 with 70% homology. Comparison of the promoter sequences of nah and sal with the analogous sequences of the xylABC and xylDEFG operons of the TOL plasmid showed little homology between the 5' regions of these two sets of positively regulated hydrocarbon degradation operons. In addition, the transcription start site of the nahR regulatory gene was located and its promoter sequence was determined. The nahR promoter overlapped at the -35 position of the sal promoter; however, the nahR gene is transcribed in the opposite direction. Sequences similar to the consensus sequences of Escherichia coli promoters (at -35 and -10) were found in nah, sal, and nahR at the appropriate positions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M. Plasmids in Pseudomonas. Annu Rev Genet. 1976;10:7–30. doi: 10.1146/annurev.ge.10.120176.000255. [DOI] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973 Jun;114(3):974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A. I., Chenchik A. A., Aivasashvilli V. A., Beabealashvilli R. Sh, Nikiforov V. G. Escherichia coli and Pseudomonas putida RNA polymerases display identical contacts with promoters. Mol Gen Genet. 1984;195(3):511–515. doi: 10.1007/BF00341455. [DOI] [PubMed] [Google Scholar]
- Grund A. D., Gunsalus I. C. Cloning of genes for naphthalene metabolism in Pseudomonas putida. J Bacteriol. 1983 Oct;156(1):89–94. doi: 10.1128/jb.156.1.89-94.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz A. H., Miyada C. G., Wilcox G. Functional limits of the araIc promoter suggest an additional regulatory site for araBAD expression. J Bacteriol. 1984 Apr;158(1):141–147. doi: 10.1128/jb.158.1.141-147.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Ebina Y., Nakazawa A., Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981 Nov;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 1984 Nov;3(11):2461–2466. doi: 10.1002/j.1460-2075.1984.tb02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso R., Di Lauro R., Rosenberg M., de Crombrugghe B. Nucleotide sequence of the operator-promoter region of the galactose operon of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Jan;74(1):106–110. doi: 10.1073/pnas.74.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Inouye S., Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombinants: analysis of insertion and deletion mutants. J Bacteriol. 1980 Oct;144(1):222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Schell M. A. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 1983 Feb;153(2):822–829. doi: 10.1128/jb.153.2.822-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene. 1985;36(3):301–309. doi: 10.1016/0378-1119(85)90185-4. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Regulation of naphthalene catabolic genes of plasmid NAH7. J Bacteriol. 1985 Jun;162(3):1008–1013. doi: 10.1128/jb.162.3.1008-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]