Abstract
Protein-protein interactions are key to function and regulation of many biological pathways. To facilitate characterization of protein-protein interactions using mass spectrometry, a new data acquisition/analysis pipeline was designed. The goal for this pipeline was to provide a generic strategy for identifying crosslinked peptides from single LC/MS/MS datasets, without using specialized crosslinkers or custom-written software. To achieve this, each peptide in the pair of crosslinked peptides was considered to be “post-translationally” modified with an unknown mass at an unknown amino acid. This allowed use of an open-modification search engine, Popitam, to interpret the tandem mass spectra of crosslinked peptides. False positives were reduced and database selectivity increased by acquiring precursors and fragments at high mass accuracy. Additionally, a high-charge-state-driven data acquisition scheme was utilized to enrich datasets for crosslinked peptides. This open-modification search based pipeline was shown to be useful for characterizing both chemical as well as native crosslinks in proteins. The pipeline was validated by characterizing the known interactions in chemically crosslinked CYP2E1-b5 complex. Utility of this method in identifying native crosslinks was demonstrated by mapping disulfide bridges in RcsF, an outer membrane lipoprotein involved in Rcs phosphorelay.
INTRODUCTION
Protein-protein interactions play a major role in many cellular processes such as signal transduction, metabolic pathways, regulation of gene expression, and transcriptional regulation. Alterations in these interactions have been shown to lead to diseases such as Huntington's, Creutzfeld-Jacob and Alzheimer's disease 1–3. Given the importance of protein-protein interactions to human health, a multitude of physical, biochemical and genetic approaches have been employed for their detection and characterization4, 5. Conventional techniques such as nuclear magnetic resonance (NMR) and X-ray crystallography, however, are limited by their stringent sample requirements such as protein amount, purity, concentration, size, and homogeneity6, 7. Biochemical methods such as affinity chromatography and immunoprecipitation are only suitable for studies in vitro8, 9. They can only be used to study one protein complex per experiment and require some prior knowledge of the interaction in order to target it. In addition, for most of these methods to be effective, the interactions under investigation have to be stable. Studying transient complexes using these methods can therefore be challenging. Chemical crosslinkers can be used to stabilize weak interactions and facilitate their detection by creating non-native covalent bonds between interacting proteins that may then be characterized10–12.
Chemical crosslinking in combination with mass spectrometry (CXMS) is a useful technique for investigating these transient interactions at molecular interfaces in protein complexes. Studying protein interactions using CXMS methods has many advantages over X-ray crystallography or NMR methods, such as 1) Protein complexes may be studied in vitro and in vivo13, 14; 2) Analytical throughput is high, making experimental turn-around times short; 3) Relative to X-ray or NMR analysis, sample quantities required for CXMS are an order of magnitude smaller; 4) There is no restriction on protein size that can be studied because analysis is based on peptides; 5) Components of the crosslinked complex, their stoichiometry and relative juxtapositions may be determined; and 6) Availability of crosslinking reagents with different lengths and amino acid specificities allows for a greater flexibility in experimental design15, 16.
Despite the excellence of mass spectrometry as an analytical tool, unambiguous identification of crosslinked peptides resulting from proteolytic digestion of crosslinked proteins has proven to be technically challenging. The problem is similar to detecting phosphopeptides in a protein digest, because crosslinked peptides are often present at sub-stoichiometric levels, which leads to failure in detecting them during data-dependent LC/MS/MS analysis. Once the tandem mass spectra of cross-linked peptides have been identified, it remains challenging to assign sequences and map sites of crosslinking. This is because tandem mass spectra of crosslinked peptides are complicated by the presence of two unique sets of fragment ions. In this sense their interpretation is similar to interpretation of tandem mass spectra containing two peptides fragmented in parallel17. Furthermore, while the masses of the two individual peptides (that constitute the crosslinked peptide) are independently present in a sequence database, the mass of the crosslinked peptide is not. Additionally, the tandem mass spectra of the crosslinked peptides contain two sets of fragment ions, which precludes direct spectral interpretation of the tandem mass spectrum using database search algorithms such as SEQUEST, Phenyx or Mascot.
In order to improve identification of protein crosslinks using mass spectrometry, several advances in the crosslinking strategy and crosslinker design have been implemented over the last few years. Affinity purification strategies have been employed to aid the purification of crosslinked protein complexes13, 18, 19. Crosslinkers with defined isotope tags have been used to distinguish crosslinker-modified peptides from unmodified peptides as well as for quantitative analysis of crosslinked peptides15, 16, 20, 21. Alternatively, chemically22 or MS2 cleavable crosslinkers (containing bonds that fragment during low-energy activation) have been utilized to facilitate the identification of crosslinked products by detection of marker ions23–25. Another technique for selectively identifying crosslinked peptides by mass spectrometry is the incorporation 18O-label into peptides during proteolysis16, 26, 27. However, most of these CXMS methods involve high overhead in cost and effort, and require custom software to interpret data28–33.
While some protein associations are transient and weak, others are stabilized via covalent bonds such as disulfide bridges. Other examples of covalent interactions that occur biologically include crosslinks formed by transglutaminase or lysyl oxidase. These covalent interactions are important for structural integrity and function of proteins and protein complexes. Characterization of these native crosslinks is complicated by the same problems as chemical crosslinks. We sought to design a method that could be used to map both chemically-introduced as well as naturally-occurring crosslinks in proteins. Towards this goal we developed a crosslink identification pipeline based on mass spectrometric analysis of crosslinked peptides followed by open-modification searches. In this pipeline, each peptide in the pair of crosslinked peptides is treated as if it were post-translationally modified with an unknown mass at an unknown amino acid, which allows use of any available open-modification search engine for data analysis. The resulting open-modification CXMS pipeline also uses accurate mass of parent and fragment ions to increase database selectivity and decrease false positives. This new, direct method is general-purpose enough to be used for characterization of chemical and native crosslinks without the necessity for specialized reagents or software. Here, we describe the implementation of this simple method and its effectiveness in characterizing protein complexes by identifying crosslinked peptides and determining putative protein-protein interaction sites. We also demonstrate the utility of this method in identifying native disulfide bridges in proteins, without reduction or derivatization of disulfide bonds.
EXPERIMENTAL SECTION
Materials
Crosslinking reagent 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) was purchased from Pierce Biotechnology, Inc. (Rockford, IL). Sequencing grade modified trypsin was from Roche Applied Science (Indianapolis, IN). Escherichia coli expressed recombinant human cytochrome b5 (b5) and cytochrome P450 2E1 (CYP 2E1) were purchased from Invitrogen (Carlsbad, CA). All other chemicals were from Sigma-Aldrich unless otherwise stated.
Crosslinking reaction and proteolytic digestion of crosslinked complex
CYP2E1 and b5 were mixed in 1:3 molar ratio (10 μM CYP2E1 and 30 μM b5) in 50mM potassium phosphate buffer (pH 7.5) containing 20% glycerol. The mixture was by gently stirred for 10 min at room temperature and then held at room temperature for 2 h. EDC was added to the solution to 8 mM final concentration from a 100 mM stock solution. Crosslinking reaction was allowed to proceed at room temperature for 2 h. Reaction was quenched by removal of EDC through dialysis against dialysis buffer A (50mM potassium phosphate buffer containing 20% glycerol). Glycerol was removed by a second dialysis against dialysis buffer B (50mM potassium phosphate buffer, pH 7.5) and the sample was dried completely in a Speed Vac. Crosslinked sample was denatured using 6M urea, reduced with DTT and alkylated with iodoacetamide. Trypsin was added to the final mixture in 30:1 ratio and the digestion was allowed to proceed at 37 C overnight.
Expression, purification and digestion of RcsF
Plasmid expressed pPagC-RcsF-6His was transformed into an ΔrcsC histidine kinase strain. An rcsC null background was necessary to prevent growth inhibition caused by strong RcsF mediated activation of the Rcs Regulon. An overnight culture was back-diluted 1:100 and expression was induced by 0.2% arabinose. At an OD600 of 0.8, cells were harvested and periplasmic fractions were prepared by osmotic shock. Periplasmic fractions containing unanchored RcsF were dialyzed in PBS overnight. 6-His tagged RcsF was purified through a Nickel column and further purified through a Superdex 200 size exclusion column. 200ug of purified RcsF was digested with trypsin without prior reduction/alkylation (to keep the disulfide bonds intact).
Mass spectrometry and HPLC
Peptide digests were analyzed by electrospray ionization in the positive ion mode on a hybrid linear ion trap-Oribtrap instrument, known as the LTQ-Orbitrap (Thermo Fisher, San Jose, CA). The LTQ-Orbitrap was equipped with a nanoflow HPLC system (NanoAcquity; Waters Corporation, Milford, MA) fitted with a home-built helium-degasser. Peptides were trapped on a home-made 100 μm i.d. x 18 mm long pre-column packed with 200Ǻ C18 stationary phase (5 μm, C18AQ; Michrom) and subsequently separated on a home-made gravity-pulled 75 μm i.d. × 150 mm long analytical column packed with 100Ǻ C18 stationary phase (5 μm, C18AQ; Michrom), coupled to the mass spectrometer.
For each injection, an estimated amount of 0.5 μg of peptide mixture was loaded onto the pre-column at 4 μl/min in water/acetonitrile (95/5) with 0.1% (v/v) formic acid. Peptides were eluted using an acetonitrile gradient flowing at 250 nl/min using mobile phase consisting of: A, water, 0.1% formic acid; B, acetonitrile, 0.1% formic acid. A linear gradient program was used as follows: 0 min: A (95%), B (5%), 55 min: A (65%), B (35%), 60 min: A (15%), B (85%), 65 min: A (5%), B (95%), 75–90 min: A (95%), B (5 %); (stop). The electrospray voltage was applied via a liquid junction using a gold wire inserted into a micro-tee union (Upchurch Scientific, Oak Harbor, WA) located in between the pre-column and analytical column. Ion source conditions were optimized using the tuning and calibration solution recommended by the instrument provider. All MS survey scans were performed in the Orbitrap from m/z 400–2000, at resolution of 60,000 (m/z 400) and ion populations of 5*105. For tandem mass spectrometry with ion detection in the Orbitrap, the ion population was set to 2*105, resolution to 7,500 and the precursor isolation width to 4 m/z units. Collision energy was set to 40% for CID in the LTQ. All data-dependent analyses were performed using MS survey scans followed by data-dependent selection of the 5 most abundant precursors for tandem mass spectrometry. Singly, doubly and triply charged precursors were rejected from data-dependent selection in all the runs except for the targeted analysis of intra-chain disulfide bridge in RcsF. Data redundancy was minimized via the Dynamic Exclusion feature, by excluding the previously selected precursor ions (−0.1 / +1.1 Da) for 45 seconds before being selected again for fragmentation.
Data processing and database searching
After data-dependent acquisition, tandem mass spectral data were converted into peaklists (.dta files) using the instrument vendor's software (extract_msn.exe; Thermo Fisher, San Jose, CA). Tandem mass spectra of higher charge state precursors were deconvoluted to 2+ charge state precursors and 1+ charge state fragments, with an in-house written Perl script (http://goodlett.proteomics.washington.edu). Deconvoluted spectra were searched by Phenyx (GeneBio SA, Geneva, Switzerland), to identify and filter-out the linear peptides. Trypsin specificity and three missed cleavage sites were specified for these searches. Methionine residues were considered as being present in reduced and oxidized form. Cysteine residues were considered alkylated with iodoacetamide. Spectra not matched by the first-pass Phenyx searches were searched using Popitam in an open-search mode. Briefly, any mass modification between −50 and +3000 Da was searched using trypsin specificity and 1 missed cleavage, against a database containing protein sequences of interest. The smallest possible tolerance (0.01 Da) was used for fragment ion matching.
Popitam
Popitam is an algorithm designed to associate amino acid sequence with tandem mass spectra of chemically modified peptides without a priori knowledge of the modification34. A “spectrum graph”, commonly used by de novo sequencing algorithms, is used to transform tandem mass spectral information into a graph with nodes and edges. Nodes in the graph represent m/z values from the tandem spectrum. Between nodes Popitam creates edges each time a mass difference between m/z values corresponds to one amino acid (single edge, represented by upper case letters) or two amino acids (double edges, represented by lower case letters). Next, for each candidate peptide in a protein database, Popitam extracts all sequence tags (according to a user-specified minimum length) that are consistent with the candidate sequence and the m/z pattern of the spectrum. Sequence tags are then combined according to logical rules to provide best-fit tandem mass spectral interpretation scenarios34. Typically, a scenario is composed of one or several sequence tags and gaps (information missing between sequence tags). By comparing each gap in the scenario and its expected value deduced from the peptide sequence under consideration, Popitam evaluates if the gap arises from a lack of information in the tandem mass spectrum due to too missing peaks (in which case, the gap is represented by dashes or -) or if it contains a modification (in which case the gap is indicated by stars or *) (http://www.expasy.ch/tools/popitam/). Finally, each scenario is scored using a function that is generated by Genetic Programming. The Popitam output displays the highest scoring candidate peptides together with the proposed scenarios.
THEORETICAL BASIS
Rationale for using an “open-modification” search tool based on sequence tag extraction
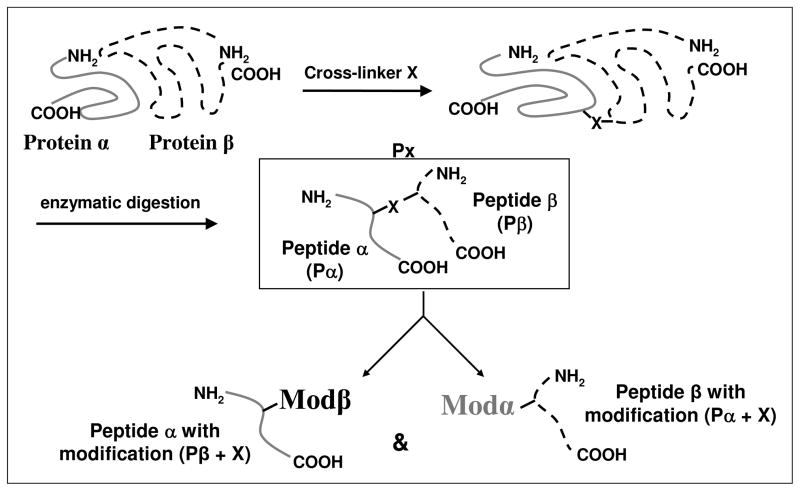
Standard MS database search engines cannot match the observed masses of crosslinked peptides to theoretical masses of peptides in a database. However, if the crosslinked peptides are considered as peptides with unknown modifications at unknown amino acid residues, the problem can be solved by using an open-modification search engine. For example, consider a crosslinked peptide Px that is composed of peptides Pα and Pβ that are covalently linked to each other by a crosslinker X (Figure 1). In this case, identification of Px can be simplified to identifying "peptide Pα with an unknown modification (Pβ +X)” and/or "peptide Pβ with an unknown modification (Pα +X)”. Since the tandem mass spectrum of Px is likely to contain fragment ions from both Pα and Pβ, a tag-extraction based method can be used to match sequence tags from Pα and Pβ one at a time. In the open-modification CXMS pipeline, an existing open-access software Popitam34 (http://www.expasy.ch/tools/popitam/) was used to perform open-modification searches. Designed to search for peptides with unknown modification masses, Popitam can detect any biological post-translational modifications (PTM), chemical artifact, or, as in our case, unknown crosslinked peptides. Because of the branched structure of crosslinked peptides, fragmentation of intact crosslinked peptides is often not very extensive, yielding less informative and hard-to-interpret tandem mass spectra than the corresponding linear peptides. The tag-extraction algorithm used by Popitam is useful in such scenarios because it can match sequences in a database by identifying short sequence-specific tags for individual peptides. In addition, Popitam allows users to select the fragment ion tolerance as a variable parameter, allowing one to take advantage of the tandem mass spectra acquired with high mass accuracy. While the open-modification CXMS pipeline utilizes Popitam, other sequence tag-extraction based algorithms, for example InsPecT35 or GutenTag36, designed to identify spectra from modified and/or mutated peptides should also be effective in the pipeline.
Figure 1.
The rationale for using an open-modification search strategy for identifying crosslinked peptides is illustrated. A crosslinked precursor Px can be treated as “peptide Pα with a modification (Pβ +X)” and/or "peptide Pβ with a modification (Pα +X)”, where X is the mass added by the crosslinker and depends on the type of crosslinker used. An open-modification search engine can thus be used to identify the peptides Pα and Pβ with unknown modifications.
Description of the open-modification CXMS pipeline
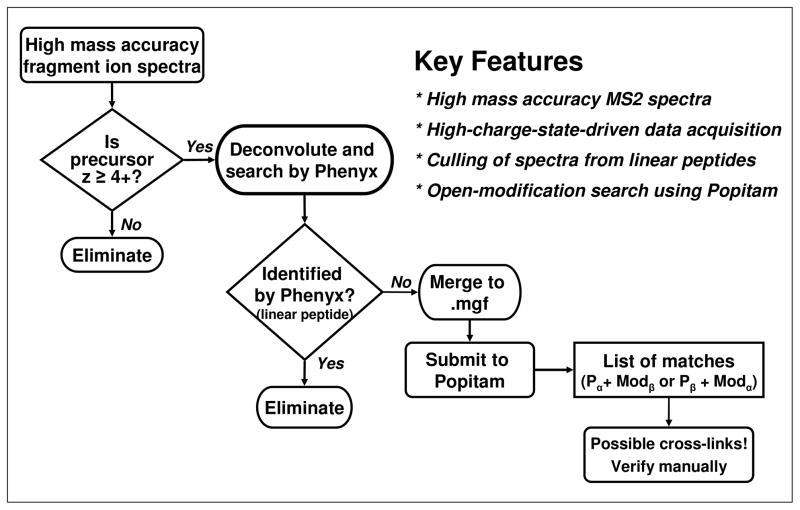
For facile identification of MS2 spectra of crosslinked peptides in the pool of tandem mass spectra consisting of mostly linear peptides, we designed a short and relatively simple data acquisition/processing work-flow named the open-modification CXMS pipeline (Figure 2). Both parent and fragment ions were acquired in the Orbitrap mass analyzer of an LTQ-Orbitrap for high mass accuracy37. Use of high measured mass accuracy on fragment ions is very important when analyzing crosslinked peptides, because of the combinatorial explosion of possible candidate sequences. High mass accuracy provides greater specificity by limiting false identifications37, 38. The combinatorial complexity inherent in analyzing large sets of MS2 spectra from crosslinked samples also requires optimization of computing time and resources. This issue was addressed in the open-modification CXMS pipeline by incorporating a series of steps described below. Orbitrap resolution was reduced from 60,000 for precursor-ion to 7,500 for fragment-ion acquisition in order to accomplish a high instrument duty cycle, comparable to linear ion trap acquisitions. Data-dependent enrichment of MS2 spectra from crosslinked peptides was achieved by targeting only high-charge-state precursors (≥ [M + 4H]4+) for collision-induced-dissociation (CID) during HPLC electrospray ionization tandem mass spectrometry (LC/ESI-MS/MS) acquisition. The high-charge-state driven acquisition was based on the prevalence of more highly charged precursors among the crosslinked peptides, because they have two tryptic termini and often carry twice the number of protons as unmodified peptides when ionized by ESI32, 39. While such a targeted data-acquisition scheme helps to enrich the acquired MS2 spectra for crosslinked peptides, it does not exclude tandem mass spectra from high-charge-state linear peptides. To eliminate spectra from linear peptides and to further enrich the dataset for crosslinked peptide tandem mass spectra, a standard database search was performed. Typically, the information contained in MS2 spectra from peptides of charge states higher than 3+ is not well utilized, because most search engines fail to either adequately identify or statistically validate these high-charge-state peptides. To solve this problem all acquired MS2 spectra were deisotoped and deconvoluted prior to the standard database search analysis. Spectra thus deisotoped and deconvoluted were searched against a database consisting of the proteins of interest, using Phenyx, a database search engine which allows use of high mass accuracy encoded in parent and fragment ions40. Charge state reduction of the MS2 spectra by deisotoping and deconvolution allowed matches to tandem mass spectra with precursor charge states ≥4+. All spectra matching linear peptides were then removed from further consideration, leaving a set of spectra that did not match sequences in the database. These remaining spectra, which presumably represented crosslinked peptides, some high charge state linear peptides (with and without modifications) and non-peptide analytes, were searched using Popitam to generate a list of peptide identifications with the corresponding modification masses. Finally, spectra from crosslinked peptide candidates were identified by querying the modification masses generated from Popitam against masses of tryptic peptides (generated in silico) from proteins of interest. Identified candidates were validated manually by examining the measured mass error of the precursor, presence of residues required for the crosslinking reaction, and quality of the spectral annotation. As shown next, this approach allows direct and unambiguous characterization of crosslinked peptides. While most CXMS methods require acquisition and analysis of multiple tandem mass spectrometry datasets, the open-modification CXMS pipeline characterizes crosslinked peptides from a single LC/MS/MS data set without the need for specialized crosslinkers or software.
Figure 2.
Flow chart depicting the key steps of the open-modification CXMS pipeline.
RESULTS AND DISCUSSION
Pipeline validation by identifying crosslinked peptides in CYP2E1-B5 complex
To assess the accuracy of the open-modification CXMS pipeline, the CYP2E1-b5 protein complex was chosen as a model system. The molecular interactions in this protein-protein complex were recently characterized by an 18O-labeling based CXMS method27. Test samples were prepared as per authors’ protocol with omission of the 18O-labeling step. Data was acquired and analyzed using the open-modification CXMS pipeline described above. The list of crosslinked candidates identified with this approach is shown in Table 1. As judged by measured mass accuracies of the identified precursor ions (within 5 ppm of predicted values) the two inter-protein crosslinked peptides previously identified by the 18O-labeling based strategy were confidently identified. Assignment of fragment ions, as discussed in the next section, further confirmed the existence of a crosslink between the peptide candidates. In addition to the two known interactions, four new interactions, which were not detected by the 18O- labeling strategy, were also identified from this analysis (Table 1). Presence of these new crosslinks in CYP2E1-b5 complex was supported by the accurate measured masses of the precursors and spectral assignments of the corresponding tandem mass spectra.
Table 1.
List of crosslinked peptides and the corresponding sequence tags (where applicable) identified from CYP2E1-b5 complex using the open-modification CXMS pipeline†.
| # | Measured precursor mass | Charge state (z) | Peptide / Sequence tag (Cytochrome b5) | Amino acid Coordinate (b5) | Peptide / Sequence tag (CYP2E1) | Amino acid Coordinate (CYP2E1) | MMA (ppm) | Method of Identification |
|---|---|---|---|---|---|---|---|---|
| 1 | 3781.70 | 4, 5 | EQAGGDATENFEDVGHSTDAR *********NF ---------- |
48–68 | YSDYFKPFSTGKR YSdy**************** |
423–435 | 1.0 | O18-labeling & CXMS pipeline |
| 2 | 3625.60 | 4, 5 | EQAGGDATENFEDVGHSTDAR ******ATENF---------- |
48–68 | YSDYFKPFSTGK ******pfSTgk |
423–434 | 2.5 | O18-labeling & CXMS pipeline |
| 3 | 2540.36 | 4, 5 | FLEEHPGGEEVLR *********EVlr |
35–47 | YKLCVIPR ****VIpr |
485–492 | 2.1 | CXMS pipeline only |
| 4 | 2491.37 | 4, 5 | FLEEHPGGEEVLR ****HPG---------- |
35–47 | VIKNVAEVK ***NVaevk |
235–243 | 5.3 | CXMS pipeline only |
| 5 | 1849.03 | 4 | LYMAED lymaE* |
124–129 |
KVIKNVAEVK - |
234–243 | 4.6 | CXMS pipeline only |
| 6 | 3781.70 | 4, 5 |
EQAGGDATENFEDVGHSTDAR **aggdatENFEDVgh---------- |
48–68 | YSDYFKPFSTGKR - |
423–435 | 2.7 | CXMS pipeline only |
According to the convention followed for Popitam’s sequence tag representation, upper case letters correspond to one amino acid edges, lower case letters correspond to two amino acid edges, ‘*’ indicates presence of modification and ‘-‘ indicates lack of information. Please see the experimental section for more details on Popitam.
Like other isotope tagging methods, the 18O-labeling strategy uses the mass difference between an un-labeled peptide and the corresponding heavy-labeled peptide as a “filter” to select crosslinked peptide candidates. However, analysis of mass spectrometric data generated from 18O-labeling is complicated due to 1) incomplete labeling or back-exchange with concomitant loss of the isotope label, 2) the presence of peptides with multiple charge states resulting from the electrospray ionization process, which can lead to loss of the mass tag, 3) shifts in retention time between the 16O-labeled and 18O-labeled LC/MS data sets and 4) the need to acquire multiple LC/MS/MS data sets that must be analyzed and cross-correlated to detect the crosslinked peptides. In addition, like most CXMS methods, this 18O-labeling strategy involves expensive and labor-intensive steps, and requires custom-written software for data analysis. In comparison, our new approach did not require any isotope labeling of peptides or any additional MS1 scans to select the crosslinked candidates. More importantly, no custom-written software was required to analyze the data.
Spectral assignment and determination of site of crosslinking
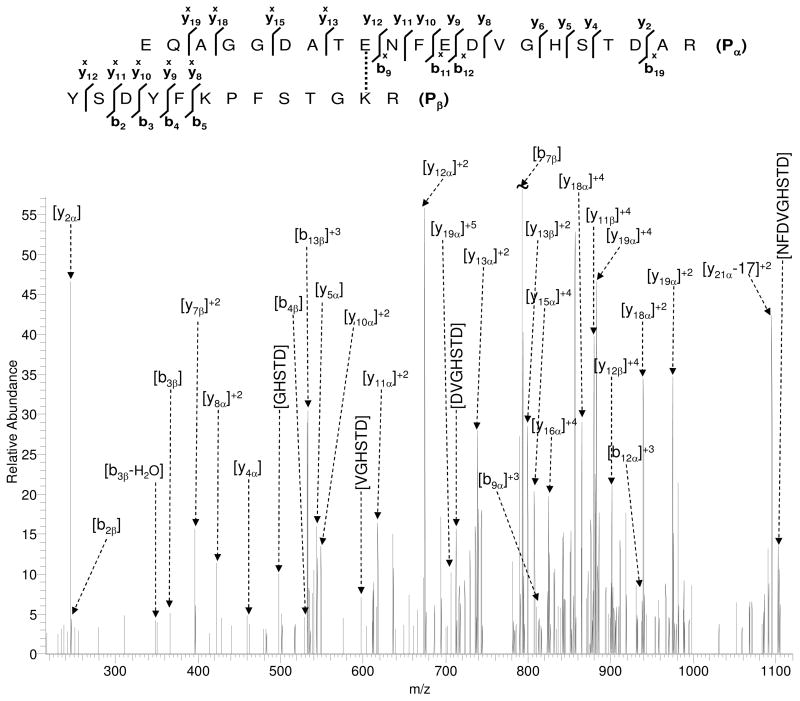
Tandem mass spectra from all six of the putative crosslinked peptides were annotated using MS2Assign41. MS2Assign is a web based application designed to assign peaks in a tandem mass spectrum of modified or crosslinked peptides. To limit the number of possible assignments, only b, y and internal cleavage ions were considered for assignment. Interpretable tandem mass spectral data were obtained for all six candidates identified in CYP2E1-b5 complex. Acquisition of fragment ions at high mass accuracy provided increased confidence in the spectral assignment. Tandem mass spectrum for crosslinked precursor with putative sequences EQAGGDATENFEDVGHSTDAR-YSDYFKPFSTGKR is shown in Figure 3. More than 40% of the fragment ions in the spectrum above the specified threshold (5% of the base peak) were identified by the program. Sequence specific b and y ions were observed for both peptides within 10 ppm measured mass accuracy. In addition, fragment ions containing crosslinked sequences from both peptide chains, e.g. [y18α-YSDYFKPFSTGKR]4+ or [y18x]α and [y11β-EQAGGDATENFEDVGHSTDAR]4+ or [y11x]β, were also observed. Presence of these fragment ion masses in the spectrum (indicated by superscript ‘x’ in Figure 3) could not be explained by fragment ions from either of the linear peptides alone, and thus further supported the presence of crosslinks. The interpretation of these high-charge-state fragment ions also underscores the importance of charge state reduction of tandem mass spectra in general and particularly for analysis of crosslinked peptide tandem mass spectra.
Figure 3.
Tandem mass spectrum of a crosslinked peptide (m/z = 757.35, candidate #1 in Table 1) identified from CYP2E1-b5 complex. All labeled fragment ions were identified with more than 10 ppm mass accuracy. Ions marked with subscript α are from the larger peptide (EQAGGDATENFEDVGHSTDAR) and ions with subscript β are from the smaller peptide (YSDYFKPFSTGKR). This nomenclature is consistent with that proposed in the recent literature (ref. 41 in the manuscript). Fragment ions with superscript ‘x’ represent the crosslinked fragment ions.
Accurate assignment of fragment ions in the tandem mass spectrum of a crosslinked peptide is important not only for identification of correct peptide sequence, but also for identifying the residues that are modified or crosslinked. Exact locations of the crosslinks in the six candidates identified in the CYP2E1-b5 complex are depicted in bold letters in Table 1. Most MS based methods rely on spectral assignment from fragmentation patterns to identify the location of the crosslink. This assignment is possible because fragment-ion intensities around the site of crosslink are generally diminished relative to other amino acid stretches of the two crosslinked peptides. In the open-modification CXMS pipeline proposed here, the sequence tag approach used by Popitam is very useful in validating, and in some cases identifying, the exact location of the crosslinks. For example, for crosslinked candidate #2 shown in Table 1, peptide YSDYFKPFSTGKR from CYP2E1 has two potential crosslinking sites--Lys428 and Lys434. However, the sequence tag “pfSTgk” obtained from Popitam search results indicated a modification site to the left of the Proline residue. This allowed confident assignment of Lys428 as the site of the crosslink, which was consistent with the previously published results20. In fact, out of the twelve crosslink-sites in the six candidates, nine could be predicted simply by examining the corresponding sequence tags (Table 1). Thus, using this new direct approach, the sites of modification may often be directly located from the sequence tag output of Popitam. Subsequent manual spectral assignment of the high mass accuracy fragment ion spectra allows the putative crosslinks proposed by Popitam to be confirmed with added confidence. Of the three peptides for which crosslinked sites could not predicted directly by Popitam, peptides KVIKNVAEVK and YSDYPFSTGKR in candidates #5 and #6 respectively did not yield any sequence-tag information, because of low fragmentation of the peptides. Sequence tag for peptide YSDYFKPFSTGKR in candidate #2 was obtained; however, it was not sufficient for unambiguous assignment of the site of crosslinking. MS2 spectra from these candidates were annotated using MS2Assign in order to identify the sites of crosslinking indicated in the table.
Use of the open-modification CXMS pipeline to map disulfide bonds
To demonstrate the utility of the open-modification CXMS pipeline in identifying native crosslinks, the disulfide bonds in a standard peptide, BNP-32 (Brain Natriuretic Peptide-32) were analyzed. In order to mimic a complex sample, a known amount of BNP-32 peptide was spiked in a bacterial whole cell lysate that was subsequently proteolyzed with trypsin. Because of the intact disulfide bond which holds BNP-32 peptide in a loop conformation (Supporting Figure S1), tryptic digestion of un-reduced peptide results in many missed cleavages. Tryptic fragments thus generated contain both inter- and intra-chain disulfide bonds depending on whether the peptide is cleaved in the loop region or not. In spite of this added complexity, peptides with inter-chain as well as intra-chain (without an actual cleavage site between the two linked cysteine residues) disulfide bonds were successfully identified from this complex mixture (Supporting Information Table S1), indicating that the open-modification CXMS pipeline can be used for detecting different disulfide geometries in proteins.
Next, the disulfide bonds in a bacterial protein, RcsF, were characterized. RcsF is an outer membrane lipoprotein involved in regulation of Rcs phosphorelay42, a pathway used to sense and respond to membrane stress in Salmonella typhimurium43. In E. coli, RcsF has been shown to be the upstream sensor for the Rcs regulon and to be required for activation in rfa envelope mutant backgrounds42, 44. However, the mechanism by which RcsF transmits the activation signal to the inner membrane constituents is unknown. RcsF is a substrate of DsbA, a periplasmic dithiol oxidase, in the absence of which RcsF is fully reduced45. It has been reported that mutations in dsbA lead to activation of the Rcs system44. Mutation experiments have also shown that point mutants of RcsF with cysteine-to-alanine (Cys-->Ala) substitution fail to activate the Rcs phosphorelay46. However, a direct correlation between redox state of RcsF and regulation of the Rcs system has not been established. Also, despite the plausible role of thiol groups in signaling state on RcsF, the arrangement of disulfide bonds in RcsF has not been characterized yet.
The final processed version of RcsF contains five cysteines. The first cysteine (Cys16) is lipidated to form an N-acyl-diacylglycerylcysteine, making it unavailable for disulfide bond formation42. This leaves four cysteines in the protein that may be combined to form two disulfide bridges—Cys74, Cys109, Cys118 and Cys124. Analyzing the tandem spectra from ≥4+ charge-state precursors acquired from a non-reduced RcsF sample, an inter-chain disulfide bond was identified between Cys74-Cys124 (Table 2a). However, any tandem mass spectrum containing Cys109 or Cys118 was not identified in either the first-pass Phenyx search or the second-pass Popitam open-modification search. The amino acid sequence of RcsF does not contain any tryptic sites (Arg or Lys residues) between Cys109 and Cys118. This means that if there were a disulfide bond connecting these two cysteines, it would manifest itself as an intra-peptide bond. It is likely that no information on the redox states of Cys109 and Cys118 was obtained from the initial analysis because the precursor with the intra-chain disulfide bond was not highly charged due to its small size and thus was not selected for fragmentation during the data-dependent selection. With the hypothesis that a tandem mass spectrum for this peptide was not acquired due to the >4+ data acquisition scheme, a second targeted tandem MS acquisition was carried out on 2+ and 3+ charge states of the calculated precursor mass of the putative intra-disulfide bond linked peptide. Indeed, this follow-up experiment resulted in tandem mass spectra that identified a peptide sequence containing Cys109 and Cys118. Additionally, Popitam was able to extract sequence tags from both N- and C-terminus of the peptide and indicated that the peptide carried a modification mass of -2.02 u, corresponding to the loss of two protons, clearly indicating an intra-chain disulfide bridge (Table 2b). Overall, these results accounted for all of the disulfide bonds in RcsF. While these results demonstrate that our crosslink identification pipeline can be used for mapping disulfide links in proteins, they also illustrate the complexities inherent in different disulfide geometries. Note that these complexities may necessitate inclusion of lower charge states in the analysis of disulfide bonds.
Table 2.
(a) Inter- and (b) intra-chain disulfide bonds identified in RcsF. Value of X for disulfide crosslinks is −2.01 u, corresponding to loss of one proton from each cysteine residue during disulfide bond formation.
| Measured precursor mass | Peptide Mass | Protein Id (AA seq) | Peptide / Scenario (shifts) | Modification mass |
|---|---|---|---|---|
| (a) Mr = 3956.91 |
2585.17 (Pα) |
RcsF 65–89 |
DLGEVSGESCQATNQDSPPNIPTAR ***************DSPpnI-- |
1371.74 (Pβ + X) |
| 1373.74 (Pβ) |
RcsF 121–134 |
QAVCIGSALNISAK ****IGSAL --- |
2583.16 (Pα + X) |
|
| Cross-linked Precursor (Px) |
|
Measured mass accuracy = 4.5 ppm | ||
| (b) Mr = 2086.97 |
2088.98 (Pα) | RcsF 101–120 |
ANAVLLHSCEITSGTPGCYR ANAVLL***************YR |
− 2.01 (X) |
| Cross-linked Precursor (Px) |
|
Measured mass accuracy = 4.2 ppm | ||
CONCLUSIONS
Need for specialized crosslinkers and/or reagents and lack of robust and open-access computer programs for crosslink analysis are major factors that limit routine use of CXMS for studying protein structures and protein-protein interactions. Herein we report a new method that allows use of readily available chemical crosslinkers and open-access MS data analysis software for characterizing crosslinked peptides. We bypassed laborious biochemical purification of crosslinked peptides and the use of isotope tagging to identify them, relying instead on direct, systematic use of high measured mass accuracy, high-charge-state-driven data acquisition, and culling of linear peptide tandem mass spectra. The open-modification CXMS method offers several advantages over previous techniques. First, it is versatile enough to unequivocally identify chemical, as well as native crosslinks, in an efficient and cost-effective manner. While we tested the method on disulfide bond mapping, other native crosslinks in proteins, e.g. isopeptide bonds47, 48 and pyridinium crosslinks49, can also be detected with this new approach. Second, besides identifying inter-protein crosslinks, as illustrated in this study, this method can also be used for identifying intra-protein crosslinks, intra-peptide crosslinks and dead-end products from chemical-crosslinking experiments. Third and finally, a distinct advantage of this approach is its potential application to rapid analysis of multi-protein complexes. Together, these advantages make our approach very attractive for routine analysis of protein-protein interactions using chemical crosslinking and mass spectrometry.
Supplementary Material
Supplementary Figure S1. Disulfide bond and tryptic sites in Brain Natriuretic peptide-32. Tryptic sites in BNP-32 are indicated by black lines. The disulfide bridge between Cys10 and Cys26 in BNP-32 forms a loop structure. Cleavage at any of the tryptic sites in this loop region (Arg13, Lys14 and Arg17) results in peptides with an inter-chain disulfide bond, and failure to cleave in the loop region results in peptides with an intra-chain disulfide bond.
Supplementary Table S1. Inter- and intra-chain disulfide bonds identified in BNP-32. Examples of disulfide-linked peptides identified from BNP-32. Grey lines indicate a disulfide bridge and black lines indicate an un-cleaved peptide sequence. Amino acid sequence derived from the sequence tags is underlined in each peptide.
Acknowledgments
We are grateful to S. Nelson, Q. Gao and A. Roberts for stimulating discussions. We thank B. Gallis for critical review of the manuscript. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID 1U54 AI57141-01) and the National Cancer Institute (R33CA099139-01).
References
- 1.Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S. Trends in Neurosciences. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 2.Parsons RB, Austen BM. Biochem Soc Trans. 2007;35:974–979. doi: 10.1042/BST0350974. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt-Ulms G, Legname G, Baldwin MA, Ball HL, Bradon N, Bosque PJ, Crossin KL, Edelman GM, DeArmond SJ, Cohen FE, Prusiner SB. Journal of Molecular Biology. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 4.Berggård T, Linse S, James P. PROTEOMICS. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 5.Phizicky E, Fields S. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 7.Zuiderweg ERP. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 8.Shen Z, Cloud KG, Chen DJ, Park MS. J Biol Chem. 1996;271:148–152. doi: 10.1074/jbc.271.1.148. [DOI] [PubMed] [Google Scholar]
- 9.Howell JM, Winstone LT, Coorssen JR, Turner RJ. PROTEOMICS. 2006;6:2050–2069. doi: 10.1002/pmic.200500517. [DOI] [PubMed] [Google Scholar]
- 10.Sinz A. Mass Spectrometry Reviews. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 11.Huang BX, Dass C, Kim HY. Biochem J. 2005;387:695–702. doi: 10.1042/BJ20041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ethier M, Lambert J-P, Vasilescu J, Figeys D. Analytica Chimica Acta. 2006;564:10–18. doi: 10.1016/j.aca.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero C, Tagwerker C, Kaiser P, Huang L. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Orlando V, Strutt H, Paro R. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 15.Petrotchenko EV, Olkhovik VK, Borchers CH. Mol Cell Proteomics. 2005;4:1167–1179. doi: 10.1074/mcp.T400016-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Collins CJ, Schilling B, Young M, Dollinger G, Guy RK. Bioorganic & Medicinal Chemistry Letters. 2003;13:4023–4026. doi: 10.1016/j.bmcl.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 17.Scherl A, Tsai YS, Shaffer SA, Goodlett DR. PROTEOMICS. 2008 doi: 10.1002/pmic.200800045. In Press. [DOI] [PubMed] [Google Scholar]
- 18.Sinz A, Kalkhof S, Ihling C. Journal of the American Society for Mass Spectrometry. 2005;16:1921–1931. doi: 10.1016/j.jasms.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Hurst GB, Lankford TK, Kennel SJ. Journal of the American Society for Mass Spectrometry. 2004;15:832–839. doi: 10.1016/j.jasms.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Ihling C, Schmidt A, Kalkhof S, Schulz DM, Stingl C, Mechtler K, Haack M, Beck-Sickinger AG, Cooper DMF, Sinz A. Journal of the American Society for Mass Spectrometry. 2006;17:1100–1113. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Muller DR, Schindler P, Towbin H, Wirth U, Voshol H, Hoving S, Steinmetz MO. Anal Chem. 2001;73:1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 22.Bennett K, Kussmann M, Bjork P, Godzwon M, Mikkelsen M, Sorensen P, Roepstorff P. Protein Sci. 2000;9:1503–1518. doi: 10.1110/ps.9.8.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Back JW, Hartog AF, Dekker HL, Muijsers AO, de Koning LJ, de Jong L. Journal of the American Society for Mass Spectrometry. 2001;12:222–227. doi: 10.1016/S1044-0305(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, Munske GR, Siems WF, Bruce JE. Anal Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 25.Soderblom EJ, Bobay BJ, Cavanagh J, Goshe MB. Rapid Communications in Mass Spectrometry. 2007;21:3395–3408. doi: 10.1002/rcm.3213. [DOI] [PubMed] [Google Scholar]
- 26.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 27.Gao Q, Doneanu CE, Shaffer SA, Adman ET, Goodlett DR, Nelson SD. J Biol Chem. 2006:20404–20417. doi: 10.1074/jbc.M601785200. [DOI] [PubMed] [Google Scholar]
- 28.Hojrup P, Hedin A, et al. Ion Formation from Organic Solvents. John Wiley & Sons; 1990. pp. 61–66. [Google Scholar]
- 29.Koning Leo J, PTK, Back Jaap Willem, Nessen Merel A, Vanrobaeys Frank, Beeumen Jozef, Gherardi Ermanno, Koster Chris G, Jong Luitzen. FEBS Journal. 2006;273:281–291. doi: 10.1111/j.1742-4658.2005.05053.x. [DOI] [PubMed] [Google Scholar]
- 30.Gao Q, Xue S, Doneanu CE, Shaffer SA, Goodlett DR, Nelson SD. Anal Chem. 2006;78:2145–2149. doi: 10.1021/ac051339c. [DOI] [PubMed] [Google Scholar]
- 31.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. J Proteome Res. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 32.Rinner O, Seebacher J, Walzthoeni T, Mueller L, Beck M, Schmidt A, Mueller M, Aebersold R. Nat Meth. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Y, Chen Y, Lichti C, Hall R, Raney K, Jennings S. BMC Bioinformatics. 2005;6:S9. doi: 10.1186/1471-2105-6-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez P, Gras R, Frey J, Appel RD. PROTEOMICS. 2003;3:870–878. doi: 10.1002/pmic.200300402. [DOI] [PubMed] [Google Scholar]
- 35.Tanner S, Shu H, Frank A, Wang L-C, Zandi E, Mumby M, Pevzner PA, Bafna V. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 36.Tabb DL, Saraf A, Yates JR. Anal Chem. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherl A, Shaffer SA, Taylor GK, Hernandez P, Appel RD, Binz PA, Goodlett DR. J Am Soc Mass Spectrom. 2008;19:891–901. doi: 10.1016/j.jasms.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Mol Cell Proteomics. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Jacques Colinge AM, Giron Marc, Dessingy Thierry, Magnin Jérôme. PROTEOMICS. 2003;3:1454–1463. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 41.Schilling B, Row RH, Gibson BW, Guo X, Young MM. Journal of the American Society for Mass Spectrometry. 2003;14:834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 42.Castanie-Cornet M-P, Cam K, Jacq A. J Bacteriol. 2006;188:4264–4270. doi: 10.1128/JB.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laubacher ME, Ades SE. J Bacteriol. 2008:JB.01740–01707. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majdalani N, Gottesman S. Annual Review of Microbiology. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 45.Kadokura H, Tian H, Zander T, Bardwell JCA, Beckwith J. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 46.Holman C. Personal Communication. University of Washington; 2008. [Google Scholar]
- 47.Lorand L. FASEB J. 2007;21:1627–1632. doi: 10.1096/fj.07-0602ufm. [DOI] [PubMed] [Google Scholar]
- 48.Gao Y, Mehta K. J Biochem. 2001;129:179–183. doi: 10.1093/oxfordjournals.jbchem.a002830. [DOI] [PubMed] [Google Scholar]
- 49.Barnard K, Light ND, Sims TJ, Bailey AJ. Biochemical Journal. 1987;244:303–309. doi: 10.1042/bj2440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Disulfide bond and tryptic sites in Brain Natriuretic peptide-32. Tryptic sites in BNP-32 are indicated by black lines. The disulfide bridge between Cys10 and Cys26 in BNP-32 forms a loop structure. Cleavage at any of the tryptic sites in this loop region (Arg13, Lys14 and Arg17) results in peptides with an inter-chain disulfide bond, and failure to cleave in the loop region results in peptides with an intra-chain disulfide bond.
Supplementary Table S1. Inter- and intra-chain disulfide bonds identified in BNP-32. Examples of disulfide-linked peptides identified from BNP-32. Grey lines indicate a disulfide bridge and black lines indicate an un-cleaved peptide sequence. Amino acid sequence derived from the sequence tags is underlined in each peptide.