Abstract
We introduce a toolset of program libraries collectively titled MATCH (Multipurpose Atom-Typer for CHARMM) for the automated assignment of atom types and force field parameters for molecular mechanics simulation of organic molecules. The toolset includes utilities for the conversion from multiple chemical structure file formats into a molecular graph. A general chemical pattern-matching engine using this graph has been implemented whereby assignment of molecular mechanics atom types, charges and force field parameters is achieved by comparison against a customizable list of chemical fragments. While initially designed to complement the CHARMM simulation package and force fields by generating the necessary input topology and atom-type data files, MATCH can be expanded to any force field and program, and has core functionality that makes it extendable to other applications such as fragment-based property prediction. In the present work, we demonstrate the accurate construction of atomic parameters of molecules within each force field included in CHARMM36 through exhaustive cross validation studies illustrating that bond increment rules derived from one force field can be transferred to another. In addition, using leave-one-out substitution it is shown that it is also possible to substitute missing intra and intermolecular parameters with ones included in a force field to complete the parameterization of novel molecules. Finally, to demonstrate the robustness of MATCH and the coverage of chemical space offered by the recent CHARMM CGENFF force field (Vanommeslaeghe, et al., JCC., 2010, 31, 671–690), one million molecules from the PubChem database of small molecules are typed, parameterized and minimized.
Keywords: Force Fields, Atom Typing, Molecular Graph, Partial Charges, Parameterization
Introduction
The increasing availability of computing resources is reshaping the researcher’s approach in the utilization of molecular simulations for the modeling of proteins, nucleic acids, their ligands and inhibitors. It is now feasible, with the growth of large libraries of drug-like compounds, to investigate receptor-ligand binding poses and other properties using molecular mechanics force fields in a high throughput manner1. A significant barrier in this process, however, is the accurate generation of explicit inter- and intra-molecular parameters for novel potential drugs that are consistent with the biomolecular force field utilized in modeling the other components of the system 2,3. Modern force fields such as CHARMM4,5, AMBER6,7 and OPLS8 rely on empirical parameters and have been developed to yield accurate modeling of conformational changes and non-covalent interaction energies for proteins and nucleic acids9,10. However, these force fields do not contain all the required parameters to represent drug-like molecules for studying receptor-ligand interactions.
Developers of the biomolecular force fields have adopted different strategies to optimize the bonded and nonbonded parameters and attempt to reproduce experimental data or quantum mechanical properties of model compounds. Therefore, it is unlikely that the combination of a particular biomolecular force field with an arbitrary ligand force field would yield properly balanced intermolecular interactions. Rather, it is crucial that the small molecule parameters follow a similar parameterization scheme to that which was used to develop the biomolecular force field11. The most straightforward parameters that can be generalized to novel compounds are those associated with the intra-molecular energy terms (e.g., equilibrium values and force constants for bond lengths and angles as well as optimal torsion angles and the respective barrier heights). Similarly, van der Waals parameters, the atomic radii ri and energy well-depth εi, are often successfully transferred among analogous atom types12. While small changes in these parameters might significantly impact the energy, they are not particularly sensitive to the bonded environment of the molecule. In contrast, the partial charges that are associated with each atom are the primary components in the electrostatic energy terms and are significantly challenging to transfer from one molecule to another due to their dependence on both bonded and nonbonded chemical environment.
Two main strategies have been suggested for generating partial charge assignments that are compatible with current biomolecular force fields. In one fixed-charge strategy, charges are adopted for an entire molecule, often based on ab initio calculations or parameterized methods that mimic these charge distributions. A restrained electrostatic potential (RESP) charge fitting procedure is advised for assigning partial charges to novel ligands in a manner that is consistent with the Generalized AMBER force field (GAFF)3. Antechamber13, an auxiliary program in the AMBER molecular modeling suite of programs that uses coordinate and connectivity information to assign atom and bond types for ligands based on atom and bond type definition tables, can generate charges using RESP, AM1 Mulliken, AM1-BCC, CM2 or Gasteiger charge methods.
In an alternative fixed-charge strategy, used by the CHARMM and OPLS family of force fields, charge distributions of a molecule are built-up from charges assigned to the component fragments of the molecule. Halgren14, in developing the MMFF94 force field, proposed bond charge increment “rules” in which optimal charges are determined for fragments of molecules and these fragments are then pieced together to construct charge distributions for novel compounds. Several programs exist to assign atom types and atomic partial charges based on the bonded environment of the atom. These automated assignment programs convert a three-dimensional structure file into a representation of the bonded environment, such as a connectivity table of atoms and bonds. Patterns within this connectivity table are identified as fragments for which atom-types and partial charges are associated. These programs differ in how the bonded environment is determined, how the specific “rules” are defined for matching the fragments in the new molecule with those of “known” fragments, and how the partial charges are distributed throughout the molecule. For example, the molecular modeling package IMPACT15 accepts a PDB file format and automatically assigns atom types and parameters for a wide range of organic molecules that are consistent with the OPLS_2003 force field. For the entire molecule, the partial atomic charges are assigned by distributing any formal ionic charges over one or more atoms and then adding contributions from the bond charge increment (BCI) parameters associated with the chemical bonds. PRODRG16,17, through its web interface, generates molecular topologies from a coordinate file and assigns partial charge distributions from a molecule’s constitutive fragments for use with the GROMOS force field18.
Recent developments in the CHARMM community have led to the generation of small molecule parameters in a novel force field denoted CHARMM General Force Field (CGENFF). Although a notable step in the right direction, the chemical space covered by the ~400 molecules in CGENFF is limited and still requires manual efforts to extend it to new molecules. Our preliminary goal with this work was to develop a publicly available solution for generating parameters for novel molecules that are consistent with the CHARMM parameterization scheme. We were interested not only in creating a way to process molecules en masse within CHARMM, but also to develop a tool to investigate the merits of different types of parameterization choices and strategies. We developed a general approach that extracts rules for both charging and parameterization based on a library of topology and parameter files for an existing biomolecular force field. This scheme then allows for the fragments comprising existing parameters to be applied or extrapolated to novel molecules in a fashion consistent with the parameterization strategy or philosophy within a given biomolecular force field. We have focused our efforts on CHARMM; however, this approach and the MATCH toolset can be used to extract rules for charging and parameterization based on any biomolecular force field.
A fundamental feature of the MATCH algorithm is the representation of molecular structures as mathematical graphs. Chemoinformatics has benefited greatly from the representation of chemical structure as graphs, such as in ring identification and characterizing chemical connectivity19. In particular, structure and substructure searching of chemical databases, such as those performed on inventory or patent databases, automated retrosynthetic analyses, property prediction, quantitative structure-activity/toxicity/property relationship analyses, visualization, and similarity/diversity analyses are applications with chemical pattern recognition solutions20–23 The unique chemical environment defining an atom type can also be depicted in graph form, enabling a chemical characteristic comparison between a library of known atom type definitions and atoms in a novel compound. Furthermore, additional operations are vastly simplified while functioning within a graph reference frame, including: quantification of the similarity of chemical environment between atom types within a given force field, atomic ring identification, and identification of atoms requiring improper angles to enforce accurate geometry. Much of the next section will be devoted to discussing the implementation of mathematical graphs within MATCH.
At present, we will demonstrate the utility of MATCH and discuss the primary components comprising the software package. MATCH supports every force field currently implemented in CHARMM36 (i.e., force fields specific for proteins, nucleic acids, lipids, carbohydrates, ethers and model small molecules). Here, we demonstrate two primary functions of the MATCH toolset. First, we show how MATCH is used to extract fragment-based atom types and associated bond charge increment rules. More specifically, we discuss how MATCH constructs libraries that contain definitions for the chemical environments described by the force field topology files for a given force field as well as the schemes for assigning partial charges to these atom type definitions. Second, we illustrate how MATCH is used to generate force-field specific MATCH libraries. These libraries are shown to be self-consistent with existing CHARMM force fields by their ability to reproduce atomic charges contained in the force field that was used to infer the rules. In addition, the viability of parameter substitution to determine missing parameters and thereby enabling complete parameterization will be demonstrated through a leave-one-out substitution study. Our benchmark for the transferability of rules learned from a force field will be the charging and parameterization of the molecules in other existing force fields. We then directly compare computed values to existing ones and measure how well the results are correlated. This exhaustive exercise, using each existing CHARMM force field to charge and parameterize the other, demonstrates the ability of the methods implemented in MATCH in generalizing a force field representation. Finally, the parameterization of one million small molecules from the PubChem database24 with the implementation of the CGENFF-based libraries within MATCH illustrates the scope and potential of MATCH in real world usage.
MATCH Strategies and Components
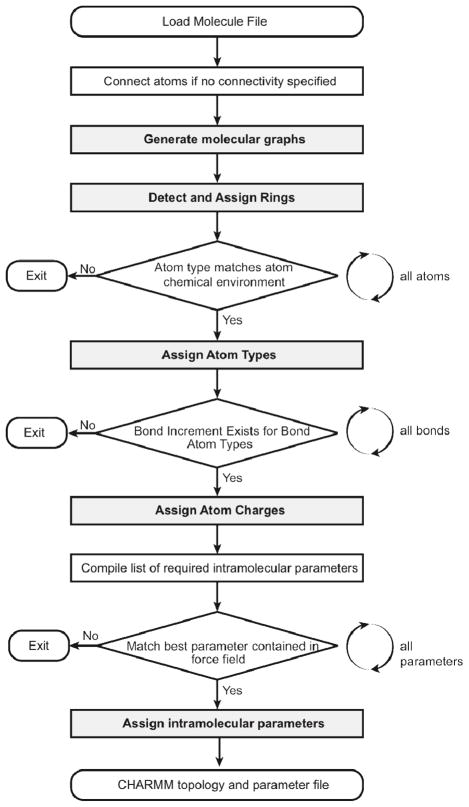
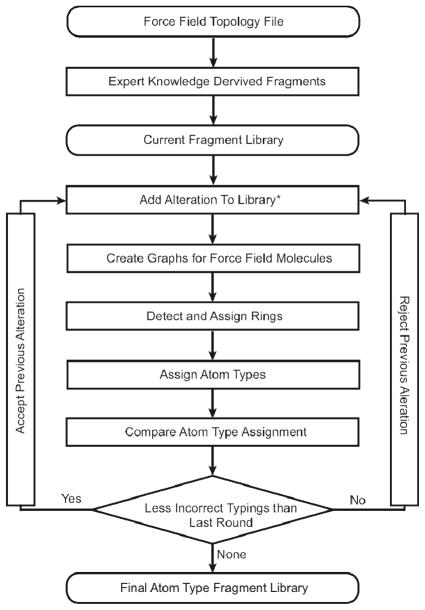
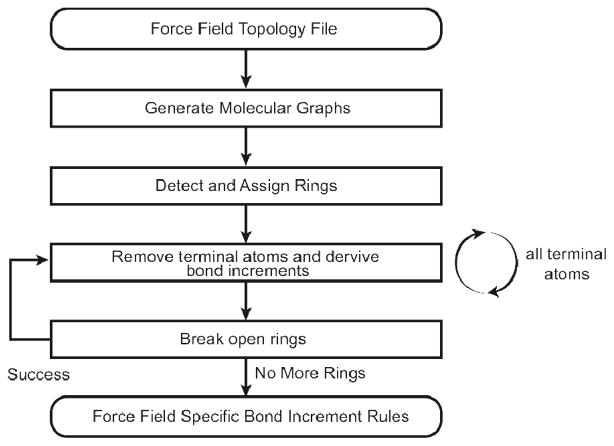
MATCH is a suite of tools that has been developed for constructing molecular fragment-based libraries and BCI rules to be utilized for the extension of a given biomolecular force field. There are two distinct applications of the MATCH toolkit: i) the utilization of atom-type molecular fragment and BCI rule libraries in the charging and parameterization of novel molecules and ii) the tools required to assemble these libraries as well as the generation of rules to allow substitution of parameters to assist in the parameterization of new molecules. The procedure in which MATCH extends a force field to a novel molecule is illustrated in Figure 1. Development of the MATCH libraries of fragments for atom typing and bond increment rules are illustrated in Figures 2 and 3, respectively. Here, we explore the ability of MATCH, with some expert intervention, to effectively construct force field specific MATCH libraries, which is to “learn” atom type definitions and bond charge increment rules from multiple CHARMM force fields. We also investigate the ability of MATCH to use these libraries and the substitution rules to parameterize molecules in different force fields.
Figure 1.
Overview of the MATCH algorithm. All major algorithm components discussed in the paper appear in bold.
Figure 2.
Overview of the process of developing atom type molecular fragments for a given force field, which is the basis of MATCH’s atom typing engine.
Figure 3.
Overview of the process of extracting the bond charge increment rules for a given force field.
Molecular graphs
Molecular graphs are assembled using the supplied connectivity information (CONECT lines in a PDB file, a CHARMM RTF file, a bond list for MOL2, MOL, and SDF, etc) or predicted using the atomic coordinates and bonding rules based on atomic radii. As described by Downs and coworkers25, molecular graphs are constructed as labeled, directed, connected graphs, where each atom is represented by a vertex and stores information about itself including element, number of bonds, ring membership, and pointers to neighboring atoms. For small molecules (less then 10 atoms) a graph represents the entire molecule; however, due to the computational expense of constructing these graphs for larger molecules, a limitation was imposed such that atoms greater than 10 bonds away are not included in the definitions of the chemical environment of a given atom. This limitation was arbitrarily set to 10 as a compromise between accuracy and efficiency. In fact, the chemical space characterization of all atom types in this study does not extend further than 3 to 4 bonds away (see atom type declarations in MATCH). The molecular graph is then expanded following a breadth-first algorithm26. Starting from one atom, each atom to which it is bonded in then added to the tree; atoms to which they are bonded and are not yet included in the tree are then added and so on until all atoms in the molecule are either represented in the tree or 10 bonds away. The end result is a branched data structure that allows for tree comparisons and other operations, which are crucial to the workings of MATCH. The process is repeated for each atom as the head vertex. While a bond is not normally considered directional, an artificial directionality is imposed by this representation and is harnessed in algorithms that will be discussed later. For clarity, atoms occurring higher in the depth of the tree are considered the parent, while bonded atoms that are added beneath are considered to be children. The only exception to this definition is in cyclic compounds, where two connected atoms may be positioned such that they are at the same depth. In this case the first atom to be traversed is considered to be the parent of the other.
Calculating whether one molecular graph is similar enough to another to be considered a “match” is a fundamental process in atom typing. The procedure to do this is straightforward: For two graphs to be considered a match, each node of the smaller graph (i.e., the atom type fragment) must exist within the larger graph (i.e., the molecular graph within the new molecule with the current atom being the head node) with the same connectivity. The procedure is analogous to a typical tree data-structure comparison in which the comparison is initiated at the head nodes. Confirming a match is a two-step process: first, features such as ring membership, aromaticity, etc., of the nodes of the smaller graph must be contained in the nodes of the larger graph. Second, the element and number of bonds of each node must be consistent. This process continues until the smaller graph has all of its nodes matched or until one node is unable to be matched to a node in the other graph. Occasionally, there are two possible matches for a node; when this occurs, the children of both potential matches are compared to the node’s children in a a recursive manner until a difference is identified or the graphs are found to be identical.
Ring Detection
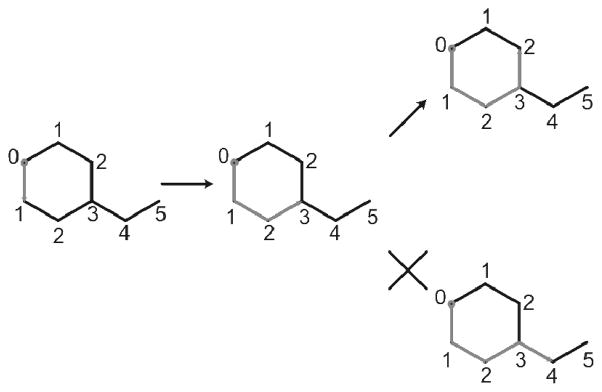
Identification of ring membership is crucial in the atom typing process of MATCH due to the specificity of atom types that are only found in rings. Ring discovery has received considerable attention in the literature because of its computational demands19. Much of the algorithmic development in this area has focused on the identification of subsets of rings that have particular meaning in some applications, for example in the analysis of synthetic pathways27. In this situation, exhaustive enumeration of rings is required for accurate atom typing due to the fact that atom types are ring specific. The algorithm developed here relies heavily on the use of molecular graphs discussed earlier and is based on the works described by Tiernan28 on mathematical graph circuit detection. The elements of our ring detection algorithm are as follows: each heavy atom with more than one bond is considered in turn unless the atom has already been detected as being part of a ring. The ring detection algorithm is a breadth-first search26 that traverses the molecular graph of the atom being considered. During each iteration of the search, each current path is extended to new heavy atoms, the path that is currently the closest in level to the starting point will always be selected to be followed first (see Figure 4). Upon reaching the start atom with the path containing more than two atoms, successful termination is reached and all atoms that were traversed along the path are marked with ring membership. Failed termination is reached when each path covers more than 50% of the molecule and is more than 50% away (by depth) from the start, making it impossible to successfully return to the starting point.
Figure 4.
Depiction of the usage of the molecule graphs to our advantage in determining rings. Here we see that given one possible path: when given the choice between two directions the ring detection algorithm will always follow the path of lowest level.
This algorithm is very fast, requiring only one atom in a ring to be searched. It also prevents duplicate identification of rings. While other ring detection algorithms have been shown to be more efficient19, this algorithm was selected because of its reliance on molecular graphs. In fact, most of the computational efforts in MATCH are for the construction of the molecular graph and the typing, charging and parameterization of novel molecules. Therefore, ring detection is not the computational bottleneck of MATCH and, thus, the decision to implement this ring detection algorithm does not hinder the performance.
Molecular Fragment-based Atom Typing
Converting molecules into mathematical graph form enables the direct comparison of the local chemical environment of one atom to another and the quantitative evaluation of the similarity between the two structures. Continuing with this ideology, the chemical space that defines an atom type can be represented as a molecular fragment. We define a molecular fragment as a group of connected atomic nodes that contain the required atomic features that describe the chemical space of the atom type (i.e., atomic element, number of bonds, ring membership, etc). These molecular fragments have the same properties as the graphs that are built for actual molecules and thus can be compared in a similar procedure. Adopting this philosophy of representing distinct chemical space as a molecular fragment reduces the atom typing process to one of tree comparison, in which the largest molecular fragment that completely matches an atom’s molecular graph is assigned to that atom. The library of atom type molecular fragments is preserved in super smiles string format with similarities to the implementation by Bone et al29.
In super smiles string notation, each atom is represented by its chemical element plus its number of bonds. More specific information is appended to the end of the string. Examples of super smiles format are displayed in Table 1. The “!” attribute denotes no ring membership whereas the “%” attribute indicates ring membership and is followed by the ring size and aromaticity. Connectivity between atoms is denoted by parentheses, where atoms within a parenthesis are bound and considered to be children to the one outside. The decision to describe atom type molecular fragments in this representation is to allow for effortless management of MATCH atom type force field libraries. It is a straightforward process to modify or add new atom types to an existing library or to create entirely new ones. In this study, we demonstrate how atom types in the CHARMM force fields can be represented by molecular fragments. Certainly, this strategy can be extended to represent other biomolecular force fields.
Table 1.
Examples of the syntax of the super smiles strings used to represent atoms within MATCH encoded molecular fragments.
| Smiles String | Atoms Matched |
|---|---|
| * | Any Atom |
| C.4 | Carbon atoms with 4 bonds |
| !N.3 | Aliphatic Nitrogens atoms with 3 bonds |
| O | Any Oxygen atom |
| [^C] | Not a Carbon atom |
| S.2% | Sulfur ring atoms with 2 bonds |
| C.3%6,6 | Carbons atoms with 3 bonds in 2 6 |
| N.3%6A | Nitrogen atoms with 3 bonds in a 6 membered aromatic ring |
| C.4%5N | Carbon atoms with 4 bonds in a 5 membered non-aromatic ring |
Bond Charge Increment Rules
Inspection of atomic charges in commonly used force fields suggest that they often follow bond charge increment rules5. Bond increments are a description of the magnitude and direction of charge of the covalent bonding of two atoms. Decomposing the atomic charges of a molecule into these increments yields a set of generalized rules based on the type of the atoms in the bond. Once these rules are identified they can be extended and applied to new molecules.
Development of bond charge increment rules has been implemented in the past for other force fields such as in MMFF94. In their approach they globally optimized a set of rules through an iterative process that best fit the training set14. While this is a valid approach and was considered when investigating charging rules in MATCH, it was discarded due to the inability to precisely reproduce the training charges in force fields such as CGENFF (data not shown). In addition, our goal is consistency: to preserve the charging rules found in the protein and nucleic acid force fields as much as possible. Our approach is consistent with our fragment-based atom typing procedure and accurately reproduces partial charge assignments in all CHARMM force fields.
Empirical force fields generally reuse atom types for bonded parameter assignments despite slightly different charge distributions. For example, in the CHARMM protein force field most methylenes transfer −0.09 electron units of charge from each of the two aliphatic hydrogens onto the adjacent carbon; however, for methylenes adjacent to a primary ammonium this increment is −0.05 electron units, despite the fact that identical atom types are used to describe the respective carbon and hydrogen atoms. This discrepancy is dealt with in MATCH by conducting a secondary level of atom typing. During the development of BCI from a force field, if there are multiple solutions exist for a given pair of bonded atom types, the most frequently used BCI is stored as the default increment rule and the infrequent ones are stored separately as refining increment rules. These refining increments are associated with a molecular fragment as found in the atom typing process. This fragment is the description of the chemical environment that is correlated with the divergent increment rule. During the charging procedure MATCH considers the refining increment rules and checks to see that the corresponding chemical environment matches the current local connectivity of the molecule: if so, the refining increment rule is applied instead of the default rule.
The simplest case to describe the magnitude and direction of charge in the covalent bonding of two atoms involves a terminal atom, that is when one of the atoms has only one covalent bond. In a neutral system, it is straightforward to determine the bond increment between a terminal atom and its bonding partner: it must be the terminal atom’s assigned atomic charge balanced by an equal and opposite signed charge assigned to its bonding partner. For example, most aliphatic hydrogen atoms bound to aliphatic carbons have a charge of 0.09 in the CHARMM force field, and this yields charge increments of +0.09 and −0.09 to the hydrogen and carbon atoms, respectively. Unfortunately, as the number of bonds increase it becomes increasingly difficult to deconvolute the charge relationship in each bond. The solution we adopted is to disassemble a molecule by removing terminal atoms and subtracting their respective bond charge increments. Returning to the example of the aliphatic hydrogen atoms, the procedure would involve removing the terminal hydrogen atoms and subtracting the charge from the BCI (i.e., subtracting −0.09 e−) from the remaining carbon atom. This process effectively nullifies the bond between the hydrogen and carbon and reduces the number of bonds in which the carbon atom participates. If the carbon atom is a methyl carbon, it would now be rendered a terminal atom with a charge reduced by −0.27 e−. This iterative procedure of nullifying bonds and adjusting the charges allows the parameterization of a large portion of the BCIs. However, atom types that are exclusively contained in rings fail to yield BCIs through this method. Thus, two follow-up algorithms have been implemented to deal with ring atom types. The first algorithm is used for rings with “symmetry” points in which one atom is bonded to two atoms with the same type. In this case it is possible to break the ring at this “symmetry” point and establish the bond charge increment rules by assuming that each of the bonds contributes exactly half of the charge of the atom bonded to both of them. The second algorithm is used for rings in which no symmetry exists for any ring atom; in this case, a previously determined BCI is used to break the ring. If all the charge is accounted for by existing rules then it is accepted as the correct increment. Using the methods described above it is possible to delineate the vast majority of bond charge increment rules for a given force field. The minority of compounds in a force field whose BCIs can not be deconvoluted with these methods can be examined on a case-by-case basis. In these situations, it is usually possible to take existing increment rules and apply them to these molecules checking to see if all the charge is accounted for.
Parameter Generation
For a molecule to be successfully represented by a force field, it requires intramolecular parameters for the bond, angle and dihedral energy contributions and intermolecular parameters: atomic partial charges and van der Waals parameters describing the nonbonded energy contributions. Assignment of atomic charges was covered in the previous section; we will now discuss generation of the remaining parameters. Producing all required bond parameters for a novel compound in MATCH is trivially accomplished by removing duplicates from the list of bonded atom types that was already acquired during the process of assigning atomic partial charges and by identifying the corresponding parameters for these bonded atom types in the parameter file. To produce the required angle parameters, each bond is traversed and the neighboring bonded atom is added to each side, growing out a bond into an angle. The same procedure is repeated with angles to obtain all required dihedral angles. The required parameters are then added to the new compound’s own parameter file. Parameters that do not exist in the parent force field parameter file are generated via substitution of the best-fit parameter.
No force field contains parameters of all chemical space; therefore, means to “interpolate” within or “extrapolate” beyond the parameterized chemical space are necessary. Our solution is to identify existing parameters that “best fit” the required parameter through a form of parameter substitution. Upon examination of atom types in a given force field it is apparent that some types are more similar than others. For example, investigating the CHARMM36 protein force field, it is evident that a correlation exists between atom types. CT1, an aliphatic carbon bonded to one hydrogen, shares many of the same bond, angle, and dihedral parameters as CT2, also an aliphatic carbon but bonded to two hydrogen atoms. This is unlikely coincidental: atom types that share a similar chemical space should also have similar bonded parameters. From careful analysis of the chemical space of atom types basic rules can be derived. First, aliphatic types behave more similarly to other aliphatic types then they do to atoms that have ring membership. Second, the number of bonds a type has also affects how similar parameters are to each other: types that share the same number of bonds have more similar angle and dihedral parameters. Keeping these basic observations in mind, it is possible to create a score describing how one type is related to another based on comparison of the molecular fragment representations used for typing. The use of this substitution method vastly increases the number of molecules that can be assimilated into the working force field. A brief overview of how the relatedness between types is built is: the molecular fragment of each atom type is compared and the overlap between the two is computed. Special penalties are put in place to distinguish the score of atom types only found in rings from atom types that are only found in aliphatic chains and the reverse case. These scores are preserved in text format that may also be altered by users if desired.
Substitution is available during both the atom charging and parameter generation stages in MATCH. In both cases the procedure is equivalent. For example, if there is a bond between atom type A and B, but the corresponding bonded parameters do not exist in the force field parameter file, the relation matrix will be queried. Each existing bond parameter is scored in the simple fashion of how closely its first atom type is related to A and how closely its second is related to B; the reverse is also considered. The relatedness is 1 if the atom types are the same and is 0 if the two atom types have neither the same element nor the same bond number; the summation of the relatedness of each pair is the score. The bond parameters with the highest score are selected as the substitution parameters for this new pair of bonded atom types.
Program Organization
MATCH supports a wide variety of molecular formats, (PDB, MOL2, MOL, SDF, RTF) and exports CHARMM formatted PDBs for files supplied in non-PDB formats. The core algorithms of the MATCH toolset have been implemented in Perl. Perl was chosen to maximize portability. MATCH is a small package that utilizes PerlChemistry, another package for more general applications of identifying similar connectivity between molecules, renaming atoms given a chemical environment (e.g., the naming convention used in RESP7), or averaging charges over atoms with identical connectivity. PerlChemistry is a set of Perl packages that provides object representations such as atom, bond and molecule. This distinction was planned so PerlChemistry can exist independently from MATCH and allows users to have access to the molecular graph API. Several examples of applications of molecular graphs are provided as part of the distribution.
The key properties of the MATCH package are contained in a single Perl package called MATCHer.pm, which allows users to write additional scripts to facilitate any of the algorithms discussed in this paper in the context of other force fields. The default script, MATCH.pl, provides the core functions of atom typing, charging and parameter determination that are associated with the processes depicted in Figure 1. All the MATCH force field libraries discussed in this paper are included in the current version of the MATCH package. The MATCH package is supplied together with basic usage instructions under a GNU license and can be downloaded at http://brooks.chem.lsa.umich.edu/software.
Methods
Constructing force field-specific MATCH libraries via MATCH
Force field-specific MATCH libraries were constructed via MATCH based on the CHARMM36 topology files: top_all22_prot, top_all27_na, top_all35_carb, top_all35_ethers, top_all36_cgenff and top_all36_lipid. For each force field the molecular fragments for each atom type were constructed through an iterative optimization procedure. Using a given force field the goal is to correctly assign types for all the atoms within the force field. The main concern in this process is to avoid mistyping by incorrectly making one type cover the space of another. To avoid this, atom types were grouped together by the atom element and bond number and were developed simultaneously. That is, each time there was a modification of a fragment, each atom that was of the group’s element and number of bonds was typed and if there were fewer mistypings this change was accepted. This was repeated until there were no mistypings. Most aliphatic atom types have rather distinct chemical space and, thus, required a few rounds of optimization. On the other hand, it was more difficult to create the optimal set of fragments for atom types that are exclusively based in rings and, thus, these atom types required multiple rounds of optimization. The Perl script TestBuildTypeStrings.t that is required for this optimization is provided in the MATCH package distribution for future optimizations and development of atom-type fragments for new force fields. Another challenge in this optimization scheme is keeping the atom-type fragments as general as possible while preserving their unique chemical environment.
For each force field that contained residue patches, each patch was applied if it increased the chemical space of the set (i.e., added new atom types or bond increment rules) or was necessary to correct polymer connectivity. By default, the NTER and CTER patches were applied to the protein force field residues and the 5TER and 3TER patches were applied to the nucleic acid force field residues. With the exception of CGENFF, all molecules in the topology files were included in the process of constructing the force field-specific MATCH libraries. In total, 53 of the 415 molecules in the CGENFF topology file were eventually excluded. There were 3 primary categories of molecules that were excluded: molecules containing a fused ring that would require all bond increments to be refined as a result of charge smearing; molecules containing a conjugated alkene chain which has alternating CG2DC1 and CG2DC2 atom type designations but the same chemical environment; and molecules that have a connectivity of two atom types A and B such that A – B – A – B – A, which would require simultaneous refinement of the A–B bond increment. The latter two categories of molecules have been incorporated into the most recent version of the CGENFF MATCH libraries, but were not used in this study.
Bond increments were extracted from each force field topology file in an automated fashion as discussed in the previous section, and can be reproduced in MATCH using GenerateBondIncrementRules.pl. Refinement bond increments were added to fix obvious exceptions to the BCIs, e.g., where the default BCIs could not reproduce the charge distributions in the molecules, and were usually small in number, with exception of CGENFF. In addition to the compounds that were excluded when constructing the CGENFF-specific MATCH libraries, several other compounds in the CGENFF topology file do not obey clear bond increment rules. With additional refinement rules, however, it was possible to reliably reproduce charges for these compounds.
Extrapolating and interpolating force field parameters via MATCH libraries
Both the self-validation and cross-validation of atomic charge was conducted with the same procedure (TestIncrements.pl). To assess the ability of the MATCH libraries to extrapolate and interpolate to new contexts, MATCH libraries of force field A were used to assign charges to the atoms of each molecule in force field B. Molecular graphs of each molecule in B were constructed and each molecule object was duplicated, but with all atom types and charges removed. Each molecule copy was then typed using the MATCH libraries based on force field A’s atom type molecular fragments. If any of the atoms could not be typed, the algorithm proceeded to the next molecule. Upon successful completion of the atom type assignments, BCIs were applied to assign atomic charges. The differences between the original and assigned charges for atoms in molecules that were successfully charged were computed. For the self-validation analyses, A and B were the same force field. A similar procedure is in place for comparing the atomic parameters of one force field compared to another and is performed using TestParameters.pl. Analysis was also completed on atoms that could be completely charged/parameterized regardless of whether its entire molecule could be (Table S1 and S2).
High-throughput small molecule parameterization
PubChem small molecules were obtained from the PubChem database in MOL2 format. Since submission of molecules is random and we are interested in the chemical space that can be covered using MATCH we took the first million of the ~26 million molecules that met the following criteria: molecular weight <600 Da and contained exclusively elements H, C, N, O, F, P, S, Cl, Br, and/or I. Molecules that fit these criteria were processed by MATCH using the CGENFF force field-based MATCH libraries. If force field generation was successful, the molecule was minimized in CHARMM using steepest descent minimization for 100 steps with nonbonded cutoffs that are defined in the protein force field. Finally, the RMSD between the minimized structure and original structure was calculated.
Results and Discussion
Recapitulating Bond Charge Increment Rules
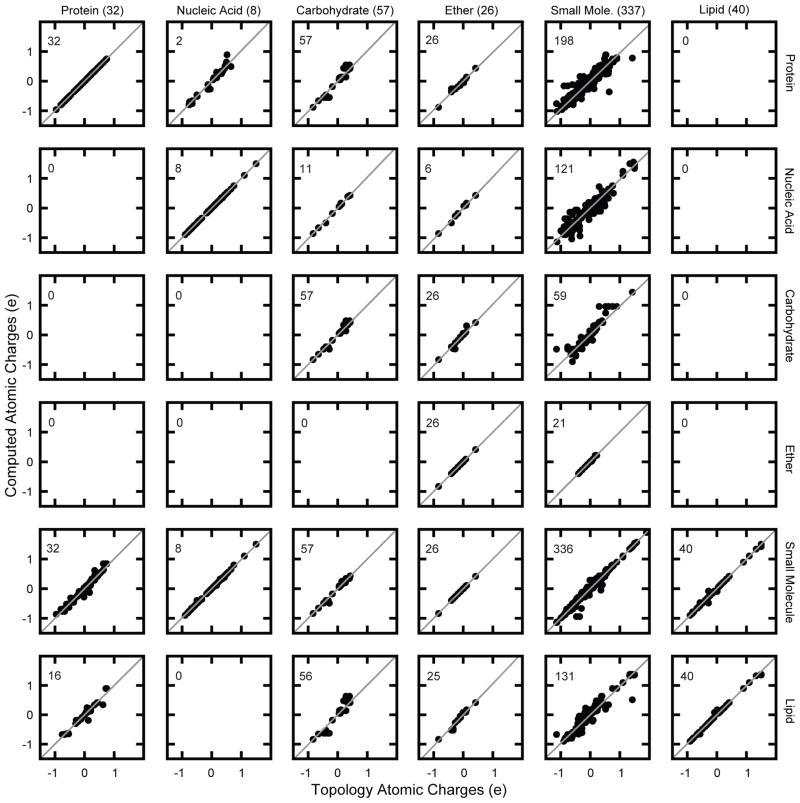
This novel suite of MATCH tools includes facilities to aid in developing force field-specific MATCH libraries that are learned from a given biomolecular force field, and to generate sets of parameters for novel compounds that are consistent with this force field in a short amount of time. Here, we explore the ability of MATCH with some expert intervention to effectively construct force field specific MATCH libraries, which is to “learn” atom type definitions and bond charge increment rules from multiple CHARMM force fields. We also investigate the ability of MATCH to use these libraries and the substitution rules to parameterize molecules in different force fields. Figure 5 summarizes the results for the MATCH-assigned partial charges for each atom compared to that in the original CHARMM topology file. The plots along the diagonal in Figure 6 represent the results for the self-consistency study in which MATCH re-predicts the properties of the force field on which the MATCH libraries were based. The off-diagonal plots represent the results from the cross validation study in which MATCH interpolates and extrapolates parameters and atom type assignments from a different force field.
Figure 5.
The x-axis denotes the reference force fields while the y-axis is the force field libraries within MATCH. The numbers in the top left corners of each graph indicate the number of molecules that were successfully charged using a given MATCH library. Additional information can be found in Tables S1 and S2 of the Supplemental Material.
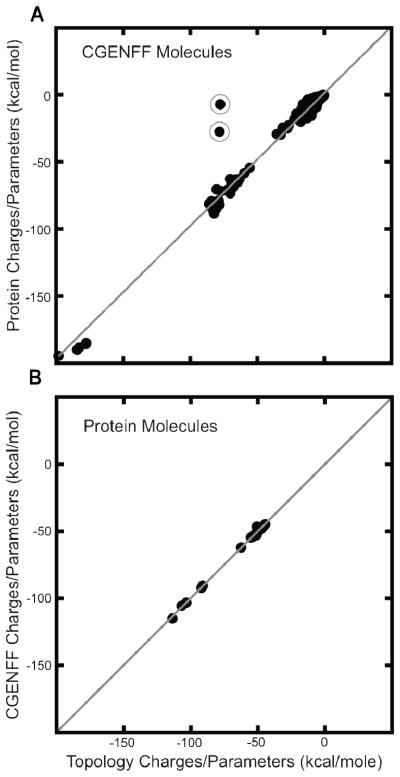
Figure 6.
A) The correlation between the solvation energy calculated using the charges and parameters found in the CGENFF topology and parameter files compared to the solvation energy calculated using the MATCH computed protein charges and parameters. There are two distinct outliers, for which MATCH computed the incorrect formal charge. Removing these outliers yields an average error of 2.2 kcal/mole. B) The correlation between the solvation energy calculated using the charges and parameters found in the protein topology and parameter files compared to the MATCH computed CGENFF charges and parameters. Excellent agreement is achieved in this test: an average unsigned error of 0.6 kcal/mole.
First, the results from the self-consistency study demonstrate that MATCH successfully recapitulates the atomic charges in the CHARMM topology files. It would be difficult to get such excellent agreement without also capturing the correct atom types. All force fields were flawlessly reproduced with the exception of the carbohydrate force field and CGENFF. In the carbohydrate force field there is a small discrepancy for the increments between atom types: CC2O3 and OC2D3, which exists in both D-psicose and ketose. In the original topology file, the assigned charges for these atoms are quite different from each other leading MATCH to learn radically different bond increment rules. Upon inspection the chemical connectivity appears identical but the stereochemistry is D compared to L, which alters the proximity between the ketone and the hydroxyl groups and, thus, may affect the charge distribution assigned to the ketone. This makes it impossible to create a refining bond increment rule to assign different increments in each case as the application requires a unique chemical environment to discriminate. As mentioned in the Methods section, CGENFF molecules whose partial charge assignments were clearly not consistent with bond increment rules were omitted from the “learning” phase in which the CGENFF-based MATCH libraries were constructed. After removal of these molecules there remained a few that had a bond increment that could not be refined with a fragment, as it was not possible to isolate a unique chemical environment; this is the cause of the relatively few outliners that appear and primarily involved fused ring systems. Consequently, charges for these molecules are not reproduced exactly, but are still of very high quality.
Second, the cross-validation study illustrates how MATCH libraries can be extended to generate parameters for novel compounds. Starting with the protein force field, which contains only atom types and bond increment rules designed for the amino acids, the rules are successfully generalized out to a significantly larger chemical space. This is illustrated primarily in typing and charging CGENFF molecules with the top_all22_prot force field, in which over 59% of the molecules in CGENFF were successfully processed in MATCH. The R2 correlation between the MATCH-assigned atomic charges and those found in the CGENFF topology file is 0.94. The average unsigned error is 0.024 electron units while the percentage unsigned error is 16.0%. While the ability of the MATCH libraries based on the CHARMM protein topology file to be successfully extended to other small molecules is very promising, it is worth considering why certain CGENFF molecules were unable to be processed in MATCH. The most significant contributor is the lack of necessary atom types for atoms of elements that are not included in the protein force field. Almost 19% of CGENFF, that is 64 molecules, contain elements P, F, Cl, Br, I, and these elements are not present in the protein force field. The remaining CGENFF molecules that could not be processed with MATCH failed because it was not possible to construct a substitution for a necessary bond increment rule to complete the atomic charges. While the substitution rules can be further generalized, the quality of both atomic charges and parameters will suffer.
The MATCH libraries based on the protein force field successfully processed all of the molecules contained in the carbohydrate and ether force fields. The R2 correlation between the MATCH-assigned and original atomic charges was 1.0 and 0.99 for the carbohydrate and ether force fields respectively. The average percentage error in atomic charge for molecules in the carbohydrate force field was 2.6% while that for molecules in the ether force field was 14.2%. The high quality of these results is not surprising since both of these force fields represent a narrow chemical space and it is mostly covered by the chemical space found in the side chains of the amino acids. However, the MATCH libraries based on the protein force field had more limited success in extending coverage to the nucleic acid and the lipid force fields. Only adenine and cytidine in the nucleic acid force field were successfully parameterized with MATCH: though the R2 correlation and average unsigned error between the computed and existing charges is excellent at 0.97 and 0.035 electron units, respectively. None of the molecules in the lipid force field were successfully parameterized with MATCH because every lipid molecule has a phosphate head group and the protein force field does not contain any atom types for phosphorus.
CGENFF is the most chemically diverse force field; this is partly due to the inclusion of the model compounds from each of the other force fields. This diversity suggests that a large portion of the chemical space of the other force fields could be covered when generating molecular fragments for the atom types and the bond increment rules based on CGENFF. This hypothesis is supported by the results of typing and charging molecules from the other force fields with the MATCH libraries developed from CGENFF. MATCH was able to successfully parameterize all compounds from the other force fields and reproduced the partial atomic charges very reliably. In fact, for the charges assigned to the 7570 atoms there is an average unsigned error of 0.0013 charge units, an average percentage error of 1.0%, and an R2 correlation with the existing charges of 1.0 This level of agreement has profound implications for further development of MATCH. As mentioned earlier, there are model compounds from each of the other force fields within CGENFF. These compounds have lead to accurate generation of the bond charge increment rules that are shared with all the other force fields, suggesting that extending the chemical space can be accomplished by adding few model compounds that represent the desired novel chemical connectivity. For example, a huge library of novel scaffolds can be parameterized by performing quantum chemical optimizations on the simplest representations of new connectivity and then extracting the necessary bond increments to develop the rules necessary to parameterize the entire set.
The lipid force field is the second largest in terms of the number of atoms next to CGENFF and is the only other force field that has the atom types and bond increments that are necessary to type and parameterize some of the protein force field. Using the lipid force field libraries within MATCH, 50% of the protein force field could be parameterized with an average percentage error for atomic charges of 6.19% and an R2 of 0.99. The majority of the error comes from attempting to parameterize phenylalanine without aromatic atom types. It is interesting that there is an overlap of chemical space between the head groups of lipids and amino acid backbone and side chains. For the carbohydrate force field near complete parameterization was possible, with an average percentage error of 3.0% and an R2 of 0.99. This agreement is excellent and further illustrates the power of extrapolating the bond increment rules. Lipids contain no ring-specific atom types and yet MATCH is still able to correctly recapitulate the atomic charges of the carbohydrate force field, which are primarily sugar rings. With the lipid-force field-based MATCH libraries, all but one molecule in the ether force field could be parameterized. The error of 27% and R2 of 0.97 indicates that the atomic charges were not computed flawlessly. Most of the error stems from the lack of an atom type that is specific for ether chemical space; the closest one that exists is for an ester oxygen. Similarly, only 40.4% of the CGENFF molecules were successfully parameterized with the lipid-based MATCH libraries, with an average percentage error of 27% and an R2 of 0.96 for atomic charges. As with the protein force field, attempts to parameterize aromatic rings lead to error. However, the largest errors come for ribose atoms. C3′ has some of the largest error with an original charge of 0.01 and a computed charge of 0.16 a percentage error of 1600%. This further reiterates the need for a quality control method, or cut off where parameter substitutions may be too unreliable.
The CHARMM nucleic acid, carbohydrate and ether force fields cover significantly less chemical space than the protein and CGENFF force fields and, thus, far fewer compounds were successfully processed with the MATCH libraries based on these force fields. For example, the nucleic acid force field-based MATCH libraries were able to type and parameterize ~36% of the molecules within the CGENFF force field and 11 molecules in the carbohydrate force field.
However, the partial charge assignments for the successfully processed molecules is very high: with R2 of 1.0 and 0.93 for the carbohydrate and CGENFF molecules, respectively, and with an average percentage error of 5.0% and 28.5% respectively. In addition the nucleic acid force field was also able to parameterize 6 of the ether molecules with an average percentage error of 11.7% and R2 of 0.99. The decrease in coverage as compared to the protein and CGENFF force fields is not surprising as the nucleic acid force field does not include as many aliphatic atom types. In fact, all of the aliphatic groups in the nucleic acid based MATCH libraries come from select patches that modulate the purine and pyrimidine groups. Similarly, the carbohydrate force field, although interesting for the parameterization of 5 and 6 membered sugar rings, has a very narrow chemical space. The MATCH libraries based on the carbohydrate force field could only parameterize 59 out of the 336 CGENFF molecules, which included all the ether molecules. Similarly, the ether force field, much like the carbohydrate force field, is very specific and contains simple aliphatic and ring ether molecules and the associated MATCH libraries were only able to type and parameterize 21 of the CGENFF molecules. The R2 correlation for the atomic charges is 1.0 and the average percentage error is 0.5%, though all but one molecule had charges that were exactly reproduced.
To quantify the relationship between the percentage unsigned error in charge and physical properties of a molecule, we calculated the solvation energy using an implicit solvent model: Generalized Born with Molecular Volume (GBMV)30 for both the CGENFF and protein molecules with their original charge/parameters and their MATCH generated parameters. Figure 6A displays the correlation of solvation energy for CGENFF molecules using their topological charges and parameters compared to the ones calculated using the protein MATCH libraries. As mentioned prior there was a 16% unsigned error when calculating the atomic charges of the molecules found in CGENFF using the protein MATCH libraries, yet there remains a very strong correlation (R2 = 0.99) between the solvation energy calculated using both charging/parameterization schemes. It should be noted that there were two outliers GUAN and PHEO, in both cases the protein force field lacked the necessary chemical space to correctly calculate the formal charge. These were the first instances of observing this type of malfunction and appear to be very rare. An additional feature was added that allows the user to specify the formal charge of a molecule as a precaution to future occurrences. When removing these the average unsigned error in solvation energy is 2.2 kcal/mole. In Figure 6B the reverse case is examined, which is the calculation of the solvation energy of protein molecules with their native CHARMM charge and parameters compared to the CGENFF MATCH scheme of parameters. In this case the cross validation study yielded only a 6% error in the charges, consequently the average difference in solvation energy is around 0.6 kcal/mole with and R2 of 1.0, which is very promising.
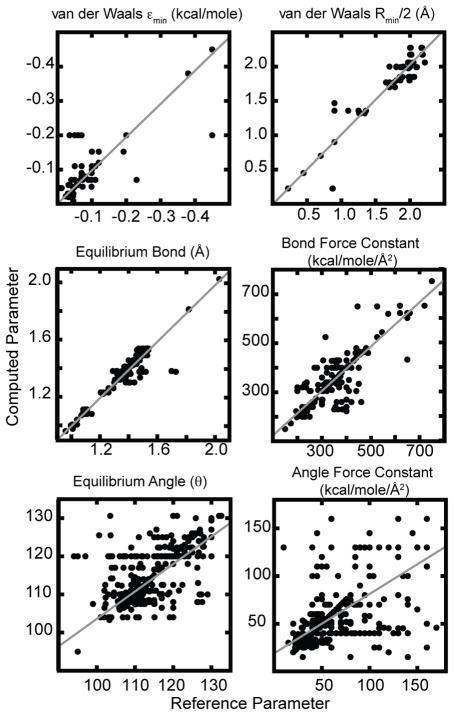
Cross Validation of Parameters
The quality of the parameters that are generated through this cross validation study are further investigated for the case in which the MATCH libraries from the protein force field are used to parameterize the CGENFF molecules. This combination requires the most extensive extrapolation of parameters from the MATCH libraries and, thus, should give a realistic scenario of our parametization procedure (Figure 7). The R2 correlation between the predicted and actual van der Waals well-depth (ε) and radius (Rmin) parameters are 0.79 and 0.96 respectively. It is not entirely surprising that there is a relative decrease in the correlation for the well-depth parameters compared to the radius due to the fact that the protein force field does not contain any nitrogen atoms within a 6 membered ring. Removal of the 50 data points that correspond to this nitrogen type results in an R2 of 0.91. The quality of the parameters is quite high for the equilibrium bond length and force constants with an R2 correlation of 0.98 and 0.87, respectively, and an average unsigned error of 0.72% and 5.4% respectively. The quality of the angle parameters show a deterioration with an R2 for equilibrium angle and force constant of 0.57 and 0.41 respectively and average unsigned errors of 1.9% and 20.9% respectively. The low error for equilibrium angles with the much higher error in the force constant suggest that the geometry is being reproduced but the rigidity of the angles is not reproduced with the same level of accuracy. For dihedral parameters it is more difficult to evaluate how similar two sets of dihedrals are to each other due to the possibility of multiple declarations of the same dihedral with different multiplicity values. Thus, we investigated how often there was the same number of declarations as this would be the major contributor to differences in behavior of the dihedrals. We find that 94.6% of the dihedrals shared the same number of declarations and of these 87% had the same multiplicity and 85% had the same multiplicity and identical reference angles. These results demonstrate that a large number of dihedral angles are correctly represented as taking part in the same number of declarations with the same multiplicity. This is an important observation because the dihedral terms largely determine the shape of the energy landscape of the small molecule with the force constants giving the height the energy barriers.
Figure 7.
Quality of the CGENFF force field parameters that were extrapolated from the protein force field libraries in MATCH.
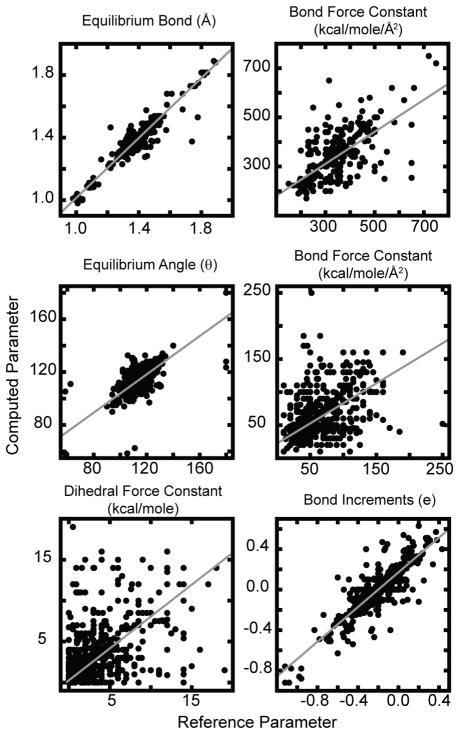
Parameter Substitution
To demonstrate that our atom-type substitution procedure is able to yield accurate results, we systematically removed one of the bond, angle or dihedral parameters within the CGENFF parameter file or a bond increment rule from the CGENFF-based MATCH libraries and identified the “best fit” or “nearest neighbor” parameter given the remaining parameters. Figure 8 summarizes the results for this leave-one-out substitution study on bond, angle, dihedral parameters and bond charge increment rules. Substitutions are not applicable to van der Waals parameters since CGENFF includes van der Waals parameters for each atom type. For equilibrium bond lengths and angles there is good agreement between the original and the substituted parameters with an R2 of 0.95 and 0.56, respectively, yielding average percentage errors of 1.6% and 2.4%, respectively. This ability to accurately preserve the geometry of a novel molecule without prior knowledge of the parameters is critical. It should be noted that for the equilibrium angles there is a small subset that contain infrequently used atom types. Due to their rarity, finding a suitable substitute does not exist in the CGENFF parameter set. Out of ~1500, about 20 fall into this category, which primarily include atoms that bridge fused rings. R2 greatly improves upon removal of these points. Indentifying these poor substitutions is currently being investigated.
Figure 8.
Quality of the parameter that was predicted from the “best fit” to the remaining parameters in the leave one out substitution study.
The results for substitutions involving the bond and angle force constants display a decrease in accuracy with an R2 of 0.51 and 0.34, respectively. However, correctly computing the force constants is of less importance than the equilibrium values as they impact the flexibility of the molecule, but not its lowest energy conformation. In many high-throughput drug design scenarios, a rough estimate of the force constants suffices in producing a reasonable parameter set for modeling the structure of a novel molecule. For researcher’s who plan to do extensive dynamic simulations using MATCH parameterized ligands, further validation may be required. As mentioned in the previous section, there is a difficultly in assessing similarity in dihedral parameters. We examined whether the substituted dihedral had the same number of declarations as the original value: 88% of the time the substituted dihedral shares the same number of declarations as the original. Of these 88%, 95% had the same multiplicity value and 92% had the same multiplicity and identical reference angles. Lastly, the bond-increment substitution is promising with an R2 of 0.76 and average unsigned error of 0.055 electron units. Not all parameters will be generated by substitutions, rather just the minority that are not explicitly defined in a given force field. This leave-one-out study along with the results from our cross-validation studies demonstrate that the atom type substitution strategy greatly increases the chemical space that a force field can cover by extrapolation or interpolation without a significant sacrifice in accuracy.
PubChem Database Screen
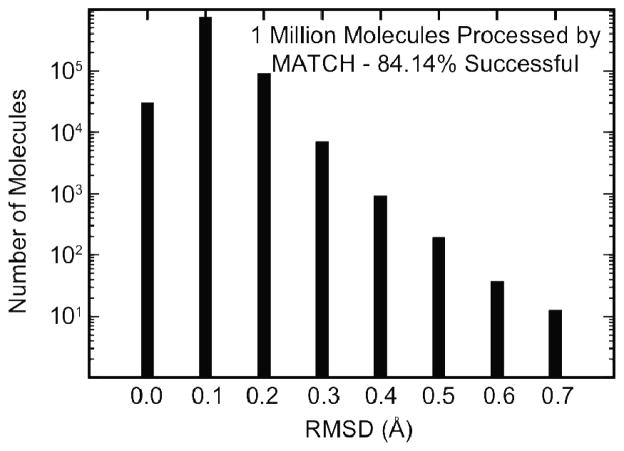
CGENFF is the most diverse CHARMM force field and provides the most extensive coverage of chemical space. As observed in the previous section it encompasses the chemical space that is spanned by all of the other CHARMM force fields. Here, we assess the ability of the CGENFF-specific MATCH libraries to generate topology and parameter files for one million drug-like molecules from the PubChem database and estimate the upper limit of the extensibility of the MATCH libraries based on the current generation of the CGENFF force field 2. The overall success rate is 84.14%, where success is defined as MATCH’s ability to generate CHARMM rtf and param files followed by minimization of the energy of the molecule with CHARMM. Only ~2 seconds were required to process each compound suggesting that MATCH can be incorporated into high-throughput drug design strategies. Additionally, each molecule only has to be processed once to be included in any number of molecular mechanics calculations. Furthermore, as illustrated by the results summarized in Figure 9, the quality of the final minimized structures is very high. After energy minimization, all molecules are within 0.75 Å RMSD of their initial pdb structure. The PubChem pdb structures are not necessarily crystallographic structures; therefore, reproducing the exact PubChem structure is not a goal of this exercise, but rather the reasonably low RMSD indicates that the geometry of the molecule is retained when the structure is minimized with the parameters assigned from MATCH parameterization based on the CGENFF libraries Although these are encouraging results, it is important to investigate why some of the molecules we examined were not successfully processed by MATCH. Because we removed all molecules containing elements not represented in the CGENFF force field, all atoms will be typed. This is clear from the innate hierarchy for the CGENFF atom types. However, due to the large number of types, there are many combinations that do not have known bond charge increment rules so no satisfactory substitution increment rule is identified. Also, corresponding bond, angle and dihedral parameter substitutions may not be identified. It is possible to increase the combination of allowed bond increments (and other parameter substitutions) by allowing a less strict substitution criterion for unknown increments (and parameters). However, this generally leads to overall lower quality parameterization (results not shown). A more practical approach would be to increase the chemical space of the bond increment rules (and parameters) by indentifying distinct model compounds or fragments that lie outside of the space encompassed by the CGENFF libraries, determine the associated charge distributions from quantum chemical calculations2 and then construct the additional bond charge increment rules, as well as other force field parametes. Future goals are to census the entire PubChem database for drug-like molecules and look for the chemical space that is prevalent yet is not included in the CGENFF-specific MATCH libraries. Learning the BCIs and parameters for a select number of new chemical groups will greatly expand the total chemical space covered by MATCH.
Figure 9. Quality of the minimized structures for the.
PubChem drug-like molecules that were successfully processed using the CGENFF libraries within MATCH to generate their respective topology and parameter files. RMSD was computed by comparing conformations found in the PubChem database to the ones after minimization.
Conclusion
We have presented a library of functions and data structures, collectively called MATCH, which is designed to facilitate the automated selection of appropriate atom types, partial charges, and molecular parameters for common molecular mechanics force fields. The toolset is customizable and extensible, such that it can act both as a solution for extrapolating and interpolating from the known chemical space to novel molecules and as a tool to study the effects of specific parameter choices and parameterization strategies. Through cross validation studies we have shown that it is possible to accurately replicate atomic charges and parameters using rules derived from another force field. This strategy has significant potential; however, the ability of MATCH to successfully generate the new parameter and topology files and the quality of the results are directly dependent on the chemical diversity that exists in the original force field topology file that is used to generate the force-field specific MATCH libraries. Given the ability of the CGENFF-derived MATCH libraries to construct physically meaningful parameters and partial charge assignments for 84% of the randomly selected drug-like compounds in the PubChem Database, MATCH with its current CHARMM-based libraries is a promising tool for high-throughput drug design applications based on the biomolecular CHARMM force field.
Future work will focus on the development of an automated procedure for generating the molecular fragments of atom types and the development of a measure of the quality of both the atomic charges and parameters to understand when a substitution of a parameter or a bond increment is likely to be too detrimental to be included. In this study we actively participated in defining the molecular fragments to ensure that the simplest representation of an atom type was generated. Automated procedures were investigated, but ultimately they produced suboptimal results compared with strategies that incorporated expert knowledge. With further research, the automated fragment generation feature will enable the MATCH strategy to be even more generalizable and facilitate the seamless integration of additional force field topology files into force-field specific MATCH libraries. Finally, MATCH parameter extrapolation and interpolation can be useful in identifying the shortcomings of a particular force field and focus optimization efforts on the parameters of specific chemical groups that can lead to the greatest improvement in overall quality of charges and parameters.
Supplementary Material
Acknowledgments
This work was supported by the NIH (Grant GM037554). We are grateful to lab members for all their thoughtful discussion.
References
- 1.Reddy AS, Pati SP, Kumar PP, Pradeep HN, Sastry GN. Curr Protein Pept Sc. 2007;8(4):329–351. doi: 10.2174/138920307781369427. [DOI] [PubMed] [Google Scholar]
- 2.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 4.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Journal of Computational Chemistry. 1983;4(2):187–217. [Google Scholar]
- 5.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. Journal of Computational Chemistry. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. J Am Chem Soc. 1984;106(3):765–784. [Google Scholar]
- 7.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1996;118(9):2309–2309. [Google Scholar]
- 8.Jorgensen WL, Maxwell DS, TiradoRives J. J Am Chem Soc. 1996;118(45):11225–11236. [Google Scholar]
- 9.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Curr Opin Struc Biol. 2009;19(2):120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 10.MacKerell AD, Nilsson L. Curr Opin Struc Biol. 2008;18(2):194–199. doi: 10.1016/j.sbi.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackerell AD. Journal of Computational Chemistry. 2004;25(13):1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 12.Halgren TA. Journal of Computational Chemistry. 1996;17(5–6):616–641. [Google Scholar]
- 13.Wang JM, Wang W, Kollman PA, Case DA. J Mol Graph Model. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Halgren TA, Bush BL. Abstr Pap Am Chem S. 1996;212:2-COMP. [Google Scholar]
- 15.Murphy RB, Philipp DM, Friesner RA. J Comput Chem. 2000;21(16):1442–1457. [Google Scholar]
- 16.Schuttelkopf AW, van Aalten DMF. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 17.Kleywegt GJ. Acta Crystallogr D. 2007;63:94–100. doi: 10.1107/S0907444906022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF. J Comput Chem. 2004;25(13):1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 19.Downs GM, Gillet VJ, Holliday JD, Lynch MF. J Chem Inf Comp Sci. 1989;29(3):172–187. [Google Scholar]
- 20.Holliday JD, Downs GM, Gillet VJ, Lynch MF. J Chem Inf Comp Sci. 1992;32(5):453–462. [Google Scholar]
- 21.Welford SM, Lynch MF, Barnard JM. J Chem Inf Comp Sci. 1984;24(2):57–66. [Google Scholar]
- 22.Gillet VJ, Welford SM, Lynch MF, Willett P, Barnard JM, Downs GM, Manson G, Thompson J. J Chem Inf Comp Sci. 1986;26(3):118–126. [Google Scholar]
- 23.Holliday JD, Downs GM, Gillet VJ, Lynch MF. J Chem Inf Comp Sci. 1993;33(3):369–377. [Google Scholar]
- 24.Bolton EE, Wang Y, Thiessen PA, Bryant SH. In: Annual Reports in Computational Chemistry. Wheeler RA, Spellmeyer DC, editors. Elsevier; 2008. pp. 217–241. [Google Scholar]
- 25.Downs GM, Gillet VJ, Holliday JD, Lynch MF. J Chem Inf Comp Sci. 1989;29(3):215–224. [Google Scholar]
- 26.Moore EF. The shortest path through a maze. 1959 [Google Scholar]
- 27.Schreiber SL. Science. 2000;287(5460):1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 28.Tiernan JC. Commun Acm. 1970;13(12):722. [Google Scholar]
- 29.Bone RGA, Firth MA, Sykes RA. J Chem Inf Comp Sci. 1999;39(5):846–860. [Google Scholar]
- 30.Lee MS, Salsbury FR, Brooks CL., III J Chem Phys. 2002;116(24):10606–10614. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.