Summary
Dominant mutation in two DNA/RNA binding proteins, TDP-43 and FUS/TLS, are causes of inherited Amyotrophic Lateral Sclerosis (ALS). TDP-43 and FUS/TLS have striking structural and functional similarities, implicating alterations in RNA processing as central in ALS. TDP-43 has binding sites within a third of all mouse and human mRNAs in brain and this binding influences the levels and splicing patterns of at least 20% of those mRNAs. Disease modeling in rodents of the first known cause of inherited ALS – mutation in the ubiquitously expressed superoxide dismutase (SOD1) – has yielded non-cell autonomous fatal motor neuron disease caused by one or more toxic properties acquired by the mutant proteins. In contrast, initial disease modeling for TDP-43 and FUS/TLS has produced highly varied phenotypes. It remains unsettled whether TDP-43 and FUS/TLS mutants provoke disease from a loss of function or gain of toxicity or both. TDP-43 or FUS/TLS misaccumulation seems central not just to ALS (where it is found in almost all instances of disease), but more broadly in neurodegenerative disease, including frontal temporal lobular dementia (FTLD-U) and many examples of Alzheimer’s or Huntington’s disease. (182 words)
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive adult-onset neurodegenerative disorder that leads to paralysis and ultimately death within 2–5 years. The disease is characterized by the selective loss of motor neurons in the brain and spinal cord leading to fatal paralysis. Most cases of ALS are sporadic, but 10% are inherited in a dominant manner (familial ALS). Mutations in eight genes have now been identified to cause typical ALS (Table 1). The most common cause of inherited ALS, which accounts for twenty percent of the familial cases, has been shown to be associated with missense mutations in the gene encoding cytoplasmic Cu/Zn superoxide dismutase (SOD1), which is involved in intracellular detoxification of superoxide. Much work on transgenic mice constitutively expressing ALS-linked mutations in SOD1, which develop late onset motor neuron death and muscle atrophy like that seen in ALS, has led to two pivotal discoveries in mutant SOD1 mediated toxicity: (1) death of motor neurons is not due to a loss of SOD1 activity since mice expressing dismutase inactive mutant SOD1 develop ALS like symptoms [1] and (2) mutant SOD1 synthesis within the motor neurons contributes to disease onset whereas damage within the glial cells expressing mutant SOD1 accelerates disease progression [2–5]. With nine prominent hypotheses emerging for how SOD1 mutations lead to selective death of motor neurons (reviewed in [6]), it seems likely now that for SOD1 mutants, ALS pathogenesis may come from the convergence of several toxic pathways acting in different cell types.
Table 1.
Genetics of human ALS.
| Locus | Gene | Protein | Mutations | Proportion of inherited ALS |
Discovery date | Reference |
|---|---|---|---|---|---|---|
| 21q22.1 | SOD1 | Cu/Zn superoxide dismutase | >150* | 20% | 1993 | [65] |
| 9p13.2–21.3 | Unknown | Unknown | Unknown | 20%** | Unknown | [66–68] |
| 1q36 | TARDBP | TDP-43 | >40* | 5% | 2008 | [9,10••,11••] |
| 16p11.2 | FUS | FUS/TLS | >40* | 4% | 2009 | [13••,14••] |
| 9p13.3 | VCP | Valosin-containing protein | 5 | 1–2%+ | 2010 | [70] |
| 10p15-p14 | OPTN | Optineurin | 1 | 1–2%+ | 2010 | [71] |
| 6q21 | FIG4 | PI(3,5)P(2)5-phosphatase | 5* | 1%+ | 2009 | [72] |
| 12q24 | DAO | D-amino acid oxidase | 1 | 1%+ | 2010 | [73] |
| 14q11 | ANG | Angiogenin | >10* | <1% | 2006 | [74] |
Only classical adult onset ALS cases with autosomal dominant inheritance are included. Each gene, except for SOD1, accounts for <1% of all (familial + sporadic) ALS cases.
Including sporadic ALS cases.
The largest remaining cohort of inherited ALS, estimates of proportion of inherited ALS are up to 20% (reported to account for 38% of familial ALS in Finland [69]).
To date, reported only in a single publication.
TDP-43 and FUS/TLS: a paradigm shift in ALS
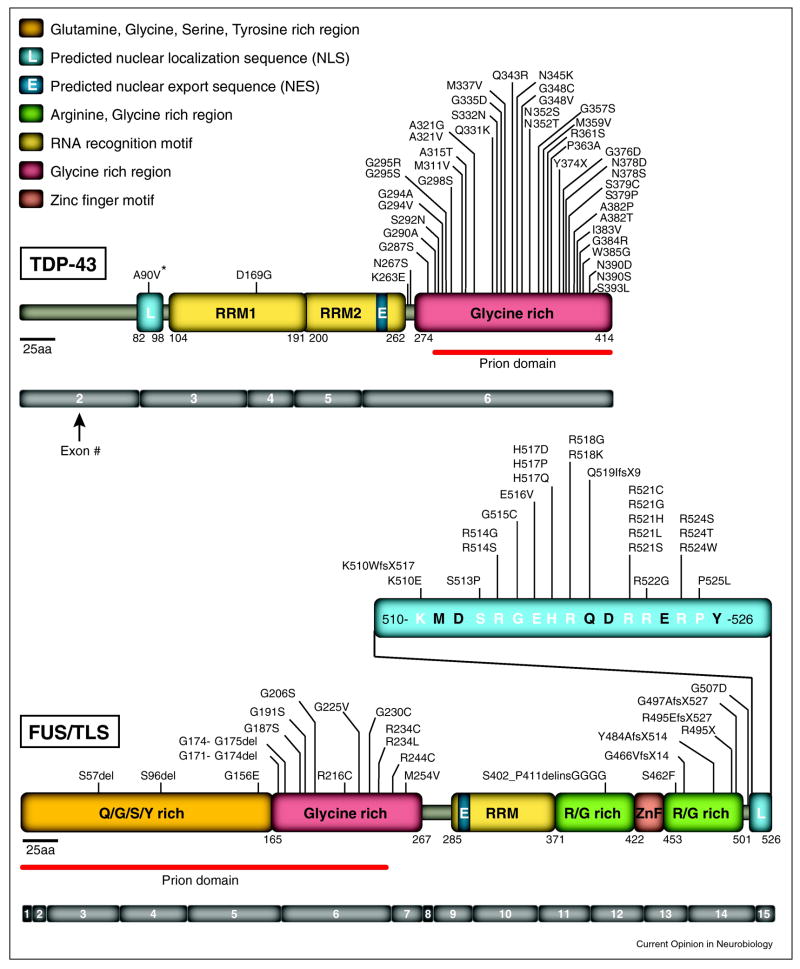
In 2006 [7,8], the beginnings of a paradigm shift in ALS emerged with identification of the 43 kDa transactive response DNA-binding protein (TDP-43) as a major component of ubiquitinated protein aggregates found in the central nervous system (CNS) of patients with sporadic ALS or in the most common form of frontotemporal dementia called FTLD-U (Frontotemporal Lobar Degeneration with Ubiquitinated inclusions). This quickly prompted direct sequencing of the gene encoding TDP-43 (TARDBP) as a means to identify mutations in TDP-43 as causative of cases of sporadic and familial ALS, as well as rare cases of FTLD [9–11]. To date, at least 44 mutations in TDP-43 (Figure 1) now account for ~5% of ALS (combining familial and sporadic ALS cases) [12]. Rapidly thereafter, mutations in another RNA/DNA binding protein, known by two names (Fused in Sarcoma or Translocated in liposarcoma; referred to here as FUS/TLS), were identified as a primary contributor to familial ALS [13,14]. A total of 43 mutations (Figure 1) have now been described in patients with or without apparent familial history, corresponding to ~4% of familial (and rare sporadic) ALS cases [12].
Figure 1. TDP-43 and FUS/TLS mutations in ALS and FTLD patients.
(Upper panel) Forty-four mutations have been identified in TDP-43 in sporadic and familial ALS patients and in rare FTLD patients, with most lying in the C-terminal glycine-rich region. The putative prion domain comprises amino acids from 277–414. (Lower panel) Forty-three mutations have been identified in FUS/TLS in familial and sporadic ALS cases and in rare FTLD patients. Most mutations are clustered in the last 17 amino acids and in the glycine-rich region and the putative prion domain comprises amino acids 1–239.
TDP-43 and FUS/TLS proteinopathies
Although mutations in TDP-43 and FUS/TLS have been reported only in ALS and FTLD patients, TDP-43 and FUS/TLS immunoreactive inclusions have been observed in the nucleus and cytoplasm of both neurons and glial cells. This is true not just in ALS and FTLD, but also in a range of other neurodegenerative diseases, including Huntington’s, Alzheimer’s and Parkinson’s disease [15]. The widespread presence of TDP-43 or FUS/TLS inclusions in so many disorders has led to a new nomenclature for the set of such diseases: TDP-43 or FUS/TLS proteinopathies [12,16].
A striking characteristic of TDP-43 pathology that is now well established in ALS, as well as in other neurodegenerative diseases (although the frequency and when it occurs during disease course remains unclear), is the nuclear clearance of TDP-43 concomitantly with its cytoplasmic mislocalization [12]. Pathogenesis is thus likely driven, at least in part, by loss of normal nuclear TDP-43 function(s). Although FUS/TLS is also a predominantly nuclear protein and is structurally close to TDP-43 (Figure 1), the redistribution of FUS/TLS from the nucleus to the cytoplasm is apparently less complete [12,17]. FUS/TLS and TDP-43 proteinopathies also differ by the absence in FUS/TLS of most of the post-translational modifications (such as hyperphosphorylation, ubiquitination and cleavage) that have been reported for TDP-43 [15,18]. Thus, if common mechanisms underlie toxicity mediated by TDP-43 and FUS/TLS, this divergence in the biochemical properties of the proteins may suggest that these alterations are not key contributors to pathogenesis.
Identifying TDP-43 RNA targets
Immunoprecipitation of RNAs interacting with TDP-43, followed by high-throughput sequencing has now been reported by three groups [19–21]. By crosslinking the RNAs to their bound proteins prior to immunoprecipitation (known as CLIP-seq or a modified version called iCLIP), two teams identified TDP-43 binding to more than third of all mouse [19] and probably an equivalent number of human [21] brain mRNAs. Immunoprecipitation of TDP-43 without crosslinking (known as RIP-seq, a technique that allows the identification of RNA binding partners but not the binding sites) also identified more than 4,000 RNAs bound by TDP-43 in primary cortical rat cells [20]. Finally, using conventional cloning and sequencing approaches [22], TDP-43 immunoprecipitation after UV-crosslinking in human neuroblastoma cell lines has led to the identification of >120 target RNAs. A comprehensive comparison of these newly reported data sets to identify conserved TDP-43 RNA targets is now eagerly anticipated.
A consensus has emerged from in vitro [20,22,23] and in vivo [19,21] approaches that the primary in vivo binding site for TDP-43 is a GU-rich motif, corroborating in vitro findings from a decade earlier [24]. Nevertheless, GU-rich motifs are neither necessary nor sufficient for TDP-43 binding [19]. Most of the TDP-43 binding sites lie deep within introns – far from splice junctions [19–22], many of which are downstream of silenced exons [21]. A broad influence of TDP-43 on mature mRNA levels and splicing was established following depletion of TDP-43 either by siRNA in cell culture [21] or from the adult mouse nervous system (by infusion of a TDP-43 complementary antisense oligonucleotide) [19]. The combination of high-throughput sequencing and splicing-sensitive microarrays has identified changes in abundance of >600 mRNAs and altered splicing patterns of 965 mRNAs in the adult mouse brain upon depletion of TDP-43 [19].
Among the mRNAs whose levels or processing is affected by TDP-43 in both mouse [19] and human [21] settings are those encoding proteins related to neuronal function and development and/or that have been implicated in neurological diseases. Indeed, RNAs whose levels are most depleted by reduction of TDP-43 are derived from genes with the longest introns (average size >100 kb) and that encode proteins involved in synaptic activity (including a N-methyl-D-aspartate receptor, an ionotropic glutamate receptor, and neurexins 1 and 3) [19]. Since genes with significantly longer introns are preferentially expressed in human and mouse nervous systems relative to other tissues, this requirement provides at least one component of selective vulnerability to neurons from the loss of nuclear TDP-43 that has been universally reported in ALS.
TDP-43 also binds and/or regulates expression and splicing patterns of several additional disease-related pre-mRNAs [19–21] including those encoding FUS/TLS, progranulin, and tau (mutations in which cause ALS and/or FTD), parkin and MEF2D (which are involved in Parkinson’s disease), as well as huntingtin and the ataxins. The last of these RNA targets has become of high interest with report that an intermediate length CAG expansion in ataxin 2 may be a contributor to ALS [25]. Among the many now identified TDP-43 target mRNAs are two previously implicated: histone deacetylase 6 (HDAC6), [26,27]) and the NF-L neurofilament subunit [28], both of which are reduced in motor neurons of ALS patients. TDP-43 neither binds to nor does its reduction affect the level or splicing of the mRNA encoding superoxide dismutase (SOD1) in mice [19] or humans [29].
Overall, TDP-43 binds to and affects procession of a very long list of RNA targets with it probably acting in multiple steps of RNA processing.
TDP-43 RNA targets in FTLD and ALS
iCLIP and RNA sequencing analyses directly from brains of FTLD patients identified significantly increased expression of, and therefore increased TDP-43 binding to two non-coding RNAs: the nuclear paraspeckle assembly transcript 1 (NEAT1) and the metastasis associated lung carcinoma (MALAT1)), both of which are themselves thought to function in splicing and regulation of gene expression). In contrast, expression of transcripts involved in synaptic activity such as neurexin 3 (NRXN3) and the glial excitatory amino acid transporter -2 (EAAT2), accompanied by diminished TDP-43 binding, were significantly decreased in FTLD brains compared to those from healthy patients [21]. Comparison of TDP-43-RNA complexes in nuclear versus cytoplasmic fractions of healthy and FTLD brain tissues revealed that more than 90% of TDP-43 interactions were with nuclear pre-RNAs both in healthy and FTLD patients. Similarly, isolation of spinal cord motor neurons from sporadic ALS patients (by laser capture microdissection) coupled with genome exon splicing arrays has revealed alterations in expression and splicing of hundreds of transcripts, albeit no significant modifications in the mRNAs encoding TDP-43 or FUS/TLS [30].
Auto-regulation of TDP-43 synthesis
Multiple teams have identified an auto-regulatory mechanism that determines the level of TDP-43 synthesis: TDP-43 controls its own expression at least in part by direct binding to the 3′untranslated region (UTR) of its own RNA transcript [19,21,23]. Auto-regulation has been documented not only in cell culture [19,21,23] but also in mice, with expression of a TDP-43-encoding transgene without the regulatory 3′UTR driving significant reduction of endogenous TDP-43 mRNA and protein [19,31,32,48]. The detailed mechanism underlying auto-regulation is unsettled. One team has argued that TDP-43 auto-regulates its synthesis, at least in part by directly binding to and enhancing splicing of an intron in the 3′ UTR of its own transcript, thereby triggering nonsense-mediated RNA degradation of the spliced mRNA [19]. Another group has argued that TDP-43 binding to its own 3′UTR promotes RNA instability and degradation by the exosome machinery [23]. Regardless, self-regulation of TDP-43 expression seems all but certain to represent an important contributor to ALS pathogenesis following an initiating event that yields reduction in nuclear TDP-43 levels (by mutation and/or cytoplasmic aggregation), thereby driving elevated TDP-43 synthesis.
FUS/TLS RNA targets
To date only few FUS/TLS RNA binding partners have been reported and a sequence binding motif(s) has not been identified. RNAs apparently bound include actin-stabilizing protein (Nd1-L), thought to play a role in actin reorganization in spines [33] and HDAC6, which has also been shown to be bound by TDP-43 [27], thus suggesting that both FUS/TLS and TDP-43 proteins can function in common biological pathways. Use of high-throughput approaches (such as those described above for TDP-43) is now essential to identify the spectrum of FUS/TLS RNA targets and establish normal function of FUS/TLS.
Identifying TDP-43 and FUS/TLS protein partners
Various interacting proteins have been reported for either TDP-43 or FUS/TLS or both (as reviewed in [15]). Most of these have been found by coupling immunoprecipitation of normal or ALS-linked mutants of TDP-43 to mass spectrometry. Despite using different starting biological tissues, numerous hnRNPs, splicing factors and RNA binding proteins have been consistently identified as TDP-43 binding partners [18,20,27,34,35]. In addition to previous identification of Drosha as a TDP-43 binding partner [36], several other components involved in microRNA processing were established as TDP-43 interacting proteins using two-step tandem-affinity purification coupled to quantitative mass spectrometry analysis [35] (using stable isotope labeling amino acids in cell culture or SILAC, an approach that allows identification of proteins of low abundance while eliminating abundant contaminants [37]).
Interaction of TDP-43 and FUS/TLS
The most striking finding of the search for TDP-43 partners is association between FUS/TLS and TDP-43. This was detected in human cell lines expressing either transiently [27,34] or stably [35] a tagged form of wild-type TDP-43 and further validated for proteins expressed at endogenous levels [27,35]. TDP-43 and FUS/TLS were found to be present in two major high molecular weight complexes: a smaller complex (of 300–400 KDa) which was enriched in nuclei and a higher molecular weight (>1 MDa) complex in the cytoplasmic fraction [27]. Proteomic analysis of the higher molecular weight complex revealed the presence of multiple ribosome and translation factors [34]. No consensus has yet emerged about the association between FUS/TLS and ALS-linked TDP-43 mutants [27,35]. While FUS/TLS binding was found to be significantly enhanced by two TDP-43 mutants (Q331K and M337V) in isogenic HeLa cell lines expressing a GFP fused transgene at near endogenous levels [35], no clear evidence for increased interactions of TDP-43 mutants (M337V, A315T, D169G and R361S) to FUS/TLS was observed in transiently transfected HeLa cells expressing a HA tagged transgene at high levels [27]. The proposed increased association of mutants of TDP-43 with FUS/TLS may be due to the surprising observation that the stability of the mutants is significantly enhanced compared to unmutated TDP-43, with mutant protein half-lives twice that of wild-type [35]. This striking finding may at least in part account for the accumulation of TDP-43 aggregation reported in ALS patients. Furthermore, if an increased interaction of FUS/TLS with TDP-43 ALS-linked mutants is confirmed in a more disease-relevant setting such as autopsied ALS brain tissues, it will suggest that this aberrant association may generate alterations of the normal function(s) of both TDP-43 and FUS/TLS, thus indicating a possible convergence of pathogenic pathways by both proteins in ALS. It will now be of high interest not only to identify the interacting partners of FUS/TLS, but also to quantitatively compare complexes containing normal TDP-43 or FUS/TLS versus ALS-linked mutants of both proteins.
Animal modeling of TDP-43 and/or FUS/TLS mediated toxicity
Multiple attempts to model ALS-like disease in fruit flies, worms, zebrafish, mice and rats from altered TDP-43 levels or mutation have now been reported [32,38–50] (Table 2). Two have been reported for FUS/TLS [51,52]. Despite the many modeling studies, no consensus has emerged on two key mechanistic questions concerning mutant TDP-43 mediated toxicity: 1) is toxicity from a gain of toxic property(ies), loss of function, or both and 2) in which cell types does the mutant act to drive toxicity (i.e., is the disease mechanism non-cell autonomous)?
Table 2.
Summary of genetic mimics of TDP-43 and FUS/TLS in disease.
| Species | Promoter | Transgene | Mutations | # lines | Fold over expression |
Phenotype | Death (days) |
Overall pathology | Loss of endogenous TDP-43 |
Nuclear clearance |
Aggregation | CTFs of TDP-43 |
Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. elegans | Synaptobre-vin (snb-1) | hTDP-43 cDNA | WT | 1 | n.r. | Unco-ordinated movement | n.r. | Abnormal MN synapses | n.r. | n.r. | n.r. | n.r. | Ash et al., 2010 [38•] |
| ΔRRM1 or −2 mutants | 1 | n.r. | Normal | n.r. | None | n.r. | n.r. | n.r. | n.r. | ||||
|
| |||||||||||||
| C. elegans | Synaptobre- vin (snb-1) | hTDP-43 cDNA | WT | 2 | n.r. |
|
None | None | n.r. | n.r. | NII TDP-43+ | Yes | Liachko et al., 2010 [43•] |
| G290A | 1 | n.r. |
|
Reduced lifespan | Degeneration of neurons in all the mutants | n.r. | n.r. | NII TDP-43+ | Yes | ||||
| A315T | 1 | n.r. | |||||||||||
| M337V | 1 | n.r. | |||||||||||
|
| |||||||||||||
| Drosophila | GMR-Gal4 (eye) D42-Gal4 (MN) | hTDP-43 cDNA | WT | 1 | n.r. | Eye depigmentation | n.r. | Degeneration of photoreceptor cells | n.r. | n.r. | Ubiquitination | n.r. | Hanson et al., 2010 [39•] |
| WT | 3 | n.r. |
|
Reduced lifespan (50%) | n.r. | n.r. | n.r. | PhosphoTD P-43 localization | No | ||||
|
| |||||||||||||
| Drosophila | GMR-Gal4 | hTDP-43 cDNA | WT | 1 | n.r. | Abnormal eye morphology | n.r. | Degeneration of photoreceptor cells | n.r. | n.r. | n.r. | n.r. | Li et al., 2010 [42•] |
| OK107-Gal4 (MB) or OK371- Gal4 (MN) | ΔNLS+ RRM1- mutant | 1 | n.r. | Normal | n.r. | None | n.r. | n.r. | n.r. | n.r. | |||
| WT | 1 | n.r. | Reduced movement | n.r. | MN loss Aggregates in cell bodies and axons Reduced NMJ | n.r. | n.r. | Rare cyto TDP-43 localization | n.r. | ||||
|
| |||||||||||||
| Drosophila | GMR-Gal4 | hTDP-43 cDNA | WT | 5 | n.r. | Modest abnormalities in eyes | n.r. | n.r. | n.r. | n.r. | n.r. | No | Ritson et al., 2010 [45•] |
| M337V | 6 | n.r. | Modest to severe abnormalities in eyes | Degeneration of photoreceptor cells | n.r. | Yes | |||||||
| ΔNLS-Mut | 6 | n.r. | Severe abnormalities in eyes | Severe degeneration of photoreceptor cells | Cyto TDP-43 localization | No | |||||||
| ΔNES-mut | 5 | n.r. | Normal | ||||||||||
|
| |||||||||||||
| Drosophila | Gal4 221 (sensory neurons) | hTDP-43 cDNA | WT Q331K M337V | n.r. | n.r. | n.r. | n.r. | Increased number of small terminal dendritic branches (higher in WT) | n.r. | n.r. | n.r. | n.r. | Lu et al., 2009 [54•] |
|
| |||||||||||||
| Drosophila | GMR-Gal4 | hTDP-43 cDNA | WT | 1 | n.r. | Mild rough eye | n.r. | Loss of pigmentation | n.r. | n.r. | Nuclear and cyto TDP-43 | n.r. | Miguel et al., 2010 [55•] |
| Elav-Gal4GS (neurons) | ΔNLS-Mut | 1 | Severe rough eye | Severe loss of pigmentation | Cyto TDP-43 | n.r. | |||||||
| ΔNES-mut | 1 | Normal | None | TDP-43 + nuclear foci | n.r. | ||||||||
| WT | 1 | Reduced lifespan | 16 | Yes | |||||||||
| ΔNLS-Mut | 1 | Reduced lifespan | 21 | Yes | |||||||||
| ΔNES-mut | 1 | Reduced lifespan | 25 (/47) | Yes | |||||||||
|
| |||||||||||||
| Zebrafish | n.a. | hTDP-43 mRNA | WT | n.a. | n.r. | Motor defects | n.r. |
|
n.r. | n.r. | Nuclear TDP-43 | n.r. | Laird et al., 2010 [41•] |
| A315T | n.a. | n.r. | Motor defects | n.r. |
|
||||||||
|
| |||||||||||||
| Zebrafish | n.a. | hTDP-43 mRNA | WT A315T | n.a. | n.r. | Mild swimming deficits | n.r. |
|
n.r. | n.r. | n.r. | n.r. | Kabashi et al., 2010 [40•] |
| A382T G348C | n.a. | n.r. | Swimming deficits | n.r. |
|
||||||||
|
| |||||||||||||
| Zebrafish | n.a. | hFUS/TLS mRNAs GFP tag | WT H517Q R521G | n.a. | n.r. | None | n.a. | None | n.r. | n.r. | Nuclear TDP-43 | n.r. | Bosco et al., 2010 [51•] |
| R495X G515X | n.a. | n.r. | n.r. | n.r. | n.r. | n.r. | Cyto TDP-43 | n.r. | |||||
|
| |||||||||||||
| Mice (C57Bl6/JxCBA, bred with C57Bl6/J F1+F2 | Prnp | hTDP-43 cDNA Flag tag | A315T | 1 het (/8 F0) | 3× |
|
155 |
|
Yes | Yes | NCI Ubi+, TDP-43− | Yes | Wegorzewska et al., 2009 [48••] |
|
| |||||||||||||
| Mice (Bl6/SJL, bred with C57Bl6/J) | Thy1 | hTDP-43 cDNA | WT | 2 het 2 homo (/23 F0) |
<1× <2× |
Normal
|
n.a. 24–200 |
None
|
n.r. n.r. |
No Yes |
No Rare NII and NCI Ubi+, TDP-43+ |
No Yes |
Wils et al., 2010 [49••] |
|
| |||||||||||||
| Mice (FVB/N) | CaMKII | mTDP-43 cDNA | WT | 3 homo (/10 F0) | 2× |
|
495 (/632 in non tg) |
|
n.r. | Rare | NCI Ubi+, TDP-43+ | Yes | Tsai et al., 2010 [47•] |
|
| |||||||||||||
| Mice (B6SJLF1, bred with CD1 or mixed bkg) | Prnp | hTDP-43 cDNA | WT | 2 het (/12) | 3× | Normal | n.a. |
|
n.r. | No | Diffuse ubi+ neurons | No | Stallings et al., 2010 [46••] |
| A315T | 3 het (/8 F0) | 4× |
|
75 |
|
n.r. | n.r. | NCI Ubi+, TDP-43+ | Yes | ||||
| M337V | 8 F0 | >10× |
|
16–42 |
|
n.r. | n.r. | NCI Ubi+, TDP-43+ | Yes | ||||
|
| |||||||||||||
| Mice (C57BL/6) | Prnp | hTDP-43 cDNA | WT | 1 het | 1.9× | Normal | n.a. | None | Yes | n.r. | n.r. | Yes | Xu et al., 2010 [32••] |
| 1 homo (/6 F0) | 2.5× |
|
30–60 |
|
Yes | rare |
|
Yes | |||||
|
| |||||||||||||
| Mice (C57BL/6;SJL, bred with C57BL/6) | Thy1 | hTDP-43 cDNA | WT | 1 het (/3 F0) | 2.3–4.6× |
|
Normal (1 line was lethal) |
|
n.r. | n.r. |
|
No | Shan et al., 2010 [45•] |
|
| |||||||||||||
| Mice (C57BL/6JxC3HeJ) | CaMKII-tTA | tetO-hTDP-43 cDNA (Tet-off inducible) | WT | 1 het (/4 F0) | 1.7× | Abnormal limb clasping (1w*) | n.r. |
|
Yes | Yes (mTDP-43) | Rare NCI ubi+, TDP-43+ | No | Igaz et al., 2010 [31••] |
| (NLS mutant | 1 het (/2 F0) | 8–9× | Abnormal limb clasping (1–3 mo*) | n.r. |
|
Yes | Yes (mTDP-43) | Rare NCI ubi+, TDP-43+ | No | ||||
|
| |||||||||||||
| Rat (Sprague- Dawley) | TARDBP | 22kb hTDP-43 gene | WT | 2 het | 2–4× | Normal | None | None | n.r. | n.r. | 3 Cyto Phospho- TDP-43 | Yes | Zhou et al., 2010 [50••] |
| CAG-tTA | TRE-miniCMV- hTDP-43 | M337V | 3 F0 | 3–6× | Loss of mobility | 10–29 | n.r. | n.r. | n.r. | n.r. | n.r. | ||
| cDNA (Tet-off inducible) | M337V | 2 het | 2–4× |
|
35–49 |
|
n.r. | n.r. |
|
Yes | |||
|
| |||||||||||||
| Rat (Sprague- Dawley) | CAG-tTA | TRE-miniCMV- hFUS cDNA (Tet-off inducible) | WT | 1 het (/12 F0) | 3–6× |
|
None |
|
n.r. | None | Ubi +, FUS− | n.r. | Huang et al., 2011 [52••] |
| R521C | 2 het (/14 F0) | 3–6× |
|
30–70 |
|
n.r. | None | Ubi +, FUS− | n.r. | ||||
Fold overexpression of transgene above endogenous levels assessed by qRT-PCR either in cortex or spinal cord; CTFs: C-terminal fragments; n.r.: not reported; n.a.: not applicable; NLS: Nuclear localization signal; NES: Nuclear export signal; RRM1: RNA Recognition Motif; SC: Spinal Cord; Het: Heterozygous; Homo: Homozygous; F0: Founder; Ubi+: Ubiquitin positive; NII: Neuronal Intranuclear Inclusions; NCI: Neuronal Cytoplasmic Inclusions; w: weeks; mo: months; d: days; NMJ: Neuromuscular junction; MN: motor neurons; SkM: Skeletal muscle; DCT: Dorsal cortical tract; LC: Lateral column; CL5: Cortical Layer 5; DG: Dentate Gyrus; MB: Mushroom Bodies; hTDP-43: human TDP-43; mTDP-43: mouse TDP-43; WT: Wild-type.
Disease modeling in Drosophila, C. elegans and zebrafish
Drosophila, Caenorhabditis elegans (C. elegans) and zebrafish all have a homologue of TDP-43. Since these models have been proved to be very powerful genetic tools to study disease mechanisms, a number of studies have reported the effects of TDP-43 disruption or elevated expression of either wild-type or mutants of human TDP-43 in these organisms.
Loss of TDP-43
Complete loss of TDP-43 function produced by either partial or complete deletion of the Drosophila homologue TBPH and down-regulation of TBPH (using small interference RNAs or siRNA) is deleterious in flies, leading to semi-lethal [42,53,54] or lethal [26] phenotypes. The surviving null flies display defects in locomotive behavior, neuromuscular junctions, reduced lifespan [53], reduced dendritic branching [54] as well as axonal loss and neuronal death [42]. Similar knock-down of TDP-43 in zebrafish using antisense morpholino oligonucleotides causes a motor phenotype consisting of shorter motor axons, excessive disorganized branching as well as swimming deficits but without clear lethality [40]. Interestingly, the motor abnormalities observed in both flies and zebrafish can be partially rescued by wild-type human TDP-43 [40,53]. Altogether these findings highlight how TDP-43 is essential both in invertebrates and vertebrates.
Increased levels of wild-type or mutant TDP-43
A consensus from multiple reports of expression of wild-type human TDP-43 in neurons of worms [38,43], flies [39,42,44,54,55] and zebrafish embryos [40,41] reveals neurodegeneration accompanied by decreased locomotive activity [38,42,43], motor deficits [40–42], motor neuron loss [42], paralysis and reduced lifespan [39,55]. Not surprisingly, the severity of the phenotypes correlates with the levels at which the transgene was accumulated in the neurons [39]. Unsolved is how the toxicity mediated by wild-type TDP-43 in such organisms relates to the situation in human disease. To date copy number variation of the TARDBP gene has not been found in humans nor are brain mRNA levels changed in most patients with various TDP-43 proteinopathies [15].
Nevertheless, in zebrafish, worms and flies, expression of human TDP-43 carrying ALS-linked mutations uniformly produces greater toxicity than wild-type TDP-43. For example, compared with zebrafish embryos expressing wild-type TDP-43, mutant TDP-43 expressing ones develop shorter motor axons, more aberrant branching and more severe motor impairments [40,41]. Similarly, expression of mutant forms of TDP-43 in the neurons of worms or flies causes a more profound neurotoxicity than expression of the wild-type protein [43,44]. No conclusion can be drawn in the fly and worm models about selective neuronal vulnerability in these examples, as toxicity simply followed the transgene expression pattern. Widespread expression in zebrafish larvae has been reported. However, toxicity has been quantified only for motor axons, leaving it unclear whether abnormalities are restricted to motor neurons. Lastly, in all the three systems, no clear insights have emerged concerning the contributions to disease from ubiquitinated aggregates, TDP-43 cleavage, cytoplasmic mislocalization or nuclear clearance. More detailed review of these models can be found in reference [56].
Increased levels of FUS/TLS
To date one study in zebrafish has described expression of wild-type or ALS-linked mutants of FUS/TLS [51]. Unlike similar expression of wild-type or ALS-linked mutant TDP-43, no significant alterations in morphology or motor axon outgrowth were found with either wild-type or mutant FUS/TLS.
Disease modeling in rodents
Loss of TDP-43 in mice
TDP-43 is essential for early mouse embryogenesis as mice with homozygous disruption of the TARBP gene were found to die embryonically due to defective outgrowth of the inner cell mass [57–60]. Heterozygous TARBP gene disruption mice are phenotypically undistinguishable from control littermates, although one of the three reports describes modest motor behavioral abnormalities in aged mice [58]. To test a physiological role by systemic deletion of TDP-43 in adult mice, wide spread post-natal deletion of TDP-43 was achieved through inactivation of a conditional (floxed) TDP-43 allele by induction with tamoxifen of a Cre recombinase (produced from insertion of Cre coding sequences into the Rosa26 gene that is thought to be ubiquitously expressed). This did not lead to ALS- or FTLD-like symptoms, but instead produced dramatic loss of body fat and lethality within 9 days [57].
Loss of FUS/TLS in mice
Disruption of the FUS/TLS gene has been achieved by two independent groups by the insertion of “gene-trap” constructs inserted into exons 8 [61] or 12 [62]. Both insertions lead to complete loss of full length FUS/TLS protein. Heterozygous FUS/TLS gene disruption mice do not display any overt phenotype. The consequence of homozygous loss of FUS/TLS produces striking differences depending on the mouse genetic background. In the inbred C57BL/6 or 129 mouse strains, absence of FUS/TLS causes perinatal death [61,62], accompanied by major defects in B-lymphocyte development [61]. In an outbred background, mice with complete loss of FUS/TLS survive until adulthood, albeit with male sterility and reduced female fertility [62]. Interestingly, murine embryonic fibroblasts derived from the two sets of mice reveal high chromosomal instability and radiation sensitivity, suggesting that FUS/TLS plays a key role in the maintenance of genome integrity. That loss of FUS/TLS from outbred adult mice apparently does not lead to neurodegeneration offers strong evidence against loss of function(s) as a major contributor to FUS/TLS proteinopathies.
Consequences of increasing wild-type TDP-43 in mice
In the past two years, eight reports in mice or rats have described elevated levels of either wild-type or mutants of TDP-43 and one report for FUS/TLS (Table 2) [31,32,45–50,52]. All of the mouse models express a human TDP-43 cDNA (either wild-type or carrying ALS-linked mutations) under the control of heterologous promoters (mouse prion, mouse Thy1 or calcium/calmodulin-dependent kinase II (CaMKII)) which are known to drive expression mostly in neuronal and non-neuronal cells of the central nervous system (CNS).
Expression of wild-type human TDP-43 in the CNS of mice has consistently been found to be toxic in a dose and threshold-dependent manner [31,32,45–47,49]. Reported phenotypes are divergent, including motor abnormalities [31,32,45,46,48–50], growth retardation [45,56,63] and cognitive impairments [47] and in some cases lethality [32,46,48–49]. The variability of the phenotypes observed does not always correlate with the promoter driving expression of the transgene. For example, a wide range of motor dysfunctions (such as gait abnormality or limb reflex impairment) were reported in mice expressing human wild-type TDP-43 under the control of Thy1 [45,49], prion [32] or CAMKII [31] promoters. On the other hand, prion-driven expression of comparable levels of TDP-43 (2.5–3 fold above that of the endogenous in total spinal cord extracts) either caused initial gait abnormalities and death by 60 days of age [32] or did not produce any overt phenotype [46]. The opposing results may be due to (1) divergence in the cells in which the transgene is expressed, (2) different genetic backgrounds (a mixed B6SJLF1xCD1 or C57BL/6xCD3heJ background or an inbred C57BL/6 or FVB strain) and/or (3) disruption of an endogenous locus by the random integration of the transgene. It is noteworthy that the severity of the phenotype was significantly increased in homozygous TDP-43 animals, compared to heterozygotes with similar total levels of transgene in the CNS. Homozygous expression of wild-type human TDP-43 driven either by Thy1 or prion promoters produced a severe motor phenotype and lethality, respectively [32,49], while no death was reported in heterozygous TDP-43 mice with apparently equivalent total levels of accumulated human TDP-43 in the CNS [45,46].
Early cognitive impairments accompanied with late motor defects, both of which are features of FTLD and primary lateral sclerosis, were found in transgenic mice expressing elevated levels of mouse TDP-43 under the control of the CaMKII promoter [47]. These transgenic mice developed learning and memory deficits at 2 months of age with motor dysfunctions (including gait abnormalities and reduced performance in rotarod) and reduction in brain mass observed at 6 months of age, concomitantly with cortical astrogliosis and neuronal loss. Untested is whether such cognitive defects develop in mice expressing comparable levels of wild-type human TDP-43 from the same promoter (such mice have so far only been reported to develop a late clasping phenotype [31]).
Hallmarks of neurodegeneration were also reported in some sets of transgenic mice (with varying promoters) to produce accumulated levels of wild-type TDP-43, estimated to range 1.7 to 4 fold above endogenous levels). Loss of cortical and/or anterior horn spinal cord neurons [47,49], or dentate gyrus neurons [31] was reported in mice where expression of wild-type TDP-43 accumulation was driven by either Thy-1 or CaMKII promoters, respectively. The neurons that degenerate are in most cases the ones in which expression was expected to be driven at highest levels by the specific transgene promoters. Astrogliosis was also found in the CNS of most of the transgenic mice [31,32,47,49]. However, axonal degeneration was only described in three sets of mice expressing TDP-43 through Thy1 [45,49] or prion [32] promoters. Altogether, expressing either wild-type human, or increased levels of mouse, TDP-43 predominantly in the CNS is toxic in mice, both when expression of the transgene is driven early during development (through the prion promoter) or post-natally (through either Thy1 or CaMKII promoters).
TDP-43 mutant expression in mice
Two studies have reported consequences in mice of expressing ALS-linked mutants of TDP-43 [46,48]. Accumulation of mutant human TDP-43 carrying the A315T mutation (TDP-43A315T) to 3–4 fold above that of endogenous TDP-43 in the CNS (using the prion promoter) caused gait abnormalities and ultimately death within 150 [46] or 75 days [48]. Despite wider expression of the transgene in the CNS when directed by the prion promoter, accumulation to 3 fold above the endogenous level of TDP-43A315T led to a loss of cortical layer V pyramidal neurons in the frontal cortex, spinal cord motor neurons as well as upper motor axons, all of which are features that are reminiscent of both ALS and FTLD [48]. Not established is whether this loss is mutant selective, as loss of these neurons was also found in mice expressing human wild-type TDP-43 predominantly in neurons [49]. No reduction in the number of neurons was reported in transgenic mice accumulating higher levels of mutant TDP-43A315T in the CNS [46]. The simplest explanation for this discrepancy is differences in cell types in which the transgene is accumulated at the highest levels, even though the expression of the transgene was driven by the same promoter (the same promoters in different lines of transgenic mice can lead to different patterns of transgene expression [63]). Settling what can be concluded from these efforts will require additional mice.
Two studies have proposed that expression of ALS-linked mutants (TDP-43A315T [46,48]; TDP-43M337V [46]) causes progressive motor neurodegeneration in mice. This is not established. Motor defects and/or loss of neurons have been reported only at one late time point of disease stage. With the exception of grip strength and gait posture, measurements have been reported at varying ages between 5 and 20 weeks; these findings reveal that 5 week old transgenic animals already have reduced motor capacities compared to their non-transgenic littermates [46]. Therefore, to determine age-dependent loss of motor neurons will require assessing motor defects at earlier ages.
Another key question not yet established is whether ALS-linked mutants of TDP-43 trigger higher toxicity than the wild-type protein in mice. Only one study has so far directly attempted to address this question. Analysis of transgenic mice expressing human wild-type, TDP-43A315T or TDP-43M337V under the same promoter (mouse prion) [46] led the authors to conclude that while expression of low levels of human ALS linked mutants of TDP-43 give rise to motor dysfunction, mice expressing the human wild-type protein do not display an overt phenotype. Nevertheless, although a large number of lines were obtained, no lines with matching levels of transgene accumulation in the CNS were identified for lines expressing mutant and wild-type TDP-43, thus preventing determination of whether TDP-43 toxicity is significantly enhanced in mice by ALS-causing mutations.
Transgenic mice with tetracycline-inducible expression in the forebrain of a human TDP-43 variant with a defective nuclear localization signal (TDP-43ΔNLS) and additional mice expressing wild-type protein (using CaMKII as a promoter) were reported in an effort to model the cytoplasmic mislocalization of TDP-43 reported in human disease [31]. High accumulation (~8–9 fold above the endogenous!) of TDP-43ΔNLS in the forebrain led to an abnormal motor clasping phenotype (a month after inducing the expression of the transgene) and a progressive loss of most dentate gyrus neurons (reaching a decrease of 90% after 6 months of induction of the transgene expression), as well as a decreased number of upper motor axons. Accumulation at much lower levels (1.7 fold above the endogenous) of wild-type TDP-43 in the forebrain caused similar neuronal and motor defects (although a longer period of induction of expression of the transgene was required). Whether either of these two transgenic mice also display cognitive/behavioral impairments or develop seizures (since most of the dentate gyrus neurons are lost) has not been reported.
A substantial down-regulation of endogenous mouse TDP-43 was found in mice expressing human wild-type TDP-43 [31]. This finding, which is also apparent in the analyses of transgenic TDP-43 mice built by other groups [19,32,48], confirms in an in vivo setting the auto-regulation mechanism for TDP-43 discussed above. Similarly, down-regulation of the endogenous TDP-43 was also found in mice expressing TDP-43ΔNLS. Although TDP-43ΔNLS was designed to be exclusively in the cytoplasm, ~50% of the transgene product was actually found in nuclear cortical extracts. It is therefore likely that the decrease in the levels of the mouse endogenous TDP-43 is also caused by the nuclear pool of the TDP-43ΔNLS regulating the mRNA encoding endogenous TDP-43.
Mechanisms of TDP-43-mediated proteinopathies
Most of the features described in TDP-43 proteinopathies, including nuclear clearance and/or cytoplasmic mislocalization, ubiquitinated aggregates, phosphorylation or cleavage of TDP-43 [15] have been examined in the multiple transgenic mice outlined above. Surprisingly, although cytoplasmic TDP-43 inclusions have systematically been reported in neurons of ALS and FTLD-U (FTLD with ubiquitinated inclusions) patients, cytoplasmic ubiquitinated inclusions that are TDP-43 or phosphorylated-TDP-43 immunopositive were rarely [31,32,46,49] or never [45,48] found in the mouse models, indicating that at least in mice cytoplasmic inclusions are not required for neurodegeneration. In independent sets of mice expressing wild-type TDP-43 under the control two different promoters [32,45], TDP-43 negative cytoplasmic aggregates have been reported to be comprised of large perinuclear accumulation of mitochondria. With the implication of mitochondria in the mutant SOD1 rodent models of ALS [64], mitochondrial dysfunction may be a common feature of ALS pathogenesis occurring in both TDP-43 and SOD1-mediated disease. In the human context, TDP-43 cytoplasmic aggregation has been shown to be accompanied by nuclear clearance of the protein. Similarly, loss of nuclear localization of TDP-43 has been reported in neurons of most of the mouse models [31,32,47,48,49]. However, the frequency of nuclear TDP-43 depletion has not been reported, so it is not yet possible to conclude that loss of nuclear TDP-43 is a principal contributor to disease. Similarly, down-regulation of endogenous mouse TDP-43 has also been observed in multiple sets of transgenic mice [31,32,48]. Whether neuronal vulnerability results from perturbations of the normal mouse TDP-43 function is not established. The presence of carboxy-terminal truncated TDP-43 fragments (CTFs) has also been implicated in neurodegeneration from their appearance in human disease. In the mouse models, several studies have reported 25kDa and 35kDa CTFs in CNS extracts [32,46–49], but not in three other mice [31,45] despite a significant loss of neurons mediated by the expression of TDP-43 in one of these latter mice [31]. Therefore, how and why these fragments are generated (and their relationship to neurodegeneration) remains unclear and their direct toxic role in TDP-43 proteinopathies has not been established by the experimental evidence available to date.
Two studies using high-throughput sequencing (RNA-seq) or microarray approaches have identified changes occurring at the mRNA level in mice expressing wild-type TDP-43 [31,45] or TDP-43ΔNLS [31], all of which developed motor abnormalities. Alterations in both differential expression and alternative splicing of hundreds of transcripts, many encoding proteins involved in cellular architecture [45] or macromolecular complex organization (in particular DNA-protein assemblies) [31] have been reported. Neurofilament mRNAs are among those significantly down-regulated in some TDP-43 transgenic mice [31,45], similar to what happens after reduction of TDP-43 in an otherwise normal adult mouse CNS [19]. Since the mice expressing human wild-type or TDP-43ΔNLS down-regulate endogenous mouse TDP-43, it is likely that the reported decreased expression of neurofilaments is caused by loss of nuclear TDP-43 function. Finally, increased levels of wild-type TDP-43 in mice was also reported to lead to an increased number, in neuronal nuclei, of an intranuclear structure called GEMs (Gemini of coiled bodies) [45]. GEMs are nuclear structures enriched in SMN (Survival Motor Neuron) complex proteins which are widely thought to play a role in the biogenesis of small ribonucleoproteins required for mRNA splicing. (Reduction in SMN is causative of the juvenile recessive disease spinal muscular atrophy (SMA), in which spinal motor neurons degenerate, causing progressive paralysis and muscular atrophy). It remains now to be determined if, and if so how, elevated numbers of these nuclear structures may contribute to damaging motor neurons.
TDP-43 model rats
Transgenic rats expressing human wild-type or an ALS-linked mutant form of TDP-43 (M337V) constitutively have also been generated [50]. To date, this is the only rodent model expressing human wild-type or mutant TDP-43 from the authentic human gene and its promoter (instead of TDP-43 cDNA). Wild-type TDP-43 expressing rats do not develop any overt phenotype. However, comparable levels of expression of the same construct but carrying the M337V substitution (TDP-43M337V) were lethal and no lines could be established. This is the only evidence today in rodents for toxicity mediated in a TDP-43 mutant selective manner. An even more attractive model emerged from additional rats ubiquitously expressing TDP-43M337V cDNA in a tetracycline dependent manner. While transgene expression when induced during development is lethal, induction at postnatal day 10 produces widespread neurodegeneration primarily in the motor system with fatal paralysis within 45–50 days of age. Of note, it is not known whether motor neuron disease from induced expression is selective for mutant TDP-43, as expression of wild-type TDP-43 using a similar promoter has not been examined.
FUS/TLS transgenic rats
Transgenic rats expressing human wild-type or an ALS-linked mutant form of FUS/TLS (R521C) conditionally have been generated [52]. Accumulation of mutant protein, but not wild-type, levels at three to six times above that of endogenous in the CNS led to degeneration of motor axons, loss of cortical and hippocampal neurons, increased denervation of neuromuscular junctions and ultimately paralysis in 30–70 days after induction of the transgene. Expression of wild-type FUS/TLS was accompanied by a mild loss of neurons in the cortex and hippocampus, as well as cognitive deficits at one year of age. Altogether, accumulation at high levels of mutant FUS/TLS in the CNS is more toxic than wild-type protein in rats.
Conclusion & Perspectives
It is early days in deciphering how mutations in the RNA/DNA binding proteins TDP-43 and FUS/TLS cause inherited ALS. TDP-43 normally functions in the maturation and splicing of thousands of pre-mRNAs. TDP-43 synthesis is controlled by an auto-regulatory mechanism mediated at least in part by direct binding to the 3′untranslated region (UTR) of its own RNA transcript. Modeling in rodent, zebrafish and invertebrate systems has produced imperfect replicas of ALS-like disease. Many of the transgenic animals reported so far develop motor and/or cognitive impairments at very young ages and progressiveness has in most cases not been established. As yet left unanswered are two of the most central questions concerning mutant TDP-43 (or FUS/TLS) mediated toxicity: 1) is pathogenesis caused by the loss of normal function(s), gain of one or more toxic properties or both, and 2) what are the crucial cell type(s) in which mutant TDP-43 or FUS/TLS acts to provoke toxicity?
Acknowledgments
We are grateful to Dr. Philippe Parone for critical reading of the manuscript and Dr. Magda Polymenidou for fruitful discussion. We acknowledge Drs. Bryan Traynor and Clotilde Lagier-Tourenne for their expert advice on the genetics of ALS. S.D.C. was funded by the Milton-Safenowitz post-doctoral fellowship from the Amyotrophic Lateral Sclerosis Association. Work has been supported by a grant to D.W.C. from the NIH (NS27036).
References
- 1.Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, Borchelt DR, Price DL, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 2.Beers DR, Henkel JS, Xiao Q, Zhao W, Wang J, Yen AA, Siklos L, McKercher SR, Appel SH. Wild-type microglia extend survival in PU. 1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Gutmann DH, Roos RP. Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum Mol Genet. 2010;20:286–293. doi: 10.1093/hmg/ddq463. [DOI] [PubMed] [Google Scholar]
- 5.Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **7.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. See reference 8. [DOI] [PubMed] [Google Scholar]
- **8.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. Along with Ref. [7], biochemical and immunohistochemical approaches are used to identify TDP-43 as a major protein component of ubiquitin-positive neuronal inclusions in sporadic ALS patients and in patients with FTLD. [DOI] [PubMed] [Google Scholar]
- **9.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. See reference 11. [DOI] [PubMed] [Google Scholar]
- **10.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. See reference 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **11.Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. Along with Ref. [9,10], these three studies represent the first reports identifying dominant mutations in TDP-43 as a primary cause of ALS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- **13.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. See Reference 14. [DOI] [PubMed] [Google Scholar]
- **14.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. Along with Ref. [13], these studies represent the first reports identifying mutations in FUS/TLS as a primary cause of familial ALS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geser F, Martinez-Lage M, Kwong LK, Lee VM, Trojanowski JQ. Amyotrophic lateral sclerosis, frontotemporal dementia and beyond: the TDP-43 diseases. J Neurol. 2009;256:1205–1214. doi: 10.1007/s00415-009-5069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seyfried NT, Gozal YM, Dammer EB, Xia Q, Duong DM, Cheng D, Lah JJ, Levey AI, Peng J. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol Cell Proteomics. 2010;9:705–718. doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **19.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011 doi: 10.1038/nn.2779. Using UV-crosslinking and immunoprecipitation (CLIP-seq) of TDP-43 from mouse brain, the authors use high-throughput sequencing to identify more than 6,000 TDP-43 RNA targets, as well as their binding sites. Furthermore, the combination of high-throughput sequencing and splicing-sensitive microarrays reveal changes in abundance and altered splicing patterns of >1000 mRNAs in the adult mouse brain upon in vivo depletion of TDP-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2010;286:1204–1215. doi: 10.1074/jbc.M110.190884. Immunoprecipitation of TDP-43 from rat cortical neurons, followed by deep sequencing (RIP-seq), led to the identification of more than 4,000 TDP-43 RNA targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011 doi: 10.1038/nn.2778. This report is the first to identify TDP-43 RNA targets in post-mortem human brain samples from FTLD patients using individual-nucleotide resolution UV-crosslinking and immunoprecipitation (iCLIP) of TDP-43 coupled with high-throughput sequencing. Furthermore, TDP-43 is found to affect expression levels and splicing patterns of mRNAs in brain samples as well as in TDP-43 knock-down human neuroblastoma cell lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao S, Sanelli T, Dib S, Sheps D, Findlater J, Bilbao J, Keith J, Zinman L, Rogaeva E, Robertson J. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol Cell Neurosci. 2011 doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- **23.Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D’Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. Embo J. 2010;30:277–288. doi: 10.1038/emboj.2010.310. Along with Ref. [19, 21], TDP-43 is found to auto-regulate its own synthesis by binding to its 3′ UTR region. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **25.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. This study reports intermediate CAG trinucleotide expansion in ataxin-2, a gene known to be mutated in spinocerebellar ataxia type 2, as an ALS susceptibility gene. Furthermore, ataxin 2 is found to be a potent modifier of TDP-43 mediated toxicity in yeast and Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiesel FC, Voigt A, Weber SS, Van den Haute C, Waldenmaier A, Gorner K, Walter M, Anderson ML, Kern JV, Rasse TM, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. Embo J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Kim SH, Shanware NP, Bowler MJ, Tibbetts RS. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. Along with Ref. [34], the authors report an interaction between TDP-43 and FUS/TLS in human cell lines. In addition TDP-43 and FUS/TLS are found to associate with histone deacetylase 6 mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- *29.Belzil VV, Daoud H, Dion PA, Rouleau GA. No Effect on SOD1 Splicing by TARDP or FUS Mutations. Arch Neurol. 68:395–396. doi: 10.1001/archneurol.2011.1. The authors report that mutations in TDP-43 or FUS/TLS do not affect the splicing of SOD1 in ALS patients. [DOI] [PubMed] [Google Scholar]
- **30.Rabin SJ, Kim JM, Baughn M, Libby RT, Kim YJ, Fan Y, Libby RT, La Spada A, Stone B, Ravits J. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet. 2010;19:313–328. doi: 10.1093/hmg/ddp498. Using laser capture microdissection to isolate spinal cord motor neurons from sporadic ALS patients coupled with genome exon splicing arrays, alterations in expression and splicing of hundreds of transcripts are identified, including enrichment in aberrantly spliced mRNAs encoding products involved in cell-matrix adhesion, but no significant modifications in the mRNAs encoding TDP-43 or FUS/TLS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **31.Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2010;121:726–738. doi: 10.1172/JCI44867. Inducible expression in mouse forebrains of either wild-type or human TDP-43 with a defective nuclear localization signal is shown to cause neuronal loss, axonal degeneration and motor spasticity. Along with Ref. [19,32], human TDP-43 expression in mice is accompanied by down-regulation of the endogenous mouse TDP-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32.Xu YF, Gendron TF, Zhang YJ, Lin WL, D’Alton S, Sheng H, Casey MC, Tong J, Knight J, Yu X, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. Mice expressing wild-type human TDP-43 under the control of the prion promoter develop axonal degeneration, gait abnormalities with ultimately early lethality. Along with Ref. [45], TDP-43 expression in neurons leads to an accumulation of neuronal mitochondria in TDP-43 negative cytoplasmic aggregates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- *34.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. Using immunoprecipitation and proteomic approaches in human cells expressing either wild-type or ALS-linked mutants of TDP-43, more than a hundred proteins are identified as potential TDP-43 interacting partners. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **35.Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. Using tandem-affinity purification and quantitative mass spectrometry in human isogenic cell lines stably expressing wild-type human TDP-43, TDP-43 is found to interact with FUS/TLS, heterogeneous nuclear ribonucleoproteins, as well as components of Drosha microRNA processing complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 37.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- *38.Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. Along with Ref. [43], the authors report that expressing wild-type TDP-43 in neurons is toxic in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Hanson KA, Kim SH, Wassarman DA, Tibbetts RS. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J Biol Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. Along with Ref. [42,44,55], expression of wild-type TDP-43 in eyes or neurons of Drosophila is toxic, leading to loss of photoreceptor cells or movement deficits, respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, Rochefort D, Bel Hadj S, Durham HD, Vande Velde C, et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. See Reference 41. [DOI] [PubMed] [Google Scholar]
- *41.Laird AS, Van Hoecke A, De Muynck L, Timmers M, Van den Bosch L, Van Damme P, Robberecht W. Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS One. 2010;5:e13368. doi: 10.1371/journal.pone.0013368. Along with Ref. [40], expression of wild-type or ALS-linked mutants of TDP-43 in zebrafish is shown to cause motor defects and axonal shortening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *42.Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, 3rd, Fushimi K, Wu JY. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. Along with Ref. [53,54], the authors report that partial or complete deletion of the TDP-43 homologue in Drosophila is toxic leading to defects in locomotive behavior, reduced lifespan as well as axonal loss and neuronal death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Liachko NF, Guthrie CR, Kraemer BC. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J Neurosci. 2010;30:16208–16219. doi: 10.1523/JNEUROSCI.2911-10.2010. See Reference 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. See Reference 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci U S A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. Transgenic mice expressing wild-type human TDP-43 predominantly in neurons are reported to develop gait abnormalities, growth retardation and reduction in large caliber axons. In addition, TDP-43 expression in neurons is found to cause an accumulation of mitochondria in cytoplasmic aggregates along with Ref. [32], as well as increased numbers of gemini bodies, nuclear structures involved in RNA splicing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **46.Stallings NR, Puttaparthi K, Luther CM, Burns DK, Elliott JL. Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol Dis. 2010;40:404–414. doi: 10.1016/j.nbd.2010.06.017. This study is the first to report mice expressing wild-type or ALS-linked mutants of TDP-43 under the control of the same promoter. The authors find that expression predominantly in the CNS of human TDP-43 harboring mutations associated with ALS is more toxic than wild-type and leads to gait abnormalities and spasticity. [DOI] [PubMed] [Google Scholar]
- *47.Tsai KJ, Yang CH, Fang YH, Cho KH, Chien WL, Wang WT, Wu TW, Lin CP, Fu WM, Shen CK. Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med. 2010;207:1661–1673. doi: 10.1084/jem.20092164. This effort reports that mice expressing mouse TDP-43 in the forebrain exhibit early impaired learning/memory, motor dysfunctions and reduced brain weight. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. This study is the first to describe mice expressing an ALS-linked mutant form of human TDP-43 under the control of the prion promoter. The mice develop gait abnormalities, weight loss and are characterized by loss of motor axons as well as cortical and spinal motor neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **49.Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. This study represents the first report of mice expressing wild-type human TDP-43 predominantly in neurons. The mice were found to develop gait and motor abnormalities as well as loss of motor axons, cortical and spinal motor neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50.Zhou H, Huang C, Chen H, Wang D, Landel CP, Xia PY, Bowser R, Liu YJ, Xia XG. Transgenic rat model of neurodegeneration caused by mutation in the TDP gene. PLoS Genet. 2010;6:e1000887. doi: 10.1371/journal.pgen.1000887. This report is the first study describing transgenic rats expressing constitutively either wild-type or an ALS-linked mutant of human TDP-43. While rats with elevated levels of wild-type TDP-43 are normal, those with elevated levels of mutant TDP-43 are lethal. Furthermore, transgenic rats ubiquitously expressing, in a tetracycline inducible manner, mutant TDP-43 display motor dysfunctions, paralysis and widespread neurodegeneration, with a predominant loss of motor neurons and motor axons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr, Sapp P, McKenna-Yasek D, Brown RH, Jr, Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. This study is the first to report that increased expression of wild-type or ALS-linked mutants of FUS/TLS in zebrafish eggs does not alter the morphology of the embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **52.Huang C, Zhou H, Tong J, Chen H, Liu YJ, Wang D, Wei X, Xia XG. FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration. PLoS Genet. 2011;7:e1002011. doi: 10.1371/journal.pgen.1002011. This study represents the first report of transgenic rats ubiquitously expressing, in a tetracycline inducible manner, either wild-type or an ALS-linked mutant of human FUS/TLS. The authors find that mutant FUS/TLS is more toxic, than wild-type protein, since rats expressing mutant FUS/TLS display early motor dysfunction, paralysis and widespread neurodegeneration, while rats expressing similar levels of normal FUS/TLS only exhibit deficits in spatial learning/memory and neuronal loss at advanced ages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *53.Feiguin F, Godena VK, Romano G, D’Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. Along with Ref. [42,54], the authors report that partial or complete deletion of TDP-43 homologue in Drosophila is toxic, leading to defects in locomotive behavior, reduced lifespan as well as axonal loss and neuronal death. [DOI] [PubMed] [Google Scholar]
- *54.Lu Y, Ferris J, Gao FB. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. See Reference 53. In addition, expression in drosophila sensory neurons of wild-type or mutant forms of human TDP-43 leads to increased dendritic branching. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *55.Miguel L, Frebourg T, Campion D, Lecourtois M. Both cytoplasmic and nuclear accumulations of the protein are neurotoxic in Drosophila models of TDP-43 proteinopathies. Neurobiol Dis. 2011;41:398–406. doi: 10.1016/j.nbd.2010.10.007. Along with Ref. [39,42,44], expression of wild-type TDP-43 in eyes or neurons of Drosophila is toxic, leading to loss of photoreceptor cells or movement deficits, respectively. [DOI] [PubMed] [Google Scholar]
- 56.Wegorzewska I, Baloh RH. TDP-43-Based Animal Models of Neurodegeneration: New Insights into ALS Pathology and Pathophysiology. Neurodegener Dis. 2010 doi: 10.1159/000321547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **57.Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. By generating a conditional Tardbp knock-out mouse model, the authors find that post-natal deletion of TDP-43 leads to loss of body fat and rapid death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58.Kraemer BC, Schuck T, Wheeler JM, Robinson LC, Trojanowski JQ, Lee VM, Schellenberg GD. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. Along with Ref. [59, 60], the authors show that homozygous disruption of the Tardbp gene in mice is lethal and that TDP-43 is essential for early mouse embryogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *59.Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P, 3rd, Herz J, Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. See Reference 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *60.Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. See Reference 58. [DOI] [PubMed] [Google Scholar]
- 61.Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White EK, Koury MJ, Oltz EM, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 62.Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, de Rooij DG, Akhmedov A, Ashley T, et al. Male sterility and enhanced radiation sensitivity in TLS(−/−) mice. Embo J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 64.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 65.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 66.Momeni P, Schymick J, Jain S, Cookson MR, Cairns NJ, Greggio E, Greenway MJ, Berger S, Pickering-Brown S, Chio A, et al. Analysis of IFT74 as a candidate gene for chromosome 9p-linked ALS-FTD. BMC Neurol. 2006;6:44. doi: 10.1186/1471-2377-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morita M, Al-Chalabi A, Andersen PM, Hosler B, Sapp P, Englund E, Mitchell JE, Habgood JJ, de Belleroche J, Xi J, et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 68.Vance C, Al-Chalabi A, Ruddy D, Smith BN, Hu X, Sreedharan J, Siddique T, Schelhaas HJ, Kusters B, Troost D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2–21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 69.Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wideassociation study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 72.Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitchell J, Paul P, Chen HJ, Morris A, Payling M, Falchi M, Habgood J, Panoutsou S, Winkler S, Tisato V, et al. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci U S A. 2010;107:7556–7561. doi: 10.1073/pnas.0914128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]