Multisite phosphorylation of proteins has been proposed to transform a graded protein kinase signal into an ultrasensitive switch-like response 1–4. Although many multiphosphorylated targets have been identified, the dynamics and sequence of individual phosphorylation events within the multisite phosphorylation process have never been thoroughly studied. In budding yeast, the initiation of S phase is thought to be governed by complexes of Cdk1 and Cln cyclins that phosphorylate six or more sites on the Clb5-Cdk1 inhibitor Sic1, directing it to SCF-mediated destruction 1, 5, 6, 7, 8. The resulting Sic1-free Clb5-Cdk1 complex triggers S phase 9. Here, we demonstrate that Sic1 destruction depends on a more complex process in which both Cln2- and Clb5-Cdk1 act in processive multiphosphorylation cascades leading to the phosphorylation of a small number of specific phosphodegrons. The routes of these phosphorylation cascades are shaped by precisely oriented docking interactions mediated by cyclin-specific docking motifs in Sic1 and by Cks1, the phosphoadaptor subunit of Cdk1. Our results suggest that Clb5-Cdk1-dependent phosphorylation generates positive feedback that is required for switch-like Sic1 destruction. Our evidence for a docking network within clusters of phosphorylation sites uncovers a new level of complexity in Cdk1-dependent regulation of cell cycle transitions, and has general implications for the regulation of cellular processes by multisite phosphorylation.
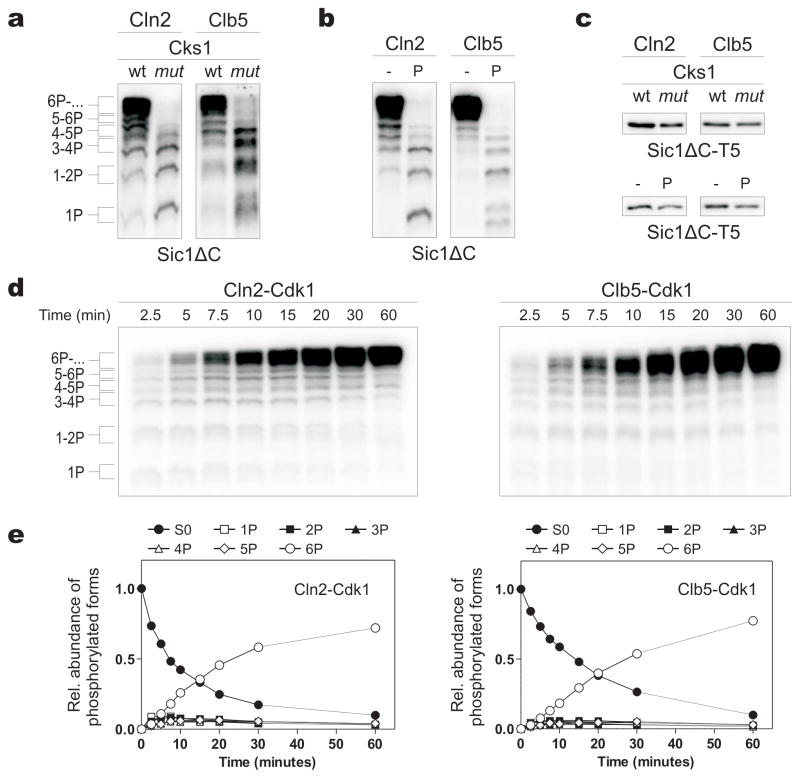
To study the multiphosphorylation of Sic1, we used a non-inhibitory truncated version of Sic1 (Sic1ΔC) as a substrate for purified Cln2- and Clb5-Cdk1 complexes (Supplementary Fig. 1a). Intriguingly, both Cln2- and Clb5-Cdk1 generated phosphorylation patterns with abruptly accumulating hyperphosphorylated species (Fig. 1a, b, d, e, Supplementary Fig. 1b). This pattern depended on Cks1, the Cdk1 subunit that binds phosphate groups 10. Mutation of the phosphate-binding site of Cks1 reduced the accumulation of multiphosphorylated forms (Fig. 1a, Supplementary Fig. 2a). Similarly, a phosphorylated competitor phosphopeptide reduced phosphorylation (Fig. 1b). Cks1 mutation and the peptide had little effect on the phosphorylation of a Sic1ΔC version containing a single Cdk site (Fig 1c).
Figure 1.
The phosphoadaptor subunit Cks1 provide processivity for the multiphosphorylation of Sic1 by Cln2-Cdk1 and Clb5-Cdk1. (a) Cln2- and Clb5-Cdk1 complexes were incubated with Sic1ΔC and 32P-ATP. The reactions also included wild-type Cks1 (wt) or a version with a mutated phosphate-binding site (mut; see Supplementary Methods). Phosphorylated substrates were separated using Phos-Tag SDS-PAGE gels. (b) Reactions were performed in the presence of a phosphopeptide competitor (P) based on the sequence surrounding T45 in Sic1. (c) The phosphorylation of a Sic1ΔC version containing a single Cdk site (Sic1ΔC-T5, with other Cdk consensus sites mutated to alanines) was not affected by Cks1mut or the phosphopeptide. The standard SDS-PAGE was used. (d) Time courses of Sic1ΔC multiphosphorylation were followed by Phos-Tag SDS-PAGE. (e) The quantified data from (d). The intensities of 32P-labeled proteins were divided by the number of phosphates as indicated to obtain the levels of different phosphoforms. In the experiments presented in Fig. 1 the enzyme concentrations were chosen to obtain roughly equal substrate labeling.
Cks1 is essential for Cdk1 function 11, 12, with roles at the G1/S and G2/M transitions 13, 14. We found that the Cks1:Cdk1 stoichiometry in vivo was about 1:1 for Cln2-Cdk1 and at least 0.5:1 for Clb5-Cdk1, confirming that Cks1-dependent multiphosphorylation is the prevalent mode of Cdk1 action in vivo (Supplementary Fig. 1a, b, c). An isothermal calorimetry binding assay of fully phosphorylated Sic1ΔC (pSic1ΔC) and Cks1 revealed a KD of 11 ± 2 μM, while the non-phosphorylated version showed no detectable binding (Supplementary Fig. 1d). Approximately 3–4 molecules of Cks1 bound each molecule of pSic1ΔC, suggesting that several phosphorylated sites can bind Cks1 independently. Finally, we found that the phospho-binding pocket of Cks1 is required for phosphorylation and degradation of Sic1 in vivo (Supplementary Fig. 1e, f).
To understand the Cks1-dependent mechanism, we analyzed Sic1ΔC multiphosphorylation over time (Fig. 1d, e). We did not observe significant accumulation of intermediate phosphorylated forms, suggesting that phosphorylation was processive. When we performed kinase reactions at Sic1ΔC concentrations higher than apparent KM, multiphosphorylation patterns remained constant despite the increase in the inhibition term 1+[S0P]/KM (Supplementary Fig. 2b–e). Thus, the enzyme displays processivity: that is, it is able to transfer two or more phosphates to the substrate during a single association event. This conclusion was additionally confirmed using different enzyme concentrations in the assay (Supplementary Fig. 2f) and in mathematical simulations (Supplementary Fig. 3). This processive pattern argues against the current model of ultrasensitivity in the Sic1 phosphorylation switch, which is based on the assumption of a distributive mechanism with equal specificity of different sites 1, 15.
To dissect the mechanism of the processive multiphosphorylation cascade, we first studied the impact of potential docking interactions between Sic1 and cyclins. In previous studies, we found that rapid Sic1 phosphorylation by Clb5-Cdk1 depends on an interaction between RXL motifs in Sic1 and the hydrophobic patch docking site (hp) in Clb5; a triple mutation in this site (Clb5hpm) decreases the net phosphorylation rate (Supplementary Fig. 4a–h) 16. We further found here that a version of Sic1ΔC with mutations at its four RXL motifs (Sic1ΔC-1234rxl) showed less abrupt production of multiphosphorylated species by Clb5-Cdk1, revealing that processive multiphosphorylation requires both Cks1-dependent and hp-dependent docking (Supplementary Fig. 4i). Cln2-Cdk1 exhibited only a mild RXL effect on the phosphorylation pattern, probably because Cln2 does not contain a conventional hp like that in the B-type cyclins. In recent studies, we also located in Sic1 a ten-amino-acid stretch, 136VLLPPSRPTS145, which confers Cln2 specificity16. Here we found that a five-alanine mutation of the first five hydrophobic residues in this stretch, or a synthetic competitor peptide containing the docking site, reduced the abrupt multiphosphorylation pattern for Cln2 (Supplementary Fig. 4j). In conclusion, both Clb5- and Cln2-Cdk1 use docking mechanisms, in addition to Cks1, to achieve processive multiphosphorylation of Sic1.
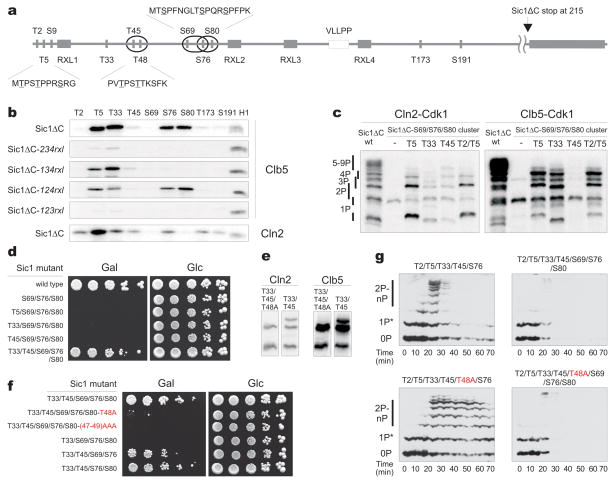
Using Sic1ΔC mutants carrying only one Cdk site (Fig. 2a, b), we found that Clb5-Cdk1 rapidly phosphorylated just four sites (T5, T33, S76, and S80), and this specificity depended on the RXL2 and RXL3 docking sites in Sic1 (Fig. 2b; note that in all figure labels, the indicated Cdk sites are those left unmutated, unless otherwise indicated). Cln2-Cdk1, on the other hand, exhibited a preference for the N-terminally located site T5 (Fig. 2b). Thus, docking interactions direct the associated kinase to a small number of primary phosphorylation sites. We speculate that these primary sites interact with Cks1 to drive processive phosphorylation of additional sites.
Figure 2.
Phosphorylated priming sites provide docking interactions for efficient phosphorylation of suboptimal sites in phosphodegrons. (a) Schematic view of phosphorylation sites, docking motifs (Clb5- and Cln2-specific), phosphodegrons (ovals 17) in Sic1. (b) Phosphorylation specificity of Clb5- and Cln2-Cdk1 towards different Cdk sites was studied using Sic1ΔC constructs containing a single fixed Cdk site. For Clb5-Cdk1, the dependence of the site specificity profile on RXL docking sites was assessed using Sic1ΔC constructs containg a single Cdk site and a single fixed RXL motif. (c) The impact of different priming phosphorylation sites on cooperative phosphorylation of the degron cluster S69/S76/S80. Phospho-site mutants of Sic1ΔC carrying the intact S69/S76/S80 cluster and the indicated sites left unmutated were used in a kinase assay with Cln2-Cdk1 and Clb5-Cdk1 using Phos-Tag SDS-PAGE. (d) Full-length Sic1 versions containing the combination of sites described in (c) were overexpressed under the galactose promoter to assay the ability of cells to degrade Sic1. (e) Comparison of the in vitro phosphorylation profiles of Sic1ΔC versions containing only the phosphorylation sites T33/T45 or T33/T45 with mutation T48A. (f) The nonconsensus Cdk1 site T48 is important for viability of cells overexpressing Sic1. The same assay as (d) was used. In panels d and f, the labels indicate unmutated amino acids, and all other consensus Cdk sites are mutated; mutations in the nonconsensus Cdk sites are highlighted in red. (g) The phosphorylation and degradation dynamics of Sic1 were followed after the release of cells from α-factor in a system constitutively expressing mutated versions of noninhibitory Sic1ΔC-3HA. The asterisk indicates a G1-specific phosphorylation by an unknown kinase.
With these primary specificities in mind, we set out to map the pathways along which Cln2- and Clb5-Cdk1 catalyze the phosphorylation of the critical sites required for Sic1 degradation. The original model of Sic1 regulation proposed that six or more sites must be simultaneously phosphorylated in vivo to facilitate binding of phospho-Sic1 to the SCF subunit Cdc4 1. On the other hand, later binding studies revealed that closely positioned pairs of phosphorylation sites (pT5/pS9, pT45/pT48, or pS76/pS80; see Fig. 2a) each present separate entities with a strong affinity for Cdc4, suggesting that just two phosphorylation sites, in the right positions, might be sufficient for Sic1 degradation 17. Our results provided a way to reconcile these findings: we hypothesized that the requirement for six or more sites in vivo reflects a requirement for priming phosphorylation events that direct processive phosphorylation of critical phosphodegrons. To test this possibility, we first measured phosphorylation of a Sic1ΔC mutant with all Cdk sites changed to alanine except for the triple cluster S69/S76/S80, which contains two potential paired degrons (S69/S76 and S76/S80). There was no processive multiphosphorylation of the cluster S69/S76/S80 (Fig. 2c, lane 2 in each panel), but processivity could be induced by adding back single Cdk1 sites to the N-terminal side of the cluster. The rate of the appearance of multiphosphorylated species correlated with the site specificity data for Cln2 and Clb5 (Fig. 2b). Addition of the most Cln2-specific site, T5, caused a much greater effect in Cln2 reactions than the less Cln2-specific sites, T33 and T45. Addition of the Clb5-specific sites T5 and T33, but not the poor Clb5 site T45, greatly increased processivity in Clb5 reactions. These results suggest that sites T5, T33, and T45 are able to serve as priming sites for Cks1-dependent phosphorylation of the S69/S76/S80 degron cluster. The phosphopeptide-dependence was confirmed for these mutants as described for Sic1ΔC-wt in Figure 1b (data not shown).
Cells overexpressing Sic1 containing only the triple cluster S69/S76/S80 were inviable (Fig. 2d). Addition of T5, T33, or T45 did not prevent this lethal effect, suggesting that phosphorylation of the S69/S76/S80 cluster alone is not sufficient for degradation of Sic1. However, viability improved when both T33 and T45 were added. Notably, the addition of T45, together with a nonconsensus Cdk site T48 (Fig. 2a), creates a double degron, T45/T48, as predicted previously 17. T48 is known to be phosphorylated in vivo 7. We hypothesized that T33 serves as a docking site for both the T45/T48 and S69/S76/S80 clusters, and that T45 serves both as a constituent site of the T45/T48 degron as well as a Cks1-dependent docking site for the degron cluster S69/S76/S80. Indeed, T48 alone (the Sic1-9A mutant with all Cdk sites changed to alanine) was a very poor substrate for Clb5 and Cln2 (data not shown), but the addition of T33 or the T33/T45 pair made it a specific site (Fig. 2e, Supplementary Fig. 5, Supplementary Table 1), implying that T33 phosphorylation allows bypass of the consensus motif requirement of a +1 proline at T48. Strikingly, we found that the alanine mutation in T48 had a strong growth-suppressing effect in the galactose assay within the context of sites T33/T45/S69/S76/S80 and also had a weaker effect in the background containing all the Cdk sites (Fig. 2f, Supplementary Fig. 4k). Our results indicate that the benefit of multisite phosphorylation of Sic1, compared to a system with a single phosphorylated site with high affinity for Cdc4 (e.g. based on an optimal degron site of Cyclin E 1), is likely to be the ability of this mechanism to provide docking-dependent kinase specificity for paired degrons. Thus, phosphorylation sites in Sic1 can be divided into three categories: (i) paired degron sites that are spaced 3–7 amino acids from each another; (ii) N-terminally positioned priming sites for each paired degron; and (iii) sites that serve as both priming and degron sites (e.g. T45).
It is unclear why a single paired degron with a priming site is insufficient for degradation of Sic1, as suggested by our observation that viability in the galactose plate assay requires both T45/T48 and either S69/S76 or S76/S80 (Fig. 2f). To test if the simultaneous presence of both paired degrons is required for degradation of Sic1, we used western blotting to measure phosphorylation and degradation of mutated versions of Sic1ΔC. Remarkably, a Sic1ΔC construct (equivalent to the Sic1-5p mutant of Nash et al. 1) containing only the degron T45/T48, but missing the intact degron around S76, was rapidly degraded after the release of cells from G1 (Fig. 2g). This degradation was abolished by mutation of the single nonconsensus Cdk site at T48. A construct also containing the S69/S76/S80 degron (Sic1-7p of Nash et al. 1) was more rapidly degraded, and mutation of T48 in this background did not influence the degradation rate. We conclude that the T45/T48 degron is sufficient to promote some degradation of Sic1 in vivo, but this rate of degradation is not sufficient to prevent the lethality of overexpressed Sic1.
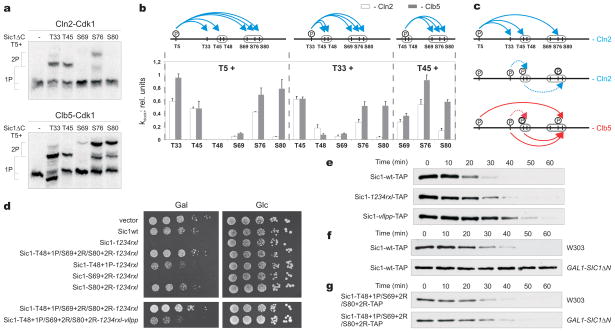
Our model assumes differential roles of Cln2 and Clb5 in the order of Sic1 phosphorylation events. To explore this possibility, we developed methods for determining the apparent rate constants, which we termed kdock, for individual Cks1-enhanced phosphorylation steps (Fig. 3a, b; Supplementary Table 1). The results revealed considerable differences between Clb5 and Cln2. Clb5-Cdk1 was much more effective than Cln2-Cdk1 in taking shortcuts to the critical degron pair of S76/S80, using T5 and T33 as priming sites for Cks1, and with assistance from RXL-mediated docking (Supplementary Tables 1 and 2; Fig. 3a–c; Supplementary Fig. 6). Notably, in the case of Clb5, different RXL motifs supported different Cks1-dependent docking events (Supplementary Table 2).
Figure 3.
Differential roles of Cln2 and Clb5 in Sic1 multiphosphorylation and degradation. (a) Pairwise mapping of the docking connections underlying Sic1 multiphosphorylation, using purified Sic1ΔC mutants containing just two of the Cdk phosphorylation sites per mutant. Representative examples of autoradiographs of phosphorylation assays, showing different docking specificities between Cln2-Cdk1 and Clb5-Cdk1. (b) The specificity profiles for different pair-wise docking connections. The error bars indicate standard errors of the means of at least two independent experiments. (See Supplementary Information and Supplementary Table 1). (c) Schematic view of fast and slow docking-dependent phosphorylation steps for Cln2- and Clb5-Cdk1. (d) Sic1 mutants with improved Cdk recognition determinants in suboptimal phosphodegrons rescue the inviability of cells overexpressing Sic1-1234rxl. The Cln2-dependent docking site becomes essential under these conditions. (e) Cells carrying SIC1wt-TAP or versions with docking site mutations at the endogeneous SIC1 locus were released from an α-factor arrest and the degradation pattern of Sic1-TAP protein was followed by western blotting, using standard SDS-PAGE. (f) A wild-type strain (SIC1wt-TAP) and a strain also expressing the nondegradable inhibitory domain of Sic1 (SIC1ΔN) under the GAL1 promoter were arrested in α-factor, followed by the addition of galactose. After 45 minutes, the cells were washed into galactose media lacking α-factor and Sic1-TAP levels were followed by western blotting (see also Supplementary Fig. 7). (g) Sic1 mutants with improved Cln-Cdk recognition determinants in suboptimal phosphodegrons trigger rapid degradation of Sic1 in the presence of SIC1- ΔN. Experiment was performed as in panel (f).
We propose that in late G1, Clb5-Cdk1 is inhibited by Sic1, and the cascade of phosphorylation events begins with T5 phosphorylation by Cln2-Cdk1. This priming event is followed by docking-enhanced phosphorylations leading to a phosphorylated chain of sites pT5/pT33/pT45/pS76 but no fully phosphorylated paired degrons, as phosphorylation by Cln2-Cdk1 of suboptimal sites in the degrons (T48 and S69, or S80) is slow (Fig. 3a, b). However, the phosphorylated cluster pT5/pT33/pT45/pS76 serves as a powerful Cln2-Cdk1-dependent docking platform for emerging Clb5-Cdk1. As Cln2 levels rise, such priming forces would create a synergistic effect between Cln2 and Clb5, greatly amplifying the impact of low emerging levels of free Clb5-Cdk1 complexes and defining the point of no return for Clb5-dependent positive feedback. A prediction of this model is that changing the limiting suboptimal degron sites to optimal Cdk sites will rescue the lethality of Sic1-1234rxl (Supplementary Fig. 4f), as the degradation in this case should be driven primarily by Cln2. Indeed, changing T48 to a Cdk1 site by introducing a proline at position +1 partially rescued the lethal phenotype of Sic1-1234rxl (Fig. 3d). A similar rescue was attained by introducing a positive determinant for Cln2-Cdk1, an arginine at position +2 16, to the site S80. Optimization of the S69 site had no effect. Almost complete rescue was gained by a triple mutation with all three limiting degron sites (T48, S69, and S80) changed to optimal Cdk sites. Importantly, these effects are unlikely to be due to improved binding of phosphodegrons to Cdc4, as the basic residues on the C-terminal side of pS/pT are known to disrupt the Cdc4 interaction 1. Finally, in order to confirm that degradation of these Sic1 mutants is driven by Cln-Cdk1, instead of Clb5-Cdk1, we additionally mutated the Cln2-specific docking site VLLPP in the triple mutant background (Fig. 3d, lower panel). The vllpp mutation abolished the rescue effect of the triple mutant. These data indicate that Cln2 alone does have the potential to drive Sic1 degradation, but the Cln2-driven phosphorylation cascade is terminated at the rate-limiting final steps. However, this mechanism allows creation of the Clb5 docking platform containing the chain of optimal sites pT5/pT33/pT45/pS76.
Finally, to compare further the functions of Clb5 and Cln2, we analyzed the degradation of endogeneous Sic1. We found that mutation of either the Cln-specific vllpp docking motif or Clb5-specific RXL docking sites delayed Sic1 degradation (Fig. 3e), confirming that both Cln2 and Clb5 have a role in the timing of Sic1 degradation. However, when all Clb-Cdk1 activity in the cell was specifically inhibited by overexpression of nondegradable Sic1, endogenous Sic1 was completely stabilized (Fig. 3f, Supplementary Fig. 7), arguing that the key trigger for Sic1 degradation and the G1/S transition is the emerging free Clb5-Cdk1, after its levels exceed those of the inhibitory complex.
The inability of Cln alone to cause Sic1 degradation could be attributed to the slow phosphorylation rate of sites in the S76/S80 degron. Indeed, by introducing optimal Cln consensus motifs into the slow degron sites, analogously to the experiment described in Figure 3d, the Sic1 degradation pattern was restored to normal despite the absence of Clb-Cdk1 activity (Fig. 3g). The processive multiphosphorylation cascades, composed of a set of fast and slow steps and different docking specificities, enable this discrimination between the signal outputs of different cyclin-Cdk1 complexes. Furthermore, the Cln output state acts as a primer state for the second complex, creating a potential ‘AND gate’ in which Cln2 is not allowed to trigger the G1/S transition until sufficient levels of Clb5 activity accumulate.
In conclusion, our model provides novel insights into the multisite phosphorylation mechanism of Sic1 and, potentially, of Cdk1 targets in general. The multiple sites create a network of docking connections that exploit Cks1-dependent and cyclin-specific docking interactions to process Cdk1 signals to achieve proper tuning of the timing of the G1/S transition (Supplementary Fig. 8). As most Cdk1 targets in the cell contain clusters of multiple sites 18, the regulation of cell cycle switchpoints by Cdk1-dependent multiphosphorylation might prove to be far more complex than generally anticipated, and it is possible that beneath the seemingly random constellations of phosphorylation sites, an intricate signal processing logic may be hidden.
Methods Summary
Yeast strains were in the W303 background and are listed in Supplementary Table 3. Plasmid constructs are listed in Supplementary Table 4. The Phos-tagTM Acrylamide AAL-107 19 was purchased from NARD Institute Ltd, Japan.
Methods
Protein purification
TAP-purification of cyclin-Cdk1 complexes was performed as described previously 20, 21 using C-terminally TAP-tagged cyclin constructs cloned into 2 micron vectors and overexpressed from the GAL1 promoter. For purification of 3HA-Cln2-Cdk1, a yeast strain (a kind gift from Dr. Doug Kellogg, UCSC) with the GAL1 promoter introduced along with the N-terminal 3HA tag in the chromosomal locus of the CLN2 gene was used. The overexpressed 3HA-Cln2-Cdk1 complex was purified as described 22, using immunoaffinity chromatography with a rabbit polyclonal antibody against the HA epitope (purchased from Labas, Estonia). Purification of N-terminally 6His-tagged recombinant Sic1 constructs was performed using cobalt affinity chromatography. Cks1 was purified as described previously 12. The optimal working concentration for purified Cks1 was taken as 500 nM based on optimization performed for cyclin-Cdk1 preparations and Sic1ΔC as a substrate.
Phosphorylation assays
For quantitative phosphorylation assays, the substrate concentration was kept in the range of 0.5–2 μM (in the linear [S] vs v0 range, several-fold below estimated KM) and the initial velocity conditions were defined as a substrate turnover ranging up to 10%/N of the total concentration of N Cdk sites. The general composition of the assay mixture was as follows: 50 mM Hepes pH 7.4, 100 mM NaCl, 0.1% NP-40, 20 mM imidazole, 2% glycerol, 2 mM EGTA, 0.2 mg/ml BSA, 500 nM Cks1 and 500 μM ATP (with added γ-32P-ATP (Perkin Elmer)). Around 1–10 nM of the purified kinase complex was used, the amount depending on the setup of the experiment. The optimal working concentration for purified Cks1 was taken as 500 nM according to the optimization performed for cyclin-Cdk1 preparations using Sic1ΔC as a substrate. For the phosphorylation assay with mutant Cks1 (Cks1mut), purified kinase complexes were preincubated for 45 minutes with Cks1wt or Cks1mut to compensate for differences in the amounts of Cks1 already present in the preparations. The composition of the preincubation mixture was: 50mM Hepes pH 7.4, 300 mM NaCl, 0.2 mg/ml BSA, 500 μM ATP. Kinase assay was initiated by adding preincubation mixture and γ-32P-ATP to the substrate. Aliquots were taken at least at two different time points and the reaction was stopped by SDS-PAGE sample buffer. For separating the phosphorylated versions of Sic1, 10% SDS-PAGE was used, supplemented with the Phos-tagTM reagent according to the instructions from the manufacturer.
For the phosphorylation assay of full-length Sic1 as part of the inhibitory complex, Clb5-TAP-Cdk1 was isolated from yeast cell extract containing overexpressed Clb5-TAP with IgG beads (Supplementary Fig. 4b). The stoichiometric Sic1-Clb5-Cdk1 complex was formed by incubating an excess amount of purified Sic1 with the beads, and unbound Sic1 was removed by washing (50 mM Hepes pH 7.4, 0.5 M NaCl, 0.1% NP-40). The phosphorylation reaction was performed according to the standard kinase assay protocol and was initiated by adding purified cyclin-Cdk1 complexes to the washed beads. The Clb5-dependent phosphorylation of full-length Sic1 as part of the stoichiometric inhibitory complex revealed a similar RXL specificity profile (Supplementary Fig. 4b). This result also suggests that Sic1ΔC is a valid model substrate, which was chosen for large scale analysis instead of the hard-to-adjust assay with the stoichiometric complex. It is reasonable to consider the N-terminal region of Sic1 as an independent polypeptide entity, given that only a short C-terminal part is required for high-affinity inhibition and that Sic1 is an intrinsically disordered protein 23.
The mutant Cks1 used in our experiments was designed to disrupt the phosphate-binding site and contained the combination of mutations R33E,S82E,R102A. The triple mutants in the hydrophobic patch (hpm) of Clb5 and Clb2 were described previously 24, and the hpm of Clb3 (F201A, L205A, T208A) was designed according to sequence homology with other B-type cyclins.
The relative rate constants for different phospho-docking enhanced steps (kdock) were determined using mutated Sic1ΔC versions containing different pairwise combinations of Cdk1 sites (the rest of the serine and threonine residues in S/TP motifs were mutated to alanines). It is impossible to estimate the effects of docking on the reaction rates directly, as it is hard to produce a version of substrate protein where the primed docking sites are fully phosphorylated and the secondary sites are unphosphorylated. Therefore, we used an indirect approach by estimating the relative formation rate of doubly phosphorylated species. The kdock was defined as the ratio of the observed fraction of the doubly phosphorylated form and the estimated kinase activity towards the single N-terminal priming site present in the pair-wise Sic1ΔC construct. The latter parameters were estimated using quantified values for single site specificities from the experiment in Figure 2b (in the absence or presence of different docking sites). Thus, the kdock values are independent of the rates of the priming steps and reflect only the rates of the secondary steps. The phosphorylation of the substrate was followed in a conventional kinase assay and singly and doubly phosphorylated species were resolved using Phos-Tag SDS-PAGE and quantified by PhosphorImager. The kdock was calculated from two consecutive time points (i.e. 8 and 16 minutes, in the low initial range of total substrate consumption) from at least two independent experiments. We applied a condition of a minimal ratio of 0.7 for the kdock values from these two time points, to ensure that the singly phosphorylated species had not reached the temporary quasi steady state. The obtained values are ‘apparent constants’ as it is impossible to precisely determine the relative contributions of processive and distributive mechanisms. Nevertheless, the obtained kdock values provide very good estimates of how much the phosphorylation of a site is enhanced when another site is present.
Western blottings and viability assays
For viability assay, log-phase cultures were equalized in density and spotted as serial dilutions on selective synthetic complete (SC) plates. The plates were incubated for 48–60h at 30°C. For the western blotting experiments, cells were grown to OD600=0.3 and treated for 2,5h with 1μg/ml α-factor or for 2,5h with 15mg/ml hydroxyurea and released by washing. In the experiments presented in Fig. 3f, g and Supplementary Fig. 7 the GAL1-SIC1ΔN was integrated into the URA3 locus. In Fig. 2g the Sic1ΔC-3HA versions were cloned into the pRS315 vector and constitutively expressed under the ADH promoter. In the experiments presented in Fig. 3e, f, g the endogeneous SIC1 was C-terminally TAP tagged. The cells were lysed by bead-beating in lysis buffer containing urea. Blotting of Phos-Tag SDS-PAGE gels was performed using a dry system iBlot (Invitrogen). The antibody used for western blotting of 3HA-tagged proteins was HA.11 Clone 16B12 from Covance, USA and the antibodies used for western blotting of Cdk1 (Cdc28 (yC-20)) and of TAP-tagged proteins (c-Myc (A-14)) were from Santa Cruz Biotechnology, USA. The rabbit anti-Cks1 was from Labas, Estonia.
Isothermal calorimetry
Recombinant Sic1ΔC was phosphorylated with purified Clb5-Cdk1, and complete phosphorylation was confirmed by following the phosphorylation shift by Phos-Tag SDS-PAGE. Recombinant Cks1 was expressed in E. coli from a pET vector and purified with anion exchange and size exclusion chromatography. Calorimetry experiments were performed with a VP-ITC system (MicroCal). 0.3–0.4 mM Cks1 was titrated into a 30 μM solution of phospho-Sic1ΔC. Experiments were carried out at 25°C in a buffer containing 25 mM Tris and 150 mM NaCl (pH 8.0). Data were analyzed with the MicroCal Origin software package. The reported binding constant and stoichiometry (n) are the average from 2 experiments, and the reported errors are the standard deviation of these measurements.
Quantitative mass-spectrometry
For quantitative determination of Cks1-dependent phosphorylation of T48, equal amounts of Sic1ΔC-wt protein were phosphorylated by Clb5-Cdk1 supplemented with normal isotopic ATP [(16O)-ATP] or heavy ATP [(18O)-ATP] (Cambridge Isotope Laboratories, Inc.). Kinase assays were incubated at room temperature for 60 min and after pooled together in a 1:1 ratio (v/v) in SDS-PAGE sample buffer. The proteins were separated by 10% SDS-PAGE and the gels were stained with Coomassie briliant blue G-250 (Sigma) and protein bands were excised from the stained gels. Proteins were in-gel digested by LysC/P (10 ng μl−1) (Wako) and peptides were purified by using C18 StageTips.
Peptides were separated by Agilent 1200 series nanoflow system (Agilent Technologies) connected to a LTQ Orbitrap classic mass-spectrometer (Thermo Electron) equipped with nanoelectrospray ion source (Proxeon). Purified peptides were loaded on a fused silica emitter (75 μm x 150 mm) (Proxeon) packed in-house with Reprosil-Pur C18-AQ 3 μm particles (Dr. Maisch). Peptides were separated with 30 minute 3–40% B gradient (A: 0.5% acetic acid, B: 0.5% acetic acid/80% acetonitrile) at a flow-rate of 200 nl/min, eluted peptides were sprayed directly into LTQ Orbitrap mass-spectrometer with a spray voltage of 2.2 kV. The MS scan range was m/z 300–1800 and the top 5 precursor ions were selected for subsequent MS/MS scans. A lock-mass was used for the LTQ-Orbitrap to obtain constant mass-accuracy during the gradient analysis. Peptides were identified with Mascot 2.3 (www.matrixscience.com) search engine. Peptide mass tolerance of 7 ppm was used and fragment ion mass tolerance of 0.6 Da. Two missed cleavage site for LysC/P was allowed. The oxidation of the methionine and the phosphorylation of serine, threonine were set as variable modifications.
Supplementary Material
Acknowledgments
We thank Doug Kellogg for strains and Lauri Peil for advice on MS. This work was supported by International Senior Research Fellowship No. 079014/Z/06/Z from the Wellcome Trust (ML), an installation grant from EMBO and HHMI (ML), No. 1253, Grant No. 6766 from the Estonian Science Foundation (ML), EMP grant No. 08071N from the Norwegian government, and a grant from the National Institute of General Medical Sciences (DOM).
Footnotes
Author Contributions
The first five authors and M. Loog designed and performed the experiments, except for the isothermal calorimetry experiments, performed by E.R.M.B. and S.M.R. M. Loog coordinated the project and wrote the manuscript with assistance from D.O.M. and S.M.R.
References
- 1.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 2.Borg M, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SY, Ferrell JE., Jr Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang X, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci U S A. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma R, et al. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 8.Cross FR, Schroeder L, Bean JM. Phosphorylation of the Sic1 inhibitor of B-type cyclins in Saccharomyces cerevisiae is not essential but contributes to cell cycle robustness. Genetics. 2007;176:1541–1555. doi: 10.1534/genetics.107.073494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 10.Arvai AS, Bourne Y, Hickey MJ, Tainer JA. Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain. J Mol Biol. 1995;249:835–842. doi: 10.1006/jmbi.1995.0341. [DOI] [PubMed] [Google Scholar]
- 11.Hadwiger JA, Wittenberg C, Mendenhall MD, Reed SI. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol Cell Biol. 1989;9:2034–2041. doi: 10.1128/mcb.9.5.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynard GJ, Reynolds W, Verma R, Deshaies RJ. Cks1 is required for G(1) cyclin-cyclin-dependent kinase activity in budding yeast. Mol Cell Biol. 2000;20:5858–5864. doi: 10.1128/mcb.20.16.5858-5864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Reed SI. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 14.Patra D, Wang SX, Kumagai A, Dunphy WG. The xenopus Suc1/Cks protein promotes the phosphorylation of G(2)/M regulators. J Biol Chem. 1999;274:36839–36842. doi: 10.1074/jbc.274.52.36839. [DOI] [PubMed] [Google Scholar]
- 15.Deshaies RJ, Ferrell JE., Jr Multisite phosphorylation and the countdown to S phase. Cell. 2001;107:819–822. doi: 10.1016/s0092-8674(01)00620-1. [DOI] [PubMed] [Google Scholar]
- 16.Koivomagi M, et al. Dynamics of Cdk1 Substrate Specificity during the Cell Cycle. Mol Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita E, Yamada A, Takeda H, Kinoshita-Kikuta E, Koike T. Novel immobilized zinc(II) affinity chromatography for phosphopeptides and phosphorylated proteins. J Sep Sci. 2005;28:155–162. doi: 10.1002/jssc.200401833. [DOI] [PubMed] [Google Scholar]
- 20.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 21.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 22.McCusker D, et al. Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol. 2007;9:506–515. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- 23.Mittag T, et al. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.