Summary
Bacillus pseudofirmus OF4 is an extreme but facultative alkaliphile that grows non-fermentatively in a pH range from 7.5 to above 11.4 and can withstand large sudden increases in external pH. It is a model organism for studies of bioenergetics at high pH, at which energy demands are higher than at neutral pH because both cytoplasmic pH homeostasis and ATP synthesis require more energy. The alkaliphile also tolerates a cytoplasmic pH > 9.0 at external pH values at which the pH homeostasis capacity is exceeded, and manages other stresses that are exacerbated at alkaline pH, e.g. sodium, oxidative and cell wall stresses. The genome of B. pseudofirmus OF4 includes two plasmids that are lost from some mutants without viability loss. The plasmids may provide a reservoir of mobile elements that promote adaptive chromosomal rearrangements under particular environmental conditions. The genome also reveals a more acidic pI profile for proteins exposed on the outer surface than found in neutralophiles. A large array of transporters and regulatory genes are predicted to protect the alkaliphile from its overlapping stresses. In addition, unanticipated metabolic versatility was observed, which could ensure requisite energy for alkaliphily under diverse conditions.
Introduction
Extremely alkaliphilic Bacillus species grow optimally at pH values above 9.5 and often exhibit growth above pH 11(Horikoshi & Akiba, 1982, Krulwich et al., 2011b, Yumoto, 2002). They are found in soil as well as in naturally alkaline ecological niches, such as alkaline soda lakes, and in enrichments that result from industrial processes, such as indigo dye plants (Grant, 2003, Horikoshi, 1999, Yumoto, 2007, Krulwich & Guffanti, 1989). Alkaliphilic bacteria are of applied interest because of the usefulness of many of their alkali-tolerant enzymes and the potential that some of them have for bio-remediation (Fujinami & Fujisawa, 2010, Horikoshi, 1998, Horikoshi, 1999, Sarethy et al., 2011). In addition, adaptations of alkaliphile physiology as well as adaptations of specific proteins provide insights into stress responses and central physiological functions such as oxidative phosphorylation and cytoplasmic pH homeostasis that have importance beyond alkaliphiles (Hicks et al., 2010, Kitada et al., 2000, Padan et al., 2005).
Growth at high pH is costly relative to neutral pH because of the energy demands of cytoplasmic pH homeostasis and the greater energy required for ATP synthesis under alkaline conditions (Hicks et al., 2010, Krulwich et al., 2007, Krulwich et al., 2011a). Yet the bulk chemiosmotic driving force, the bulk protonmotive force (PMF) is much lower than in neutralophilic bacteria (Krulwich et al., 2007, Krulwich et al., 2011a). This is the driving force proposed by Mitchell to be required for ATP synthesis and energization of sodium/proton antiporters on which alkaliphile pH homeostasis depends (Mitchell, 1961, West & Mitchell, 1974). Bioenergetic studies on oxidative phosphorylation in alkaliphilic Bacillus species led to the proposal that a surface-associated PMF that is larger than the bulk PMF is used for oxidative phosphorylation even in non-alkaliphilic settings (Brändén et al., 2006, Cherepanov et al., 2003, Heberle et al., 1994, Krulwich, 1995, Mulkidjanian et al., 2006). Studies of ATP synthesis at high pH in alkaliphilic B. pseudofirmus OF4 further demonstrated a requirement for specific adaptations of the ATP synthase that provided structure-function insights into this centrally important enzyme (Fujisawa et al., 2010, Liu et al., 2009, Preiss et al., 2010, Wang et al., 2004). Similarly, studies of alkaliphile pH homeostasis led to discovery of a novel category of hetero-oligomeric cation/proton antiporters, the Mrp (Cation Proton Antiporter-3) family of antiporters, which has important roles in non-alkaliphiles as well (Hamamoto et al., 1994, Kitada et al., 2000, Swartz et al., 2005). Studies of alkaliphilic Bacillus species also identified a family of voltage-gated sodium channels and the Na+-coupled MotPS type of flagellar stator, which have roles both in alkaliphilic and in non-alkaliphilic bacteria (Fujinami et al., 2009, Ito et al., 2004a, Ito et al., 2004b, Ren et al., 2001).
Here we report the complete genome sequence of extremely alkaliphilic Bacillus pseudofirmus OF4, a facultative aerobic alkaliphile isolated from soil that grows non-fermentatively in a pH range from ~7.5 to above 11.4 and also grows fermentatively (Guffanti & Hicks, 1991, Sturr et al., 1994, Wang et al., 2004). This alkaliphilic capacity somewhat exceeds that of closely related alkaliphilic Bacillus halodurans C-125 (Ito, 2002), whose genome was sequenced over a decade ago (Takami et al., 2000). Because of its broad alkaline pH range and genetic accessibility, B. pseudofirmus OF4 has become a model system for bioenergetic studies of alkaliphily (Ito et al., 1997, Ito et al., 2004a, Ito et al., 2004b, Liu et al., 2009, Wang et al., 2004). However, major unanswered questions remain about oxidative phosphorylation, pH homeostasis and stress responses of this and other extremely alkaliphilic Bacillus species. The questions include the strategies for meeting high energy costs, the basis for growth when the cytoplasmic pH is as high as 9.5 and the strategies for managing other stresses that are exacerbated at high pH. The genome sequence data for the B. pseudofirmus OF4 chromosome and two resident plasmids suggest a built-in capacity for genomic rearrangements that could foster adaptability. The genomic data further reveal unanticipated versatility in the metabolic and energy storage capacities of B. pseudofirmus OF4 that may account in part for its robust alkaliphily. The data also provide information that will lead to testable hypotheses about “overlapping stress management” and about further adaptations that contribute to oxidative phosphorylation and pH homeostasis at high pH.
Results and discussion
General features of the genome, including two resident plasmids
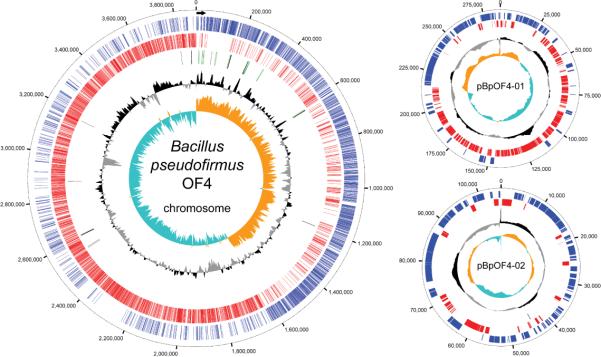
The genome of Bacillus pseudofirmus OF4 consists of a 3,858,997 bp circular chromosome and two plasmids; pBpOF4-01 (285,222 bp) and pBpOF4-02 (109,029 bp) (Table 1). The sizes of the main chromosome and the small plasmid are consistent with earlier chromosomal mapping (Gronstad et al., 1998) whereas the presence of the large plasmid has not been previously reported. The average GC content of the chromosome is 40.4% whereas the large and small plasmids have an average GC content of 36% and 35.5% respectively. Graphical representations of the chromosome and plasmids are shown in Fig. 1. There are seven ribosomal RNA operons on the chromosome. These are clustered within .65 Mb downstream of the origin of replication (Fig. 1) as is observed in Bacillus halodurans C-125 (Nakasone et al., 2000). Each operon contains a 16S, 23S and 5S rRNA except for the 5th operon which possesses two 5S sequences. The annotation data, obtained as described under Experimental Procedures, are summarized in Table 1. Phylogeny data shown in Table 1 indicate that a very large proportion of top BLASTP hits for B. pseudofirmus OF4 chromosomal genes are from alkaliphilic Bacillus halodurans C-125, followed by two other alkaliphilic Bacillus strains. The top BLASTP hits for the genes in the two plasmids are distinct from those chromosomal sources and are also distinct from each other.
Table 1.
Bacillus pseudofirmus OF4 genome statistics
| Chromosome (Ctg1) | pBpOF4-01 (Ctg2) | pBpOF4-02 (Ctg3) | |
|---|---|---|---|
| Accession # | CP001878 | CP001879 | CP001880 |
| Size (bp) | 3,858,997 | 285,222 | 109,029 |
| GC Content (%) | 40.4 | 36 | 35.5 |
| ORFs | 3,922 | 292 | 122 |
| ORF Coding percentage | 85.63% | 79.59% | 71.63% |
| ORFs with functional assignment | 2,235 | 117 | 30 |
| Hypothetical ORFs | 1,686 | 175 | 92 |
| Proteins with TM domains | 1105 | 87 | 46 |
| Proteins with Enyzme Classification (EC) | 921 | 13 | 4 |
| Proteins with Gene Ontology (GO) | 997 | 28 | 4 |
| rRNA operons | 7 | 0 | 0 |
| tRNAs | 75 | 0 | 2 |
| Mobile element related | 59 | 33 | 12 |
| Species with most top BLASTP hits* | Bacillus halodurans (1510) | Bacillus sp. (26) | Bacillus cereus (11) |
| Bacillus sp. (418) | Lysinibacillus sphaericus (20) | Bacillus cellulosilyticus (6) | |
| Bacillus clausii (234) | Paenibacillus sp. (13) | Bacillus sp. (6) | |
| Bacillus cellulosilyticus (180) | Bacillus halodurans (12) | Bacillus halodurans (4) | |
| Bacillus megaterium (101) | Bacillus cellulosilyticus (10) | Bacillus thuringiensis (3) |
Numbers in parentheses indicate the number of top BLASTP hits
Fig. 1.
Circular representation of the B. pseudofirmus OF4 genome. Black arrow – dnaA gene (oriC). Blue bars – genes on the forward strand, red bars – genes on the reverse strand, green bars – ribosomal RNA, black bars – tRNAs. Black ring – GC plot, Orange and blue ring – GC skew.
Resident plasmids
Many of the genes of the resident plasmids are predicted to encode mobile elements. Apart from those genes, which will be discussed in the next section, both plasmids have a large number of hypothetical proteins along with a mix of genes predicted to encode regulatory proteins, signaling proteins, enzymes, transporters, redox proteins and chaperones. For both plasmids, especially the larger pBpOF4-1, one theme of the genes with proposed functions is metal acquisition and heavy metal resistance. The pBPOF4-01 plasmid has genes predicted to encode a ferritin Dps protein (BpOF4_19979), a P-type ATPase (BpOF4_20074) as well as a CadD type (BpOF4_20084) cadmium transporter, a P-type ATPase for copper (BpOF4_20189) as well as a copper chaperone protein (BpOF4_20194) and copper resistance protein (BpOF4_20289), a manganese ABC transporter (BpOF4_20504/_20509/_20514), and a mercury resistance locus including an efflux protein (BpOF4_20579). The small plasmid, pBpOF4-02, has genes predicted to encode: a P-type ATPase (BpOF4_21834) as well as CadD type (BpOF4_21839) cadmium resistance transporter, but also metabolic proteins including a locus with a TRAP-T dicarboxylate uptake system (BpOF4_21499_21504_21509), malate/lactate dehydrogenase (BpOF4_21514), alcohol dehydrogenase (BpOF4_21519) and aldehyde dehydrogenase (BpOF4_21524). Some of the plasmid genes have chromosomal genes as their closest homologues, suggesting gene transfers between plasmids and chromosome as has been shown in Bacillus megaterium strains (Eppinger et al., 2011).
During the sequencing effort, independent preparations of DNA were used. One of them lacked the large pBpOF4-01 plasmid, as had the earlier sample used in a physical mapping study (Gronstad et al., 1998). Like the mapping study sample, the cells from which this sample was isolated had been shipped from one collaborating laboratory to another, suggesting that particular manipulations or stresses might lead to loss of the large plasmid of B. pseudofirmus OF4. DNA and/or colony PCR, with primer sets for regions of the chromosome and each plasmid, were used to survey the plasmid content of different strains that have been used as “wild type strains” in earlier studies: (i) B. pseudofirmus OF4, the object of this genome sequencing project (6 stocks, 3 of which had been shipped between collaborating laboratories); two stocks of B. pseudofirmus OF4-811M, which is a derivative of B. pseudofirmus OF4 with a methionine auxotrophy marker (Clejan et al., 1989); and (iii) a single stock of Bacillus pseudofirmus RAB (formerly called Bacillus firmus RAB), a more obligately alkaliphilic strain that is closely related to B. pseudofirmus OF4 (Guffanti et al., 1980). Four stocks of these strains that had been stored in the laboratory of origin were found to have both plasmids, while two stocks of B. pseudofirmus OF4 that had been shipped to other laboratories had lost the large plasmid (Table 2, top section). The energetic burden associated with introduction of multi-copy plasmids did not cause loss of endogenous plasmids from B. pseudofirmus OF4-811M as shown for four distinct transformants, one of which had an insert (Table 2, middle section). Analyses of diverse mutant strains constructed in B. pseudofirmus OF4-811M and kept as stocks in the laboratory of origin, revealed that six of the ten mutant strains made directly from B. pseudofirmus OF4-811M lacked the large pBpOF4-01 plasmid (Table 2, bottom), Loss of the small pBpOF4-02 plasmid was only observed in two mutants that also had lost the large plasmid. Strains that lacked both plasmids were isolates with mutations in the ctaD gene of cytochrome c oxidase, and five out of twelve isolates of a double mutant made in the ATP synthase atpE gene using the pBpOF4-01-minus and Fo-ATP synthase-minus (ΔatpB-F) background. The analyses as a whole suggest that the loss of the large pBpOF4-01 plasmid may be stochastic. Loss of the small plasmid was only observed in strains without the large plasmid and there may be some dependence of its loss on a defect in oxidative phosphorylation. No effects of plasmid loss on growth for 14 hr in Malate-Yeast Extract (MYE) medium at pH 7.5 and 10.5 were observed in comparisons, respectively, of the atpB silent mutant vs. parent strain B. pseudofirmus OF4-811M. Nor were growth deficits observed in comparisons of strains of the double atpE mutant, ala18/22gly, that had no plasmids vs. strains of the same mutant that had the small plasmid only. Perhaps loss of one or both plasmids affects growth on heavy metals, where the plasmid resistance loci may have a significant impact, as they might under rapidly changing or unusually extreme environmental conditions, where the large number of mobile elements may become assets as agents of change.
Table 2.
Plasmids present in Bacillus pseudofirmus OF4, closely related strains and derivative transformants and mutant strainsa.
| Strains | # of independent stocks | Large plasmid | Small plasmid | Source |
|---|---|---|---|---|
| "Wild-type" strains | ||||
| Bacillus pseudofirmus OF4 | 6 | 4Yes/2No | Yes | (Guffanti et al.., 1986) |
| Bacillus pseudofirmus OF4-811M | 2 | Yes | Yes | (Clejan et al., 1989) |
| Bacillus pseudofirmus RAB | 1 | Yes | Yes | (Guffanti et al., 1980) |
|
| ||||
| Transformant s of B. pseudofirmus OF4-811M | ||||
| pBK15 | 1 | Yes | Yes | (Ito et al., 1997) |
| pYH56 | 1 | Yes | Yes | (Hicks et al., 2003) |
| pHP13-clsA | 1 | Yes | Yes | Hicks, unpublished |
| pYM24 | 5 | Yes | Yes | (Ito et al., 2004a) |
|
| ||||
| Mutant strains of B. pseudofirmus OF4-811M | ||||
| Δ amhT | 1 | Yes | Yes | (Wei et al., 2003) |
| Δ nhaC | 1 | No | Yes | (Ito et al., 1997) |
| Δ ncbA | 1 | No | Yes | (Ito et al., 2004b) |
| Δ atpZ | 1 | No | Yes | (Hicks et al., 2003) |
| Δ atpI | 1 | Yes | Yes | (Hicks et al., 2003) |
| Δ atpZI | 1 | Yes | Yes | (Hicks et al., 2003) |
| atpD,his6-tagged | 2 | Yes | Yes | (Preiss et al., 2010) |
| Δ atpB-F | 1 | No | Yes | (Wang et al., 2004) |
| atpB silent mutation | 2 | No | Yes | (Wang et al., 2004) |
| atpE, pro51ala | 2 | No | Yes | (Wang et al., 2004) |
| atpE, TMH1 | 1 | No | Yes | (Wang et al., 2004) |
| atpE, ala18/22gly | 12 | No | 7Yes/5No | (Liu et al., 2011) |
| atpE, ala16/18gly | 2 | No | Yes | (Liu et al., 2011) |
| atpE, ala20/22gly | 2 | No | Yes | (Liu et al., 2011) |
| Δ ctaC | 1 | No | Yes | This work |
| Δ ctaD | 1 | No | No | (Gilmour & Krulwich, 1997) |
The presence of the large resident plasmid, pBpOF4-01 and/or small resident plasmid, pBpOF4-02, were detected by colony PCR and/or sequencing efforts of DNA samples for the indicated number of independent stocks of the indicated strains of B. pseudofirmus OF4, its derivatives and closely related B. pseudofirmus RAB.
Mobile elements
B. pseudofirmus OF4 contains mobile and repeated genetic elements including DNA transposons (insertion sequences), group I and group II introns, and interspersed DNA repeats. In total, 104 ORFs (open reading frames) are related to mobile element functions, including transposases, resolvases, integrases, reverse-transcriptases (RT), and homing endonucleases (Table S1). While these ORFs represent only 1.5% of the chromosomal ORFs, they constitute ~10% of the gene content of each plasmid.
A remarkable feature of the B. pseudofirmus OF4 genome is the presence of a group I intron located in the gene encoding the tmRNA, a special RNA molecule with tRNA-like and mRNA-like domains involved in translation regulation ((Karzai et al., 2000); Fig. S1). Whereas the tmRNA gene has been identified in almost all sequenced bacterial genomes, the distribution of the group I intron is very limited, as less than 1% of the hundreds of known tmRNA sequences are interrupted by an intron (according to data in the tmRNA website, tmRDB, and Rfam databases (Williams, 2002, Zwieb et al., 2003, Gardner et al., 2009)). Intron-containing tmRNAs are mostly found in species from the Clostridium genus, such as C. botulinum and C. tetani, and in the uncharacterized Bacillus sp. SG-1. Indeed, the tmRNA-associated group I introns from these organisms and B. pseudofirmus OF4 are inserted at the same site within the tRNA-like domain (P12) of the tmRNA and share 68–78% nucleotide sequence identity over their entire length, indicating a common origin (Fig. S1).
The B. pseudofirmus OF4 genome also harbors six group II introns, one of which is inserted in the chromosome while five are inserted in the pBpOF4-01 plasmid (Table S1). Noticeably, two of the plasmidic introns are nested and form a twintron structure, where the innermost intron is inserted at the site corresponding to the catalytic site of the RT encoded by the outermost intron (Fig. S2). Twintrons are uncommon in prokaryotes and have only been found in very few specific genomes of Bacteria and Archaea (Dai & Zimmerly, 2003, Dai et al., 2003). The innermost intron of the B. pseudofirmus OF4 twintron is related to particular group II introns of Bacillus thuringiensis, Photorhabdus luminescens, and Methanosarcina acetivorans, which specifically target RTs of group II introns and other retroelements (Tourasse & Kolstø, 2008, Dai & Zimmerly, 2003). The other striking group II intron feature of B. pseudofirmus OF4 is that one of the pBpOF4-01 introns harbors an unusual structural extension at the 3' end which is similar to that found in introns from bacteria belonging to the Bacillus cereus group (Stabell et al., 2009, Tourasse et al., 2010).
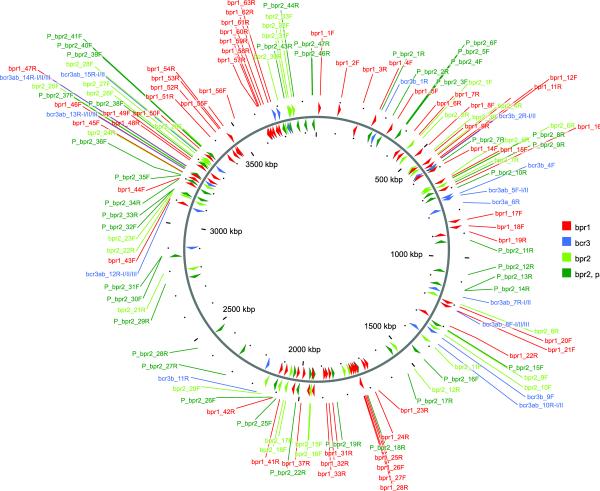
In addition to known mobile elements, B. pseudofirmus OF4 contains three sets of intergenic, interspersed, non-coding repeated DNA elements of length 100–400 bp, which are exclusively found in the chromosome (Fig. 2). The B. pseudofirmus OF4 chromosome encodes 35 copies of the bcr3 repeat elements that were originally identified and analyzed in bacteria from the B. cereus group, and which are also present in some Bacillus and Geobacillus species (Tourasse et al., 2006, Kristoffersen et al., 2011). These elements are ~140 bp long and exhibit characteristics of transposable elements, including terminal inverted repeat motifs and target site duplication. As in B. cereus group chromosomes, bcr3 repeats can form arrays containing two or three overlapping copies, and are over-represented near the 3' end of genes, some of them having housekeeping or important functions such as NADH dehydrogenase and the GroEL chaperone. In contrast, the major repeat element of B. pseudofirmus OF4, tentatively named bpr1 (for Bacillus pseudofirmus repeat), is unique to that strain. It is ~170 bp in length and is present in 63 copies. bpr1 itself exhibits a repetitive modular structure made up of CGCAATAA or CGCATAAA motifs repeated seven times in the sequence. In four chromosomal loci, adjacent copies of bpr1 and bcr3 elements were found. The third repeat of B. pseudofirmus OF4, bpr2, also exhibits a peculiar structure. bpr2 is ~400 bp long and consists of two inverted regions of ~90 bp separated by a linker region of ~150 bp. While there are 33 full-length bpr2 copies in B. pseudofirmus OF4, in some chromosomal regions partial copies corresponding to the left (5') or right (3') end are found, and particular loci contain three or four adjacent ends, giving a total number of 80 bpr2-like sequences in B. pseudofirmus OF4. Altogether, the bpr1, bpr2, and bcr3 repeat elements are distributed throughout the B. pseudofirmus OF4 chromosome, with higher densities in regions close to the origin and terminus of replication, and do not exhibit a strand bias (Fig 2). Alignments of the three groups of repeat elements are shown in Figs. S3–S5.
Fig. 2.
Distribution of interspersed DNA repeats of 100 bp or more in the chromosome of B. pseudofirmus OF4. Each repeat element (bpr1, bpr2 and bcr3) is drawn in a different color. Repeat copies are numbered consecutively. Copies located on the forward DNA strand are drawn above the thick grey line representing the chromosome and their names contain the letter F, whereas copies located on the reverse strand are drawn below the grey line and their names contain the letter R. Partial copies of the bpr2 repeat element are indicated with names starting with the letter P. The figure was generated using CGview (Stothard & Wishart, 2005).
Origin of replication of the chromosome
The origin of replication in B. pseudofirmus OF4 chromosome is comparable to that in B. halodurans C-125. The web-based tool Ori-Finder was used to locate the region of the genome where DNA asymmetry was at a maximum as well as the location of putative 9 bp dnaA boxes (TTWTNCACA). The region identified was 2 kilobases in length (280nt – 2,280nt) and the location of OriC associated genes such as DnaA and DnaN were located nearby. DNA replication has been shown to require two regions of dnaA boxes up and downstream of the DnaA gene in Bacillus subtilis (Moriya et al., 1992). These two dnaA box regions (Box region C and Box region R) are also found in B. halodurans C-125 (Takami et al., 1999). Searching in the location identified by ORI-finder for the perfect Escherichia coli dnaA box TTATCCACA with a maximum of 1 mismatch identified these dnaA clusters in B. pseudofirmus OF4 as well (Fig. S6). Unwinding of the DNA during initiation in B. subtilis occurs when DnaA binds to the dnaA box 14bp upstream of an AT-rich region in the intergenic region between DnaA and DnaN. This event unwinds 28 bp in the AT-rich region with an additional 16–18bp being unwound in the presence of single-stranded DNA-binding protein (Krause & Messer, 1999). A comparison of the nucleotide sequences of the oriC at the AT-rich region of B. subtilis 168, B. halodurans C-125 and B. pseudofirmus OF4 is shown in Fig. S6.
Isoelectric point (pI) profiles of proteins from B. pseudofirmus OF4 and other Bacillus species
Analyses of diverse bacterial genome sequences have revealed that the profile of the pI values for the overall genome proteins and/or proteins with particular localizations exhibit fall into different ranges (Kennedy et al., 2001, Schwartz et al., 2001). Among alkaliphilic Bacillus proteins, it was noted even before genome sequences were available that particular excreted enzymes (van der Laan et al., 1991) and segments of membrane proteins that are exposed on the outside surface had a significantly higher content of acidic residues than their neutralophilic homologues (Krulwich et al., 2007, Krulwich et al., 2011a, Quirk et al., 1993). A comparison of the average predicted pI of proteins in different compartments was carried out for four alkaliphilic Bacillus species, B. pseudofirmus OF4, B. selenitireducens MLS10, B. halodurans C-125 and B. clausii KSM-K16, and three neutralophilic Bacillus species, B. subtilis 168, B. cereus ATCC 14579, and B. pumilis SAFR-032. As shown in Fig. 3, a lower pI for the alkaliphile proteins was observed in all compartments, with the difference being most pronounced among the cell wall and extracellular proteins. Acidic residues, which will be charged under highly alkaline conditions, may be required for proper folding or retention of function by some proteins or protein segments (Quirk et al., 1993, van der Laan et al., 1991). For fixed elements on the cell wall or outer membrane surface, the negative charges may also contribute to attracting sodium ions and protons near the surface (Krulwich et al., 2007, Krulwich et al., 2011a). The small but significantly more acidic character to cytoplasmic proteins from alkaliphilic vs. neutralophilic Bacillus species may be part of a broader set of global adaptations of cytoplasmic proteins that underpins retention of function when at cytoplasmic pH > 9. Cytoplasmic enzymes from alkaliphilic Bacillus alcalophilus and Bacillus circulans ssp. alkalophilus exhibit distinctive structural and sequence features that are likely to be adaptations for function at elevated pH (Dubnovitsky et al., 2005, Kapetaniou et al., 2006).
Fig. 3.
Predicted amino acid sequences from 7 complete Bacillus genomes were submitted to the Compute pI / Mw Tool at the Swiss Institute of Bioinformatics ExPASy Proteomics Server (http://ca.expasy.org/tools/pi_tool.html). The same sequences were also submitted to PSORTb v3.0 (Yu et al., 2010) for determination of subcellular location. Comparison of the means between B. pseudofirmus OF4 and the six other Bacilli was performed with unpaired 2-sample T-tests. ** p<0.0001, * p<0.05
Synteny with Bacillus halodurans C-125 and some of the genomic differences
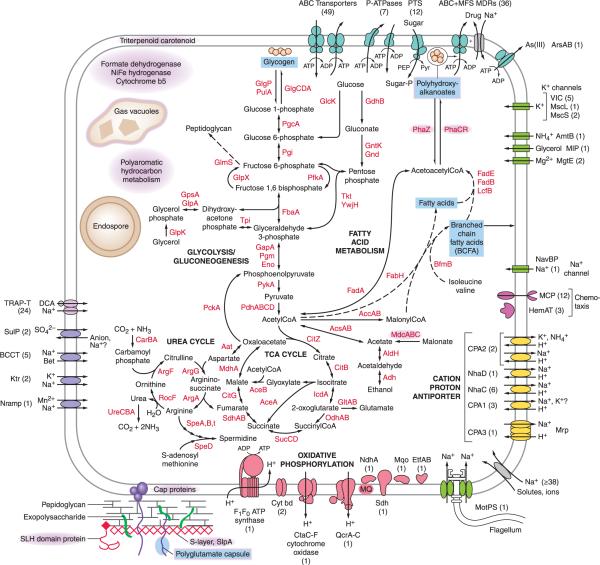
The chromosome of B. pseudofirmus OF4 was compared with its closest taxonomic sequenced neighbor, B. halodurans C-125 using annotations obtained from RAST (Rapid Annotations with subsystems). First, the chromosomes were rearranged so that origin and orientation were the same. Then, each gene on the B. pseudofirmus OF4 chromosome was compared to the B. halodurans C-125 genome with BLASTP searches. About 80% of the genes gave positive hits using a cutoff of 50% protein identity and the remaining 20% were not considered to be orthologous. A dot plot was generated using the B. pseudofirmus OF4 query and B. halodurans C-125 BLAST hit gene identifier numbers as coordinates. This dot plot illustrates the genic co-linearity of the two species identifying several regions of insertions or deletions interspersed throughout the genome but no significant inversions (Fig. S7). Specific differences between the two alkaliphiles that might account for the somewhat more robust alkaliphily of B. pseudofirmus OF4 were sought among the chromosomal genes in B. pseudofirmus OF4 for which there were “no hits” from BLASTP searches of B. halodurans C-125; but for which there was a suggested function. Four large gene clusters were identified, including those accounting for the largest gaps in the synteny plot (Fig. S7). These are loci encoding: a Ni/Fe hydrogenase/cytochrome b5 complex (BpOF4_01290 to _01355); gas vacuoles (BpOF4_07620 to _07690); PAH (polyaromatic hydrocarbon) breakdown enzymes (BpOF4_12555 to _12625); proteins with SLH (S-layer homology) domains together with assembly enzymes for SCWPs (Secondary Cell Wall Polymers) (BpOF4_05780 to _05940). Smaller loci found in B. pseudofirmus OF4 but not in B. halodurans C-125 include: Cap proteins for a polyglutamate capsule (BpOF4_06650 to _06660); malonate transport and utilization enzymes (BpOF4_11955 to _11980); enzymes for selenocysteine synthesis and incorporation into proteins (BpOF4_19305 to _19345); synthetic enzymes for a membrane triterpenoid carotenoid (BpOF4_14550 to _14575); bacillithiol spermidine synthetase (BpOF4_01225 to _01235); and enzymes for PHA (polyhydroxyalkanoate) synthesis (BpOF4_07445 to _07450). Many of these loci are indicated in the diagram of a B. pseudofirmus OF4 cell shown in Fig. 4. A comprehensive examination of B. halodurans C-125 genes that are absent from B. pseudofirmus OF4 was not undertaken. However, it is of interest to note that B. pseudofirmus OF4 lacks the tuaG gene (BH3661) and has no close homologue of the tuaA gene (BH3650) that are required for synthesis of acidic cell wall-associated teichuronic acids that have a vital role in cytoplasmic pH homeostasis in B. halodurans C-125 (Aono et al., 1999, Aono & Ohtani, 1990). This is consistent with divergent patterns of SWCPs and their contributions to alkaliphily in the two closely related alkaliphiles.
Fig. 4.
Features of the Bacillus pseudofirmus OF4 genome that are proposed adaptations to the energy demands and overlapping stresses related to alkaliphily and reveal structural and metabolic properties not found in closely related Bacillus halodurans C-125. The selected metabolic pathways shown focus on energy-generation and potential stored energy forms (light blue). Dashed arrows represent multi-step pathways for which only representative enzymes are shown. The primary transporter groups (shown in aqua) are predicted to have roles in antibiotic resistance/cell envelope stress, nutrient uptake, and ion homeostasis. The ABC-type MDRs (multi-drug transporters) included in the 36 total MDRs are also counted in the ABC transporter total of 49. The selected groups of secondary transporters that are shown have potential or established roles in: cytoplasmic pH homeostasis (yellow); sodium-dependent efflux of toxins and uptake of nutrients, ions or compatible solutes (gray); and the TRAP-T transporters, with DCA (dicarboxylic acids as a substrate), whcih are highlighted in purple because the 24 loci significantly exceeds the 16 loci found in the B. halodurans C-125 genome. See text for full names of transporters abbreviated here. The representative channels (green) that are shown have roles in motility, pH homeostasis, osmo-regulation, cation and nutrient uptake. In the square-shaped figure that represents a rod-shaped bacterium, the top/bottom represent the long sides, where the flagellum is found, and the sides represent the poles, where the sodium channel and chemotaxis receptors and unreleased spores are localized. The other localizations are not known. Outside bottom left corner: peptidoglycan and additional cell wall associated polymers and proteins are shown. Examples of elements not found in Bacillus halodurans C-125 (highlighted or outlined in purple) are: two of the putative energy storage forms, polyhydroxyalkanoates and polyglutamate capsule; cell wall S-layer and SLH-domain proteins; cell membrane triterpenoid carotenoid; gas vacuoles; a NiFe hydrogenase/cytochrome b5 locus; enzymes and regulators of polyaromatic hydrocarbon metabolism; and a malonate utilization locus.
Cell division, transcription and translation
The B. pseudofirmus OF4 chromosome has full complements of genes related to cell division, transcription and translation that are comparable to those in B. subtilis and B. halodurans C-125. As expected from the pI data above, several divisome proteins that are integral membrane proteins exhibit much lower pI values in B. pseudofirmus OF4 and other alkaliphilic Bacillus species than in neutralophilic Bacillus species. For example, the integral membrane protein MreC, which functions together with MreB and MreD in cell shape determination and has a domain just on the outside of the membrane (Kruse et al., 2005), has a range of pI values from 4.2 to 4.58 in the four alkaliphiles shown in the analyses in Fig. 3, with the pI for B. pseudofirmus OF4 MreC (BpOF4_02815) at 4.54. By contrast, the range for the neutralophiles shown in Fig. 3 is 6.79–8.75. Differences of similar magnitude and direction are observed for other divisome proteins with externally exposed segments such as RodA (BpOF4_04055), which also has a role in shape determination (Henriques et al., 1998, Real et al., 2008), and FtsX (BpOF4_05485) that is part of an ABC transporter (Reddy, 2007). With respect to transcription, the genome revealed 21 sigma factors that are all encoded in the chromosome (Table S2). Eleven of them are likely to be Extracytoplasmic Function (ECF) type sigma factors that are part of a response system to external environmental signals (Helmann, 2002).
Cell surface, secretion and chaperones
Cell surface. As part of a full complement of synthetic enzymes for the membrane phospholipids of B. pseudofirmus OF4, three cardiolipin synthase genes were identified. The clsA gene (BpOF4_ 01900) had been identified earlier (Guo & Tropp, 1998) and two additional genes, clsB (BpOF4_07070) and clsC (BpOF4_03205) were identified here. It has been hypothesized that the relatively high levels of membrane cardiolipin in the alkaliphile (Clejan et al., 1986) support oxidative phosphorylation by facilitating rapid proton transport along the membrane surface (Haines & Dencher, 2002, Haines, 2009). Identification of the cls genes will facilitate tests of this hypothesis. The neutral membrane lipids of B. pseudofirmus OF4 contain a carotenoid component that is presumed to account for the yellow color of the alkaliphile colonies. It is likely to be a triterpenoid carotenoid of the general type noted in other alkaliphilic Bacillus spp. (Aono & Horikoshi, 1991) and characterized detail in Staphylococcus aureus (Pelz et al., 2005) and Bacillus indicus HU36 (Perez-Fons et al., 2011). B. pseudofirmus OF4 genes (BpOF4_14550 to _1480) resemble those involved in staphyloxanthin pigment production in S. aureus. The B. pseudofirmus OF4 pigment probably differs significantly from staphyloxanthin, however, since only some of the genes are strongly homologous, e.g. ctrM (BpOF4_14560), while others are not, e.g. putative crtQ (BpOF4_14570) and crtO (BpOF4_14575), and the gene arrangement is distinct, with two apparent sub-clusters in B. pseudofirmus OF4 that are transcribed in opposite directions.
The secondary cell wall polymers (SCWP) that form the cell wall layer of B. pseudofirmus OF4 together with the peptidoglycan are similar to those of the B. cereus group of Bacillus species in featuring an S-layer, encoded by slpA (BpOF4_ 06075) in B. pseudofirmus OF4. SlpA was identified as a major externally exposed protein in a proteomics study of cell-membrane associated proteins (Gilmour et al., 2000). That study showed that SlpA is expressed at high levels during growth of B. pseudofirmus OF4 at both pH 7.5 and 10.5 even though its production is detrimental to growth at pH 7.5, as assessed in studies of an slpA deletion mutant. Like the low pI profiles of externally exposed proteins, such constitutive expression of slpA is evidence of “hard-wiring” in support of alkaliphily (Gilmour et al., 2000, Krulwich et al., 2011a). The genome sequence reveals approximately a dozen SLH domain proteins that are bound outside the cell as shown schematically in Fig. 4 (Desvaux et al., 2006). The SLH domain proteins include a hydrolase (BpOF4_05835), 5'-nucleotidase (BpOF4_ 05925), and N-acetylmuramoyl-L-alanine amidase (cell wall lytic enzyme, LytC, BpOF4_06105) in the chromosome and a β-lactamase on the large plasmid (BpOF4_20104). Several of these SLH proteins are in a gene cluster that also contains genes for: “cell surface attachment” proteins CsaA (BPOF4_ 05790) and CsaB (BpOF4_05780), which respectively are a putative oligosaccharide transporter and a pyruvyltransferase that are critical for SCWP assembly (Mesnage et al., 2000); TagO (BpOF4_05775) and TagA (BpOF4_05795), glycosyltransferases that have been shown to be required for SCWP synthesis in B. anthracis (Kern et al., 2010); and a SecA (BpOF4_05800) secretion protein.
Another SCWP of B. pseudofirmus OF4 predicted by the genome sequence is a poly-γ-glutamate capsule that is anchored in the cell wall (Fig. 4). There are two anchoring capD genes, capD1 (BpOF4_05215), which is next to another lytR gene (BpOF4_05210), and capD2 (BpOF4_06640), which is in a larger group of cap (capsule) genes that include other synthetic genes for the polyglutamate capsule. The synthetic capACB genes (BpOF4_06650,_06655, and _06660) are close to capD2, but no capE was identified. CapE is required for synthesis (Candela & Fouet, 2006). We hypothesize that the needed CapE protein is found as a domain of the EpsX protein (product of epsX, BpOF4_06635), which is predicted to have a role in extracellular polysaccharide (EPS) synthesis. BLASTP analyses showed that the EpsX of B. pseudofirmus OF4 is longer than most other EpsX homologues; the top hit of the B. thuringiensis CapE protein in the B. pseudofirmus OF4 genome is the first 60 amino acid residues of EpsX and the top hit for that domain of the alkaliphilie EpsX in the Bacillus subtilis genome is CapE. The cap locus is epsX capD2 capH capA capC capB. The presence of capH (BpOF4_06645), which is transcribed in the opposite direction of the synthetic cap genes, is consistent with the capsule being a potential energy source since CapH is predicted to be a polyglutamate hydrolase (Suzuki & Tahara, 2003). In addition, a polyglutamate capsule could play a role, as proposed for other bacteria, in binding metal ions (Candela & Fouet, 2006); at very high external pH, the bioavailability of important ions of metals such as iron is very low (McMillan et al., 2010). Additional chromosomal gene products are predicted to be regulators or enzymes with roles in production of extracellular polysaccharides and capsular polysaccharides. They include a tyrosine kinase of the BY kinase type (BpOF4_06120,_06125) and capsular polysaccharide biosynthesis proteins CpsD and CpsC (BpOF4_06615,_06620).
Secretion and chaperones
The complement of secretory systems and chaperones in B. pseudofirmus OF4 are similar to those in both alkaliphile and non-alkaliphile Bacillus genomes. The Sec system (Driessen & Nouwen, 2008) components include: two SecA proteins, SecA1 and A2 (BpOF4_05510,_05800), SecC (BpOF4_15960), SecD (BpOF4_11730), SecE (BpOF4_08475), SecF (BpOF4_11735), SecG (BpOF4_05265) and SecY (BpOF4_08650). There is one pair of twin-arginine targeting (Tat) system genes of the type found in Gram-positive bacteria, TatA (BpOF4_09290) and TatC (BpOF4_09295) (Barnett et al., 2008). B. halodurans C-125 has two such systems (Yen et al., 2002). A Type VII secretion system (ESAT-6/WXG100 protein superfamily) of the type found among Firmicutes is the predicted product of a cluster of genes (BpOF4_09430 to BpOF4_09450) (Pallen, 2002). The protein secretion chaperonin PrsA (BpOF4_11455) has a pI of 3.63. Two Oxa/YidC family chaperone/insertion proteins (Bp_11410;_07940) and three CtaG assembly proteins, one in the large plasmid pBpOF4-01 (BpOF4_20324) and two in the chromosome (BpOF4_02205; _10060), support membrane protein assembly, stability and/or ion co-factor incorporation. Cytoplasmic chaperones include GroES and GroEL (BpOF4_09355;_09360).
Bioenergetics: transporters, motility and chemotaxis, oxidative phosphorylation
Interconnected cycles of proton efflux/proton uptake and sodium ion efflux and uptake are essential for cytoplasmic pH homeostasis of B. pseudofirmus OF4. The alkaliphile can maintain a cytoplasmic pH that is 2.3 pH units more acidic than the external pH at very high pH (Krulwich, 1995, Sturr et al., 1994). Under respiratory conditions, the proton-pumping respiratory chain (highlighted in pink in Fig. 4) generates the PMF, an electrochemical proton gradient, which drives the major energy-consuming processes that result in net proton entry into the cytoplasm. Enzymes that provide electrons to the respiratory chain by reduction of menaquinone (MQ) include the membrane-embedded succinate dehydrogenase (Sdh) complex (BpOF4_03110 to _03120), as well as membrane-associated malate:quinone oxidoreductase (Mqo) (BpOF4_07745), electron transfer flavoprotein EtfAB (BpOF4_03150/_03155) and Ndh2 (non-proton pumping) NADH dehydrogenase, NdhA (BpOF4_04810). The products of two other Ndh2-encoding genes (BpOF4_06965, BpOF4_04855) are likely to have roles in thioredoxin reduction and nitrate assimilation. The two proton-translocating respiratory chain complexes are the quinone:cytochrome c oxidoreductase (QcrA-C) (BpOF4_15525 to _15535) and the Cta, cytochrome caa3-type terminal oxidase (CtaC-F, BpOF4_00900 to _00915). There are two alternate cytochrome bd-type terminal oxidases, cydABI and II (BpOF4_09310/_09315 and BpOF4_10585/_10590). Unlike their homologues in other bacteria (Puustinen et al., 1991, Winsted & von Wachenfeldt, 2000), they cannot replace the proton-pumping Cta in supporting non-fermentative growth (Gilmour & Krulwich, 1997). In addition to the cytochrome c subunits found in each of the proton-pumping respiratory chain complexes, QcrC and CtaC, the B. pseudofirmus OF4 genome encodes cccB (BpOF4_05495) and cccA (BpOF4_13740), respectively lipid-anchored and trans-membrane span-anchored cytochrome c551 proteins that have homologues in other Bacillus species (Bengtsson et al., 1999).
The most critical PMF-consuming process for cytoplasmic pH homeostasis at alkaline pH is sodium/proton antiport, which is catalyzed by most of the 13 cation/proton antiporters shown in Fig. 4 (highlighted in yellow), a number significantly higher than the 5 such antiporters found in B. halodurans C-125. The hetero-oligomeric Mrp antiporter (BpOF4_ 13210 to _13180) of the Cation:Proton Antiporter (CPA3) family is a necessary sodium/proton antiporter for pH homeostasis in both B. pseudofirmus OF4 and B. halodurans C-125 (Hamamoto et al., 1994, Swartz et al., 2005). The additional sodium/proton antiporters are likely to make contributions under particular conditions (Krulwich et al., 2009). The only one studied to date, apart from Mrp, is NhaC5 (BpOF4_13965) which plays a role at both pH 7.5 and 10.5 under conditions of low sodium ion concentration (Ito et al., 1997). The other process that is energized by the PMF is proton-coupled ATP synthesis by the F1Fo-ATP synthase that is encoded by the atp operon (BpOF4_06845 to _06890); it supports pH homeostasis while producing ATP (Krulwich et al., 2011b, Wang et al., 2004). Two of the membrane-embedded synthase proteins, the a-subunit and rotor-forming c-subunits, have alkaliphile-specific sequence motifs that are required for ATP synthesis at high pH (Hicks et al., 2010).
Ongoing Mrp-mediated sodium ion efflux in exchange for protons is essential for B. pseudofirmus OF4, especially at high pH. Therefore, completion of the sodium cycle by transporters that bring sodium ions back into the cytoplasm is also critical for pH homeostasis. Two channels contribute to sodium ion re-entry, the voltage-gated NavBP sodium channel (BpOF4_14445), which is located at cell poles and plays a role in chemotaxis, and the MotPS sodium channel (BpOF4-03820,_03815), which powers flagellar rotation (Fujinami et al., 2009, Ito et al., 2004a, Ito et al., 2004b). However, transporters that catalyze sodium-coupled solute uptake, sodium/solute symporters, constitute the major part of the sodium uptake-limb of the B. pseudofirmus OF4 sodium cycle (Krulwich et al., 1985). The evidence to date indicates that essentially all the ion-coupled solute symporters of B pseudofirmus OF4 are sodium-coupled (Padan et al., 2005, Slonczewski et al., 2009). The genome sequence reveals that this alkaliphile has a particularly large number of ion (presumably sodium ion)-coupled symporters (Fig. 4), even relative to some other extreme alkaliphiles. For example B. pseudofirmus OF4 has a much lower number of sodium-independent, ATP-energized ABC transporters than B. halodurans C-125, 49 versus 111, but has significantly higher numbers of presumably sodium-coupled transporters, including Tripartite ATP-independent periplasmic transporters, TRAP-T (24 versus 16), which facilitate high affinity solute binding (Rabus et al., 1999), along with many other sodium-coupled symporter types (Fig. 4).
Metabolic versatility and energy storage
Energy storage
As noted earlier, there are special energy costs associated with growth at external pH values well above 9.5, that are especially acute under non-fermentative conditions, where the burden of energy-dependent pH homeostasis is not significantly offset by metabolic acid production (Krulwich et al., 2011a, Slonczewski et al., 2009). The B. pseudofirmus OF4 genome sequence suggests that its ability to sustain growth at high pH is fostered by a capacity to store multiple types of compounds that can be catabolized as needed: the polyglutamate capsule polymer described earlier; polyhydroxyalkanoates; glycogen; and fatty acids, including branched chain fatty acids (Fig. 4). An earlier proteomic study of membrane associated enzymes showed that catabolic enzymes for branched chain fatty acids (BpOF4_05110; _01410; and _01415) were increased during growth at pH 10.5 relative to 7.5 (Gilmour et al., 2000).
Energy sources
In addition to its diverse forms of stored energy, the B. pseudofirmus OF4 genome reveals versatility in the sources for its energy-yielding pathways. The 24 TRAP-T systems provide a large capacity for uptake of dicarboxylates, Tripartite Tricarboxylate Transporter (TTT) (BpOF4_12495 to _12505) and CitMHS (BpOF4_09395) family transporters take up tricarboxylates and three GntP type transporters (BpOF4_09335,_09395,_13435) are likely to take up either gluconate or citrate. In addition, there are: multiple transporters for uptake of readily metabolizable aspartate and glutamate (BpOF4_05190;_07190; _11710;_14810); an ABC uptake system for acetoin (BpOF4_19110, _19115); phosphotransferase (PTS) components for uptake and phosphorylation of over a half dozen mono- and di-saccharides; a MIP channel (BpOF4_10315) predicted to take up glycerol; and a MadLM transporter (BpOF4_11950,_11955) for malonate uptake and enzymes for its catabolism (BpOF4_11965 to _11990) (Dimroth & Hilbi, 1997).
Further metabolic versatility is evidenced by the locus for catabolism of polyaromatic hydrocarbons (BpOF4_12555 to _12625), a capacity that could also confer bioremediation potential (Peng et al., 2008). In addition, a 15-gene cluster encodes putative gas vacuoles (BpOF4_07620 to _07690), which are likely to be similar to those found in strains of B. megaterium (Eppinger et al., 2011, Li & Cannon, 1998). Such vacuoles could facilitate access of the organism to oxygen under conditions of soil flooding so that PMF-dependent pH homeostasis remains optimal. The NiFe-hydrogenase gene cluster, which also contains a formate dehydrogenase (BpOF4-1290 to _01350), might further support growth under oxygen limiting conditions as hypothesized for the mycobacterial homologue of the hydrogenase (Berney & Cook, 2010)..
Signaling related to energy sources
Analysis of the 33 chromosomal and 2 plasmid Two-Component Signaling systems (TCSs) (Table S3) predicted that 4 of them are involved in dicarboxylate and/or tricarboxylate metabolism, consistent with the patterns of transport and metabolism described above. Many of the signaling kinases that are involved in chemotactic responses, i.e. 12 MCPs (Methylated Chemotaxis Proteins), mediate positive taxis toward energy sources or negative taxis from metabolic uncouplers or inhibitors. The 3 HemAT sensors of oxygen support movement toward optimal conditions of aeration (Table S4). B. pseudofirmus OF4 has genes encoding homologues of the B. subtilis CheW, CheY, CheA, CheV, CheR, CheB, CheC, and CheD chemotaxis proteins but, like B. halodurans C-125, it also has a second phosphatase, CheX (BpOF4_11185), and two genes instead of single genes encoding CheY and CheV proteins (Fujinami et al., 2009). It has been suggested that the serine/threonine protein kinase PrkC of B. subtilis may have a role in metabolic regulation (Pietack et al., 2010); the genome sequence identified the PrkC (BpOF4_00425) of B. pseudofirmus OF4 and its companion PrpC, a serine/threonine phosphoprotein phosphatase (BpOF4_00430).
Stress responses
Overview
The B. pseudofirmus OF4 genome sequence reveals gene complements for managing several well-characterized stress responses that are comparable to those found in non-alkaliphiles, without evidence for greater robustness in the form of greater numbers of effectors. These stress responses include genes associated with the SOS response, heat- and cold-shock and general stress proteins, as well as the sigma B gene locus that starts with toxin-antitoxin genes mazE mazF (BpOF4_09165, _09170) and extends through rsbRSTUW sigB rsbX (the last gene tag being BpOF4_09210). Additionally, although reactive oxygen species (ROS) generation is increased at high pH (Selivanov et al., 2008), the gene complement identified as related to ROS stress is entirely comparable to that of B. subtilis (Zuber, 2009). Perhaps distinctive properties for some of these gene products will eventually be found to relate to alkaliphily. For some other stresses that are exacerbated by alkaline pH, the genome analysis suggests that there are augmented countermeasures in the alkaliphile. These are described below.
Sodium and high osmolarity stress
As already noted, B. pseudofirmus OF4 has a relatively high number of cation/proton antiporters that are predicted to catalyze efflux of sodium ions, whose cytotoxicity is increased at alkaline pH (Padan et al., 2005) (Fig. 4). The contribution of antiporters to sodium-resistance is augmented by an ABC-type sodium ion efflux system (BpOF4_11390 to _11400). The interacting challenge of osmoregulation is predicted by the genome sequence to be ameliorated by a large number of different channels, transporters and pathways for synthesis or uptake of compatible solutes. Potassium and glutamate uptake are part of an early response to elevated osmolarity that could be supported in part by potassium channels and transporters (Booth et al., 2005, Wood, 2011), which include: two Ktr-type potassium uptake systems, KtrAB (BpOF4_07080/_07075) and KtrCD (BpOF4_01110/_03125), which are similar to those described in B. subtilis (Holtmann et al., 2003) although the alkaliphile Ktr pairs are likely to couple potassium uptake to sodium uptake as in cyanobacteria (Matsuda et al., 2004); 5 putative voltage-gated (BpOF4_01220,_04575,_07435,_10900,_14445), one large-conductance mechanosensitive potassium channel (BpOF4_01755) and two small-conductance ones (BpOF4_01120/_1205); glutamate uptake could be mediated by 5 putative sodium coupled glutamate transporters (BpOF4_05190,_07190,_09415_11710, _14810). The compatible solute synthesis and uptake systems include those for: synthesis of ectoine and hydroxyectoine (BpOF4_10765 to _10775, and BpOF4_12395), which are part of a conserved response system of demonstrated physiological importance (Bursy et al., 2007); uptake of glycine betaine, coupled to sodium by 5 OpuD transporters (BpOF4_01050, _01200, _14175, _18090, _18915), transporters whose importance has been shown in B. subtilis (Kappes et al., 1996), as well as an ABC system (BpOF4_16115/_16120); and uptake of proline by 6 sodium-coupled PutP transporters (BpOF4_06790, _09890, _10120, _10245, _12885, _16425), a homologue of which has also been shown to be regulated by osmotic stress in B. subtilis (von Blohn et al., 1997).
Cell envelope-antibiotic-alkaline stress intersection
Evidence from non-alkaliphilic bacteria, including B. subtilis, indicates that there is an intersection between cell envelope stress and alkaline stress as well as antibiotic stress (Cao et al., 2002a, Cao et al., 2002b, Wiegert et al., 2001). Alkaliphiles like B. pseudofirmus OF4 are particularly susceptible to inhibition by membrane permeable cations during growth at high pH, because of their large transmembrane electrical potential, inside negative relative to outside. They are also sensitive to basic antibiotic compounds during growth at high pH because of the large transmembrane pH gradient, inside acidic relative to outside (Krulwich et al., 2011b, Slonczewski et al., 2009). Five of the 35 annotated TCSs (Two-Component Signaling systems) of B. pseudofirmus OF4 are predicted, by findings in non-alkaliphiles, to have roles in cell envelope stress: WalKR (BpOF4_07825, _07830); BceSR (BpOF4_09980, _09985); LiaRS (BpOF4_11560, _11565); YycF/YvrG (BpOF4_12970, _12975); and CssSR (BpOF4_11425, _11420) (Table S3) (Mascher et al., 2003, Mascher et al., 2004, Dubrac et al., 2008, Rietkotter et al., 2008, Hyyryläinen et al., 2001, Gusarov et al., 2009). Thirty six drug exporting transporters of the Major Facilitator and ABC families (Fig. 4) and the nitric oxide synthase (BpOF4_10365) are predicted to directly contribute to antibiotic resistance (Gusarov et al., 2009). Further, B. pseudofirmus OF4 has 2 paralogous FabH genes (BpOF4_02090, BpOF4_03935) that may, as found in B. subtilis (Kingston et al., 2011), decrease membrane fluidity in response to sigma W signaling by increasing the relative content of long straight chain fatty acids. However, that pattern is already found in B. pseudofirmus OF4 relative to B. subtilis (Clejan et al., 1986). Perhaps the alkaliphile FabH paralogues are both more like FabHb, which is activated by the sigma W signal in B. subtilis, or the FabHb of the alkaliphile is hard-wired for substantial basal activity at near neutral pH and expressed even more upon sigma B signaling after exposure to high pH.
Methylglyoxal toxicity
Another stress that is increased by alkaline conditions is the toxicity of methylglyoxal, which is produced when phosphate is limiting in cells that are metabolizing sugars (Ozyamak et al., 2010). In addition to genes encoding putative glyoxalases, GloA (BpOF4_14285/_02030) and GloB (BpOF4_01695), the B. pseudofirmus OF4 genome encodes 5 sodium-coupled Npt type phosphate transporters (BpOF4_0270, _04330, _13520, _13885, _21614) and one Pit family phosphate transporter (BpOF4_1485), which could reduce the susceptibility to phosphate insufficiency.
Low iron bioavailability
At high pH, B. pseudofirmus OF4 must overcome the problem of low solubility and hence low bioavailability of critically important metals such as iron, whose bioavailability at pH 10 is estimated at about 10−23 M (Drechsel & Jung, 1998). The B. pseudofirmus OF4 genome revealed the 6-gene gene cluster asbABCDEF (BpOF4_08925 to _08950) encoding a catecholate siderophore, whose synthesis is independent of a nonribosomal peptide synthetase (Barry & Challis, 2009). The siderophore is predicted to be similar to the Peribactin siderophore of the B. cereus group and probably corresponds to the catecholate siderophore recently detected in B. pseudofirmus OF4 (Lee et al., 2007, McMillan et al., 2010). Upstream and contiguous with this gene cluster are four more genes, FhuGB and FhuRD (BpOF4_08905 to _08920) that are predicted to encode an ABC type siderophore transporter and its regulator. This is one of 6 ABC transporters predicted to transport ferric siderophores. Genes for 2 siderophore ferric reductases (BpOF4_11075 and BpOF4_02495), which are needed to release the siderophore-bound iron by reduction of the metal, were also identified in the chromosome. One is in the same locus as a ferric dicitrate ABC uptake system, FdcPTA (BpOF4_11060, _11065, _11070), and the ferric reductase that follows is homologous to a ferric siderophore reductase recently described in B. halodurans C-125 and designated FchR (Miethke et al., 2011). The second siderophore ferric reductase (BpOF4_0249) is predicted to have 262 amino acids, and its top two BLASTP matches are in B. halodurans C-125 and thermoalkaliphilic Caldalkalibacillus thermarum TA2.A1. It will be of interest to study whether the alkaliphile Peribactin-like siderophore and siderophore ferric reductases exhibit features that are adaptive for function at high pH.
Growth and/or survival with cytoplasmic pH > 9
It is likely that the capacity of B. pseudofirmus OF4 for growth when the cytoplasmic pH is high will in part depend upon the structural impact of specific patterns of amino acid usage in cytoplasmic proteins of alkaliphiles (Kapetaniou et al., 2006). B. pseudofirmus OF4 has a chromosomal gene encoding a 134 amino acid protein, asp23 (BpOF4_01535), which is a Firmicute protein of unknown function that was first identified in S. aureus as alkaline-shock inducible (Kuroda et al., 1995), and also has a gene encoding a conserved ~59 amino acid protein (BpOF4_01690) that was annotated in B. halodurans C-125 as “alkaliphily-related” (BH2819). We note that the B. pseudofirmus OF4 sequence further reveals two small hypothetical, contiguous genes encoding highly polar proteins, BpOF4_10505 and BpOF4_10510, that are 83 and 45 amino acids respectively. Homologous proteins are found only in the alkaliphiles B. halodurans C-125 and B. clausii KSMK16 in which they are also the products of contiguous genes. Perhaps they are chaperones or proteins with other protective functions that promote tolerance to elevated cytoplasmic pH and whose functions will emerge after further investigation.
Concluding comments
B. pseudofirmus OF4 grows well at both pH 7.5 and 10.5 on both non-fermentative and fermentative carbon sources. The data from this study indicate that the genome provides impressive redundancy in key transporters. This may reflect the need to have distinct isoforms of some transporters for at near neutral and at extremely alkaline pH values. The metabolic versatility, yet to be fully elucidated, is hypothesized to be adaptive for a soil extremophile that is likely to encounter large and sudden perturbations. The upper limit of pH has not yet been defined for this extreme alkaliphile. Continuous cultures of B. pseudofirmus OF4 that were maintained on MYE medium at a constant at pH of 11.4 exhibited growth and higher pH values were not examined (Sturr et al., 1994). With the genome in hand, it would be of interest to determine the upper pH limit for growth. Preliminary, unpublished data on cells from the earlier pH 11.2 cultures suggested that these cells were better adapted for growth at pH ≥ 10.5 than the cells used to start the culture. Genome wide transcriptional data could now be gathered on variants that arise at pH ≥ 11.4 to generate specific mechanistic hypotheses that could be tested with site-directed mutants, e.g. about requirements for growth near the external alkaline limit and the requirements for tolerating the high cytoplasmic pH that accompanies growth at such external pH values. Genome rearrangements could also be tracked at extreme alkaline stress, to test the idea that the myriad of mobile elements found in the endogenous plasmids represent a “tool-kit” for producing change under conditions in which the majority of the cell population cannot not persist.
Experimental procedures
Strains and growth conditions
Bacillus pseudofirmus OF4 has been deposited in the American Type Culture Collection (ATCC BAA-2126) and the Japan Collection of Microorganisms (JCM 17055). The sources for this strain and other strains used are in Table 2. For DNA isolation and assessments of plasmid content, cells were grown on either the MYE (malate yeast extract) or GYE (glucose yeast extract) media described earlier (Ito et al., 1997). For disruption of the chromosomal B. pseudofirmus OF4 ctaC gene, a spectinomycin resistance gene was introduced into a ctaC fragment and homologous recombination-based disruption of the chromosomal gene was carried out by a strategy using pG+host4 (Biswas et al., 1993, Wang et al., 2004).
DNA isolation, sequencing and sequence assembly
Genomic DNA was prepared essentially as described by (Sambrook et al., 1989). Isolated genomic DNA from Bacillus pseudofirmus OF4 was used to generate two single stranded DNA libraries. One was a single stranded shotgun library which contained fragments of ~500bp. The second library was a Paired End Library consisting of 100–400bp of paired sequences separated by 4.5kb on average. Each library was sequenced separately on a 454 Life Sciences/Roche FLX Sequencer. The runs were assembled together with the Newbler 2.0.00.20 Assembler. The two sequencing runs combined generated an assembly of 11 scaffolds containing 76 large contigs (>500bp), representing a coverage depth of ~54X.
Finishing
Primers were designed off the ends of each intra-scaffold contig and the scaffolds themselves. PCR products were generated, cleaned and subsequently sequenced on an ABI3730xl. Multiple rounds of primer walking were performed using the sequencing results of the previous round to close each of the remaining gaps.
Annotation and analysis
The B. pseudofirmus OF4 assembly was submitted to PGAAP (Prokaryotic Genomes Automatic Annotation Pipeline) via the web-site: http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html. This annotation was combined with manually curated annotations. The final assembly and annotations can be accessed using the GenBank accession numbers CP001878, CP001879, and CP001880. A second annotation was obtained using the RAST (Rapid Annotations with subsystems) (Aziz et al., 2008) service to identify and categorize features of the genome.
Determination of the origin of replication
The web-based tool Ori-Finder was used to locate the region containing the origin of replication on the main chromosome (http://tubic.tju.edu.cn/Ori-Finder/). This tool takes into account DnaA boxes and GC, AT, RY and MK disparity. The region identified by Ori-Finder was then manually inspected and compared to corresponding locations of DnaA, DnaN and surrounding sequences in Bacillus subtilis 168 and Bacillus halodurans C-125.
Dot-Plot Co-linearity with Bacillus halodurans C-125
The ORFs on the B. pseudofirmus OF4 chromosome and B. halodurans C-125 were re-oriented such that DnaA was labeled gene #1. Each ORF on the OF4 genome was then submitted as a query in a BLASTP search against the C-125 ORFs. The gene number on C-125 was recorded if there was a hit > 50%. Hits < 50% or no hit were recorded as 0. The query gene number (x) and the BLAST hit number (y) were then used as coordinates (x,y) to generate a Dot-Plot.
Distribution of two resident plasmids in diverse strains related to B. pseudofirmus OF4
Colony PCR and PCR using complete genomic DNA were used to determine the presence of big or small plasmids in the Bacillus pseudofirmus OF4 strain or derivatives. For the colony PCR, a small amount of colony from a Petri plate was mixed with 50 μl of sterile water and used as PCR template. The PCR was conducted according to the instruction from HotstarTaq Master Mix (Qiagen). A 551 bp PCR product, designated chromosome PCR product, was amplified with the following primers: Chromosome_F 5'GCAAGCACAGACCTGAAGAAGAAG 3' and Chromosome_R 5' GGAAATTGAAACGAAGATTGAGGAG 3'; a 976 bp pcr product, designated large plasmid (pOF4-01) PCR product, was amplified with the following primers: Large_plasmid_F 5' CCTTTGACTTTAGATAACGGGTTCG 3' and Large_plasmid_R 5' GCATCCATTTTAACAACACCACATAGA 3'; a 1739 bp PCR product, designated small plasmid (pBpOF4-02) PCR product, was amplified with the following primers: Small_plasmid_F 5' GCAAAGCCAATGGAAGGACAAA 3' and Small_plasmid_R 5' TTCACTCAGTATCTGGCTTTCCAATA 3'.
Intron and repeat identification
Search for sequences and/or secondary structures homologous to known group I and group II introns were carried out using the Infernal (Nawrocki et al., 2009) and RNAweasel (Lang et al., 2007) softwares. For Infernal searches, the full structural multiple alignments of all group I and group II introns available in the Rfam database (http://rfam.sanger.ac.uk/; entry IDs RF00028 and RF00029, respectively, were used to build covariance models by means of the cmbuild program of the Infernal package (Gardner et al., 2009). The models were then calibrated using cmcalibrate, and cmsearch was used to search the B. pseudofirmus OF4 genome with the calibrated models (cmbuild, cmcalibrate, and cmsearch were run with default settings). RNAweasel searches were conducted at the RNAweasel website hosted by the University of Montreal, Canada, (http://megasun.bch.umontreal.ca/RNAweasel/), a service based on curated sets of mitochondrial and chloroplast introns.
To identify interspersed DNA repeats, intergenic regions were extracted from the B. pseudofirmus OF4 genome using Genome2D (Baerends et al., 2004) and were compared following dual iterative searches based on BLASTN (Altschul et al., 1997) as previously described (Tourasse et al., 2006, Klevan et al., 2007). For bcr3 elements, additional searches were conducted using bcr3 sequences previously identified in bacteria from the Bacillus cereus group as queries (Kristoffersen et al., 2011). Multiple sequence alignments of repeat sequences were computed using MUSCLE 3.8 (Edgar, 2004) and further edited and analyzed in SEAVIEW4 (Gouy et al., 2010).
Supplementary Material
Acknowledgements
This work was supported in part by research grant GM028454 from the National Institute of Health to TAK, support from the Allegheny Singer Research Institute/Allegheny General Hospital as well as grants DHHS/HRSA C76HF00659 and National Institutes of Health grant DC02148 to FH, grant 30621005 from the National Science Foundation of China (NSFC) and 973 programs 2007CB707801 from the Ministry of Sciences and Technology of China to YM, the Strategic Research Foundation Grant-aided for Private Universities and Grant-in-Aid for Scientific Research (B) No. 21370074 of the Ministry of Education, Culture, Sports, Science and Technology of Japan to MI.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono A, Horikoshi K. Carotenes produced by alkaliphilic yellow-pigmented strains of Bacillus. Agric. Biol. Chem. 1991;55:2643–2645. [Google Scholar]
- Aono R, Ito M, Machida T. Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphile Bacillus lentus C-125. J. Bacteriol. 1999;181:6600–6606. doi: 10.1128/jb.181.21.6600-6606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono R, Ohtani M. Loss of alkalophily in cell-wall-component-defective mutants derived from alkalophilic Bacillus C-125. Isolation and partial characterization of the mutants. Biochem. J. 1990;266:933–936. [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJ, Smits WK, de Jong A, Hamoen LW, Kok J, Kuipers OP. Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 2004;5:R37. doi: 10.1186/gb-2004-5-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JP, Eljilander RT, Kuipers OP, Robinson C. A minimal Tat system from a gram-positive organism: a bifunctional TatA subunit participates in descrete TatAC and TatA complexes. J. Biol. Chem. 2008;283:2534–2542. doi: 10.1074/jbc.M708134200. [DOI] [PubMed] [Google Scholar]
- Barry SM, Challis GL. Recent advances in siderophore biosynthesis. Curr. Opin. Chem. Biol. 2009;13:205–215. doi: 10.1016/j.cbpa.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Bengtsson J, Rivolta C, Hederstedt L, Karamata D. Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J. Biol. Chem. 1999;274:26179–26184. doi: 10.1074/jbc.274.37.26179. [DOI] [PubMed] [Google Scholar]
- Berney M, Cook GM. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0008614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Gruss A, Ehrlich SD, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 1993;175:3628–36235. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR, Edwards MD, Murray E, Miller S. The role of bacterial channels in cell physiology. In: Kubalski A, Marinac B, editors. Bacterial Ion Channels and Their Eukaryotic Homologs. ASM Press; Washington, D.C.: 2005. pp. 291–312. [Google Scholar]
- Brändén M, Sandén T, Brzezinski P, Widengren J. Localized proton microcircuits at the biological membrane-water interface. Proc. Natl. Acad. Sci. USA. 2006;103:19766–19770. doi: 10.1073/pnas.0605909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursy J, Pierik AJ, Pica N, Bremer E. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J. Biol. Chem. 2007;282:31147–31155. doi: 10.1074/jbc.M704023200. [DOI] [PubMed] [Google Scholar]
- Candela T, Fouet A. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 2006;60:1091–1098. doi: 10.1111/j.1365-2958.2006.05179.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Kobel PA, Morshedi MM, Wu MF, Paddon C, Helmann JD. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 2002a;316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 2002b;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- Cherepanov DA, Feniouk BA, Junge W, Mulkidjanian AY. Low dielectric permittivity of water at the membrane interface: effect on the energy coupling mechanism in biological membranes. Biophys. J. 2003;85:1307–1316. doi: 10.1016/S0006-3495(03)74565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan S, Guffanti AA, Cohen MA, Krulwich TA. Mutation of Bacillus firmus OF4 to duramycin resistance results in substantial replacement of membrane lipid phosphatidylethanolamine by its plasmalogen form. J. Bacteriol. 1989;171:1744–1746. doi: 10.1128/jb.171.3.1744-1746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan S, Krulwich TA, Mondrus KR, Seto-Young D. Membrane lipid composition of obligately and facultatively alkalophilic strains of Bacillus spp. J. Bacteriol. 1986;168:334–340. doi: 10.1128/jb.168.1.334-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA. 2003;9:14–19. doi: 10.1261/rna.2126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, Dumas E, Chafsey I, Hebraud M. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 2006;256:1–15. doi: 10.1111/j.1574-6968.2006.00122.x. [DOI] [PubMed] [Google Scholar]
- Dimroth P, Hilbi H. Enzymic and genetic basis for bacterial growth on malonate. Mol. Microbiol. 1997;25:3–10. doi: 10.1046/j.1365-2958.1997.4611824.x. [DOI] [PubMed] [Google Scholar]
- Drechsel H, Jung G. Peptide siderophores. J. Pept. Sci. 1998;4:147–181. doi: 10.1002/(SICI)1099-1387(199805)4:3%3C147::AID-PSC136%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Dubnovitsky AP, Kapetaniou EG, Papageorgiou AC. Enzyme adaptation to alkaline pH: atomic resolution (1.08 A) structure of phosphoserine aminotransferase from Bacillus alcalophilus. Protein Sci. 2005;14:97–110. doi: 10.1110/ps.041029805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycG) essential signal transduction pathway. Mol. Microbiol. 2008;70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M, Bunk B, Johns MA, Edirisinghe JN, Kutumbaka KK, Koenig SS, Huot Creasy H, Rosovitz MJ, Riley DR, Daugherty S, Martin M, Elbourne LD, Paulsen I, Biedendieck R, Braun C, Grayburn S, Dhingra S, Lukyanchuk V, Ball B, Ul-Qamar R, Seibel J, Bremer E, Jahn D, Ravel J, Vary PS. Genome sequences of the biotechnologically important Bacillus megaterium strains QM B1551 and DSM319. J. Bacteriol. 2011;193:4199–4213. doi: 10.1128/JB.00449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinami S, Fujisawa M. Industrial application of alkaliphiles and their enzymes -- past, present and future. Environ. Technol. 2010;31:845–856. doi: 10.1080/09593331003762807. [DOI] [PubMed] [Google Scholar]
- Fujinami S, Terahara N, Krulwich TA, Ito M. Motility and chemotaxis in alkaliphilic Bacillus species. Future Microbiol. 2009;4:1137–1149. doi: 10.2217/fmb.09.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Fackelmayer O, Liu J, Krulwich TA, Hicks DB. The ATP synthase a-subunit of extreme alkaliphiles is a distinct variant. J. Biol. Chem. 2010;285:32105–32115. doi: 10.1074/jbc.M110.165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour R, Krulwich TA. Construction and characterization of a mutant of alkaliphilic Bacillus firmus OF4 with a disrupted cta operon and purification of a novel cytochrome bd. J. Bacteriol. 1997;179:863–870. doi: 10.1128/jb.179.3.863-870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour R, Messner P, Guffanti AA, Kent R, Scheberl A, Kendrick N, Krulwich TA. Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J. Bacteriol. 2000;182:5969–5981. doi: 10.1128/jb.182.21.5969-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grant WD. Alkaline environments and biodiversity. In: Gerday C, Glansdorff N, editors. Extremophiles (Life Under Extreme External Conditions) Eolss Publishers; Oxford, U.K.: 2003. On-line publication www.eolss.net. [Google Scholar]
- Gronstad A, Jaroszewicz E, Ito M, Sturr MG, Krulwich TA, Kolstø AB. Physical map of alkaliphilic Bacillus firmus OF4 and detection of a large endogenous plasmid. Extremophiles. 1998;2:447–453. doi: 10.1007/s007920050091. [DOI] [PubMed] [Google Scholar]
- Guffanti AA, Blanco R, Benenson RA, Krulwich TA. Bioenergetic properties of alkaline-tolerant and alkalophilic strains of Bacillus firmus. J. Gen. Microbiol. 1980;119:79–86. [Google Scholar]
- Guffanti AA, Hicks DB. Molar growth yields and bioenergetic parameters of extremely alkaliphilic Bacillus species in batch cultures, and growth in a chemostat at pH 10.5. J. Gen. Microbiol. 1991;137:2375–2379. doi: 10.1099/00221287-137-10-2375. [DOI] [PubMed] [Google Scholar]
- Guo D, Tropp BE. Cloning of the Bacillus firmus OF4 cls gene and characterization of its gene product. Biochim. Biophys. Acta. 1998;1389:34–42. doi: 10.1016/s0005-2760(97)00086-6. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines TH. A new look at cardiolipin. Biochim. Biophys. Acta. 2009;1788:1997–2002. doi: 10.1016/j.bbamem.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Haines TH, Dencher NA. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- Heberle J, Riesle J, Thiedemann G, Oesterhelt D, Dencher NA. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 1994;370:379–382. doi: 10.1038/370379a0. [DOI] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Glaser P, Piggot PJ, Moran CP., Jr. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 1998;28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- Hicks DB, Liu J, Fujisawa M, Krulwich TA. F1F0-ATP synthases of alkaliphilic bacteria: lessons from their adaptations. Biochim. Biophys. Acta. 2010;1797:1362–1377. doi: 10.1016/j.bbabio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: Two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 2003;185:1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi K. Alkaliphiles. In: Horikoshi K, Grant WD, editors. Extremophiles: Microbial Life in Extreme Environments. Wiley-Liss, Inc.; New York: 1998. pp. 155–179. [Google Scholar]
- Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi K, Akiba T. Alkalophilic Microorganisms. Springer-Verlag; Heideberg: 1982. [Google Scholar]
- Hyyryläinen HL, Bolhuis A, Darmon E, Muukkonen L, Koski P, Vitikainen M, Sarvas M, Pragai Z, Bron S, van Dijl JM, Kontinen VP. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 2001;41:1159–1172. doi: 10.1046/j.1365-2958.2001.02576.x. [DOI] [PubMed] [Google Scholar]
- Ito M. Aerobic alkaliphiles. In: Bitton G, editor. Encyclopedia of Environmental Microbiology. Wiley; New York: 2002. pp. 133–140. [Google Scholar]
- Ito M, Guffanti AA, Zemsky J, Ivey DM, Krulwich TA. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J. Bacteriol. 1997;179:3851–3857. doi: 10.1128/jb.179.12.3851-3857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Hicks DB, Henkin TM, Guffanti AA, Powers B, Zvi L, Uematsu K, Krulwich TA. MotPS is the stator-force generator for motility of alkaliphilic Bacillus and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 2004a;53:1035–1049. doi: 10.1111/j.1365-2958.2004.04173.x. [DOI] [PubMed] [Google Scholar]
- Ito M, Xu H, Guffanti AA, Wei Y, Zvi L, Clapham DE, Krulwich TA. The voltage-gated Na+ channel NavBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc. Natl. Acad. Sci. USA. 2004b;101:10566–10571. doi: 10.1073/pnas.0402692101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetaniou EG, Thanassoulas A, Dubnovitsky AP, Nounesis G, Papageorgiou AC. Effect of pH on the strcuture and stability of Bacillus circulans ssp. alkalophilus phosphoserine aminotransferase: thermodynamic and cystallographic studies. Proteins. 2006;63:742–753. doi: 10.1002/prot.20935. [DOI] [PubMed] [Google Scholar]
- Kappes RM, Kempf B, Bremer E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 1996;178:5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Ryan C, Faull K, Schneewind O. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J. Mol. Biol. 2010;401:757–775. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston AW, Subramanian C, Rock CO, Helmann JD. A σw-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol. Microbiol. 2011;81:69–79. doi: 10.1111/j.1365-2958.2011.07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada M, Kosono S, Kudo T. The Na+/H+ antiporter of alkaliphilic Bacillus sp. Extremophiles. 2000;4:253–258. doi: 10.1007/s007920070010. [DOI] [PubMed] [Google Scholar]
- Klevan A, Tourasse NJ, Stabell FB, Kolstø AB, Økstad OA. Exploring the evolution of the Bacillus cereus group repeat element bcr1 by comparative genome analysis of closely related strains. Microbiology. 2007;153:3894–3908. doi: 10.1099/mic.0.2007/005504-0. [DOI] [PubMed] [Google Scholar]
- Krause M, Messer W. DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene. 1999;228:123–132. doi: 10.1016/s0378-1119(99)00009-8. [DOI] [PubMed] [Google Scholar]
- Kristoffersen SM, Tourasse NJ, Kolstø AB, Økstad OA. Interspersed DNA repeats bcr1-bcr18 of Bacillus cereus group bacteria form three distinct groups with different evolutionary and functional patterns. Mol Biol Evol. 2011;28:963–983. doi: 10.1093/molbev/msq269. [DOI] [PubMed] [Google Scholar]
- Krulwich TA. Alkaliphiles: `basic' molecular problems of pH tolerance and bioenergetics. Mol. Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Federbush JG, Guffanti AA. Presence of a nonmetabolizable solute that is translocated with Na+ enhances Na+-dependent pH homeostasis in an alkalophilic Bacillus. J. Biol. Chem. 1985;260:4055–4058. [PubMed] [Google Scholar]
- Krulwich TA, Guffanti AA. Alkalophilic bacteria. Annu. Rev. Microbiol. 1989;43:435–463. doi: 10.1146/annurev.mi.43.100189.002251. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Hicks DB, Ito M. Cation/proton antiporter complements of bacteria: why so large and diverse? Mol. Microbiol. 2009;74:257–260. doi: 10.1111/j.1365-2958.2009.06842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich TA, Hicks DB, Swartz TH, Ito M. Bioenergetic adaptations that support alkaliphily. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. ASM Press; Washington, DC: 2007. pp. 311–329. [Google Scholar]
- Krulwich TA, Liu J, Morino M, Fujisawa M, Ito M, Hicks D. Adaptive mechanisms of extreme alkaliphiles. In: Horikoshi K, Antranikan G, Bull A, Robb FT, Stetter K, editors. Extremophiles Handbook. Springer; Heidelberg: 2011a. pp. 120–139. [Google Scholar]
- Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011b;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 2005;55 doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Hayashi H. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Lee JY, Janes BK, Passalacqua KD, Pfleger BF, Bergman NH, Liu H, Hakansson K, Somu RV, Aldrich CC, Cendrowski S, Hanna PC, Sherman DH. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J. Bacteriol. 2007;189:1698–1710. doi: 10.1128/JB.01526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Cannon MC. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J. Bacteriol. 1998;180:2450–2458. doi: 10.1128/jb.180.9.2450-2458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fujisawa M, Hicks DB, Krulwich TA. Characterization of the functionally critical AXAXAXA and PXXEXXP motifs of the ATP synthase c-subunit from an alkaliphilic Bacillus. J. Biol. Chem. 2009;284:8714–8725. doi: 10.1074/jbc.M808738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 2003;50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- Mascher T, Zimmer SL, Smith TA, Helmann JD. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 2004;48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Kobayashi H, Katoh H, Ogawa T, Futatsugi L, Nakamura T, Bakker EP, Uozumi N. Na+-dependent K+ uptake Ktr system from the cyanobacterium Synechocystis sp. PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. J. Biol. Chem. 2004;279:54952–54962. doi: 10.1074/jbc.M407268200. [DOI] [PubMed] [Google Scholar]
- McMillan DG, Vetasquez I, Nunn BL, Goodlett DR, Hunter KA, Lamont I, Sander SG, Cook GM. Acquisition of iron by alkaliphilic bacillus species. Appl. Environ. Microbiol. 2010;76:6955–6961. doi: 10.1128/AEM.01393-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 2000;19:4473–4484. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Pierik AJ, Peuckert F, Seubert A, Marahiel MA. Identification and characterization of a novel-type ferric siderophore reductase from a Gram-positive extremophile. J. Biol. Chem. 2011;286:2245–2260. doi: 10.1074/jbc.M110.192468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Moriya S, Atlung T, Hansen FG, Yoshikawa H, Ogasawara N. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol. 1992;6:309–315. doi: 10.1111/j.1365-2958.1992.tb01473.x. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Heberle J, Cherepanov DA. Protons @ interfaces: implications for biological energy conservation. Biochim. Biophys. Acta. 2006;1757:913–930. doi: 10.1016/j.bbabio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Nakasone K, Masui N, Takaki Y, Sasaki R, Maeno G, Sakiyama T, Hirama C, Fuji F, Takami H. Characterization and comparative study of the rrn operons of alkaliphilic Bacillus halodurans C-125. Extremophiles. 2000;4:209–214. doi: 10.1007/pl00010713. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyamak E, Black SS, Walker CA, Maclean MJ, Bartless W, Miller S, Booth IR. The critical role of S-lactoylglutathione formation during methylglyoxal detoxification in Escherichia coli. Mol. Microbiol. 2010;78:1577–1590. doi: 10.1111/j.1365-2958.2010.07426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ. The ESAT-6/WXG100 superfamily - and a new Gram-positive secretion system? Trends Micriobiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- Pelz A, Wieland KP, Putzbach K, Hentschel P, Albert K, Götz F. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 2005;280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- Peng R-H, Xiong A-S, Xue Y, Fu X-Y, Gao F, Zhao W, Tian Y-S, Yao Q-H. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Revs. 2008;32:927–955. doi: 10.1111/j.1574-6976.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- Perez-Fons L, Steiger S, Khaneja R, Bramley PM, Cutting SM, Sandmann G, Fraser PD. Identification and developmental formation of carotenoid pigments in the yellow/orange Bacillus spore-formers. Biochim. Biophys. Acta. 2011;1811:177–185. doi: 10.1016/j.bbalip.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Pietack N, Becher D, Schmidt SR, Saier MH, Hecker M, Commichau FM, Stulke J. In vitro phosphorylation of key metabolic enzymes from Bacillus subtilis: PrkC phosphorylates enzyme from different branches of basic metabolism. J. Mol. Microbiol. Biotechnol. 2010;18:129–140. doi: 10.1159/000308512. [DOI] [PubMed] [Google Scholar]
- Preiss L, Yildiz Ö, Hicks D, Krulwich TA, Meier T. A new type of proton coordination in an F1F0-ATP synthase rotor ring. PLoS Biol. 2010;8:e1000443. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen A, Finel M, Haltia T, Gennis RB, Wikstrom M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- Quirk PG, Hicks DB, Krulwich TA. Cloning of the cta operon from alkaliphilic Bacillus firmus OF4 and characterization of the pH-regulated cytochrome caa3 oxidase it encodes. J. Biol. Chem. 1993;268:678–685. [PubMed] [Google Scholar]
- Rabus R, Jack DL, Kelly DJ, Saier MH., Jr. TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology. 1999;145:3431–3445. doi: 10.1099/00221287-145-12-3431. [DOI] [PubMed] [Google Scholar]
- Real G, Fay A, Eldar A, Pinto SM, Henriques AO, Dworkin J. Determinants for the subcellular localization and function of a nonessential SEDS protein. J. Bacteriol. 2008;190:363–376. doi: 10.1128/JB.01482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. Role of FtsEX in cell division of Escherichia coli: viability of the ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 2007;189:98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Rietkotter E, Hoyer D, Mascher T. Bacitracin sensing in B. subtilis. Mol. Microbiol. 2008;68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor; New York: 1989. [Google Scholar]
- Sarethy IP, Saxena Y, Kapoor A, Sharma M, Sharma SK, Gupta V, Gupta S. Alkaliphilic bacteria: applications in industrial technology. J. Ind. Microbiol. Biotechnol. 2011;38:769–790. doi: 10.1007/s10295-011-0968-x. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Ting CS, King J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- Selivanov VA, Zeak JA, Roca J, Cascante M, Trucco M, Votyakova TV. The role of external and matrix pH in mitochondrial reactive oxygen species generation. J. Biol. Chem. 2008;283:29292–29300. doi: 10.1074/jbc.M801019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 2009;55:1–79. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- Stabell FB, Tourasse NJ, Kolstø AB. A conserved 3' extension in unusual group II introns is important for efficient second-step splicing. Nucleic Acids Res. 2009;37:3202–3214. doi: 10.1093/nar/gkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturr MG, Guffanti AA, Krulwich TA. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J. Bacteriol. 1994;176:3111–3116. doi: 10.1128/jb.176.11.3111-3116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Tahara Y. Characterization of the Bacillus subtilis ywtD gene, whose product is involved in γ–polyglutamic acid degradation. J. Bacteriol. 2003;185:2379–2382. doi: 10.1128/JB.185.7.2379-2382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 2005;9:345–354. doi: 10.1007/s00792-005-0451-6. [DOI] [PubMed] [Google Scholar]
- Takami H, Masui N, Nakasone K, Horikoshi K. Replication origin region of the chromosome of alkaliphilic Bacillus halodurans C-125. Biosci. Biotechnol. Biochem. 1999;63:1134–1137. doi: 10.1271/bbb.63.1134. [DOI] [PubMed] [Google Scholar]
- Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse NJ, Helgason E, Økstad OA, Hegna IK, Kolstø AB. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol. 2006;101:579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Kolstø AB. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: insights into their spread and evolution. Nucleic Acids Res. 2008;36:4529–4548. doi: 10.1093/nar/gkn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse NJ, Stabell FB, Kolstø AB. Structural and functional evolution of group II intron ribozymes: insights from unusual elements carrying a 3' extension. N Biotechnol. 2010;27:204–211. doi: 10.1016/j.nbt.2010.02.014. [DOI] [PubMed] [Google Scholar]
- van der Laan JC, Gerritse G, Mulleners LJ, van der Hoek RA, Quax WJ. Cloning, characterization, and multiple chromosomal integration of a Bacillus alkaline protease gene. Appl. Environ. Microbiol. 1991;57:901–909. doi: 10.1128/aem.57.4.901-909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blohn C, Kempf B, Kappes RM, Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hicks DB, Guffanti AA, Baldwin K, Krulwich TA. Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and non-fermentative growth at pH 10.5. J. Biol. Chem. 2004;279:26546–26554. doi: 10.1074/jbc.M401206200. [DOI] [PubMed] [Google Scholar]
- West IC, Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem. J. 1974;144:87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- Williams KP. The tmRNA Website: invasion by an intron. Nucleic Acids Res. 2002;30:179–182. doi: 10.1093/nar/30.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsted L, von Wachenfeldt C. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 2000;182:6557–6564. doi: 10.1128/jb.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JM. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Ann. Rev. Microbiol. 2011;65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- Yen M-R, Tseng Y-H, Nguyen EH, Wu L-F, Saier MH., Jr. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 2002;177:441–450. doi: 10.1007/s00203-002-0408-4. [DOI] [PubMed] [Google Scholar]
- Yumoto I. Bioenergetics of alkaliphilic Bacillus spp. J. Biosci. Bioeng. 2002;93:342–353. doi: 10.1016/s1389-1723(02)80066-4. [DOI] [PubMed] [Google Scholar]
- Yumoto I. Environmental and taxonomic biodiversities of Gram-positive alkaliphiles. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. ASM Press; Washington, D.C.: 2007. pp. 295–310. [Google Scholar]
- Zuber P. Management of oxidative stress in Bacillus. Annu. Rev.Microbiol. 2009;63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]
- Zwieb C, Gorodkin J, Knudsen B, Burks J, Wower J. tmRDB (tmRNA database) Nucleic Acids Res. 2003;31:446–447. doi: 10.1093/nar/gkg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.