Abstract
Sequential immunolocalisation of 5-methylcytosine (5-MeC) and fluorescence in situ hybridisation with chromosome-specific BAC clones were performed on Brachypodium distachyon mitotic metaphase chromosomes to determine specific DNA methylation patterns of each chromosome in the complement. In the majority of cells examined, chromosomes Bd4 and Bd5, which bear the loci of 5S and 35S ribosomal DNA, respectively, had characteristic 5-MeC patterns. In contrast, the distribution of 5-MeC along the metacentric chromosome pairs Bd1, Bd2 and Bd3 was more variable. There were numerous differences in distribution of methylated sites between homologous chromosomes as well as between chromosome arms. Some chromosome sites, such as pericentromeric regions, were highly methylated in all chromosomes. Additionally, the influence of a hypomethylating agent, 5-azacytidine, on B. distachyon chromosome methylation patterns was confirmed. It was found that some chromosome pairs underwent demethylation more easily than others, but there was no apparent regularity in demethylation of particular chromosome segments.
Keywords: 5-azacytidine, Brachypodium distachyon, Chromosome, DNA methylation, Epigenetics
Introduction
Epigenetic regulation is not mediated by changes in nucleotide sequence but may confer mitotically heritable alterations in gene activity and may play a major role in the expression of some eukaryotic genes. Mechanisms that define the epigenetic landscape of the cell include methylation of the DNA, modifications of the nucleosomal histones and remodelling of chromatin (Tariq and Paszkowski 2004; Wada 2005; Law and Jacobsen 2010). The most widely studied epigenetic modification is DNA methylation, which mediates gene silencing (Gehring and Henikoff 2007; Vaillant and Paszkowski 2007), and therefore is essential for normal plant development and reproduction (Zluvova et al. 2001; Xiao et al. 2006; Meijon et al. 2010).
Covalent DNA modification of cytosine at position 5 occurs primarily at symmetrical CG dinucleotides; in plants it is also observed in non-CG sites, like symmetrical CNG and asymmetric CNN trinucleotides (where N is A, C or T; Bender 2004; Henderson and Jacobsen 2007). Plant nuclear DNA usually possesses high levels of 5-methylcytosine (5-MeC). Recent studies of Cokus et al. (2008) revealed total methylation levels of 24% CG, 6.7% CNG and 1.7% CNN in the genome of the model dicot plant, Arabidopsis thaliana. Moreover, it was proved that 5-MeC content is higher in the genomes of monocots compared with dicot plants (Vanyushin and Ashapkin 2011). These methylation levels correlate with different repetitive DNA content, especially retrotransposons, among species. DNA methylation is considered one of the most important processes to silence transposable element transcription and movement, and probably protects the plant genome from a variety of mutations (Casacuberta and Santiago 2003). There is much evidence suggesting the significant role of DNA methylation in stabilising the plant genome. For example in A. thaliana met1 DNA methyltransferase mutants, various mobile elements and pseudogenes became activated, while in other DNA methyltransferase mutant lines, such as drm1, drm2 and cmt3, the transcriptional activity of these elements remained broadly similar to wild-type plants (Miura et al. 2001; Singer et al. 2001; Lippman et al. 2004). These results, along with the findings of Zhang et al. (2006) clearly suggest that in most cases CG methylation is sufficient for transposable element deactivation.
To date, DNA methyltransferase mutant lines such as met1 are not available for economically important grass crop species, and consequently hypomethylating agents are widely used to study the consequences of genome-wide demethylation. One of them, 5-azacytidine (5-AzaC), is a cytosine analogue in which ring carbon 5 is replaced by nitrogen (Haaf 1995; Mirouze and Paszkowski 2011). Consequences of 5-AzaC treatment have been studied at both molecular and cytological level in various grass species, including triticale and wheat (Neves et al. 1995; Castilho et al. 1999). Akimoto et al. (2007) demonstrated that manipulation of the rice methylome by hypomethylating chemicals may be a powerful biotechnological tool to increase crop quality. They used the progeny of 5-AzaC-treated seedlings to initiate lines for further propagation. Ultimately, a line was obtained whose resistance to a bacterial pathogen, Xanthomonas oryzae, was correlated with promoter hypomethylation of a resistance gene.
Although the majority of information about DNA methylation is based on biochemical and molecular methods, such as methylcytosine immunoprecipitation (Zhang et al. 2006; Zilberman et al. 2007; Gupta et al. 2010) and DNA bisulphite modification followed by ultra-high throughput sequencing (BS-Seq; Cokus et al. 2008; Lister et al. 2008), some cytological techniques have also been developed. Miller et al. (1974) described a method based on detection of highly methylated regions after incubation with antibodies raised against 5-MeC, which has been used for determination of methylation patterns in mammalian chromosomes. The first report which described the 5-MeC distribution in plant chromosomes came from the work of Frediani et al. (1986) on Phaseolus coccineus. During the last two decades, distribution of 5-MeC on metaphase chromosomes of different plant species has been frequently analysed in this way (Ruffini Castiglione et al. 1995, 2008; Frediani et al. 1996; Cremonini et al. 2003; Suzuki et al. 2010). Studies of the distribution of methylation-rich regions provide a further insight into the structure and function of the plant genome at the chromosomal level (Ruffini Castiglione et al. 2002).
Brachypodium distachyon is an internationally accepted model grass which possesses numerous, highly desirable ‘model’ biological features, such as one of the smallest (~350 Mb) nuclear genomes described to date in Poaceae with low (x = 5) chromosome number, small stature, self-fertility, short life cycle and undemanding growth requirements. In 2010, under the extensive collaborative effort of the International Brachypodium Initiative, its nuclear genome was sequenced and annotated (IBI 2010). At present, the ever-growing repertoire of experimental tools includes large collections of accessions and mutants, highly efficient transformation protocols, microarrays, large insert (BAC) libraries of genomic DNA, EST libraries from different tissues and resequencing data (Brkljacic et al. 2011; Mur et al. 2011; Vain 2011). Although the organisation of the B. distachyon genome both at the molecular and cytogenetic level is well studied, to our knowledge no analyses on its epigenetics have been done to date.
In this paper, we demonstrate for the first time the distribution of 5-MeC in mitotic metaphase chromosomes of B. distachyon using a specific monoclonal antibody raised against 5-MeC and epifluorescent visualisation. 5-MeC signals were measured and averaged along the longitudinal axes of all chromosomes. Furthermore, alterations of methylation patterns after 5-AzaC treatment were studied.
Materials and methods
Plant material and root meristem preparation
B. distachyon genotype ABR1 (2n = 10) was obtained from the collection held by Aberystwyth University (UK). Preparations of root meristems were made according to previously described procedure (Jenkins and Hasterok 2007). Seeds were grown on filter paper moistened with tap water for 3 days at room temperature in the dark. Additionally, some seeds were germinated on filter paper moistened with 5-azacytidine solution (5-AzaC; Sigma-Aldrich) at three different concentrations: 0.1, 0.01 and 0.001 mmol/L. 5-AzaC was prepared fresh every day of germination as previously described (Castilho et al. 1999). Whole seedlings with roots 2.5–3.5 cm long were collected and immersed in ice-cold water for 24 h, fixed in 3:1 (v/v) methanol/glacial acetic acid at 4°C overnight and then stored at −20°C until use.
After washing in 0.01 mmol/L citric acid–sodium citrate buffer (pH 4.8) for 15 min, fixed seedlings were digested in enzyme mixture comprising 20% (v/v) pectinase (Sigma-Aldrich), 1% (w/v) cellulase (Calbiochem) and 1% (w/v) cellulase ‘Onozuka R-10’(Serva) for 1.5 h at 37°C. After digestion, meristems were dissected out from the root tips, squashed in 45% acetic acid and frozen on dry ice. After freezing, cover slips were removed and preparations were postfixed in 3:1 ethanol/glacial acetic acid, dehydrated in absolute ethanol and air dried.
Immunodetection of 5-methyl-cytosine
Methylated cytosines were immunodetected with mouse antibodies raised against 5-methylcytosine (Abcam; 1:200 in 1% bovine serum albumin (BSA) in 1× phosphate-buffered saline (PBS)) and goat anti-mouse secondary antibody conjugated with Alexa488 (Invitrogen; 1:200 in 1% BSA in 1× PBS). Slides were denatured in 70% formamide for 2 min at 70°C and blocked with 5% BSA. Incubation with primary antibody was done at 37°C for 1 h and after washes in PBS, the secondary antibody was applied at the same conditions. Chromosomes were counterstained with 2.5 mg/ml 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) in Vectashield (Vector Laboratories).
DNA probes and fluorescence in situ hybridisation
After immunodetection of 5-MeC, a sequential fluorescence in situ hybridisation (FISH) experiment was done to identify particular chromosomes in the complement. BAC clones (ABR1-26-H1, ABR1-42-H8, ABR1-54-D7, ABR1-32-C1 and ABR1-63-E9) from the B. distachyon genomic library constructed by Hasterok et al. (2006) were used as probes to discriminate chromosomes Bd1 to Bd5, respectively. Two different combinations of BAC clones were used in different experiments (Fig. 1c–d). All BACs were labelled with tetramethyl-rhodamine-5-dUTP (Roche) by nick translation as described by Hasterok et al. (2006). Slides previously used for immunodetection of 5-MeC were washed in 4× saline sodium citrate (SSC) with 0.1% Tween 20 at 37°C to remove cover slips and then washed in 2× SSC at room temperature. Slides were postfixed in 4% paraformaldehyde in 2× SSC, washed in 2× SSC, dehydrated in ethanol series and air dried.
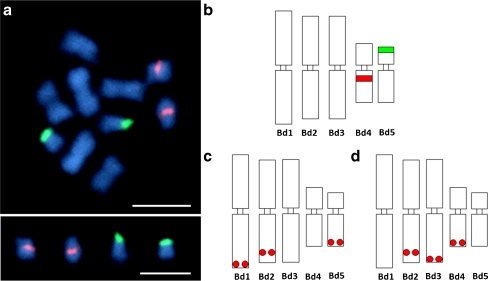
Fig. 1.
Metaphase chromosomes of B. distachyon genotype ABR1. a FISH of 5S (red fluorescence) and 25S rDNA (green fluorescence) to five pairs of chromosomes. b Idiogram of haploid set of chromosomes. The sites of rDNA loci are indicated. c–d Idiograms of B. distachyon chromosomes showing physical localisation of BAC clones used in sequential FISH reactions. The positions of BAC landing sites are marked by red dots. Bar 5 μm
The FISH procedure was adopted from Jenkins and Hasterok (2007). The hybridisation mixture consisted of 50% deionized formamide, 20% dextran sulphate, 2× SSC, 5% sodium dodecyl sulphate, salmon sperm blocking DNA in 75–100 fold-excess of labelled probe and 2.5–3.0 ng/ml of each DNA probe. The mixture was predenatured at 75°C for 10 min, applied to slides with chromosome preparations and then denatured together at 70°C for 4.5 min in Hybaid OmniSlide in situ denaturation system (Thermo Electron). Hybridisation was performed overnight at 37°C in a humid chamber. Post-hybridisation washes were performed in 10% deionized formamide in 0.1× SSC for 10 min at 42°C. Chromosomes were counterstained with DAPI in Vectashield.
Image acquisition and processing
All images were captured with a CoolSNAP cf CCD camera (Photometrics) attached to a Leica DMRB epifluorescence microscope and then processed using Photoshop CS3 (Adobe). To determine the DNA methylation pattern in chromosomes, the ‘RGB Profile Plot’ plugin for ImageJ (NIH, USA) software was used.
Results
In situ immunodetection of the 5-MeC on metacentric chromosomes Bd1, Bd2 and Bd3
The methylation patterns of B. distachyon metaphase chromosomes were studied by immunodetection of 5-MeC, followed by BAC-FISH to identify each chromosome of the complement. B. distachyon is a diploid with the basic chromosome number of x = 5 and an asymmetrical karyotype in which three out of five chromosomes can be easily distinguished based on morphometrical features alone (Fig. 1). However, unambiguous identification of metacentric chromosomes Bd2 and Bd3 requires additional markers, such as chromosome-specific BAC clones. In this paper, five clones were used both to reliably identify each of the five chromosomes and to discriminate between their short and long arms (Fig. 1c–d).
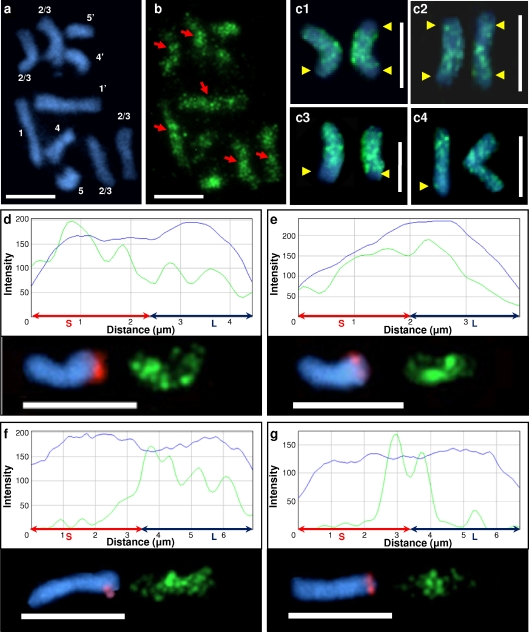
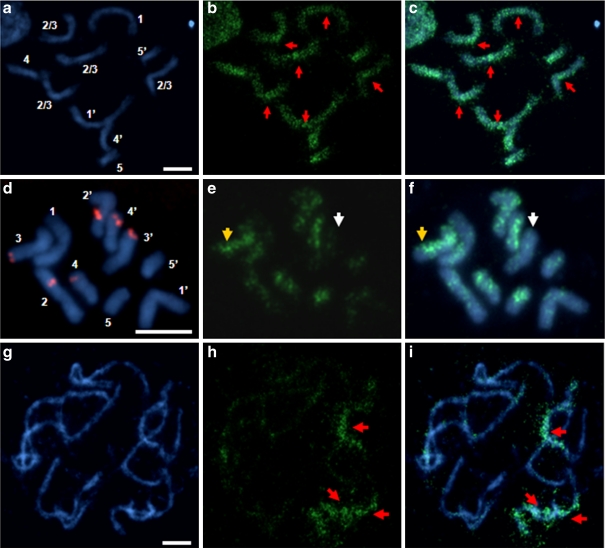
In mitotic metaphase cells from root meristems, distinct 5-MeC foci distributions were detected in all chromosomes (Fig. 2). Metacentric chromosome pairs showed a dispersed anti-5-MeC signals along the arms with some regions almost always more intensively labelled than the others. These highly methylated segments were identified as pericentromeric regions (Fig. 2a–b). The high density of anti-5-MeC signals in pericentromeric segments commonly form characteristic peaks on methylation profiles of all B. distachyon chromosomes. In contrast to pericentromeric sequences, distal regions of metacentric chromosomes were frequently unmethylated or had remarkably lower methylation levels than interstitial and proximal segments (Fig. 2c1–4). Based on the number of unmethylated terminal regions inferred from the pattern of 5-MeC fluorescent signals, all Bd1, Bd2 and Bd3 chromosome pairs were classified in one of the five distinct groups, with 4, 3, 2 or 1 region(s) unmethylated or all distal regions highly methylated (Table 1).
Fig. 2.
Distribution of the 5-MeC foci (green fluorescence) on the metacentric chromosomes of B. distachyon (Bd1, Bd2 and Bd3). a Mitotic metaphase complement stained with DAPI, b distribution of 5-MeC signals at the same chromosomes, pericentromeric regions are pointed out by red arrows. c1–c4 Bd1 homologues, which are representative of other metacentric chromosomes in the complement. Terminal regions with significantly lower methylation levels are marked by yellow arrows. d–g 5-MeC foci along the longitudinal axes of Bd1 chromosomes. Chromosomes are oriented with short arms to the left. The long arm of each chromosome is identified by the BAC clone ABR1-26-H1 (red fluorescence). Profiles of the counterstain (DAPI) are shown by blue curves, the green curves denote the distribution of methylation foci. The length of chromosomes is shown on the x-axis in microns, while the fluorescence intensity on the y-axis is presented in arbitrary units. d–e and f–g Homologous chromosomes from one metaphase complement. DAPI counterstaining, blue fluorescence. Bars 5 μm
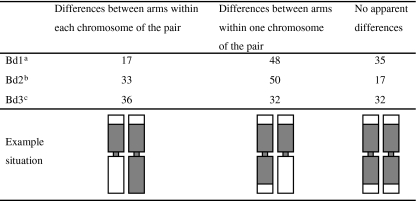
Table 1.
The percentage of examined chromosome pairs Bd1, Bd2 and Bd3 with absence of DNA methylation in distal chromosome regions of individual cells
| 4 terminal regions unmethylated | 3 terminal regions unmethylated | 2 terminal regions unmethylated | 1 terminal region unmethylated | All terminal regions methylated | |
|---|---|---|---|---|---|
| Bd1a | 32 | 18 | 23 | 16 | 11 |
| Bd2b | 30 | 13 | 33 | 17 | 7 |
| Bd3c | 20 | 12 | 28 | 36 | 4 |
Percentages in rows sum to 100%
a95 chromosome pairs examined
b30 chromosome pairs examined
c25 chromosome pairs examined
As can be seen in Fig. 2 (d–e, f–g), differences were detectable in 5-MeC foci distribution between homologous chromosomes. Variation in methylation pattern was also observed between arms of the same chromosome (Fig. 2d, f). Such dissimilarities include both distribution and signal intensity of immunofluorescence corresponding to 5-MeC in particular chromosome segments. Different distribution of anti-5-MeC signals between chromosome arms was observed in some cases in both homologues and in others in only one chromosome of the given pair. In some cases, no apparent difference between homologues was found (Table 2). Sequential FISH with BAC clones revealed that differential methylation of chromosomes Bd1, Bd2 and Bd3 occurs over both short (Fig. 2d) and long arms (Fig. 2f) at similar frequency. In addition, where significant differences in anti-5-MeC signal intensity were seen between the arms of a chromosome, its homologue showed a characteristic methylation pattern with the most prominent pericentromeric anti-5-MeC signals showing either a gradual (Fig. 2e) or a more abrupt (Fig. 2g) boundary with the distal regions.
Table 2.
The percentage of examined chromosome pairs Bd1, Bd2 and Bd3 with different 5-MeC foci (grey areas) distribution between the arms of each chromosome of the pair
Percentages in rows sum to 100%
a95 chromosome pairs examined
b30 chromosome pairs examined
c25 chromosome pairs examined
In situ immunodetection of 5-MeC on chromosomes with rDNA loci
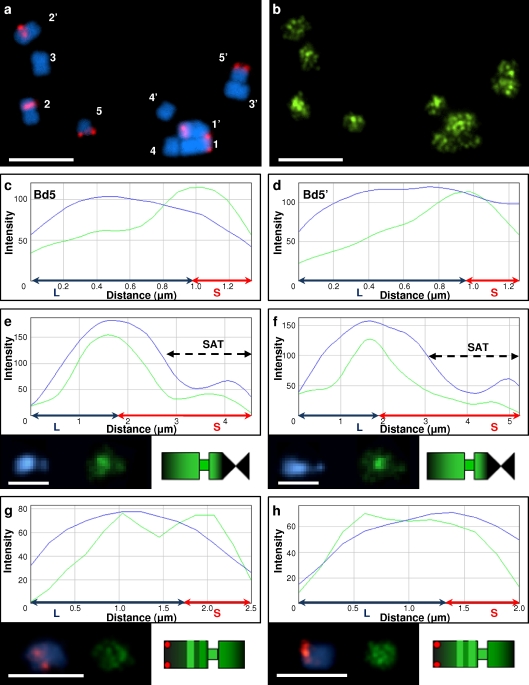
DNA methylation patterns were also analysed in the submetacentric chromosomes Bd4 and Bd5, which carry 5S and 35S rDNA loci respectively (Fig. 1a–b). The 5S rDNA site is localised proximally in the long arm of chromosome Bd4, while a nucleolar organising region (i.e. containing transcriptionally active 35S rDNA loci) is found distally in the short arm of chromosome Bd5 (Draper et al. 2001; Garvin et al. 2008). Unlike the previous group, these chromosomes demonstrate more specific patterns of DNA methylation. Two general types of 5-MeC foci distribution were apparent for chromosome Bd5, depending on condensation, one for highly condensed chromosomes (Fig. 3a–d) and another one for those with clearly visible satellite regions (Fig. 3e–f). Both were characterised by the highest levels of DNA methylation in pericentromeric regions, which abruptly decreased towards both chromosome termini. The methylation profile observed in less condensed Bd5 chromosomes showed considerably lower methylation at 35S rDNA sites (Fig. 3e–f) than in the highly condensed chromosomes (Fig. 3c–d).
Fig. 3.
DNA methylation patterns on chromosomes Bd4 and Bd5 of B. distachyon. a FISH with BAC clones ABR1-26-H1, ABR1-42-H8, ABR1-63-E9 (red fluorescence). b Distribution of 5-MeC signals on the same chromosomes. c–d 5-MeC foci distribution along the longitudinal axes of highly condensed chromosome pair Bd5 excised from the metaphase spread shown on a–b. e–f 5-MeC signal distribution of Bd5 homologues with visible satellite region. g–h 5-MeC foci arrangement of Bd4 homologues. Profiles, idiograms and chromosomes Bd4–Bd5 are oriented with their long arm to the left. Dark green tints on idiograms reflect low methylation level. Methylation profile descriptions as for Fig. 2. DAPI counterstaining, blue fluorescence. Bars 5 μm
The methylation pattern of chromosome Bd4 revealed two characteristic peaks of high-density 5-MeC foci (Fig. 3g–h). The first corresponded with the pericentromeric regions of the chromosome while the second was located interstitially on the long arm. Decrease in intensity of anti-5-MeC signals in proximal regions of chromosomes Bd4 was observed.
Effect of 5-AzaC on DNA methylation
No prominent differences in anti-5-MeC signal distribution were observed in B. distachyon chromosome complements from the material subjected to the lowest (0.001 mmol/L) concentration of 5-AzaC. Immunolocalisation of 5-MeC in metacentric chromosome pairs showed strong similarity to methylation patterns found in chromosomes of the nontreated material (Fig. 4a–c). The specific DNA methylation patterns of the smallest submetacentric pairs Bd4–Bd5 were also retained.
Fig. 4.
DNA methylation patterns on mitotic B. distachyon chromosomes after 5-AzaC treatment. a prometaphase chromosomes subjected to 0.001 mmol/L 5-AzaC. b Methylation pattern of the same chromosomes. Positions of centromeres are pointed out by arrows. d FISH with BAC clones ABR1-26-H1, ABR1-54-D7 and ABR1-32-C1 (red fluorescence) on metaphase chromosomes subjected to 0.01 mmol/L 5-AzaC. e Distribution of 5-MeC foci on the same chromosomes. g Prophase/prometaphase chromosomes after 0.1 mmol/L 5-AzaC treatment. h Methylation pattern of the same chromosomes. c, f, i Superimposed images of DAPI stained chromosomes and signals of 5-MeC residues. The arrows colour coding: red—very high; yellow—high and white—low methylation level. DAPI counterstaining, blue fluorescence. Bars 5 μm
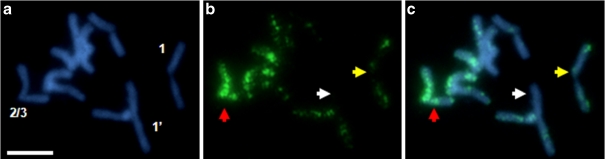
In the material subjected to 5-AzaC at 0.01 mmol/L concentration, signals corresponding to 5-MeC were weaker and their distribution in chromosome pairs was changed in comparison to their counterparts in the control material. Noticeably, some chromosome pairs seemed to undergo demethylation more easily than others, e.g. while chromosome Bd4 still showed strong signals corresponding to 5-MeC, the methylation of chromosome Bd5 was already much lower (Fig. 4d–f). In the majority of the cells examined, neither homologue of Bd5 displayed any strong immunofluorescence signals (data not shown), although in some cases one of the chromosomes showed higher methylation than its homologue (Fig. 4d–f). Moreover, there were considerable differences in methylation between homologous chromosomes Bd1, Bd2 and Bd3. In most cases, one chromosome of the pair showed stronger anti-5-MeC signals than its homologue. This pattern was particularly visible for chromosome Bd3 (Fig. 4d–f). In addition, there was no regularity in demethylation of particular chromosome regions.
Application of 5-AzaC at the highest concentration (0.1 mmol/L) led to decondensation and strong demethylation in B. distachyon chromosomes, however, there were no chromosomes completely lacking methylation foci (Fig. 4g–i). Some chromosomes were almost always found to be labelled, e.g. one homologue of the Bd4 pair (data not shown) and one homologue of either the Bd2 or Bd3 pairs (Fig. 5). Differences in methylation between the homologues were also observed, especially in the case of chromosome Bd1, where one homologue often displayed a weak anti-5-MeC fluorescence along both arms, while its counterpart had no visible methylation foci in one arm (Fig. 5). As in the material subjected to lower concentrations of 5-AzaC, there was no preferential demethylation of particular chromosome regions.
Fig. 5.
Different demethylation of particular B. distachyon chromosomes subjected to 0.1 mmol/L 5-AzaC. a DAPI-stained chromosomes. b Distribution of 5-MeC residues. c Superimposed images of DAPI stained chromosomes and 5-mC distribution. Arrow colour coding as for Fig. 4. Bar 5 μm
Discussion
The aim of this study was to analyse methylation patterns in B. distachyon mitotic metaphase chromosomes using specific monoclonal antibody against 5-MeC according to the method described by Podesta et al. (1993). The distribution of 5-MeC signals in metacentric chromosome pairs Bd1, Bd2 and Bd3 was very variable. Our results indicate differential distribution and intensity of the fluorescence signals among the chromosomes of different cells and between homologues within the same cell (Fig. 2), as well as between the arms of the same chromosome (Fig. 2d, f; Table 2). These data are consistent with the results obtained for other plant species with nuclear genomes of different size, e.g. Allium cepa (Ruffini Castiglione et al. 1995; Suzuki et al. 2010), Vicia faba (Frediani et al. 1996), Triticale (Castilho et al. 1999), Zingeria biebersteiniana (Cremonini et al. 2003) and Haplopapus gracilis (Ruffini Castiglione et al. 2008). We assume that the heterogeneity within a particular chromosome pair could be explained by distinct chromatin organisation, which can be linked with different transcriptional activity of homologous chromosomes, although the possibility that differential distributions of the 5-MeC foci between homologues are the result of variation in the accessibility to the antibody also cannot be ruled out. A well-established precedent is random inactivation of one of the X chromosomes, caused by methylation in CpG islands, which is positively correlated with transcriptional inactivation and late replication (Monk and Grant 1990). Different distribution of 5-MeC foci has also been found in plant sex chromosomes. Siroky et al. (1998) observed that in the female (XX) homozygotes of Melandrium album one of the homologues was hypermethylated, while the second displayed more intensive 5-MeC labelling on its short arm.
It was noticeable that all B. distachyon chromosomes revealed very high intensity of anti-5-MeC signals in pericentromeric regions (Fig. 2a–b), which is probably correlated with accumulation of transposable elements. Analysis of the B. distachyon genome sequence confirmed the presence of LTR elements, predominantly of the Ty3-gypsy superfamily, within these chromosome regions (IBI 2010). A high methylation level of pericentromeric segments is most likely the consequence of the genome defences against mobile element activity, which may also lead to greater condensation of these regions. It has been clearly demonstrated for many organisms, e.g. in human and fruit fly, that the functional centromeric chromatin (CEN chromatin) which is associated with a specific histone H3 variant (CENH3) is enriched with a euchromatic mark, dimethylation of histone H3 at lysine 4 (H3K4me2), while pericentromeric regions which flank CEN chromatin possess heterochromatic modifications H3K9me2 and H3K9me3 (Sullivan and Karpen 2004). Zhang et al. (2008) revealed that hypomethylated CEN chromatin in A. thaliana and maize is also flanked by heterochromatic fractions enriched with H3K9me2. Interestingly, the most recent studies on epigenetic status of active and inactive centromeres in maize B chromosomes showed that inactivation of CEN chromatin is accompanied by hypermethylation (Koo et al. 2011). In our study, methylation profiles of somatic metaphase chromosomes were analysed. The resolution of 5-MeC mapping in highly condensed metaphase chromosomes is probably too low to identify hypomethylated CEN chromatin, however, on some profiles a decrease in 5-MeC foci distribution within centromere sites was seen (Fig. 2d, e, g). Furthermore, all B. distachyon centromeres are flanked by gene-poor regions (IBI 2010; Qi et al. 2010), which is in concordance with data obtained for other grasses, like rice, wheat and barley (Moore 2000).
In contrast, distal chromosome segments in B. distachyon were often unmethylated or had significantly lower methylation level than the other regions (Fig. 2c1–4, Table 1). These data contradict the results obtained for several plant and animal species, where high density of 5-MeC was detected also in telomere-associated sequences (Frediani et al. 1996; Henderson and Jacobsen 2007). Analysis of B. distachyon genome composition revealed that coding sequences are rather evenly distributed along chromosome arms with the exception of pericentromeric and centromeric regions. This kind of gene arrangement is also typical for other small genomes, like rice (Moore 2000) and A. thaliana (AGI 2000; Zhang et al. 2006). Low methylation level at the termini of B. distachyon chromosomes suggests the localisation of transcriptionally active genes within these segments.
Our results indicate that only the smallest chromosomes in the karyotype bear specific and recurrent methylation patterns (Fig. 3). It is well known (Draper et al. 2001; Garvin et al. 2008) that the Bd4 and Bd5 chromosomes carry ribosomal RNA genes (Fig. 1a–b). It is also well documented that while plant genomes contain hundreds to thousands of rRNA gene copies, only some of them are transcriptionally active (Birch and Zomerdijk 2008). Active and silent rRNA genes can be distinguished by markers of their epigenetic status, such as DNA methylation and histone modification. The clearest demonstration of the role of DNA methylation in silencing of rRNA genes has come from studies on nucleolar dominance, the phenomenon present in some natural and resynthesised interspecific plant hybrids. It was demonstrated that treatment with hypomethylating agents like 5-AzaC can activate previously silenced rRNA genes (Neves et al. 1995; Lawrence et al. 2004). Reduction in methylation levels was observed across the B. distachyon rDNA loci (Fig. 3), however 5-MeC signals were still visible. Detection of 5-MeC foci within rDNA sites by immunocytological reaction has been reported for some other plant species, e.g. Z. biebersteiniana (Cremonini et al. 2003), H. gracilis (Ruffini Castiglione et al. 2008) and Colpodium versicolor (Ruffini Castiglione et al. 2009), and has been attributed to the repressed fraction of rRNA genes.
We also demonstrated that 5-AzaC is effective in hypomethylating the B. distachyon genome (Figs. 4, 5), as reported for rice (Sano et al. 1990), flax (Fieldes et al. 2005) and wheat (Santos et al. 2002). Inhibition of DNA methylation was observed at the two highest concentrations used, 0.1 and 0.01 mmol/L. There is no evidence for specific demethylation of particular chromosome segments in B. distachyon, however, some chromosome pairs retain strong anti-5-MeC signals after 5-AzaC treatment while the others seem to undergo demethylation more readily. Furthermore, application of 5-AzaC led to loss of reproducible methylation patterns within the chromosomes Bd4 and Bd5. This result is in agreement with the report of Siroky et al. (1998), where 5-AzaC treatment caused the loss of specific 5-MeC patterns in M. album sex chromosomes.
This study provides the first insight into the epigenetic landscape of the model grass B. distachyon at the chromosomal level. Immunocytological data revealed heterogeneity in the distribution of highly methylated DNA along the chromosomes. The variability of methylation patterns in different cells most likely reflects changes in gene activity. Moreover, analyses with the use of 5-AzaC showed that while the hypomethylating agent caused significant demethylation of B. distachyon genomic DNA, some chromosome regions remained consistently methylated. Future studies should address the question why some parts of the B. distachyon genome can be demethylated more easily than the others. It would be of interest to find a correlation between methylation of particular chromosome segments and the cell differentiation pathway.
Acknowledgements
The authors are grateful to Dr Tim Langdon (Aberystwyth University, UK) and Ms Ewa Breda (University of Silesia, Katowice, Poland) for their helpful comments on the manuscript. This work was supported by the Polish Ministry of Science and Higher Education grant no N303 570738. NB acknowledges scholarships from the UPGOW project co-financed by the European Social Fund.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- 5-AzaC
5-azacytidine
- 5-MeC
5-methylcytosine
- BAC
Bacterial artificial chromosome
- FISH
Fluorescence in situ hybridisation
- NOR
Nucleolus organiser region
- L
Long arm of the chromosome
- rDNA
Ribosomal DNA
- S
Short arm of the chromosome
References
- AGI Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Akimoto K, Katakami H, Kim HJ, et al. Epigenetic inheritance in rice plants. Ann Bot. 2007;100:205–217. doi: 10.1093/aob/mcm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R et al (2011) Brachypodium as a model for the grasses: today and the future. Plant Physiol. doi:10.1104/pp.111.179531, Epub ahead of print [DOI] [PMC free article] [PubMed]
- Casacuberta JM, Santiago N. Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene. 2003;311:1–11. doi: 10.1016/S0378-1119(03)00557-2. [DOI] [PubMed] [Google Scholar]
- Castilho A, Neves N, Rufini-Castiglione M, Viegas W, Heslop-Harrison JS. 5-Methylcytosine distribution and genome organization in triticale before and after treatment with 5-azacytidine. J Cell Sci. 1999;112(Pt 23):4397–4404. doi: 10.1242/jcs.112.23.4397. [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremonini R, Ruffini Castiglione M, Grif VG, et al. Chromosome banding and DNA methylation patterns, chromatin organisation and nuclear DNA content in Zingeria biebersteiniana. Biol Plant. 2003;46:543–550. doi: 10.1023/A:1024863511570. [DOI] [Google Scholar]
- Draper J, Mur LA, Jenkins G, et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001;127:1539–1555. doi: 10.1104/pp.010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieldes MA, Schaeffer SM, Krech MJ, Brown JC. DNA hypomethylation in 5-azacytidine-induced early-flowering lines of flax. Theor Appl Genet. 2005;111:136–149. doi: 10.1007/s00122-005-2005-9. [DOI] [PubMed] [Google Scholar]
- Frediani M, Tagliasacchi AM, Durante M, Avanzi S. Distribution of 5-methylcytosine-rich regions in the polytene chromosomes of Phaseolus coccineus embryo suspensor as shown by the immunoperoxidase technique. Exp Cell Res. 1986;167:337–342. doi: 10.1016/0014-4827(86)90174-6. [DOI] [PubMed] [Google Scholar]
- Frediani M, Giraldi E, Castiglione MR. Distribution of 5-methylcytosine-rich regions in the metaphase chromosomes of Vicia faba. Chromosome Res. 1996;4:141–146. doi: 10.1007/BF02259707. [DOI] [PubMed] [Google Scholar]
- Garvin DF, Gu YQ, Hasterok R, et al. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci The Plant Genome. 2008;48:S69–S84. [Google Scholar]
- Gehring M, Henikoff S. DNA methylation dynamics in plant genomes. Biochim Biophys Acta. 2007;1769:276–286. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gupta R, Nagarajan A, Wajapeyee N. Advances in genome-wide DNA methylation analysis. Biotechniques. 2010;49:iii–xi. doi: 10.2144/000113493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T. The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: implications for methylation-associated cellular processes. Pharmacol Ther. 1995;65:19–46. doi: 10.1016/0163-7258(94)00053-6. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Marasek A, Donnison IS, et al. Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization. Genetics. 2006;173:349–362. doi: 10.1534/genetics.105.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- IBI Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Jenkins G, Hasterok R. BAC ‘landing’ on chromosomes of Brachypodium distachyon for comparative genome alignment. Nat Protoc. 2007;2:88–98. doi: 10.1038/nprot.2006.490. [DOI] [PubMed] [Google Scholar]
- Koo DH, Han F, Birchler JA, Jiang J. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 2011;21:908–914. doi: 10.1101/gr.116202.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/S1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijon M, Feito I, Valledor L, Rodriguez R, Canal MJ. Dynamics of DNA methylation and histone H4 acetylation during floral bud differentiation in azalea. BMC Plant Biol. 2010;10:10. doi: 10.1186/1471-2229-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OJ, Schnedl W, Allen J, Erlanger BF. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14:267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Monk M, Grant M (1990) Preferential X-chromosome inactivation, DNA methylation and imprinting. Development 55–62 [PubMed]
- Moore G. Cereal chromosome structure, evolution, and pairing. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:195–222. doi: 10.1146/annurev.arplant.51.1.195. [DOI] [PubMed] [Google Scholar]
- Mur LA, Allainguillaume J, Catalán P, et al. Exploiting the Brachypodium tool box in cereal and grass research. New Phytol. 2011;191:334–347. doi: 10.1111/j.1469-8137.2011.03748.x. [DOI] [PubMed] [Google Scholar]
- Neves N, Heslop-Harrison JS, Viegas WS. rRNA gene activity and control of expression mediated by methylation and imprinting during embryo development in wheat × rye hybrids. Theor Appl Genet. 1995;91:529–533. doi: 10.1007/BF00222984. [DOI] [PubMed] [Google Scholar]
- Podesta A, Ruffini Castiglione M, Avanzi S, Montagnoli G. Molecular geometry of antigen biding by a monoclonal antibody against 5-methylcytidine. Int J Biochem. 1993;25:929–933. doi: 10.1016/0020-711X(93)90249-E. [DOI] [PubMed] [Google Scholar]
- Qi L, Friebe B, Wu J, Gu Y, Qian C, Gill BS. The compact Brachypodium genome conserves centromeric regions of a common ancestor with wheat and rice. Funct Integr Genomics. 2010;10:477–492. doi: 10.1007/s10142-010-0190-3. [DOI] [PubMed] [Google Scholar]
- Ruffini Castiglione M, Giraldi E, Frediani M. The DNA methylation patterns of Allium cepa metaphase chromosomes. Biologisches Zentralblatt. 1995;114:57–66. [Google Scholar]
- Ruffini Castiglione M, Cremonini R, Frediani M. DNA methylation patterns on plant chromosomes. Caryologia. 2002;55:275–282. [Google Scholar]
- Ruffini Castiglione M, Frediani M, Venora G, Cremonini R. Cytological investigation of Haplopappus gracilis (Nutt.) Gray: 5-methylcytosine-rich regions, fluorochrome banding and chromatin sensitivity to DNase I digestion. Protoplasma. 2008;233:107–113. doi: 10.1007/s00709-008-0296-9. [DOI] [PubMed] [Google Scholar]
- Ruffini Castiglione M, Kotseruba V, Cremonini R. Methylated-rich regions and tandem repeat arrays along the chromosome complement of Colpodium versicolor (Stev.) Schmalh. Protoplasma. 2009;237:13–18. doi: 10.1007/s00709-009-0063-6. [DOI] [PubMed] [Google Scholar]
- Sano H, Kamada I, Youssefian S, Katsumi M, Wabiko H. A single treatment of rice seedlings with 5-azacytidine induces heritable dwarfism and undermethylation of genomic DNA. Mol Gen Genet. 1990;220:441–447. doi: 10.1007/BF00391751. [DOI] [Google Scholar]
- Santos AP, Abranches R, Stoger E, Beven A, Viegas W, Shaw PJ. The architecture of interphase chromosomes and gene positioning are altered by changes in DNA methylation and histone acetylation. J Cell Sci. 2002;115:4597–4605. doi: 10.1242/jcs.00160. [DOI] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA. Robertson’s mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene decrease in DNA methylation (DDM1) Genes Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siroky J, Castiglione MR, Vyskot B. DNA methylation patterns of Melandrium album chromosomes. Chromosome Res. 1998;6:441–446. doi: 10.1023/A:1009244210622. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Shiomi M, Morihana S, Yamamoto M, Mukai Y. DNA methylation and histone modification in onion chromosomes. Genes Genet Syst. 2010;85:377–382. doi: 10.1266/ggs.85.377. [DOI] [PubMed] [Google Scholar]
- Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Vain P. Brachypodium as a model system for grass research. J Cereal Sci. 2011;54:1–7. doi: 10.1016/j.jcs.2011.04.002. [DOI] [Google Scholar]
- Vanyushin BF, Ashapkin VV. DNA methylation in higher plants: past, present and future. Biochim Biophys Acta. 2011;1809:360–368. doi: 10.1016/j.bbagrm.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Wada Y. Physiological functions of plant DNA methyltransferases. Plant Biotech. 2005;22:71–80. doi: 10.5511/plantbiotechnology.22.71. [DOI] [Google Scholar]
- Xiao W, Custard KD, Brown RC, et al. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell. 2006;18:805–814. doi: 10.1105/tpc.105.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang W, Lee HR, Koo DH, Jiang J. Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- Zluvova J, Janousek B, Vyskot B. Immunohistochemical study of DNA methylation dynamics during plant development. J Exp Bot. 2001;52:2265–2273. doi: 10.1093/jexbot/52.365.2265. [DOI] [PubMed] [Google Scholar]