Abstract
Viral infection converts the normal functions of a cell to optimize viral replication and virion production. One striking observation of this conversion is the reconfiguration and reorganization of cellular actin, affecting every stage of the viral life cycle, from entry through assembly to egress. The extent and degree of cytoskeletal reorganization varies among different viral infections, suggesting the evolution of myriad viral strategies. In this Review, we describe how the interaction of viral proteins with the cell modulates the structure and function of the actin cytoskeleton to initiate, sustain and spread infections. The molecular biology of such interactions continues to engage virologists in their quest to understand viral replication and informs cell biologists about the role of the cytoskeleton in the uninfected cell.
The shape and movement of cells, as well as phagocytosis, intercellular communication and the distribution of organelles, depend on actin1 (for history, see BOX 1). Actin persists in the cell as two different forms: monomeric globular actin (G-actin) and polymeric filamentous actin (F-actin) (FIG. 1). F-actin is composed of two parallel strands of actin monomers. The directionality of the filament is determined by the orientation of the monomers, with the positive end being that opposite the end with the ATP-binding pocket. Polymerization begins with actin monomers being stabilized by an initiation complex, of which there are many. The initiation complex that is most often described as interacting with viruses is the ARP2/3 complex2. On its own, ARP2/3 has little polymerization-stimulating activity, but this activity is enhanced through interaction with multiple polymerization induction factors, such as members of the Wiskott–Aldrich syndrome protein (WASP) family and WASP-interacting proteins (WIPs)3,4 (FIG. 2). Filament formation is promoted and stabilized through the action of proteins such as profilin and cortactin, and the filament is depolymerized through the action of proteins such as cofilin or gelsolin5,6. Actin filaments (called microfilaments) also bundle with other actin-interacting proteins, including fascins7,8, forming more substantial structures. Alternatively, the filaments can be crosslinked by branching, which is initiated by actin-nucleating proteins9, to form a meshwork such as cortical actin. F-actin fibres form the microfilament network inside the cell, varying from myosin-containing contractile stress fibres to the cortical actin network that resides beneath the plasma membrane and around intracellular organelles (FIG. 3). Actin fibres are also used to make: sheet-like extensions, such as lamellipodia, membrane ruffles and blebs; finger-like protrusions, such as microvilli and filopodia; or dot-like podosomes. These structures are modified by the action of several actin-binding and signalling proteins.
Box 1. A brief history of actin.
Actin was first observed and isolated in 1887 by Halliburton et al. as a coagulating activity associated with extracts of muscle tissue150. It was not until the 1940s that the term ‘actin’ would become associated with the filamentous material that was isolated from muscle tissue151. Under the electron microscope, actin filaments had a consistent width and varied lengths. When mixed with myosin, the filament increased in width and became studded with “nodose structures which in the myofibril may be aligned” (REF. 152), indicative of the structures that are observed in muscle tissue. It was not until 1962 that people began to understand that actin was found in every eukaryotic cell153 and not until 1973 that the first connection between actin and viruses was reported in the literature16.
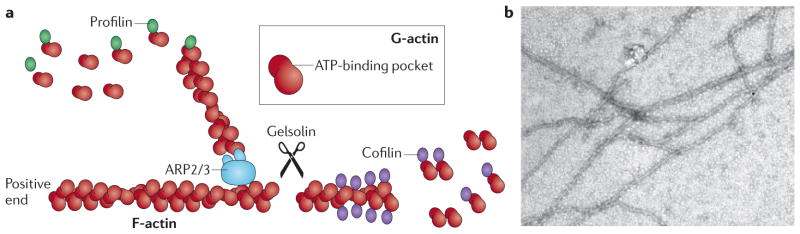
Figure 1. Actin filament dynamics.
a | Actin filaments (known as filamentous actin (F-actin)) are formed by two parallel strands of head–tail polymers of actin monomers (globular actin (G-actin)). Actin polymerization is initiated by the ARP2/3 complex and stimulated by cofactors such as profilin. The direction of actin filaments is determined by the orientation of the monomers, with the positive end being defined as opposite the ATP-binding pocket. Actin depolymerization can occur at either end of the filament. Cofilin interacts with actin dimers to promote disassembly, which can be initiated by the activity of gelsolin. b | Actin filaments that were polymerized in vitro and visualized under an electron microscope. Image is reproduced, with permission, from REF. 154 © (2009) American Society for Biochemistry and Molecular Biology.
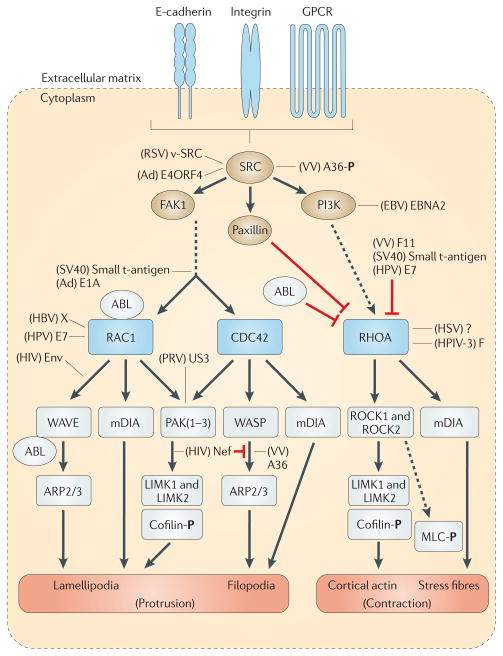
Figure 2. RHO-family GTPase-mediated modelling of the actin cytoskeleton.
E-cadherins, integrins or guanylyl-nucleotide-binding protein (G protein)-coupled receptors (GPCRs) activate SRC, which phosphorylates focal adhesion kinase 1 (FAK1). FAK1 promotes the formation of protrusive actin structures by activating RHO-family GTPases such as RAC1 and cell division cycle 42 (CDC42). The downstream effectors of RHO-family GTPases are: Wiscott–Aldrich syndrome protein (WASP)–WASP-family verprolin-homologous protein (WAVE) proteins, Diaphanous-related formins (mDIA proteins; also known as DRF or DIAPH proteins) and kinases such as PAKs and RHO-associated protein kinases (ROCKs). WASP–WAVE proteins stimulate the activation of the ARP2/3 complex. PAKs and ROCKs contribute to the formation of actin filaments by inactivating cofilin via phosphorylation of LIM domain kinases (LIMKs). mDIA stimulates the nucleation and extension of parallel actin filaments. RHOA is initially inactivated by integrin signalling via paxillin. Phosphoinositide 3-kinase (PI3K) is phosphorylated by SRC and then activates RHOA, leading to the formation of stress fibres. ABL tyrosine kinases negatively regulate the RHO–ROCK signalling pathway while activating RAC1 and WASP–WAVE (for reviews, see REFS 10,11). ROCK proteins also stimulate the phosphorylation of myosin regulatory light chain (MLC), thus contributing to the contractility of actin–myosin. Several viral proteins interact with this signalling machinery at multiple levels (see main text for details). Black arrows indicate direct phosphorylation or stimulation of the downstream molecule. Inhibitory or indirect interactions are shown as red or dotted lines, respectively. Stimulatory or modulatory interactions of viral proteins are indicated by black lines. Ad, adenoviruses; E4ORF4, early region ORF4 protein; EBNA2, EBV nuclear antigen 2; EBV, Epstein–Barr virus; F, fusion protein; HBV, hepatitis B virus; HPV, human papillomavirus; HSV, herpes simplex virus; HPIV-3, human parainfluenza virus 3; PRV, pseudorabies virus; RSV, Rous sarcoma virus; SV40, simian virus 40; VV, vaccinia virus;
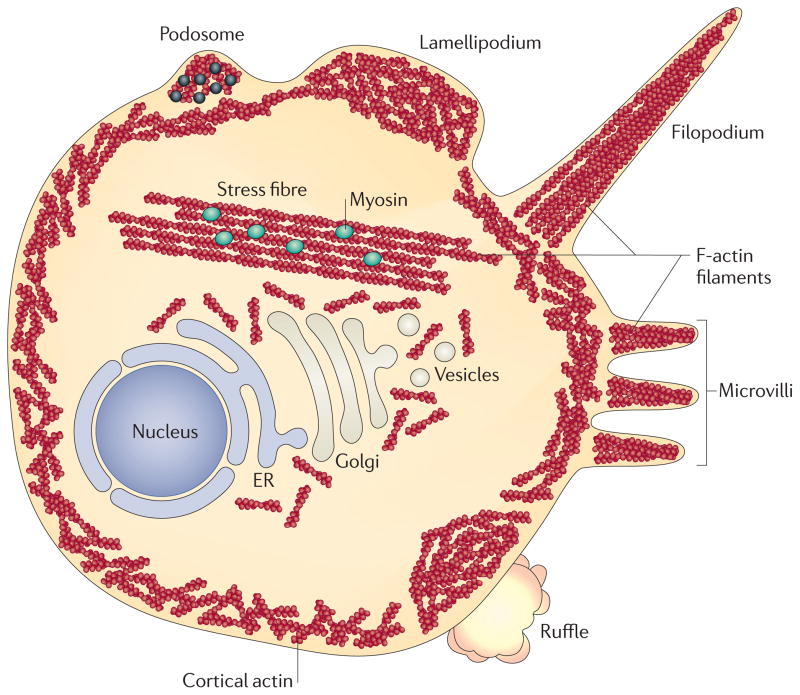
Figure 3. Manifestations of actin rearrangement.
Within the cell, actin filaments can be arranged to form multiple structures. Stress fibres are large assemblies of actin filaments that can span the length of the cell. The presence of myosin in stress fibres enables contractility. Underneath the plasma membrane is the loosely organized network of actin filaments that is termed cortical actin. Actin filaments also can be organized to produce a range of cellular extensions, including podosomes, lamellipodia, filopodia, microvilli and large membrane ruffles. Podosomes contain several actin-binding proteins, signalling molecules and metalloproteinases (black dots). ER, endoplasmic reticulum.
The actin cytoskeleton is highly dynamic and is mainly manipulated by members of the RHO-family GTPases that control signal transduction pathways linking membrane receptors to the cytoskeleton (FIG. 2). RHO-family GTPases regulate several cellular processes, including F-actin polymerization, assembly of intercellular junctions, cell polarity, cell migration and membrane trafficking (reviewed in REF. 10). More than twenty different RHO-family GTPases regulate cytoskeletal dynamics. Among these, the most ubiquitous members are: RHOA, which is responsible for the formation of stress fibres; RAC1, which induces membrane ruffles or lamellipodia; and cell division cycle 42 (CDC42), which regulates the formation of protrusive filopodia11,12. Many pathogens, including viruses, have evolved gene products to engage and subvert the actin cytoskeleton and, in particular, the RHO-family GTPase signalling system (reviewed in REFS 13–15).
In this Review, we highlight some of the interactions that are promoted by viral proteins which redirect the structure and function of the actin cytoskeleton. We attempt to connect the historic literature concerning actin with the current advances in the field. We discuss the role of actin rearrangement during the earliest known effect of viruses on cells: transformation. We continue by describing the ways in which actin is manipulated during entry, assembly and egress, the main stages of the viral life cycle.
It is important not to be anthropomorphic when discussing viruses. However, it is difficult not to be when viral infections seem to be so insidious and capable of thwarting or bypassing everything that blocks their replication and dissemination. The title of this Review uses the evocative word ‘subversion’ more to attract attention to the amazing cell biology engaged by these small entities than to ascribe any purpose to what is observed.
Viral infection and cell transformation
The first links between viral infection, cell morphology and changes in the actin cytoskeleton were realized in the early 1970s with the description of ‘transformation’ (BOX 2). The first-identified transforming viruses were: Rous sarcoma virus (RSV), an avian retrovirus16; simian virus 40 (SV40), a primate polyomavirus17; and adenoviruses18. The most prominent changes in transformed cells are the rounded cell shape, with motile blebs, bulging pseudopodia, loss of contact inhibition and lack of stress fibres. Indeed, transformed cells are immobile in cell culture and pile up on each other, with less pronounced adherens junctions, randomly distributed microtubules, diminished microfilaments and disrupted stress fibres17–19. Many of these changes also are seen in mitotic cells but are more organized temporally20. At the onset of mitosis, cells round up, matrix adhesion is lost and a contractile actin–myosin ring is formed.
Box 2. Cell transformation.
Virus-mediated cell transformation is a multistep process resulting in abnormally proliferating and morphologically altered cells in vitro which share the characteristics of tumour cells in vivo. Typically, transformed cells lack contact inhibition, are anchorage independent and form multiple layers in the culture dish, termed foci17. Key steps of virus-mediated transformation include deregulation of the cell cycle and inhibition of apoptotic pathways by the action of viral oncoproteins. To achieve such complex deregulation, oncoproteins of small DNA tumour viruses (for example, the small t- and large T-antigens of simian virus 40 (REF. 44), the E1A and E1B proteins of adenoviruses20,47 or the E6 and E7 proteins of human papillomaviruses61) work in an orchestrated manner to interfere with several steps of signal transduction pathways. Incomplete action of these oncoproteins may result in immortalized cells, which do not form foci in vitro and are not tumorigenic when injected in animals47. In the case of retroviruses, viral RNA genomes contain aberrant forms of cellular proto-oncogenes (such as c-SRC, c-FOS, c-JUN and c-MYC) encoding proteins with abnormal functions (for example, the v-SRC protein of Rous sarcoma virus25) that cause deregulation of several signalling cascades, resulting in a completely transformed cell phenotype. Cells that are infected and transformed by these viruses do not produce infectious progeny virions and therefore are not lysed as a result of cytopathic effects. Transforming viruses rather stimulate unlimited cell proliferation and persist in these immortal cells as an evolutionary strategy.
These changes are accompanied by an increase in cortical rigidity, which involves actin remodelling mediated by a signalling pathway that involves RHO proteins and RHO-associated protein kinases (ROCKs)21. The main questions that emerged from these early studies focused on how viral infection induces dramatic cytoskeletal reorganization and whether these changes are related to the in vivo neoplastic properties of these viral infections. This connection seemed obvious because cells establish themselves in a stable state in vivo by contacting adjacent cells and the extracellular matrix. The transition from this normal state to an invasive, proliferating phenotype certainly requires drastic changes in the regulation of cell signalling, the cytoskeleton, transcription and the cell cycle, and these changes are probably caused by the specific actions of viral proteins.
RSV, SV40 and adenoviruses
Work with RSV, the causative agent of chicken tumours, revealed the first viral protein that was sufficient to transform cells19. Using a temperature-sensitive RSV mutant, investigators proved that the viral src (v-src) gene product (v-SRC; also known as pp60v–src) was responsible for the actin filament reorganization and altered surface topography observed during infection. RSV v-SRC-induced morphological changes occurred in three steps. In the first 3 hours of temperature shift, membrane alterations were obvious, with the appearance of actin ruffles22. At later times after the temperature shift, cells rounded up with retraction of the peripheral cytoplasm before the final step, which was marked by the production of microvilli, microspikes and blebs19,23. Subsequent investigations revealed that v-SRC was able to ‘dissolve’ actin stress fibres when microinjected into the cytosol of 3T3 cells, confirming that the cytoskeleton and cell motility system were among the main targets of the viral oncoprotein24. In the following years, v-SRC was shown to be a membrane-associated phosphoprotein with tyrosine kinase activity25. In transformed cells, v-SRC concentrates in podosomes and rosettes, where it colocalizes with actin-modifying proteins such as vinculin, talin, α-actinin26,27 and cortactin28. Cytoskeletal targets that are phosphorylated by v-SRC include vinculin29 and cofilin30. In normal cells, cellular SRC kinases (c-SRC proteins) regulate the formation of integrin-mediated focal adhesions. Initially, researchers thought that v-SRC changed normal focal adhesions into unique dot-like podosomes in transformed cells. However, similar podosome structures were identified in normal motile cells such as macrophages, osteoclasts22 and dendritic cells31, as well as in smooth muscle cells32 and endothelial cells33. Although podosomes are often referred to as ‘actin dots’, they are in fact complex structures containing an actin core that is surrounded by several cytoskeletal proteins and matrix metalloproteinases34 that, together with signalling molecules, remodel the cytoskeleton and extracellular matrix as necessary for tissue invasion35. Myosin was observed close to the actin core of v-SRC-induced podosomes or naturally occurring podosomes of osteoclasts, but the podosomes of cancerous or transformed cells are usually not contractile (reviewed in REF. 36). Given these observations, these podosomes of cancerous or transformed cells were renamed invadopodia, referring to the invasive properties of those cells35.
v-SRC induces the loss of stress fibres, not by depolymerizing F-actin but rather by reorganizing actin polymers so that canonical stress fibres are not formed37. These F-actin aggregates, located at the adhesion areas of transformed cells, are more resistant to the cytoskeleton-disrupting drug cytochalasin B (TABLE 1) than normal F-actin, confirming that altered polymerization occurs during transformation38. This phenotype is linked to the reduction of caldesmon expression in transformed cells, which results in a reduction in calcium-regulated actin reorganization39. In addition, other calcium-dependent actin-interacting proteins — gelsolin and α-actinin — are localized at the regions of small stress fibres in RSV-transformed rat fibroblasts40. In addition to these changes in calcium-regulated, actin-binding proteins, RSV- or SV40-transformed cells exhibit a substantial decrease in their synthesis of a subset of the tropomyosin family of proteins8,41. Tropomyosin in non-muscle cells modulates actin binding to actin-severing, actin-capping, actin-crosslinking and actin-nucleating proteins42. Tropomyosin binding to F-actin stabilizes the stress fibres and protects them from disassembly43. Reduction of not only tropomyosin but also extracellular matrix proteins (for example, collagen and fibronectin) may contribute to the disruption of both cytoskeletal and extracellular architecture in cells transformed with RNA and DNA viruses41.
Table 1.
Modulation of actin filaments
| Drug | Mechanisms of action and additional notes | Reference |
|---|---|---|
| Latrunculin A and B |
|
156 |
| Cytochalasin A, B, C and D |
|
157 |
| Mycalolide B |
|
158 |
| Phalloidin |
|
157 |
| Jasplakinolide |
|
159 |
| Blebbistatin |
|
160 |
| Tat–C3 |
|
161 |
| Y-27632 |
|
162 |
| IPA-3 |
|
163 |
The small t-antigen of the small DNA tumour virus SV40 also engages the actin cytoskeleton during cell transformation. This viral protein alone is responsible for the loss of actin filaments in rat cells44. In 2003, it was shown that the SV40 small t-antigen affects protein phosphatase 2A (PP2A), resulting in the loss of tight junctions as well as the stimulated rearrangement of F-actin networks in epithelial cells45. These rearrangements include RAC-induced membrane ruffling and formation of lamellipodia, CDC42-initiated formation of filopodia, and loss of RHO-dependent stress fibres. In epithelial cells expressing small t-antigen, levels of RAC1 and CDC42 increased while RHOA levels decreased.
Some types of adenovirus efficiently transform primary rodent cells in culture (reviewed in REF. 46). Adenovirus infection stimulates the cell cycle through the action of the viral E1A proteins (both the 13S and 12S RNA-encoded isoforms)47. In addition, E1A proteins also disrupt actin stress fibres48 and interact with the RAC–CDC42 pathway in transformed rodent cells. These interactions lead to a disorganization of the actin cytoskeleton with increased filopodial and lamellipodial production, thus enhancing cellular motility and contributing to the loss of contact inhibition49, as seen in cells transformed with RSV and SV40.
EBV, HBV and HPV
Epstein–Barr virus (EBV), a member of the family Herpesviridae, infects a large proportion of the human population worldwide and establishes a long-term, quiescent infection in virally transformed B cells. EBV infections induce efficient proliferation and transformation of B cells and are associated with several types of cancer. The first indication that cells transformed with EBV have an altered actin cytoskeleton was noted in the late 1970s. Analysis of EBV-transformed lymphocytes50 and the sera from patients with Burkitt’s lymphoma or nasopharyngeal carcinoma51 revealed substantial increases in the levels of actin and tubulin in these cells compared with levels in untransformed cells and in the sera of healthy individuals. Surprisingly, actin and tubulin accumulated on the surface of EBV-transformed cells in large quantities52. Such an extracellular presence of cytoskeletal proteins has also been observed by other groups and has attracted much attention (reviewed in REFS 53,54). However, the transport route and the extracellular function of these normally intracellular molecules still await elucidation. Moreover, in EBV-transformed B cells, the expression of fascins, which are actin-bundling proteins, is increased by more than 200-fold55. This overexpression might result in the induction of membrane protrusions, such as lamellipodia, through the reorganization of F-actin at the cell periphery56 and possibly increases cytoskeletal rigidity, as evidenced by in vitro studies57. The changes in EBV-transformed B cells reflect the action of EBV nuclear antigen 2 (EBNA2), a transcription factor that induces several cellular changes which lead to cell proliferation. EBNA2 activates a signalling pathway through the induction of one of the regulatory subunits of phosphoinositide 3-kinase (PI3K), p55α (an isoform encoded by the PI3KR1 gene), presumably interfering with signalling of the RHO-family GTPases58.
Hepatitis B virus (HBV) is a hepadnavirus that causes serious liver diseases, including hepatocellular carcinoma, in humans. HBV replication in HepG2 cells activates RAC1 and induces lamellipodia formation and membrane ruffling59. The viral protein X interacts directly with RAC1 and enhances virus replication. The cytoskeletal rearrangements that are induced by protein X (for example, the redistribution of F-actin-binding proteins at pseudopodia tips) may contribute to the prominent increased migratory phenotype that is observed in HBV-infected hepatocellular carcinoma cells60.
The early oncoproteins E6 and E7 of human papillomaviruses (HPVs) trigger malignant transformation by interfering with several cell cycle and survival pathways. E7 affects both cell cycle and RHO-family GTPase signalling cascades. E7 also enhances the cytoplasmic accumulation of p27 (also known as KIP1) (a positive regulator of cell motility and tumour invasion), which in turn inhibits RHOA activity and induces cell migration61. RAC1 is also overexpressed in papilloma cells, leading to misregulation of the RHO-family GTPase signalling pathway and thus resulting in elevated cyclooxygenase 2 expression62.
Actin and the viral life cycle: entry
The first steps of every viral infection involve virions engaging the cell surface, with subsequent penetration of the cell membrane and entry into the cytoplasm. The physical barrier imposed by the cortical actin meshwork must be overcome, and this process often involves the stimulation of actin cytoskeleton remodelling. Virion binding can initiate signalling events that change the cell surface and activate endocytosis, as we discuss below.
Virion surfing
Virion surfing is the physical movement of virions on the surface of cells or cell projections. Remarkably, interactions between viral and cellular proteins stimulate virion movement to previously masked entry sites, where fusion or endocytosis occurs. Viral proteins located on the virion surface bind with high specificity and affinity to cellular receptors that are associated with actin filaments just beneath the cell surface. The virion movement (surfing) towards entry sites is mediated by internal myosin motors, which are located in the cortical actin or at the base of filopodia and which contract the actin filaments63 (FIG. 4a). In effect, the movement of the actin cytoskeleton drags the cellular receptor–virion complexes to sites of high endocytic activity. Viruses that have been observed surfing across the cell surface include murine leukemia virus, avian leukosis virus, HIV, vesicular stomatitis virus (VSV)64, HPV65 and vaccinia virus (VV)66. Surfing is sensitive to cytochalasin D and the myosin II inhibitor blebbistatin (TABLE 1). Treatment with these drugs results in the undirected wandering of virions on the cell surface, with a corresponding reduction in viral infectivity. The virus–cell interaction that initiates such motion is largely uncharacterized. For VV, surface phosphatidylserine is crucial for virion movement and subsequent endocytosis. This unusual lipid, which is normally found on extracellular apoptotic vesicles, stimulates rapid endocytosis. Virions bind and transit to the base of filopodia, where the phosphatidylserine induces signalling mediated by serine/threonine-protein kinases PAK1 and PAK2 to initiate virion engulfment in a ‘bleb’ of plasma membrane. Active actin remodelling is necessary for both steps in this process, as VV infection is inhibited by the above-mentioned drugs66.
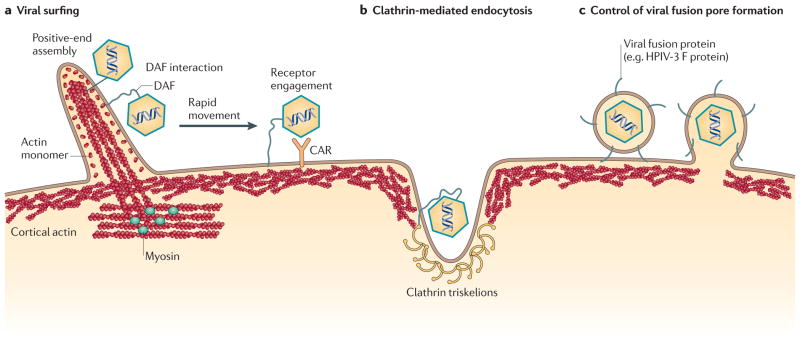
Figure 4. Models of entry.
Virions enter cells through a range of pathways, some of which are depicted here. a | Viral surfing. Myosin II at the base of the filopodium pulls the actin filaments, while the constant actin turnover at the tip pushes them. This movement makes virions ‘surf’ down the filopodium to the sites of entry. In some cases, virions bind glycophosphatidylinositol (GPI)-anchored complement decay-accelerating factor (DAF), resulting in cytoskeletal reorganization that enables the virions to rapidly reach their specific receptors. b | Actin-enhanced clathrin-mediated endocytosis. Clathrin initiates endocytosis, and actin filaments (possibly cortical actin) are recruited to increase the size of the endocytic structure. c | The suggested involvement of cortical actin in regulating formation of the fusion pore; this pore formation is mediated by viral fusion proteins such as the human parainfluenza virus 3 (HPIV-3) fusion protein (F). CAR, coxsackie virus and adenovirus receptor.
Some non-enveloped virions also move on the cell surface and induce actin remodelling for entry. Cellular entry of coxsackie viruses (which are picornaviruses) and adenoviruses relies on interactions with the same cellular receptor. Coxsackie virus and adenovirus receptor (CAR) is located at tight junctions between epithelial cells67, where it is effectively hidden from extracellular virions. To gain entry via this receptor, virions first bind the glycophosphatidylinositol (GPI)-anchored complement decay-accelerating factor (DAF), which is easily accessible on the apical cell surface. This interaction activates the tyrosine protein kinase ABL, which promotes reorganization of the cytoskeleton through the activity of RAC GTPases. DAF-bound virions move laterally to tight junctions to engage CAR68. Subsequent internalization of adenoviruses and of some coxsackievirus serotypes requires clathrin-mediated endocytosis69 (FIG. 4b).
Virion endocytosis
The engagement of endocytosis for virion entry is a subject of research and debate. Evidence for the endocytosis of infecting virions is widespread and is often actin dependent or actin stimulated (for a comprehensive review, see REF. 70). For example, entry of Kaposi’s sarcoma-associated virus, a herpesvirus, is sensitive to the action of drugs that disrupt actin filaments71 (for example, cytochalasin D, latrunculin B and jasplakinolide (TABLE 1)). Similarly, entry of Ebola virus particles and VSV pseudovirions is stimulated when RHOB and RHOC are overexpressed and decreased when RHOA is overexpressed72. These results are consistent with actinmediated macropinocytosis. Entry of VSV virions via clathrin-coated pit-mediated endocytosis is enhanced by the formation of actin filaments73 (FIG. 4b). Entry of poliovirus, an unenveloped picornavirus, is dependent on actin74 and, following entry, the rapid transit of poliovirus particles to replication centres is also dependent on an intact actin network75. In general, actin-enhanced endocytic pathways, which have previously been associated with uptake of bacteria, are often stimulated by small virus particles76.
Fusion of virion membranes and cell membranes at the cell surface
Virions that do not engage the endocytic machinery at the cell surface may gain entry by direct fusion with the cell’s plasma membrane. The involvement of actin in this process is not well understood. Viruses of the family Paramyxoviridae, such as human respiratory syncytial virus (HRSV) and human parainfluenza virus 3 (HPIV-3), encode an attachment glycoprotein G (a haemagglutinin–neuraminidase) and a fusion protein (F), both of which are required for virion entry. F protein mediates fusion of the viral membrane with the cellular membrane (FIG. 4c), subsequent transfer of the viral genome and the formation of syncytia between infected and adjacent cells. Modulation of actin filaments by cytochalasin D reduces HRSV entry as well as subsequent synctium formation, suggesting that actin filaments are involved in both processes77. The fusion and synctium-forming activity of the HPIV-3 F protein are dependent on a dynamic actin filament network and RHOA78,79. The expression of RAC1 and CDC42 promotes glycoprotein G-mediated cell–cell fusion, whereas the expression of RHOA decreases this activity80.
Receptor clustering
While some virions move on the cell surface to find an entry site, HIV virion attachment induces clustering and enrichment of the receptors CD4 and CXC-cytokine receptor 4 (CXCR4) at points of virion contact81. Receptor clustering is an actin-dependent process that uses RHO-family GTPase signalling and the actin-remodelling proteins filamin and cofilin82,83. Env-mediated fusion of virions with the plasma membrane activates RAC1, which signals to the WASP-family verprolin-homologous protein 2 (WAVE2) complex and the kinase ABL1, resulting in the initiation of ARP2/3-mediated actin polymerization and capsid entry84. Inhibition of any of these functions blocks entry at the step of fusion initiation or fusion pore expansion.
Extension induction
Virion attachment to cells can stimulate an increased uptake of virions by several mechanisms, including the extension of cell surface protrusions. Herpes simplex virus (HSV) virions engage the cytoskeleton during entry by activating RHOA-induced phagocytosis. This process involves the production of cell surface extensions and clustering of viral-entry receptors85. Formation of the extensions requires long actin filaments and is sensitive to treatment with cytochalasin D and latrunculin B. Stimulating an endophagocytic pathway through the activation of RHO-family GTPases encourages uptake of multiple virions, perhaps ensuring robust viral replication. HPV infection induces stress fibre dissolution and rapid filopodia formation within 10 minutes of infection. This activity is blocked by genistein or wortmannin treatment, suggesting that tyrosine phosphorylation and PI3K activity are behind the signalling pathways that induce these transient structures86. The production of filopodia not only allows entry of already engaged virions but also acquires virions that are near, but not in direct contact with, the target cell.
Viral replication and assembly
Viral replication requires the coordinated production, processing and assembly of viral proteins and nucleic acids to produce progeny virions that will be used to infect new cells. Depending on the cellular compartment in which replication and assembly occur, the cytoskeleton has interesting and sometimes surprising roles that are not always well understood. For example, it is known that many enveloped virions contain cellular actin87, but whether this actin has a function (for example, whether it is required for assembly or egress) or is an incidental passenger largely remains an unanswered question. Some insights have come from studying the role of actin during viral replication and assembly.
Enhancement of viral replication
Actin enhances paramyxovirus replication, but cytochalasin D has little effect on virus production77. In vitro reconstitution of HRSV RNA replication required actin (FIG. 5a) and could be further enhanced by the addition of profilin88. Furthermore, the matrix (M) protein of the related canine distemper virus does not associate with actin, but replication is sensitive to actin depolymerization89. By contrast, the M protein of influenza viruses, which are orthomyxoviruses, does associate with actin filaments90. Monomeric G-actin and profilin may enhance the replication and assembly of negative-strand RNA viruses in general, either by acting as a transcription factor or by stabilizing nucleic acids, although no clear mechanism has yet been elucidated91.
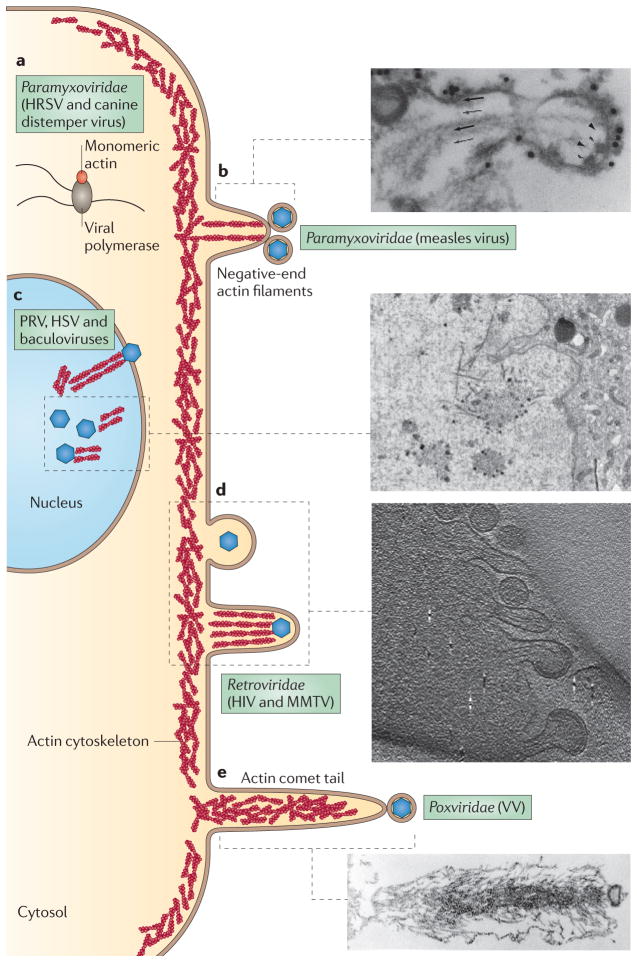
Figure 5. Actin involvement in viral replication and egress.
a | The cytoplasmic replication complexes of some negative-strand RNA viruses associate with actin. A putative replication complex of a paramyxovirus is shown, with globular actin stimulating RNA polymerase activity. b | Measles virions bud off the plasma membrane at the ends of microvillus-like structures, often associating with the ends of negative-oriented actin filaments. As seen in the myosin-labelled electron micrograph, budding virions are indicated by arrowheads, extracellular virions are dense structures at the membrane, and the directionality of the microfilament (arrows) is determined by the orientation of myosin barbs. c | Actin fibres interact with assemblies of DNA virus capsids within the nucleus. An electron micrograph of a pseudorabies virus (PRV)-infected nucleus is shown. The high-contrast spots are capsids, and the long, fibrous projections are actin fibres. d | Retroviruses also bud off the plasma membrane, presumably by stimulating the cortical actin network. As virion budding progresses, small actin-filled microvilli form underneath the virions, as observed under cryoelectron microscopy. e | Vaccinia virus (VV)-infected cells produce dense actin comet tails underneath virions. These comet tails push the virus long distances away from the cell to enhance viral dissemination and spread. HRSV, human respiratory syncytial virus; HSV, herpes simplex virus; MMTV, mouse mammary tumour virus. Part b electron micrograph is reproduced, with permission, from REF. 101 © (1986) Elsevier. Part c electron micrograph is reproduced from REF. 104. Part d electron micrograph is reproduced from REF. 112. Part e electron micrograph is reproduced, with permission, from REF. 155 © (1996) The Company of Biologists.
Movement of intracellular assemblies
The actin cytoskeleton has been suggested to have a role in moving viral genomes and proteins to cytoplasmic assembly sites. After entry, the phosphorylated HIV Gag–MA complex associates with F-actin before microtubule-based transport of virions to the nucleus92. Disruption of actin filaments with cytochalasin D not only inhibits entry of retroviral capsids but also inhibits reverse transcription. Inhibition of reverse transcription by cytochalasin D indicates that actin may regulate or enhance HIV genome synthesis, similar to the role of actin in paramyxovirus replication. Nuclear localization of Gag–MA was reduced, as was HIV infectivity, when inhibitors of myosin light chain kinases were used to inhibit myosin-dependent movement on actin filaments93. Taken together, these results show that actin affects many early steps of HIV infection, from entry through to nuclear import.
Actin may have a role in the final assembly and budding of infectious HIV virions at the cell surface94. Assembly usually occurs at the plasma membrane, where Gag assembles and buds into plasma membrane vesicles containing Env. HIV budding assemblies are associated with Triton-X100-insoluble lipid rafts, but the driving process for interactions between Gag and Env is believed to be the actin network95. Actin-depolymerizing agents such as mycalolide B (TABLE 1) inhibit HIV assembly96. HIV Gag interacts with actin filaments, and equine infectious anaemia virus Gag multimers associate with F-actin assemblies during infection97.
The interaction of Newcastle disease virus and Sendai virus M proteins with actin filaments is important during virion maturation98,99. For Sendai virus, expression of M protein is essential for the actin restructuring that is observed during infection and virion release100. The nucleocapsids of measles virus, a related paramyxovirus, associate with actin filaments in small villus-like structures at the cell periphery101 (FIG. 5b). Cytochalasin D inhibits the release of both measles virus102 and Sendai virus, implicating the formation of new actin filaments in viral assembly and release.
Actin has also been implicated in the nuclear assembly and egress of DNA viruses. Alphaherpesviruses, such as HSV and pseudorabies virus (PRV), assemble capsids in the nucleus, following which the capsids move to sites of nuclear egress along the inner nuclear membrane. The intranuclear movement of HSV capsids is sensitive to temperature, inhibition of ATP hydrolysis by sodium azide treatment, and inhibition of actin polymerization by latrunculin B treatment but not by cytochalasin D treatment103. Infection by both HSV and PRV induces nuclear actin filaments (FIG. 5c) and the accumulation of nuclear myosin V that may be necessary for movement and nuclear egress104. Further, latrunculin B and cytochalasin D treatment affects the nuclear size changes and chromatin marginalization that are prominent after HSV infection105. The mechanism that stimulates the formation of nuclear actin filaments is unknown but may involve the actin-modifying activities of the viral kinase US3 (REF. 106), as discussed below. Actin filaments in the nucleus are not unique to herpesvirus infections and may be a requirement for other viruses that replicate in the nucleus, such as baculoviruses107. After release from the nucleus, completely assembled HSV-1 virions navigate out of the cell in a process that requires the action of myosin Va motors. When infected Vero cells express a dominant-negative form of myosin Va, virion secretion is inhibited with a concomitant increase in intracellular virions108. Myosin Va may interact with virion transport vesicles to mediate intracellular movement, or may modify the cortical actin network to facilitate access of intracellular virions to the plasma membrane for subsequent release.
Viral egress and spread
Newly assembled virions leaving the cell face many of the same barriers that are encountered during virus entry, including the need to move past the cortical actin meshwork under the plasma membrane, to cover large spatial distances and to engage a new, viable host cell. Considerable evidence suggests that some viruses require the actin cytoskeleton for successful egress and spread109.
Viral budding
The first connection between virion egress and the actin cytoskeleton was made for mouse mammary tumour virus (MMTV), a retrovirus. Mammary tumour epithelial cells were observed to release virus particles from the ends of long microvilli110. This observation was extended by immunogold staining of actin in egress sites and MMTV particles111, and for other retroviruses, including HIV112 (FIG. 5d), raising the question of whether viral proteins interact with the actin cytoskeleton for the final steps of assembly and release. Disrupting actin filaments with cytochalasin D dramatically blocked virus budding and reduced virus yield by 80%113.
Extension production
Efficient retroviral spread depends on cell-to-cell contacts and does not occur readily by free virion release and serial infection. Recent evidence reveals the existence of thin, actin-based, filopodial extensions — termed cytonemes or tunnelling nanotubes — that act as bridges for the transfer of virus particles between infected and uninfected cells. Infected macrophages produce large numbers of these extensions during infection114. In MMTV-infected cells, engagement of Env and receptors on the uninfected cell filopodia results in stabilization and lengthening of the actin-based bridge115. Virus particles bud from the infected cell and move in a directed manner across the extension before entry at the base of the filopodium, similar to the viral surfing described earlier for the entry of free particles64.
The HIV Nef protein is involved in the formation of filopodial extensions in Jurkat T cells. Nef affects actin filaments by altering neural WASP (N-WASP) activity116. In addition, Nef also inhibits membrane ruffling and cell motility. The modifications of the actin network and GTPase signalling have long-term effects on cell adhesion and T cell motility in chemokine gradients117. Nef also modulates PAK2 activity, despite there being a lack of detectable interaction between these two proteins by immunoprecipitation experiments118; this modulation of PAK2 activity results in the inactivation of cofilin by hyper-phosphorylation119, and the inhibition of actin remodelling. The F195A point mutation in Nef blocks its interaction with PAK1 and PAK2, as well as the majority of its actin-modulating activity120.
Other viruses also induce actin extensions to disseminate infection over large distances in cultured cells. HSV and PRV produce filopodia or nanotube-like extensions through the activity of the viral kinase US3 (REFS 121,122). These extensions are necessary for efficient cell-to-cell spread of virions in vitro123. The activity of US3 may modify actin kinase signalling directly via phosphorylation of PAK1 and PAK2. PAK1 is necessary for the PRV-induced formation of actin projections, whereas PAK2 is necessary for the disassembly of stress fibres124. US3-induced extensions and their function in in vitro viral dissemination is also observed in cells infected with other alphaherpesvirus species125.
Rotaviruses also induce filopodial extensions for disseminated spread. The expression of rotavirus protein NSP4 and rotavirus infection both result in loss of phosphorylated cofilin in a calcium-dependent manner, whereas extended expression of NSP4 induces long, actin-based extensions from cells126. The spike protein, VP4, associates with actin filaments during infection and ectopic expression. VP4 immunoprecipitates with actin and localizes to the apical surface of polarized Caco-2 cells. Long-term VP4 expression results in actin aggregation, similar to that observed after cytochalasin D treatment127. Furthermore, VP4 modulates the polarized spread of infection, using actin treadmilling to promote virus release from apical surfaces in polarized cells128. Together, these findings suggest that two viral proteins may synergistically alter the actin architecture to promote the directional, long-distance transport of virions from the infected cell. The necessity of filopodia formation for rotavirus spread in vivo remains to be determined.
Comet tails and long-distance spread
VV infections reveal extensive interactions between the virus and the actin cytoskeleton. The first interactions involve the assembly and transport of intracellular virions. The viral protein F11 (coded by the gene F11L) blocks the regulation of RHO-family GTPase GTP cycling by binding RHOA129, thus preventing RHOA interactions with ROCK proteins and Diaphanous-related formins (mDIA proteins; also known as DRF or DIAPH proteins), and therefore resulting in a loss of actin stress fibres. Further, microtubule and actin dynamics are required for efficient viral transport and release. An F11L deletion or the expression of a mutated F11 protein that cannot bind RHOA results in the accumulation of virions beneath the plasma membrane130. Latrunculin B- or cytochalasin D-mediated dissolution of cortical actin restores viral movement to the cell periphery in the absence of functional F11, supporting the conclusion that F11 reorganizes the cortical actin network to allow virions access to the plasma membrane. Interestingly, the activity of F11 also is needed for VV-induced actin-mediated cell motility via lamellipodial extensions131 and for viral spread in vivo132.
VV-infected cells exhibit a unique phenotype involving changes in the actin cytoskeleton that are required for the spread of the infection. Hiller first reported actin extensions with considerable accumulations of actin filaments, reminiscent of the ‘comet tails’ that had been seen underneath extracellular virions133. These actin comet tails induced by VV (FIG. 5e) propel extracellular virions quickly, and for long distances, away from the infected cell134. The viral protein A36 (REF. 135) is an integral membrane protein of the outer viral envelope. When intracellular enveloped virions fuse with the plasma membrane, becoming cell-associated extracellular virions, A36 remains in the plasma membrane underneath the virion, where it undergoes c-SRC-mediated tyrosine phosphorylation136. Phosphorylated A36 recruits NCK proteins and growth factor receptor-bound protein 2 (GRB2)137, which in turn recruit N-WASP and WIPs3 to stimulate ARP2/3-mediated actin polymerization. In the absence of A36, or of other accessory viral proteins or the cellular factors required for actin polymerization, viral spread is substantially reduced (most often observed as a small plaque on cell monolayers). The association between actin tail formation and rapid plaque growth pointed directly at the process of cell-to-cell spread. On cell monolayers, cell-to-cell spread of VV is rapid — much faster than the replication cycle in a single cell. Remarkably, the proteins that mediate actin tail formation also reduce superinfection by mediating active repulsion through a similar mechanism138. Under these circumstances, A36 expressed by the newly infected cells is recruited to subsequently attaching virions, thus repelling the virion away from the cell body using actin tail polymerization. In this manner, infection travels much faster than one cell distance per replication cycle, resulting in rapid plaque formation.
Actin tails on egressing virions are a hallmark of poxvirus infections139, even when obvious A36 homologues are not present. Yaba-like disease virus, another member of the family Poxviridae, has short-lived actin tails that are dependent on the protein YL126 and on the recruitment of NCK1 and NCK2 through the tyrosine phosphorylation of YL126, similar to NCK protein recruitment by A36 (REF. 140). Further, actin-based extensions have been observed in a divergent number of large, cytoplasmically replicating DNA viruses (LCDV), including the iridovirus frog virus 3 (REF. 141).
Infection of the asfarvirus African swine fever virus (ASFV), another LCDV, induces actin-based projections, but these resemble filopodia in that they are linear actin filaments bound together with fascins. These extensions are similar to those formed during VV egress in that actin filament polymerization is induced to extend virions to distances away from the cell142, but the mechanism of viral transport in these ASFV-induced extentions is radically different from that in the VV-induced extensions. Importantly, ASFV virions remain inside the cell at the tip of the actin-based extension.
Cell disruption
In some circumstances, virions are released by destruction of the infected cell. Little is known about the mechanisms of cell disruption after viral infection in eukaryotic cells. The most informative studies have been carried out with adenovirus-infected permissive cells in vitro. Here, the expression of early region ORF4 protein (E4ORF4) leads to altered RHO-family GTPase signalling, which results in cell death143. This early viral protein triggers a p53-independent cell death pathway by accumulating on cellular membranes and the cytoskeleton, and by interacting with c-SRC family kinases as well as with PP2A143. E4ORF4 interaction with c-SRCs deregulates c-SRC signalling and activates the JUN kinase signalling pathway, resulting in dramatic changes in the actin cytoskeleton and in cell blebbing. In particular, E4ORF4 triggers de novo actin polymerization through complex interactions with CDC42, RAC1 and RHOA, which in turn promote the assembly of a contractile juxtanuclear actin–myosin network144. As a consequence, endosomes are recruited to the sites of actin assembly, leading to impaired membrane trafficking, which results in caspase-independent cell death.
Actin as an antiviral target
Cell biologists have used small-molecule modulators of actin to influence the cytoskeleton for years (TABLE 1). These same compounds also alter the outcome of viral infections, implying a function for the cytoskeleton. Such an interpretation is complicated by the involvement of actin in essential cellular processes, including cell division, polarity, motility and uptake of nutrients. The interpretation of these experiments is further complicated by the fact that actin-modulating compounds often have pleiotropic effects, disrupting more cellular processes than just those involving the actin cytoskeleton. Despite these complications, current research indicates that viral interactions with the actin cytoskeleton can be successfully targeted for new antiviral therapies and to treat virally induced disease. The small molecule Y-27632, a RHO–ROCK pathway inhibitor, has been demonstrated to block metastasis of HPV-transformed cervical cancer cells and could be developed as a novel antimetastatic therapy145. Antiviral screens using RNA interference and traditional small-molecule inhibitors will undoubtedly uncover new regulatory nodes and other potential therapeutic targets146.
Conclusion
Actin and actin-like molecules are ubiquitous in cells, from microorganisms to eukaryotes. These ancient proteins have fundamental roles in cell architecture as well as in many cell functions. It is noteworthy that every virus system studied to date has revealed interactions with the actin cytoskeleton. However, viral genomes do not usually encode actin cytoskeleton or myosin components. One exception is the reported presence of actin-like genes in the ASFV genome147, although it is unknown whether these virally encoded homologues function as cytoskeletal proteins. Therefore, the modification and modulation of actin structures in infected cells must result from the action of viral proteins that activate cell signalling pathways or directly recruit actin-remodelling proteins. The molecular biology of such interactions continues to both engage virologists in their quest to understand viral replication, and inform cell biologists about the role of the cytoskeleton in the uninfected cell.
Historically, studies of the transforming viruses provided the first appreciation that viral infection alters the host cytoskeleton. Virally transformed cells round up and cell adhesions are altered, leading to uncontrolled cell division and invasive or metastatic phenotypes. These changes are mediated by viral proteins binding to and modifying cellular signalling pathways that involve proteins such as RAC1 and CDC42. Subsequently, it was found that non-transforming viral infections alter the same pathways to promote replication and dissemination. In both cases, infection induces the disruption of densely packed actin stress fibres, resulting in the production of new actin-based structures within the cell or extending far from the cell body. Many similarities exist between transforming and non-transforming viruses, such as the effects of RHOA inhibition by VV protein F11 (R EF. 129) and the SV40 small t-antigen45. In both cases, the viral proteins cause a decrease in cell adherence to substrates and an increase in cellular motility.
Both infected and uninfected cells require a dynamic actin cytoskeleton to carry out their functions. An important caveat is that most of our current knowledge about virus–cytoskeleton interactions is derived from in vitro studies. How these findings relate to infected tissues and animals is not known. For example, stress fibre assemblies (a common feature of tissue culture cells) are often altered during viral infection of cultured cells. In vivo, these structures have been observed only in endothelial cells148,149. There are reports of novel actin structures in tissues after infection of animals. One study used serial block face scanning electron microscopy of infected rodent peripheral nervous system tissue to provide high-resolution, three-dimensional images of nuclear actin filaments that were stimulated by herpesvirus infection104. Another study demonstrated that VV dissemination in animals requires actin-modulating machinery132. Certainly, observing and manipulating the three-dimensional architecture of infected tissues is challenging experimentally but will yield large rewards. Multiphoton imaging of infected tissue and animals at the single-cell and single-molecule level is but one of many new approaches for in vivo studies. The systems biology tools of proteomics, metabolomics and lipidomics will provide much needed insights into the interface between viral proteins and the actin cytoskeleton in cells and tissues. Although studies of virus-infected cultured cells have provided many basic ideas about the molecular and cellular biology of the actin cytoskeleton, in vivo studies will undoubtedly reveal even more interactions of viruses with the cytoskeleton.
Acknowledgments
The authors acknowledge M. Way, W. Bohn, K. Gruenwald and K. DeMali for generously providing the original electron micrographs. They also appreciate the guidance and encouragement from all members of the Enquist laboratory. L.W.E. and O.O.K. are supported by the US National Institutes of Health grants R37 NS033506-16 and R01 NS060699-03. M.P.T. is supported by an American Cancer Society Postdoctoral Research Fellowship (PF-10-057-01-MPC). Competing interests statement The authors declare no competing financial interests.
Glossary
- Gelsolin
A calcium-activated protein that severs actin filaments
- Lamellipodia
Wide, thin sheets of membrane extending from the cell body; lamellipodia are often associated with the leading edge of motile cells.
- Membrane ruffles
Membrane-enclosed, densely packed actin bundles that increase during cell migration or transformation. Ruffles localize to the leading edge of the lamellipodia, giving these structures a flower-like appearance
- Filopodia
Long, thin membranous extensions of the cell with a core of actin filaments
- Podosomes
Dot-like extracellular matrix attachment sites in motile cells. In transformed cells, podosomes aggregate in the presence of serum to form ring-or crescent-shaped rosettes
- Pseudopodia
Large membranous protrusions that are used to promote the movement of highly motile cells; the name is derived from the Greek for ‘false-footed’.
- Contact inhibition
The inhibition of uncontrolled cell division by cell–cell contact through mitogen-activated protein kinase signalling. Contact inhibition is deregulated in transformed cell populations
- Adherens junctions
Epithelial cell-to-cell junctions that connect the actin cytoskeleton of one cell to the cytoplasm of the neighbouring cell via cadherins and catenins
- Vinculin
A focal-adhesion plaque protein that is associated with the microfilament ends and talin
- Talin
An actin-binding protein associated with adherens junctions, ruffling membranes and other sites of actin–membrane interaction
- α-actinin
A large family of proteins that crosslink and bundle actin filaments in a calcium-dependent manner in non-muscle cells
- Focal adhesions
Macromolecular complexes that connect the actin cytoskeleton to the extracellular matrix by the association of transmembrane integrins with extracellular proteins such as fibronectin
- Caldesmon
A calmodulin-binding and F-actin-binding protein that regulates the function of actin filaments in a calcium-dependent manner
- Clathrin-mediated endocytosis
The process of enveloping extracellular material and bringing it into the cellular cytoplasm within a membranous vesicle. Invagination of the plasma membrane and stabilization of the vesicle is carried out by triskelions of clathrin that polymerize at the membrane surface to induce curvature
- Macropinocytosis
A specialized form of endocytosis that is used by the cell to obtain soluble materials from the extracellular environment
- Actin treadmilling
The act of moving a specific actin monomer, or its position, along an actin filament using active polymerization and depolymerization processes
- Comet tails
Dense structures of actin filaments that are used to propel objects through the cytoplasm or long distances away from the cell
Footnotes
FURTHER INFORMATION
Lynn W. Enquist’s homepage:
http://www.molbio1.princeton.edu/labs/enquist/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver AM, Young ME, Lee WL, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 3.Moreau V, et al. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nature Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 4.Weisswange I, Newsome TP, Schleich S, Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. doi: 10.1038/nature07773. [DOI] [PubMed] [Google Scholar]
- 5.Carlier MF, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashiro S, Yamakita Y, Ono S, Matsumura F. Fascin, an actin-bundling protein, induces membrane protrusions and increases cell motility of epithelial cells. Mol Biol Cell. 1998;9:993–1006. doi: 10.1091/mbc.9.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumura F, Yamashiro-Matsumura S, Lin JJ. Isolation and characterization of tropomyosin-containing microfilaments from cultured cells. J Biol Chem. 1983;258:6636–6644. [PubMed] [Google Scholar]
- 9.Uruno T, et al. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nature Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz AA, Govek EE, Bottner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- 11.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 12.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nature Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. A recent and extensive review on the biology of RHO-family GTPases. [DOI] [PubMed] [Google Scholar]
- 13.Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Münter S, Way M, Frischknecht F. Signaling during pathogen infection. Sci STKE. 2006;2006:re5. doi: 10.1126/stke.3352006re5. A comprehensive review on how pathogen infection affects signalling mechanisms. [DOI] [PubMed] [Google Scholar]
- 15.Favoreel HW, Enquist LW, Feierbach B. Actin and Rho GTPases in herpesvirus biology. Trends Microbiol. 2007;15:426–433. doi: 10.1016/j.tim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Fleissner E, Tress E. Chromatographic and electrophoretic analysis of viral proteins from hamster and chicken cells transformed by Rous sarcoma virus. J Virol. 1973;11:250–262. doi: 10.1128/jvi.11.2.250-262.1973. The earliest work on RSV-induced cytoskeletal changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNutt NS, Culp LA, Black PH. Contact-inhibited revertant cell lines isolated from SV40-transformed cells. IV. Microfilament distribution and cell shape in untransformed, transformed, and revertant Balb-c 3T3 cells. J Cell Biol. 1973;56:412–428. doi: 10.1083/jcb.56.2.412. The earliest report on SV40-induced cell transformation and morphological changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman RD, Chang C, Williams JF. Properties and behavior of hamster embryo cells transformed by human adenovirus type 5. Cold Spring Harb Symp Quant Biol. 1975;39 :601–614. doi: 10.1101/sqb.1974.039.01.074. [DOI] [PubMed] [Google Scholar]
- 19.Wang E, Goldberg AR. Changes in microfilament organization and surface topogrophy upon transformation of chick embryo fibroblasts with Rous sarcoma virus. Proc Natl Acad Sci USA. 1976;73:4065–4069. doi: 10.1073/pnas.73.11.4065. The earliest demonstration of the gradual changes in cell morphology that occur as a result of cell transformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson P, Bellett AJ. Relationship between organization of the actin cytoskeleton and the cell cycle in normal and adenovirus-infected rat cells. J Virol. 1989;63:311–318. doi: 10.1128/jvi.63.1.311-318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- 22.Marchisio PC, Capasso O, Nitsch L, Cancedda R, Gionti E. Cytoskeleton and adhesion patterns of cultured chick embryo chondrocytes during cell spreading and Rous sarcoma virus transformation. Exp Cell Res. 1984;151:332–343. doi: 10.1016/0014-4827(84)90384-7. [DOI] [PubMed] [Google Scholar]
- 23.Boschek CB, et al. Early changes in the distribution and organization of microfilament proteins during cell transformation. Cell. 1981;24:175–184. doi: 10.1016/0092-8674(81)90513-4. [DOI] [PubMed] [Google Scholar]
- 24.McClain DA, Maness PF, Edelman GM. Assay for early cytoplasmic effects of the src gene product of Rous sarcoma virus. Proc Natl Acad Sci USA. 1978;75:2750–2754. doi: 10.1073/pnas.75.6.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellie S, Horvath AR, Elmore MA. Cytoskeletal targets for oncogenic tyrosine kinases. J Cell Sci. 1991;99:207–211. doi: 10.1242/jcs.99.2.207. [DOI] [PubMed] [Google Scholar]
- 26.Rohrschneider LR. Adhesion plaques of Rous sarcoma virus-transformed cells contain the src gene product. Proc Natl Acad Sci USA. 1980;77:3514–3518. doi: 10.1073/pnas.77.6.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shriver K, Rohrschneider L. Organization of pp60src and selected cytoskeletal proteins within adhesion plaques and junctions of Rous sarcoma virustransformed rat cells. J Cell Biol. 1981;89:525–535. doi: 10.1083/jcb.89.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiura K, Lim SS, Little SP, Lin S, Sato M. Differentiation dependent expression of tensin and cortactin in chicken osteoclasts. Cell Motil Cytoskeleton. 1995;30:272–284. doi: 10.1002/cm.970300405. [DOI] [PubMed] [Google Scholar]
- 29.Sefton BM, Hunter T, Ball EH, Singer SJ. Vinculin: a cytoskeletal target of the transforming protein of Rous sarcoma virus. Cell. 1981;24:165–174. doi: 10.1016/0092-8674(81)90512-2. [DOI] [PubMed] [Google Scholar]
- 30.Yoo Y, Ho HJ, Wang C, Guan JL. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin-proteasome pathway. Oncogene. 2010;29:263–272. doi: 10.1038/onc.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burns S, Thrasher AJ, Blundell MP, Machesky L, Jones GE. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood. 2001;98:1142–1149. doi: 10.1182/blood.v98.4.1142. [DOI] [PubMed] [Google Scholar]
- 32.Hai CM, Hahne P, Harrington EO, Gimona M. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp Cell Res. 2002;280:64–74. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- 33.Moreau V, Tatin F, Varon C, Genot E. Actin can reorganize into podosomes in aortic endothelial cells, a process controlled by Cdc42 and RhoA. Mol Cell Biol. 2003;23:6809–6822. doi: 10.1128/MCB.23.19.6809-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato T, et al. Identification of the membrane-type matrix metalloproteinase MT1-MMP in osteoclasts. J Cell Sci. 1997;110:589–596. doi: 10.1242/jcs.110.5.589. [DOI] [PubMed] [Google Scholar]
- 35.Saltel F, et al. Invadosomes: Intriguing structures with promise. Eur J Cell Biol. 2011;90:100–107. doi: 10.1016/j.ejcb.2010.05.011. A recent review about invadopodia, connecting earlier observations and nomenclature with recent literature. [DOI] [PubMed] [Google Scholar]
- 36.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci. 2009;122:3037–3049. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felice GR, Eason P, Nermut MV, Kellie S. pp60v-src association with the cytoskeleton induces actin reorganization without affecting polymerization status. Eur J Cell Biol. 1990;52:47–59. [PubMed] [Google Scholar]
- 38.Carley WW, Lipsky MG, Webb WW. Regulation and drug insensitivity of F-actin association with adhesion areas of transformed cells. J Cell Physiol. 1983;117:257–265. doi: 10.1002/jcp.1041170218. [DOI] [PubMed] [Google Scholar]
- 39.Owada MK, et al. Occurrence of caldesmon (a calmodulin-binding protein) in cultured cells: comparison of normal and transformed cells. Proc Natl Acad Sci USA. 1984;81:3133–3137. doi: 10.1073/pnas.81.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang E, Yin HL, Krueger JG, Caliguiri LA, Tamm I. Unphosphorylated gelsolin is localized in regions of cell-substratum contact or attachment in Rous sarcoma virus-transformed rat cells. J Cell Biol. 1984;98:761–771. doi: 10.1083/jcb.98.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendricks M, Weintraub H. Tropomyosin is decreased in transformed cells. Proc Natl Acad Sci USA. 1981;78:5633–5637. doi: 10.1073/pnas.78.9.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostap EM. Tropomyosins as discriminators of myosin function. Adv Exp Med Biol. 2008;644:273–282. doi: 10.1007/978-0-387-85766-4_20. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 44.Graessmann A, Graessmann M, Tjian R, Topp WC. Simian virus 40 small-t protein is required for loss of actin cable networks in rat cells. J Virol. 1980;33:1182–1191. doi: 10.1128/jvi.33.3.1182-1191.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nunbhakdi-Craig V, Craig L, Machleidt T, Sontag E. Simian virus 40 small tumor antigen induces deregulation of the actin cytoskeleton and tight junctions in kidney epithelial cells. J Virol. 2003;77:2807–2818. doi: 10.1128/JVI.77.5.2807-2818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endter C, Dobner T. Cell transformation by human adenoviruses. Curr Top Microbiol Immunol. 2004;273:163–214. doi: 10.1007/978-3-662-05599-1_6. [DOI] [PubMed] [Google Scholar]
- 47.Nielsch U, Fognani C, Babiss LE. Adenovirus E1A-p105(Rb) protein interactions play a direct role in the initiation but not the maintenance of the rodent cell transformed phenotype. Oncogene. 1991;6:1031–1036. [PubMed] [Google Scholar]
- 48.Bellett AJ, Jackson P, David ET, Bennett EJ, Cronin B. Functions of the two adenovirus early E1A proteins and their conserved domains in cell cycle alteration, actin reorganization, and gene activation in rat cells. J Virol. 1989;63:303–310. doi: 10.1128/jvi.63.1.303-310.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer RS, Quinlan MP. While E1A can facilitate epithelial cell transformation by several dominant oncogenes, the C-terminus seems only to regulate rac and cdc42 function, but in both epithelial and fibroblastic cells. Virology. 2000;269:404–419. doi: 10.1006/viro.2000.0232. [DOI] [PubMed] [Google Scholar]
- 50.Bachvaroff RJ, Klein G, Rapaport FT. Alterations in cell characteristics in relation to malignant transformation. Transplant Proc. 1979;11:1055–1059. [PubMed] [Google Scholar]
- 51.Lamelin JP, Williams EH, Souissi T, De-The G, Gabbiani G. Smooth muscle antibody in Burkitt’s lymphoma and in nasopharyngeal carcinoma. Clin Exp Immunol. 1977;28:157–162. [PMC free article] [PubMed] [Google Scholar]
- 52.Bachvaroff RJ, Miller F, Rapaport FT. Appearance of cytoskeletal components on the surface of leukemia cells and of lymphocytes transformed by mitogens and Epstein–Barr virus. Proc Natl Acad Sci USA. 1980;77:4979–4983. doi: 10.1073/pnas.77.8.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalheiser NR. Proteins in unexpected locations. Mol Biol Cell. 1996;7:1003–1014. doi: 10.1091/mbc.7.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnoys EJ, Wang JL. Dual localization: proteins in extracellular and intracellular compartments. Acta Histochem. 2007;109:89–110. doi: 10.1016/j.acthis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Mosialos G, et al. Epstein-Barr virus infection induces expression in B lymphocytes of a novel gene encoding an evolutionarily conserved 55-kilodalton actin-bundling protein. J Virol. 1994;68:7320–7328. doi: 10.1128/jvi.68.11.7320-7328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishikawa R, Yamashiro S, Kohama K, Matsumura F. Regulation of actin binding and actin bundling activities of fascin by caldesmon coupled with tropomyosin. J Biol Chem. 1998;273:26991–26997. doi: 10.1074/jbc.273.41.26991. [DOI] [PubMed] [Google Scholar]
- 57.Tseng Y, Fedorov E, McCaffery JM, Almo SC, Wirtz D. Micromechanics and ultrastructure of actin filament networks crosslinked by human fascin: a comparison with α-actinin. J Mol Biol. 2001;310:351–366. doi: 10.1006/jmbi.2001.4716. [DOI] [PubMed] [Google Scholar]
- 58.Spender LC, et al. Cell target genes of Epstein–Barr virus transcription factor EBNA-2: induction of the p55α regulatory subunit of PI3-kinase and its role in survival of EREB2.5 cells. J Gen Virol. 2006;87:2859–2867. doi: 10.1099/vir.0.82128-0. [DOI] [PubMed] [Google Scholar]
- 59.Tan TL, et al. Rac1 GTPase is activated by hepatitis B virus replication — involvement of HBX. Biochim Biophys Acta. 2008;1783:360–374. doi: 10.1016/j.bbamcr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 60.Lara-Pezzi E, et al. The hepatitis B virus X protein (HBx) induces a migratory phenotype in a CD44-dependent manner: possible role of HBx in invasion and metastasis. Hepatology. 2001;33:1270–1281. doi: 10.1053/jhep.2001.1270. [DOI] [PubMed] [Google Scholar]
- 61.Charette ST, McCance DJ. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene. 2007;26:7386–7390. doi: 10.1038/sj.onc.1210541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu R, Coniglio SJ, Chan A, Symons MH, Steinberg BM. Up-regulation of Rac1 by epidermal growth factor mediates COX-2 expression in recurrent respiratory papillomas. Mol Med. 2007;13:143–150. doi: 10.2119/2007-00005.Wu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nature Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 64.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. The earliest description of viral surfing as it relates to viral entry and infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schelhaas M, et al. Human papillomavirus type 16 entry: retrograde cell surface transport along actinrich protrusions. PLoS Pathog. 2008;4:e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 67.Huang KC, Yasruel Z, Guérin C, Holland PC, Nalbantoglu J. Interaction of the Coxsackie and adenovirus receptor (CAR) with the cytoskeleton: binding to actin. FEBS Lett. 2007;581:2702–2708. doi: 10.1016/j.febslet.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 68.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 69.Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2004;6:S152–S163. doi: 10.1002/jgm.553. An essential review covering virus-induced cytoskeletal changes and endocytosis. [DOI] [PubMed] [Google Scholar]
- 70.Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Ann Rev Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 71.Greene W, Gao SJ. Actin dynamics regulate multiple endosomal steps during Kaposi’s sarcoma-associated herpesvirus entry and trafficking in endothelial cells. PLoS Pathog. 2009;5:e1000512. doi: 10.1371/journal.ppat.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quinn K, et al. Rho GTPases modulate entry of Ebola virus and vesicular stomatitis virus pseudotyped vectors. J Virol. 2009;83:10176–10186. doi: 10.1128/JVI.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SPJ. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. doi: 10.1371/journal.ppat.1000394. A visually stunning analysis of VSV entry and the role of actin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brandenburg B, et al. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaughan JC, Brandenburg B, Hogle JM, Zhuang X. Rapid actin-dependent viral motility in live cells. Biophys J. 2009;97:1647–1656. doi: 10.1016/j.bpj.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veiga E, Cossart P. The role of clathrin-dependent endocytosis in bacterial internalization. Trends Cell Biol. 2006;16:499–504. doi: 10.1016/j.tcb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kallewaard NL, Bowen AL, Crowe JE., Jr Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology. 2005;331:73–81. doi: 10.1016/j.virol.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 78.Wurth MA, et al. The actin cytoskeleton inhibits pore expansion during PIV5 fusion protein-promoted cell– cell fusion. Virology. 2010;404:117–126. doi: 10.1016/j.virol.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pastey MK, Gower TL, Spearman PW, Crowe JE, Jr, Graham BS. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nature Med. 2000;6:35–40. doi: 10.1038/71503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schowalter RM, et al. Rho GTPase activity modulates paramyxovirus fusion protein-mediated cell–cell fusion. Virology. 2006;350:323–334. doi: 10.1016/j.virol.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 81.Iyengar S, Hildreth JEK, Schwartz DH. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol. 1998;72:5251–5255. doi: 10.1128/jvi.72.6.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoder A, et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T Cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jimenez-Baranda S, et al. Filamin-A regulates actin-dependent clustering of HIV receptors. Nature Cell Biol. 2007;9:838–846. doi: 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- 84.Harmon B, Campbell N, Ratner L. Role of Abl kinase and the Wave2 signaling complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog. 2010;6:e1000956. doi: 10.1371/journal.ppat.1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clement C, et al. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol. 2006;174:1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith JL, Lidke DS, Ozbun MA. Virus activated filopodia promote human papillomavirus type 31 uptake from the extracellular matrix. Virology. 2008;381:16–21. doi: 10.1016/j.virol.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cantin R, Methot S, Tremblay MJ. Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J Virol. 2005;79:6577–6587. doi: 10.1128/JVI.79.11.6577-6587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burke E, Mahoney NM, Almo SC, Barik S. Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J Virol. 2000;74:669–675. doi: 10.1128/jvi.74.2.669-675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klauschies F, et al. Viral infectivity and intracellular distribution of matrix (M) protein of canine distemper virus are affected by actin filaments. Arch Virol. 2010;115:1503–1508. doi: 10.1007/s00705-010-0737-6. [DOI] [PubMed] [Google Scholar]
- 90.Bucher D, et al. M protein (M1) of influenza virus: antigenic analysis and intracellular localization with monoclonal antibodies. J Virol. 1989;63:3622–3633. doi: 10.1128/jvi.63.9.3622-3633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harpen M, Barik T, Musiyenko A, Barik S. Mutational analysis reveals a noncontractile but interactive role of actin and profilin in viral RNA-dependent RNA synthesis. J Virol. 2009;83:10869–10876. doi: 10.1128/JVI.01271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arhel N, et al. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nature Methods. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 93.Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. A biochemical analysis of the association between actin and HIV particles at different stages post-entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fackler OT, Kräusslich HG. Interactions of human retroviruses with the host cell cytoskeleton. Curr Opin Microbiol. 2006;9:409–415. doi: 10.1016/j.mib.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 95.Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J Virol. 2007;81:5547–5560. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sasaki H, Ozaki H, Karaki H, Nonomura Y. Actin filaments play an essential role for transport of nascent HIV-1 proteins in host cells. Biochem Biophys Res Commun. 2004;316:588–593. doi: 10.1016/j.bbrc.2004.02.088. [DOI] [PubMed] [Google Scholar]
- 97.Chen C, et al. Association of Gag multimers with filamentous actin during equine infectious anemia virus assembly. Curr HIV Res. 2007;5:315–323. doi: 10.2174/157016207780636542. [DOI] [PubMed] [Google Scholar]
- 98.Tyrrell DL, Ehrnst A. Transmembrane communication in cells chronically infected with measles virus. J Cell Biol. 1979;81:396–402. doi: 10.1083/jcb.81.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106:133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 100.Miazza V, Mottet-Osman G, Startchick S, Chaponnier C, Roux L. Sendai virus induced cytoplasmic actin remodeling correlates with efficient virus particle production. Virology. 2011;410:7–16. doi: 10.1016/j.virol.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 101.Bohn W, Rutter G, Hohenberg H, Mannweiler K, Nobis P. Involvement of actin filaments in budding of measles virus: studies on cytoskeletons of infected cells. Virology. 1986;149:91–106. doi: 10.1016/0042-6822(86)90090-5. A study producing high-quality electron micrographs that demonstrate the directional alignment of actin filaments with measles virions. [DOI] [PubMed] [Google Scholar]
- 102.Stallcup KC, Raine CS, Fields BN. Cytochalasin B inhibits the maturation of measles virus. Virology. 1983;124:59–74. doi: 10.1016/0042-6822(83)90290-8. [DOI] [PubMed] [Google Scholar]
- 103.Forest T, Barnard S, Baines JD. Active intranuclear movement of herpesvirus capsids. Nature Cell Biol. 2005;7:429–431. doi: 10.1038/ncb1243. [DOI] [PubMed] [Google Scholar]
- 104.Feierbach B, Piccinotti S, Bisher M, Denk W, Enquist LW. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2006;2:e85. doi: 10.1371/journal.ppat.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Simpson-Holley M, Colgrove RC, Nalepa G, Harper JW, Knipe DM. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J Virol. 2005;79:12840–12851. doi: 10.1128/JVI.79.20.12840-12851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wagenaar F, et al. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 107.Ohkawa T, Volkman LE, Welch MD. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J Cell Biol. 2010;190:187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts KL, Baines JD. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J Virol. 2010;84:9889–9896. doi: 10.1128/JVI.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nature Rev Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. A comprehensive review about the hurdles of cell-to-cell spread of viruses. [DOI] [PubMed] [Google Scholar]
- 110.Damsky CH, Sheffield JB, Tuszynski GP, Warren L. Is there a role for actin in virus budding? J Cell Biol. 1977;75:593–605. doi: 10.1083/jcb.75.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Akiyama T, et al. Substrate specificities of tyrosinespecific protein kinases toward cytoskeletal proteins in vitro. J Biol Chem. 1986;261:14797–14803. [PubMed] [Google Scholar]
- 112.Carlson LA, et al. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maldarelli F, King NW, Jr, Yagi MJ. Effects of cytoskeletal disrupting agents on mouse mammary tumor virus replication. Virus Res. 1987;7:281–295. doi: 10.1016/0168-1702(87)90043-8. [DOI] [PubMed] [Google Scholar]
- 114.Eugenin EA, Gaskill PJ, Berman JW. Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cell Immunol. 2009;254:142–148. doi: 10.1016/j.cellimm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sherer NM, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nature Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haller C, Rauch S, Fackler OT. HIV-1 Nef employs two distinct mechanisms to modulate Lck subcellular localization and TCR induced actin remodeling. PLoS ONE. 2007;2:e1212. doi: 10.1371/journal.pone.0001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nobile C, et al. HIV-1 Nef inhibits ruffles, induces filopodia, and modulates migration of infected lymphocytes. J Virol. 2010;84:2282–2293. doi: 10.1128/JVI.02230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stolp B, Abraham L, Rudolph JM, Fackler OT. Lentiviral Nef proteins utilize PAK2-mediated deregulation of cofilin as a general strategy to interfere with actin remodeling. J Virol. 2010;84:3935–3948. doi: 10.1128/JVI.02467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stolp B, et al. HIV-1 Nef interferes with host cell motility by deregulation of cofilin. Cell Host Microbe. 2009;6:174–186. doi: 10.1016/j.chom.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 120.Rauch S, Pulkkinen K, Saksela K, Fackler OT. Human immunodeficiency virus type 1 nef recruits the guanine exchange factor Vav1 via an unexpected interface into plasma membrane microdomains for association with p21-activated kinase 2 activity. J Virol. 2008;82:2918–2929. doi: 10.1128/JVI.02185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Minnebruggen GV, Favoreel HW, Jacobs L, Nauwynck HJ. Pseudorabies virus US3 protein kinase mediates actin stress fibre breakdown. J Virol. 2003;77:9074–9080. doi: 10.1128/JVI.77.16.9074-9080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Favoreel HW, et al. Alphaherpesvirus use and misuse of cellular actin and cholesterol. Vet Microbiol. 2010;143:2–7. doi: 10.1016/j.vetmic.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 123.Favoreel HW, Van Minnebruggen G, Adriaensen D, Nauwynck HJ. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an αherpesvirus are associated with enhanced spread. Proc Natl Acad Sci USA. 2005;102:8990–8995. doi: 10.1073/pnas.0409099102. An investigation that demonstrates the direct induction of cytoskeletal extensions by a herpesvirus protein kinase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Van den Broeke Cl, et al. Alphaherpesvirus US3-mediated reorganization of the actin cytoskeleton is mediated by group A p21-activated kinases. Proc Natl Acad Sci USA. 2009;106:8707–8712. doi: 10.1073/pnas.0900436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Deruelle MJ, Favoreel HW. Keep it in the subfamily: the conserved alphaherpesvirus US3 protein kinase. J Gen Virol. 2010;92:18–30. doi: 10.1099/vir.0.025593-0. [DOI] [PubMed] [Google Scholar]
- 126.Berkova Z, Crawford SE, Blutt SE, Morris AP, Estes MK. Expression of rotavirus NSP4 alters the actin network organization through the actin remodeling protein cofilin. J Virol. 2007;81:3545–3553. doi: 10.1128/JVI.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gardet A, Breton M, Fontanges P, Trugnan G, Chwetzoff S. Rotavirus spike protein VP4 binds to and remodels actin bundles of the epithelial brush border into actin bodies. J Virol. 2006;80:3947–3956. doi: 10.1128/JVI.80.8.3947-3956.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gardet A, Breton M, Trugnan G, Chwetzoff S. Role for actin in the polarized release of rotavirus. J Virol. 2007;81:4892–4894. doi: 10.1128/JVI.02698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Valderrama F, Cordeiro JV, Schleich S, Frischknecht F, Way M. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science. 2006;311:377–381. doi: 10.1126/science.1122411. [DOI] [PubMed] [Google Scholar]
- 130.Arakawa Y, Cordeiro JV, Schleich S, Newsome TP, Way M. The release of vaccinia virus from infected cells requires RhoA-mDia modulation of cortical actin. Cell Host Microbe. 2007;1:227–240. doi: 10.1016/j.chom.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 131.Sanderson CM, Way M, Smith GL. Virusinduced cell motility. J Virol. 1998;72:1235–1243. doi: 10.1128/jvi.72.2.1235-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cordeiro JV, et al. F11-mediated inhibition of RhoA signalling enhances the spread of vaccinia virus in vitro and in vivo in an intranasal mouse model of infection. PLoS ONE. 2009;4:e8506. doi: 10.1371/journal.pone.0008506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 134.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. A seminal work on the role of actin-based motility in VV infection. [DOI] [PubMed] [Google Scholar]
- 135.Wolffe EJ, Weisberg AS, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;244:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]
- 136.Frischknecht F, et al. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 137.Scaplehorn N, et al. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr Biol. 2002;12:740–745. doi: 10.1016/s0960-9822(02)00812-6. [DOI] [PubMed] [Google Scholar]
- 138.Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science. 2010;327:873–876. doi: 10.1126/science.1183173. A report that connects the history of VV-mediated actin modulation with long-distance viral spread and the observable phenomenon of plaque formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reeves PM, et al. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on Abl and Src family tyrosine kinases. J Virol. 2011;85:21–31. doi: 10.1128/JVI.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dodding MP, Way M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe. 2009;6:536–550. doi: 10.1016/j.chom.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 141.Murti KG, Chen M, Goorha R. Interaction of frog virus 3 with the cytomatrix: III. Role of microfilaments in virus release. Virology. 1985;142:317–325. doi: 10.1016/0042-6822(85)90340-x. [DOI] [PubMed] [Google Scholar]
- 142.Jouvenet N, et al. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol. 2006;8:1803–1811. doi: 10.1111/j.1462-5822.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- 143.Champagne C, Landry MC, Gingras MC, Lavoie JN. Activation of adenovirus type 2 early region 4 ORF4 cytoplasmic death function by direct binding to Src kinase domain. J Biol Chem. 2004;279:25905–25915. doi: 10.1074/jbc.M400933200. [DOI] [PubMed] [Google Scholar]
- 144.Robert A, et al. Adenovirus E4orf4 hijacks rho GTPase-dependent actin dynamics to kill cells: a role for endosome-associated actin assembly. Mol Biol Cell. 2006;17:3329–3344. doi: 10.1091/mbc.E05-12-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Amine A, et al. Novel anti-metastatic action of cidofovir mediated by inhibition of E6/E7, CXCR4 and Rho/ROCK signaling in HPV tumor cells. PLoS ONE. 2009;4:e5018. doi: 10.1371/journal.pone.0005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 2010;6:e1000954. doi: 10.1371/journal.ppat.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ferreira C. Expression of ubiquitin, actin, and actinlike genes in African swine fever virus infected cells. Virus Res. 1996;44:11–21. doi: 10.1016/0168-1702(96)01334-2. [DOI] [PubMed] [Google Scholar]
- 148.Drenckhahn D, Wagner J. Stress fibres in the splenic sinus endothelium in situ: molecular structure, relationship to the extracellular matrix, and contractility. J Cell Biol. 1986;102:1738–1747. doi: 10.1083/jcb.102.5.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wong AJ, Pollard TD, Herman IM. Actin filament stress fibres in vascular endothelial cells in vivo. Science. 1983;219:867–869. doi: 10.1126/science.6681677. [DOI] [PubMed] [Google Scholar]
- 150.Halliburton WD. On muscle-plasma. J Physiol. 1887;8:133–202. doi: 10.1113/jphysiol.1887.sp000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Straub FB. In: Studies from the Institute of Medical Chemistry University Szeged. Szent-Györgyi A, editor. Vol. 2. Karger; Basel: 1942. pp. 3–15. [Google Scholar]
- 152.Jakus MA, Hall CE. Studies of actin and myosin. J Biol Chem. 1947;167:705–714. [PubMed] [Google Scholar]
- 153.Ohnishi T, Tomoko O. Extraction of actin- and myosin-like proteins from liver mitochondria. J Biochem. 1962;52:230–231. doi: 10.1093/oxfordjournals.jbchem.a127604. [DOI] [PubMed] [Google Scholar]
- 154.Wen KK, Rubenstein PA, DeMali KA. Vinculin nucleates actin polymerization and modifies actin filament structure. J Biol Chem. 2009;284:30463–30473. doi: 10.1074/jbc.M109.021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model system for actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 156.Spector I, Shochet NR, Blasberger D, Kashman Y. Latrunculins — novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 157.Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saito S, Watabe S, Ozaki H, Fusetani N, Karaki H. Mycalolide B, a novel actin depolymerizing agent. J Biol Chem. 1994;269:29710–29714. [PubMed] [Google Scholar]
- 159.Bubb MR, Spector I, Beyer BB, Fosen KM. Effects of jasplakinolide on the kinetics of actin polymerization. J Biol Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- 160.Limouze J, Straight A, Mitchison T, Sellers J. Specificity of blebbistatin, an inhibitor of myosin II. J Muscle Res Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- 161.Sahai E, Olson MF. Purification of TAT-C3 exoenzyme. Methods Enzymol. 2006;406:128–140. doi: 10.1016/S0076-6879(06)06011-3. [DOI] [PubMed] [Google Scholar]
- 162.Ishizaki T, et al. Pharmacological properties of Y-27632, a specific inhibitor of Rho-associated kinases. Mol Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- 163.Klussmann E, Scott J, Deacon SW, Peterson JR. In: Protein-Protein Interactions as New Drug Targets. Kass RS, Clancy CE, editors. Springer; Berlin: 2008. pp. 431–460. [Google Scholar]