Abstract
Reversible protein phosphorylation by protein kinases and phosphatases is a common event in various cellular processes. The eukaryotic protein kinase superfamily, which is one of the largest superfamilies of eukaryotic proteins, plays several roles in cell signaling and diseases. We identified 482 eukaryotic protein kinases and 39 atypical protein kinases in the bovine genome, by searching publicly accessible genetic-sequence databases. Bovines have 512 putative protein kinases, each orthologous to a human kinase. Whereas orthologous kinase pairs are, on an average, 90.6% identical, orthologous kinase catalytic domain pairs are, on an average, 95.9% identical at the amino acid level. This bioinformatic study of bovine protein kinases provides a suitable framework for further characterization of their functional and structural properties.
Keywords: ePK, kinome, aPK, bovine
The protein kinase family is one of the largest families of proteins. Protein kinases play important roles in many intracellular or intercellular signaling pathways, resulting in cell proliferation, gene expression, metabolism, motility, membrane transport, apoptosis and differentiation. Furthermore, they modulate the activity of their substrate proteins by phosphorylating serine, threonine or tyrosine residues that mediate the activation, inhibition, translocation or degradation of substrate proteins (Brognard and Hunter, 2011).
Protein kinases are subdivided into two distinct superfamilies, referred to as eukaryotic protein kinases (ePKs) and atypical protein kinases (aPKs) (Hanks and Hunter, 1995). ePKs contain a conserved catalytic domain of approximately 250 amino acids. This domain is divided into 12 subdomains with highly conserved individual amino acids and motifs (Hanks et al., 1988). Within this domain, three motifs, ‘VAIK’, ‘HRD’ and ‘DFG’, are critical for the catalytic function, even though any residue from this region is fully conserved in all family members (Manning et al., 2002b). Conservation of these typical motifs is thought to be due to selection pressure for conserving important functions, such as the interaction with ATP, and the transfer of a phosphate group to the substrate. aPKs are functionally known to have kinase activity which lack significant sequence similarity to the ePK domain.
The sequencing of several vertebrate genomes has been completed. Initial estimates of the number of protein kinases in the human genome place this at around 1000 (Hunter, 1987), with later studies identifying 518 putative protein kinases (Manning et al., 2002b). The mouse and rat genomes contain 540 and 555 protein kinases, respectively, with 509 human orthologs (Caenepeel et al., 2004; Kazi et al., 2008), thereby implying possible functional conservation across species. In the present study, and by using sensitive bioinformatics approaches we identified the near complete set of bovine protein kinases. These were further classified into groups, families and subfamilies, based on the Hanks et al. (1988) and Manning et al. (2002b) classification scheme. This classification reveals many kinases that are conserved between bovine and human, thus reflecting functional constraints of these protein kinases in the core of signaling pathway. This study provides a suitable framework for further characterization of the functional and structural properties of these protein kinases.
A search was made of bovine proteome sequences available in GenBank (Benson et al., 2010) and Ensembl (Hubbard et al., 2009) for bovine protein kinases, using various tools. A preliminary search for protein kinases was performed using PSI-BLAST (Altschul et al., 1997), against the bovine proteome with an e-value threshold of 0.0001, and an h-value of 0.1 for five iterations. Previously published human (Manning et al., 2002b), mouse (Caenepeel et al., 2004) and rat (Kazi et al., 2008) eukaryotic protein kinases and kinase catalytic domains, as well as eukaryotic protein kinase catalytic domains from a variety of organisms available at the kinase.com database were used as query sequences. A further search for protein kinases was performed using HMMER (Eddy, 1998). A Hidden Markov profile was created and validated by means of known eukaryotic protein-kinase catalytic domains. A further search was made for atypical protein kinases, using human, mouse and rat atypical protein kinases by PSI-BLAST or HMMER. Hits identified by using the different methods were combined, and duplicate records removed. Where splice variants were encountered, the variant showing either the closest proximity to the human ortholog, or the longest protein encoding variant, was recorded. All protein kinases were then evaluated for the presence of a conserved eukaryotic protein kinase domain (Hanks et al., 1988; Manning et al., 2002b). Catalytic domains were defined using RPS-BLAST in the BLAST package (Altschul et al., 1997) against the Pfam database (Finn et al., 2010), and sequence alignments carried out with AlinX implemented in the Vector-NTI package (Lu and Moriyama, 2004). Alignments were then manually edited, and all the kinases manually evaluated. Finally, 482 eukaryotic protein kinases and 39 atypical protein kinases were identified (Table S1). The primary names of the protein kinases were derived from their respective homologs in human (Manning et al., 2002b), mouse (Caenepeel et al., 2004) and rat (Kazi et al., 2008) protein kinases. On deriving a second name and synonyms from the Entrez Gene records (Maglott et al., 2005), the full protein names were retrieved thence. Representative records in Entrez Gene, corresponding to each bovine sequence, were identified, whereupon related information was included. Based upon the human protein kinases classification scheme (Manning et al., 2002b), these protein kinases were further classified into 10 groups, 129 families and 81 subfamilies (Table 1 and Table S1).
Table 1.
Protein-kinases and their distribution by groups in different genomes.
| Species | AGC | CAMK | CK1 | CMGC | Other | RGC | STE | TK | TKL | Atypical | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 63 | 74 | 12 | 61 | 83 | 5 | 47 | 90 | 43 | 40 | 518 |
| Bovine | 62 | 75 | 12 | 61 | 85 | 5 | 47 | 91 | 44 | 39 | 521 |
| Rat | 60 | 112 | 11 | 61 | 84 | 7 | 46 | 91 | 43 | 40 | 555 |
| Mouse | 60 | 97 | 11 | 60 | 84 | 7 | 47 | 90 | 43 | 39 | 538 |
| Drosophila melanogaster | 30 | 32 | 10 | 33 | 45 | 6 | 18 | 32 | 17 | 17 | 240 |
| Caenorhabditis elegans | 30 | 46 | 85 | 49 | 67 | 27 | 25 | 90 | 15 | 20 | 454 |
| Saccharomyces cerevisiae | 17 | 21 | 4 | 21 | 38 | 0 | 14 | 0 | 0 | 15 | 130 |
Previous studies have shown that almost all human protein-kinase orthologs are present in mouse and rat genomes (Caenepeel et al., 2004; Kazi et al., 2008). Thus, a search among bovine kinase sequences was conducted for the orthologous kinases using BLASTP (Altschul et al., 1997). The results were parsed, the symmetrically best hits being considered as orthologous kinases. The orthology relationships were further analyzed by CLUSTALW alignment (Thompson et al., 1997), followed by phylogenetic analysis. The latter was carried out by phylogenetic tree option incorporated into the CLUSTALW program. The NJ clustering algorithm was used for drawing bootstrap trees. As almost all bovine and human protein kinases exist as orthologous pairs (Figure S1), the similar functions in both organisms give to understand their evolutionary conservation. Human and bovine genomes contain 512 common protein kinase orthologs. Our search could not identify seven human protein kinase orthologs, likely due to the incomplete nature of bovine genome sequencing data (Table 2). In these seven human protein kinases, only TAF1L is absent in the chimpanzee genome. All are present in various higher eukaryotes, such as of orangutans and monkeys. Ten protein kinases were absent in the human genome, eight of which being bovine specific and the other two, PLK5 and TSSK5 found in other genomes (Table 2). EphB1L might be a retrotransposed copy of the EphB1 gene, with 89.2% of amino acid sequence identity with the EphB1 protein.
Table 2.
Lineage-specific protein kinases.
| Found in | Gene Name | Introns | Paralog | Other orthologs |
|---|---|---|---|---|
| Human and bovine | 512 genes | |||
| Human | CK1a2 | No | CK1a1 (91%) | Chimpanzee, Orangutan, Monkey |
| TAF1L | No | TAF1 (94%) | Orangutan, Monkey | |
| PKACg | No | PKACa (83%) | Chimpanzee, Orangutan | |
| LTK | Yes | - | Chimpanzee, Orangutan, Rat, Mouse, Horse, Chicken | |
| ROS | Yes | - | ||
| PSKH2 | Yes | - | Chimpanzee, Monkey, Dog, Orangutan, Rabbit, Horse | |
| PRKY | Yes | PRKX (92%) | Chimpanzee, Gorilla, Monkey | |
| Bovine | AURL | Yes | AurB (61%) | - |
| CK1b | Yes | CK1a (81%) | - | |
| EphB1L | No | EphB1 (89.2%) | - | |
| FERL | Yes | FER (82%) | - | |
| MARKL | Yes | - | - | |
| MASTL2 | Yes | - | - | |
| Par-1L | Yes | - | - | |
| PLK5 | Yes | - | Orangutan, Monkey, Mouse, Rat | |
| RYKL | Yes | - | - | |
| TSSK5 | Yes | - | Dog, Horse, Mouse, Rat | |
In ‘Paralog’ parentheses show % of identity (amino acid) with respective paralog.
Several proteins, such as ErbB3, SCYL1 and KSR1, have an inactive catalytic domain (Citri et al., 2003; Manning et al., 2002a). These inactive kinases, besides acting mainly as adaptor proteins, or dimerizing with active kinases, have also been shown to be involved in various cellular functions (Salerno et al., 2005; Schmidt et al., 2007; Sergina et al., 2007). Three conserved motifs, ‘VAIK’, ‘HRD’ and ‘DFG’, are important for catalytic activities. Inactive kinases lack at least one of these three conserved motifs. Fifty catalytic domains and 45 protein kinases in the human genome were predicted as catalytically inactive due to the lack of at least one of the three conserved residues (Manning et al., 2002b). The bovine complement of inactive kinases is equivalent to that of the human (Table S1).
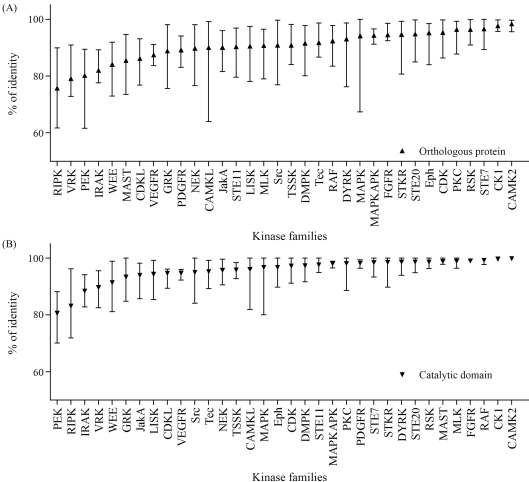
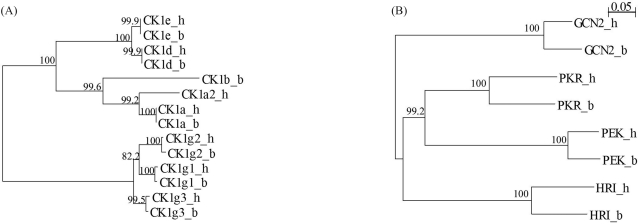
All the human and bovine orthologous protein kinase pairs and orthologous catalytic domain pairs were analyzed for the percentage of identity by AlignX incorporated into Vector-NTI (Lu and Moriyama, 2004). In protein sequence alignments of orthologous kinase pairs we observed a wide variation in local sequence conservation (Figure 1A). These were, on an average, 90.6% identical (amino acid sequence), although some were as low as 47.7%, with four pairs presenting high levels of sequence identity throughout the protein (Table S1). Although most differences between orthologs are due to amino acid substitution, many proteins contained substantial insertions or deletions (indels) between orthologs, which may account for many of the functional differences between species. Several proteins contained insertions or deletions (indels), as shown by sequence alignment (Table S1). These comparisons are also informative within the conserved domains. Although orthologous catalytic domains were, on an average, 95.9% identical, some were as low as 63.8%. Sixty two pairs were identical across the full domain, whereas 48 differed by only one amino acid (Table S1), thus indicative of strong conservative pressure throughout the catalytic domain. Catalytic domain pairs showed clearly family-dependent variability (Figure 1B). For example, of the six casein kinase 1 (CK1) family domain pairs, three were identical, and the other three differed by two residues, an average difference of only 0.4%, thereby indicating that changes in almost any amino acid within the domain destroyed some function, and thus have been eliminated by evolution (Figure 2A). At the other extreme, PEK family catalytic domain pairs are 70–88% identical, thereby implying that the core functions of this family of kinases do not greatly constrain the domain sequence (Figure 2B).
Figure 1.
Conservation within orthologous kinases (A) and catalytic domains (B) is family-dependent. Triangles indicate mean identity within selected families, and bars indicate the range.
Figure 2.
Catalytic domain conservation is family-dependent. (A) The CK1 family is highly conserved, with zero to two amino acid changes between bovine and human orthologs. (B) Catalytic domains of the PEK family are poorly conserved between human and bovine. h, Human; b, Bovine.
Our study presents a bioinformatic overview and evolutionary insight into the kinases within the bovine genome. Comparison with the human kinome revealed the evolutionary conservation of the protein kinase function. The curated kinase dataset from the bovine genome, presented here, could serve as a framework for further investigation of this important gene family.
Supplementary Material
Acknowledgments
This work was supported by a KN Biomedical Research Grant. We are grateful to Nadid Ahnaf Kazi.
Footnotes
Associate Editor: Alexandre Rodrigues Caetano
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J, Hunter T. Protein kinase signaling networks in cancer. Curr Opin Genet Dev. 2011;21:4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: Discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci USA. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Skaria KB, Yarden Y. The deaf and the dumb: The biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- Kazi JU, Kabir NN, Soh JW. Bioinformatic prediction and analysis of eukaryotic protein kinases in the rat genome. Gene. 2008;410:147–153. doi: 10.1016/j.gene.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Lu G, Moriyama EN. Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform. 2004;5:378–388. doi: 10.1093/bib/5.4.378. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002a;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002b;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Salerno M, Palmieri D, Bouadis A, Halverson D, Steeg PS. Nm23-H1 metastasis suppressor expression level influences the binding properties, stability, and function of the kinase suppressor of Ras1 (KSR1) Erk scaffold in breast carcinoma cells. Mol Cell Biol. 2005;25:1379–1388. doi: 10.1128/MCB.25.4.1379-1388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WM, Kraus C, Hoger H, Hochmeister S, Oberndorfer F, Branka M, Bingemann S, Lassmann H, Muller M, Macedo-Souza LI, et al. Mutation in the Scyl1 gene encoding amino-terminal kinase-like protein causes a recessive form of spinocerebellar neurodegeneration. EMBO Rep. 2007;8:691–697. doi: 10.1038/sj.embor.7401001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Kinase.com database, Genomics, evolution and function of protein kinases. http://kinase.com/kinbase/FastaFiles/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.