Abstract
The cloning, expression and purification of the glutathione (sulfur) import system ATP-binding protein (gsiA) was carried out. The coding sequence of Escherichia coli gsiA, which encodes the ATP-binding protein of a glutathione importer, was amplified by PCR, and then inserted into a prokaryotic expression vector pWaldo-GFPe harboring green fluorescent protein (GFP) reporter gene. The resulting recombinant plasmid pWaldo-GFP-GsiA was transformed into various E. coli strains, and expression conditions were optimized. The effect of five E. coli expression strains on the production of the recombinant gsiA protein was evaluated. E. coli BL21 (DE3) was found to be the most productive strain for GsiA-GFP fusion-protein expression, most of which was insoluble fraction. However, results from in-gel and Western blot analysis suggested that expression of recombinant GsiA in Rosetta (DE3) provides an efficient source in soluble form. By using GFP as reporter, the most suitable host strain was conveniently obtained, whereby optimizing conditions for overexpression and purification of the proteins for further functional and structural studies, became, not only less laborious, but also time-saving.
Keywords: Escherichia coli, glutathione transporter, gsiA, gene expression, green fluorescent protein
Introduction
Glutathione (γ-L-glutamyl-L-cysteinyl-glycine; GSH), a tripeptide of glutamate, cysteine and glycine, is the most important low-molecular-weight antioxidant in living cells. This tripeptide is synthesized by the sequential addition of cysteine to glutamate, followed by the addition of glycine. Reduction and conjugation reactions are usually considered as the most important functions of GSH, in which the sulfhydryl group (-SH) of cysteine is always involved.
As an antioxidant, GSH, besides playing a leading role in the protection of cells and organisms by reducing oxygen species and peroxides, also acts against xenobiotics and drugs by the formation and excretion of glutathione S conjugates (Meister, 1989). For instance, GSH can not only help in maintaining the redox environment (Schafer and Buettner, 2001), but also protects cells against environmental stress (Hayes and McLellan, 1999; Dickinson and Forman, 2002). It was reported that GSH acts in maintaining cell physiological functions under H2O2-induced oxidative stress in Saccharomyces cerevisiae (Izawa et al., 1995; Penninckx, 2000). Likewise, accumulated intracellular GSH protects cells against injuries by H2O2 stress (Pompella et al., 2003). Through these special features, glutathione plays an important role in the treatment of certain diseases caused by oxygen pressure, especially cancer (Biswas and Rahman, 2009; Yuan and Kaplowitz, 2009). Due to the great need in medicine, health-care, food and cosmetics, the commercial demand for GSH is in constant expansion.
It has been shown that glutathione and its derivatives are shifted in and out of cells by diverse transporters. Glutathione transporters themselves contain a Na+-dependent transporter (Kannan et al., 1996), an H+ symporter (Jamai et al., 1996) and glutathione S conjugate exporters (Cole et al., 1992; Li et al., 1996), which are members of the ATP binding cassette. So far, existing results regarding microbial glutathione transporters have shown that attention is mainly focused on fungi. Glutathione S conjugate transporters, such as S. cerevisiae YCF1 of the vacuolar membrane (Rebbeor et al., 1998), and yeast HGT1 of the plasma membrane, both are known as membrane-potential-dependent glutathione importers across the plasma membrane (Bourbouloux et al., 2000).
However, there are few reports on the identification of bacterial glutathione transporters. Previous studies have shown that, during transition from the exponential to the stationary phase, glutathione content in Escherichia coli cells increased significantly (Fahey et al., 1978). Furthermore, when grown in aerobic conditions, content reaches a high level.
E. coli secretes glutathione into the culture-medium during the exponential phase, its concentration reaching a maximum in the early stationary phase (Owens and Hartman, 1986; Suzuki et al., 1987). Thereafter, it is hydrolyzed by γ-glutamyltranspeptidase (GGT) in the periplasm, to liberate glutamic acid and cysteinylglycine (Suzuki et al., 1987, 1993). Cysteinylglycine is then conveyed into the cytoplasm, whereat it is cleaved into cysteine and glycine by aminopeptidases A, B, and N and dipeptidase D, this constituting a source of these two amino acids for the strain (Suzuki et al., 2001). The GGT catalyzing the initial step of extracellular glutathione cleavage, as a source of cysteine and nitrogen, was also verified by other researchers in a mammalian cell line (Hanigan and Ricketts, 1993), as well as in yeast (Mehdi and Penninckx, 1997). Nonetheless, some researchers found that even in GGT-deficient E. coli, the concentration of glutathione in the culture-medium still gradually decreased after prolonged incubation. This finding prompted a novel glutathione transporter to be reported in bacteria, providing evidence that gsiA, -B, -C, and -D encode a new type of glutathione transporter, which is also a member of the ATP-binding cassette transporter superfamily (Suzuki et al., 2005). This implies that gsiA encodes the ATP binding component, the -B periplasmic binding protein, and both gsiC and -D, two plasma membrane components (Keseler et al., 2009).
The ATPase components of ATP binding cassette transporters can provide the necessary energy by binding and hydrolyzing ATP. The transport of glutathione by GsiABCD transporters greatly depends on ATPase activity (Suzuki et al., 2005). Crystal structure analysis of the ATPase subunit of the maltose transporter, which is also an ATP binding transporter from E. coli, gives to understand it works as a dimer and has three configurations. The two nucleotide binding domains of the dimer open and close like a pair of tweezers, thus implying the regulatory mechanism for ATPase activity might be intimately related to translocation (Chen et al., 2003). Does GsiA make use of the same mechanism to power the glutathione transporter? This still remains to be seen.
The specific glutathione transport mechanism may be different from both the E. coli dipeptide transporter system and oligopeptide transporters in other bacteria. Studies of the glutathione system will not only help to clarify the mechanism involved, but also contribute to understanding the evolutionary pathway from dipeptide to oligopeptide transporters in bacteria.
Herein, the spectrum of expression conditions of gsiA from E. coli k-12 was described. The GFP reporter gene was here employed for quickly optimizing GsiA expression and purification conditions, the resultant purified protein possibly facilitating further protein structure analysis.
Materials and Methods
The expression vector pWaldo-GFPe (Waldo et al., 1999; Drew et al., 2001; Rapp et al., 2004) and the E. coli strains, C41 (DE3), C43 (DE3), Rosetta (DE3) and BL21 (DE3)plysS were kindly provided by Dr Dong Changjiang from the University of St. Andrews. The E. coli strains BL21 (DE3) and DH5α were kept in our own laboratory. rTaq DNA polymerase, T4 DNA ligase and DNA markers were purchased from TaKaRa (Dalian, China), and KOD-FX polymerase from ToYoBo (Shanghai, China). XhoI and BamHI restriction enzymes and protein size markers were obtained from Fermentas (Shenzhen, China). Mouse anti-his monoclonal antibodies and horseradish peroxidase labeled goat anti-mouse antibodies were bought from Zhongshan Goldenbridge Biotechnology Company (Beijing, China).
Construction of the expression plasmid
The sequence encoding the GsiA protein was amplified by standard PCR, using KOD-FX DNA polymerase. The forward primer was 5′-CCGCTCGAGATGCCACACAG TGATGAACTTG-3′ and the reverse 5′-CGC GGATCCTCTACGCATGAATGCGTATTCTG-3′ (the underlined sequences are XhoI and BamHI restriction sites, respectively). The PCR procedure was carried out in a Mastercycler gradient (Eppendorf, Germany) in a 50 μL reaction volume containing 25 μL of 2 × KOD-FX buffer (TOYOBO), 10 μL of dNTP (2 mmol/L), 1.5 μL of each primer (10 μM), 10 μL of PCR-grade water, 1 μL of KOD-FX polymerase (1 U μL−1) and 1 μL of genomic DNA template. The amplification protocol was as follows: 2 min predenaturing at 94 °C, followed by 30 cycles of 98 °C for 10 s, 58 °C for 30 s, 68 °C for 2 min, and a final extension step at 68 °C for 7 min.
The target product was gel-purified, digested with XhoI and BamHI, and then ligated into pre-digested pWaldo-GFPe at the molar ratio 5:1 (fragment:vector). The subsequent ligation mix was used for transforming E. coli DH5α competent cells, and the recombinant plasmid for sequencing in BGI-Shenzhen (China). The resultant plasmid, denominated pWaldo-GFP-GsiA, will be subsequently referred to as pGFP-GsiA. The gsiA sequence (GenBank accession Number JF714943) and its deduced amino acid residues were analyzed using a blast algorithm at NCBI. The composition and properties of the sequences were analyzed with DNAMAN 6.0. Clustal X2 and MEGA 4.0 were employed for generating multiple sequence alignment and a phylogenetic tree, respectively.
Recombinant expression of GsiA
Optmization of GsiA-GFP expression
pGFP-GsiA was transformed into different E. coli strains such as BL21 (DE3), C41 (DE3), C43 (DE3), Rosetta (DE3) and BL21 (DE3)plysS. The transformed cells, first grown overnight at 37 °C in 2 mL Luria-Bertani (LB) medium containing either kanamycin 50 μg/mL (BL21 (DE3), C41 (DE3) and C43 (DE3)), or kanamycin 50 μg/mL plus chloramphenicol 68 μg/mL (Rosetta (DE3) and BL21 (DE3)plysS), were then used for inoculating 10 mL LB medium supplemented with antibiotics, for posterior growth at 37 °C to an OD600 of 0.5. GsiA-GFP overexpression was subsequently induced for 4 h and 24 h at 18 °C, 25 °C and 30 °C, by the addition of isopropyl-thio --β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mmol/L, 0.4 mmol/L and 1.0 mmol/L, respectively. Cells were harvested by centrifugation at 15,800 g for 2 min and then washed with phosphate buffered saline (PBS pH 7.4). The pellet could either be stored at −20 °C for at least one week or used immediately in the following steps.
SDS-PAGE analysis of GsiA-GFP
Pellets can be variably treated for different purposes. In order to select the most suitable strains for GsiA-GFP overexpression, pGFP-GsiA was induced in five tested host-strains with 1.0 mmol/L IPTG at 18 °C and 210 rpm for 24 h. The cells, first harvested and washed by, and re-suspended in, phosphate buffered saline (PBS pH 7.4), equally mixed with SB [solubilization buffer: 200 mmol/L Tris-HCl (pH 8.8), 20% glycerol, 5 mmol/L EDTA (pH 8.0), 0.02% bromophenol blue, 4% SDS and 0.05 mol/L DTT], were then incubated for 5 min at 37 °C. SDS-PAGE was used for sample analysis. The gel was rinsed with distilled water and exposed to ultraviolet light. Images were captured with a CCD camera system to detect and analyze fluorescent bands. The desired band intensity was obtained by varying exposure time. Subsequently, the gel was stained with Coomassie blue, whereupon another picture was taken under white light with a CCD camera system. Thus, both photos could be compared. The cells was also mixed at 1:4 (v:v) with 5 × SDS loading buffer (250 mmol/L Tris-HCl, pH 6.8, 10% SDS, 50% glycerol, 0.5% bromophenol blue, 5% β-mercaptoethanol), boiled for 5 min and then separated by SDS-PAGE.
Overexpression and purication of GsiA-GFP
Rosetta (DE3) harboring pGFP-GsiA was incubated in 2 mL LB medium containing 50 μg/mL kanamycin and 68 μg/mL chloramphenicol and grown at 37 °C. The bacteria culture was then inoculated into 200 mL of LB medium and incubated at 37 °C until OD600 ≈ 0.5, whereupon 0.1 mmol/L of IPTG was added to induce recombinant protein GsiA-GFP expression. After induction at 18 °C for 24 h, the cells were harvested by centrifugation at 6.000 g, whereupon the pellet was washed in 40 mL phosphate buffered saline (PBS, pH 7.4). Subsequently the pellet was re-suspended and lysed in 30 mL PBS containing 20 mmol/L imidazole and 1 mmol/L phenylmethanesulfonyl fluoride (PMSF), for posterior sonication on ice for 12 min (5 s on, 8 s off). The lysate was clarified by centrifugation at 12,000 g for 15 min at 4 °C, whereupon the supernatant was loaded onto a Ni2+-NTA affinity column (GE Healthcare), pre-equilibrated with lysis buffer. The recombinant protein was purified through Ni2+ chelating chromatography (GE, USA), by means of a linear gradient of 20~500 mmol/L imidazole in PBS (pH 7.4), according to manufacturer’s recommendation.
Imidazole in the eluted protein was removed by means of three buffer (PBS pH 7.4) exchanges (8 h each) at 4 °C, and then the protein was concentrated by PEG 20000. Purity of recombinant GsiA-GFP protein was monitored on a 12% SDS-PAGE gel. The concentration was assayed with the Bradford method, using bovine serum albumin as standard.
Western blot analysis
Western blot analysis, at room temperature, was as follows: the protein GsiA-GFP, first resolved by 12% SDS-PAGE, was then electroblotted onto a nitrocellulose membrane with the iBlot Dry Blotting System (Invitrogen), using an iBlot Gel Transfer Device to perform dry blotting of proteins for 7 min. The membrane was first incubated for 2 h in blocking-buffer (PBS, pH 7.4 containing 0.1% Tween 20 and supplemented with 5% (w/v) nonfat dry milk), washed three times, 10 min each, and incubated 1 h with an anti-his monoclonal antibody (anti-His, 400 μg/mL, 1:1000 dilution). After washing the membrane as above, it was incubated for 1 h in a horseradish peroxidase labeled antibody (goat anti-mouse, 0.8 mg/mL, 1:5000 dilution). The membrane was then processed by the enhanced chemiluminescence method, and the protein band visualized by autoradiography.
Results and Discussion
GsiA expression plasmid construction
Many powerful vectors can be used for highly expressing various foreign genes in E. coli, the expressed products being easily detected by means of various reporter genes. The use of GFP, as the reporter in expression vector pWaldo-GFPe to optimize conditions for protein overexpression and purification in functional and structure studies, is convenient and time-saving (Drew et al., 2006). GFP facilitates direct monitoring and visualization of the protein of interest at any stage during expression, solubilization and purification. As the GFP gene is fused to the C-terminal of the foreign gene, in-gel fluorescence can be used to check for the presence of the expressed full-length protein. The GFP moiety is exceptionally stable, thus fluorescence can not only be observed during expression, solubilization and purification, but can also be detected in standard SDS polyacrylamide gels, with a detection limit of less than 5 ng GFP per protein band (Drew et al., 2006).
The coding sequence of the gsiA gene was amplified from the E. coli k-12 genome and cloned upstream of the GFP gene in pWaldo-GFPe, as described above. DNA sequencing-results confirmed the insertion. The resultant plasmid was denominated pGFP-GsiA.
Optimizing strains for gsiA expression
After transforming the E. coli strains BL21 (DE3), C41 (DE3), C43 (DE3), Rosetta (DE3) and BL21 (DE3)plysS competent cells with pGFP-GsiA, the optimal expression conditions for gsiA in the LB medium were defined, by both monitoring bacterial growth and protein fluorescence under various temperatures and IPTG concentrations, and analyzing the expressed protein in cell lysates by SDS-PAGE and Western blot, using an antibody against the His-tag.
As a proof of concept, a panel of genetically well-characterized E. coli strains with different phenotypes (Terpe, 2006) was used to screen for high-level and more-soluble expression of the GsiA-GFP protein. Actually, all the five strains above had already been successfully used as expression systems for several proteins.
The BL21 (DE3) strain, deficient in lon and ompT proteases that degrade abnormal and outer membrane proteins, respectively, provides higher levels of expression, as well as a tighter control of protein expression, by reducing proteolysis in expressed proteins. C41 (DE3), a mutant designed for membrane-protein expression, facilitates over-expression in proteins that are toxic to the BL21 (DE3) parental strain (Terpe, 2006). C43 (DE3) is a double mutant also designed for membrane-protein expression. The Rosetta (DE3) strain bears a chloramphenicol-resistant plasmid pRARE, which contributes tRNAs for codons rarely used in E. coli, viz., AUA, AGG, AGA, CGG, CUA, CCC, and GGA, thereby enhancing the expression of eukaryotic proteins. BL21 (DE3)pLysS harbors the pLysS plasmid constitutively expressing T7 lysozyme, an inhibitor of T7 RNA polymerase activity that reduces cell polymerase activity until induced.
In order to monitor GsiA-GFP expression in E. coli, cells were grown and then induced with 0.1 mmol/L, 0.4 mmol/L and 1.0 mmol/L of IPTG at 18 °C, 25 °C and 30 °C, respectively, whereupon the induced proteins were separated by SDS-PAGE. As GFP was used as reporter, GsiA-GFP fusion-protein expression could be conveniently detected by in-gel analysis (Figure 1). We found that the concentrations of IPTG had no obvious effect on GsiA-GFP expression, and the change of the induction temperatures greatly affected the expression of correctly folded fluorescent GsiA-GFP (Figure 2). The higher the induction-temperature, the less correctly folded the protein. Thus, on considering both the effects of IPTG induction-concentration and temperature, the strategies used to improve protein solubility were growth and induction at a low temperature (18 °C) and low concentration of IPTG (0.1 mmol/L).
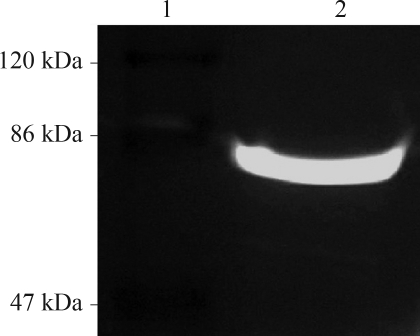
Figure 1.
In-gel fluorescence of GsiA-GFP fusion. The samples were treated with SB at 37 °C for 5 min. 1: Marker (SM0441); 2: EcoGsiA-GFP (96 kDa) induced with 1.0 mmol/L IPTG at 18 °C.
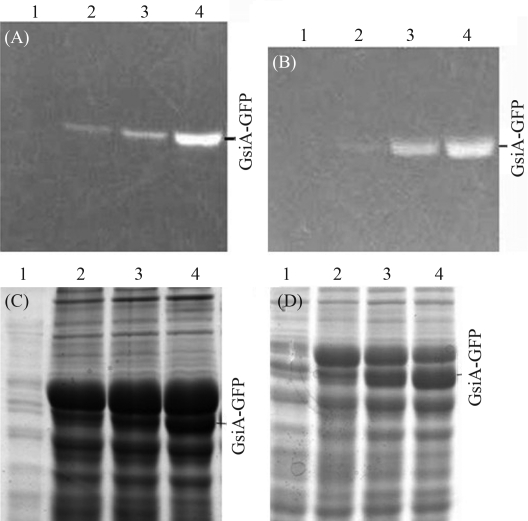
Figure 2.
Electrophoretic mobility of GFP fusion proteins expressed at different levels under various conditions. The samples were treated with SB at 37 °C for 5 min. A and B: In-gel GFP fluorescence. C and D: The same gel stained with Coomassie blue. A, C and B, D are BL21 (DE3) and Rosetta (DE3) samples respectively; Lane 1: 18 °C no IPTG; Lanes 2~4: induced with 0.1 mmol/L IPTG at 30 °C, 25 °C, 18 °C.
Variation in the extent of expression and solubility of GsiA-GFP protein in the different strains was significant. Visual inspection of the gel revealed the best GsiA-GFP productivity in BL21 (DE3) and the worst in BL21 (DE3)plysS (Figure 3A and B). Thus, on considering the maximum and minimum extent of GsiA-GFP expression, the five strains could be ranked, with BL21 (DE3) performing the best, followed by Rosetta (DE3), C41 (DE3), C43 (DE3) and BL21 (DE3)plysS.
Figure 3.
In-gel fluorescence (A) and SDS-PAGE (B) analysis of expressed GsiA-GFP fusion in several strains. The samples were treated with SB at 37 °C for 5 min. Lanes 1~5 induced with 1.0 mmol/L of IPTG at 18 °C: BL21 (DE3); C41 (DE3); C43 (DE3); Rosetta (DE3); BL21 (DE3)plysS, respectively.
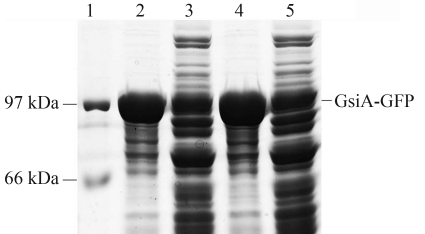
Preliminary analysis with BL21 (DE3) cell lysate showed that GsiA-GFP was mainly localized in the insoluble cellular fraction. Efforts to express in and purify recombinant GsiA-GFP from the BL21 (DE3) host strain were frustrated by this insolubility of the expressed protein (Figure 4). Due to the availability of a wide variety of specialized E. coli strains for protein expression, GsiA-GFP protein expression was re-examined.
Figure 4.
Solubility analysis of GsiA-GFP in BL21 (DE3), after induction of the cells with 0.1 mmol/L of IPTG at 18 °C. The cells were boiled in SDS loading buffer. Lane 1: Marker; Lane 2: soluble fraction; Lane 3: insoluble fraction.
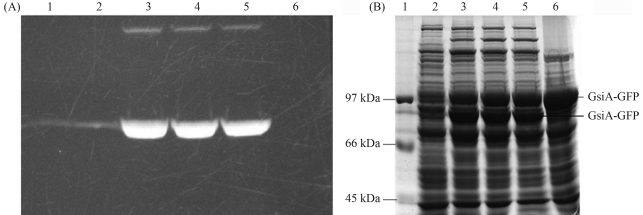
Notwithstanding, a large part of the protein produced was encountered in the insoluble fraction (Figure 5). Thus, to obtain more soluble protein for further studies of GsiA, Rosetta (DE3), by proving to be an excellent host strain for GsiA-GFP overexpression, the best among the five strains, was grown and induced at 18 °C, with 0.1 mmol/L IPTG.
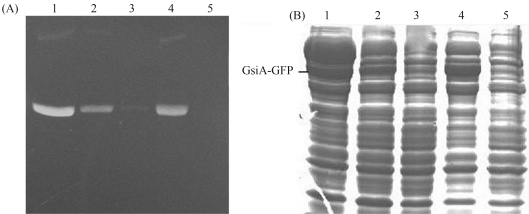
Figure 5.
Solubility analysis of GsiA-GFP expressed in Rosetta (DE3). The cells were boiled in SDS loading buffer. Lane 1: Marker; Lanes 2~5: Insoluble (lanes 2 and 4) and soluble fractions (lanes 3 and 5) after induction of the cells at 18 °C with 0.1 mmol/L and 0.4 mmol/L of IPTG, respectively.
In-gel mobility shift of GFP fusion proteins
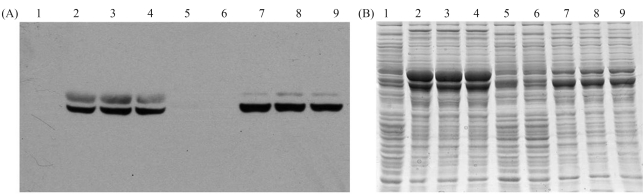
The GsiA protein was expressed in E. coli with a GFP-His8 moiety fused to the GsiA C terminus. After cell disruption, all the cellular proteins in the different expression-strains were analyzed by SDS-PAGE and in-gel detection of GFP fluorescence. A single prominent fluorescent band was observed in all the GsiA-GFP-transformed strains. The expressed GsiA-GFP protein was also analyzed by Western blot and immunodetection, with an antibody directed against the His-tag (Figure 6A). The blot not only revealed a band at the same position as the fluorescent signal, but also, surprisingly, an additional band of an apparent molecular mass that was about 10 kDa higher (Figure 6A, upper band). Two bands were observed in all tested strains that can express GsiA-GFP fusion protein (Figure 6A and B), but intensity ratios varied among the different strains and induction conditions.
Figure 6.
Western blot analysis of GsiA-GFP expression in various host strains. The samples were treated with SB at 37 °C for 5 min. A: Immunoblot of the gel decorated with anti-His tag antibody. Lower and upper bands indicate the positions of fluorescent and nonfluorescent species of the GsiA-GFP fusion proteins, respectively. B: Gel stained with Coomassie blue. Lanes1~4: BL21 (DE3) induced with 0, 1.0 mmol/L, 0.4 mmol/L and 0.1 mmol/L IPTG at 18 °C; Lanes 5–6: C41 (DE3) and C43 (DE3) induced with 0.1 mmol/L IPTG at 18 °C; Lanes 7~9: Rosetta (DE3) induced with 0.1 mmol/L, 0.4 mmol/L and 1.0 mmol/L IPTG at 18 °C.
To analyze the origin of the dual migration, we compared the electrophoretic mobility of the GFP fusion proteins treated in diverse ways (Figure 7A and B). Boiled protein samples, did not show the dual electrophoretic mobility and migrated as a single band. With the boiled protein as a reference, the apparent molecular mass of the upper band was in accordance with the predicted mass of the fusion protein of GsiA-GFP, whereas, in low temperature-treated samples, the lower fluorescent band migrated about 10~15 kDa lower than the upper one. This implies that the GFP moiety of the fusion protein in the upper band was fully denatured, thus consistent with its predicted migration in SDS-PAGE and the absence of fluorescence. The anomalous migration of the lower band could be the result of GFP moiety structure preservation, thus consistent with the observed fluorescence.
Figure 7.
In-gel analysis of the expressed GsiA-GFP in Rosetta (DE3) cells. A: In-gel GFP fluorescence. B: The gel stained with Coomassie blue. Lane 1: Marker; Lanes 2~5 treated with SB at 37 °C for 5 min: induced with 0, 0.1 mmol/L, 0.4 mmol/L and 1.0 mmol/L IPTG at 18 °C; Lane 6: induced with 0.1 mmol/L IPTG at 18 °C and the cells boiled in SDS loading buffer.
As the correct folding of the GFP moiety in GFP fusion proteins depends on the productive folding of the preceding protein domain (Waldo et al., 1999; Drew et al., 2001), the observed dual in-gel migration of GFP fusion proteins could represent the folded and aggregated protein populations produced under these expression-conditions. Clearly, methodology facilitating such rapid and simultaneous quantification of both folded and aggregated protein would be of immense value during, for instance, optimization of protein expression and purification conditions. Thus, an attempt was made to establish the suitability of dual in-gel migration of GFP fusion-proteins, as an indicator of the amounts of folded and aggregated protein present in a sample.
Intensity ratios of the two bands depend on expression levels
From the data on BL21 (DE3) (Figure 2A and C) and Rosetta (DE3) (Figure 2B and D), it appeared that the ratio between the intensities of the two bands changed, when the expression conditions were altered to produce more or less aggregated protein. Furthermore, the double bands of expressed GFP fusion proteins were not a mere artifact, but indeed originated from the different folding-states.
High expression-situations generally produce more aggregated protein in inclusion bodies than lower ones. Even so, the distribution of intensities over the two bands varied markedly. Low inducer concentrations and low inducing temperatures could contribute to the lower (fluorescent) protein bands, consistent with a high fraction of properly folded protein. Overall, low expression levels increased the relative contribution of the lower fluorescent band, whereas high levels increased the proportion of the upper band.
Noticeably, intensities of the upper bands of the proteins expressed in Rosetta (DE3) were compared with the lower in BL21 (DE3). Notably, a significant amount of recombinant GsiA-GFP protein in BL21 (DE3) was present in the pellet fraction (Figure 4). Interestingly, the expression level of GsiA-GFP in the Rosetta (DE3) strain was not higher, but more soluble, hence the likelihood of the codon context affecting GsiA-GFP folding in E. coli. We do not have an explanation for the observed behavior at this moment.
Identification of native GsiA-GFP protein in the supernatant and pellet fractions from the overproducing strains was through Western blotting with anti-His antibodies. A protein band, with a size consistent with that determined for GsiA-GFP protein (96 kDa), was detected in the IPTG-induced cell-free lysates, in both pellet and soluble fractions (Figure 6). Moreover, a significant amount of GsiA-GFP protein was present in the soluble supernatant fraction in Rosetta (DE3).
Purication of GsiA-GFP fusion proteins
Rosetta (DE3) was chosen to overexpress GsiA-GFP, since this host-strain can greatly enhance the level of expressed protein in soluble form. As the addition of imidazole during binding improves the purity of histidine-tagged proteins, 20 mmol/L imidazole was added to the lysate. Following sonication, the cell-free supernatant was loaded onto a 1 mL nickel-agarose column, previously pre-equilibrated with buffer A (PBS, pH 7.4) containing 20 mmol/L imidazole. The column was washed with 30 mL of the same buffer solution, whereupon the bound-protein was eluted with a linear gradient of imidazole (50~500 mmol/L). The Ni2+-NTA column was efficient at enriching the 8His-tagged GsiA-GFP fusion protein, which comprised > 90% of the total protein eluted with 150 mmol/L imidazole. Protein-purity was evaluated on a 12% SDS polyacrylamide gel. The purified GsiA-GFP protein was > 85% pure, as revealed by Coomassie blue staining of the gel. The molecular mass determined by SDS-PAGE was in accordance with the calculated molecular mass of 96 kDa. Fractions containing GsiA-GFP were pooled and dialyzed into storage buffer. Approximately 2.0 mg of GsiA-GFP proteins were obtained from 1L of bacteria-culture in LB.
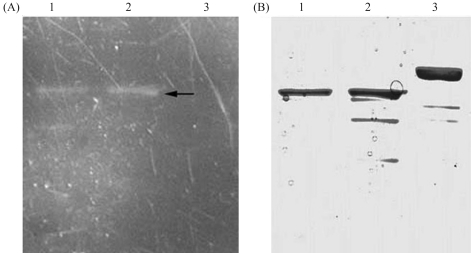
Even though the two bands may respectively represent the folded and aggregated protein, the possibility of the upper band having been denatured by the loading buffer (SB) could not be ruled out. To demonstrate the impossibility of this having occurred, the purified recombinant protein was in-gel analyzed by SDS-PAGE, whereupon it was noted that the SB buffer, incubated at 37 °C, did not affect protein-folding (Figure 8A and B).
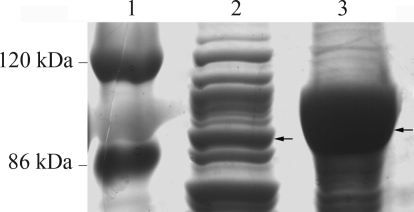
Figure 8.
In-gel analysis of purified recombinant protein. A: In-gel GFP fluorescence. B: The same gel stained with Coomassie blue. Lanes 1–2: 150 mmol/L and 100 mmol/L imidazole eluate treated with SB at 37 °C for 5 min; Lane 3: 150 mmol/L imidazole eluate boiled with SB for 5 min.
Conclusions
In general, it is difficult to decide which host and which induction situation is the more appropriate for heterologous protein production, this often depending on the target protein itself.
Herein, an efficient method for producing protein was used. Functional proteins can be easily recovered from protein-GFP fusions by cleavage with a TEV site-specific pro-tease, even though protein expression depends greatly on individual sequence of the proteins. Notwithstanding, the protocol proposed here offers a feasible alternative for the efficient and economic expression and purification of proteins for NMR and other biophysical studies.
Expression strains and induction temperatures may greatly affect both the quantity and quality of GsiA-GFP. Although higher induction temperatures favor the accumulation of insoluble proteins, extremely low temperatures (4 °C, data not shown), through suppressing cell-growth, could impede the supply of sufficient proteins for further study. As the concentration of IPTG did not visibly affect GsiA-GFP expression, low concentrations would suffice. Using GFP as a fusion strategy we could quickly optimize the expression and purification of GsiA-GFP. In-gel results showed that Rosetta (DE3) was the best strain for over-expression of GsiA-GFP at 18 °C and 0.1 mmol/L IPTG.
Upper-band intensity may represent aggregated proteins. On the contrary, lower fluorescent band intensity, besides corresponding to the GFP fluorescence observed in whole cells and the amount of active protein, also represents the fraction of GFP fusion proteins that is functionally expressed and readily solubilized upon exposure to detergents.
The availability of functionally active proteins would facilitate the application of high-resolution X-crystallography and spectroscopic analysis to structure-function studies of E. coli GsiA proteins. Finally, it should be noted that the GFP-based methodology, described in this protocol, will also greatly further the expression and purification of membrane proteins for functional and structural studies.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 30871344).
Footnotes
Associate Editor: Carlos F.M. Menck
References
- Biswas SK, Rahman I. Environmental toxicity, redox signaling and lung inflammation: The role of glutathione. Mol Aspects Med. 2009;30:60–76. doi: 10.1016/j.mam.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbouloux A, Shahi P, Chakladar A, Delrot S, Bachhawat AK. Hgt1p, a high affinity glutathione transporter from yeast Saccharomyces cerevisiae. J Biol Chem. 2000;27:513259–13265. doi: 10.1074/jbc.275.18.13259. [DOI] [PubMed] [Google Scholar]
- Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Dickinson DA, Forman HJ. Glutathione in defense and signaling: Lessons from a small thiol. Ann NY Acad Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Meth. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- Drew DE, von Heijne G, Nordlund P, de Gier JW. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- Fahey R, Brown W, Adams W, Worsham M. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanigan MH, Ricketts WA. Extracellular glutathione is a source of cysteine for cells that express γ-glutamyl trans-peptidase. Biochemistry. 1993;32:6302–6306. doi: 10.1021/bi00075a026. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McLellan LI. Glutathione and glutathione-dependent enzymes represent a coordinately regulated defence against oxidative stress. Free Radic Res. 1999;31:273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- Izawa S, Inoue Y, Kimura A. Oxidative stress response in yeast: Effect of glutathione on adaptation to hydrogen peroxide stress in Saccharomyces cerevisiae. FEBS Lett. 1995;368:73–76. doi: 10.1016/0014-5793(95)00603-7. [DOI] [PubMed] [Google Scholar]
- Jamai A, Tommasini R, Martinoia E, Delrot S. Characterization of glutathione uptake in broad bean leaf proto-plasts. Plant Physiol. 1996;111:1145–1152. doi: 10.1104/pp.111.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan R, Yi JR, Tang D, Li Y, Zlokovic BV, Kaplowitz N. Evidence for the existence of a sodium-dependent glutathione (GSH) transporter. Expression of bovine brain capillary mRNA and size fractions in Xenopus laevis oocytes and dissociation from γ-glutamyltranspeptidase and facilitative GSH transporters. J Biol Chem. 1996;271:9754–9758. doi: 10.1074/jbc.271.16.9754. [DOI] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martªnez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: A comprehensive view of Escherichia coli Biology. Nucleic Acids Res. 2009;37:464–470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Szczypka M, Lu YP, Thiele DJ, Rea PA. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- Mehdi K, Penninckx MJ. An important role for glutathione and γ-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology. 1997;143:1885–1889. doi: 10.1099/00221287-143-6-1885. [DOI] [PubMed] [Google Scholar]
- Meister A. A brief history of glutathione and a survey of its metabolism and functions. In: Dolphin D, Poulson R, Avramovic O, editors. Glutathione, Chemical, Biochemical, and Medical Aspects. John Wiley and Sons; New York: 1989. pp. 1–48. [Google Scholar]
- Owens RA, Hartman PE. Export of glutathione by some widely used Salmonella typhimurium and Escherichia coli strains. J Bacteriol. 1986;168:109–114. doi: 10.1128/jb.168.1.109-114.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx M. A short review on the role of glutathione in the response of yeast to nutritional, environmental and oxidative stresses. Enzyme Microb Technol. 2000;26:737–742. doi: 10.1016/s0141-0229(00)00165-4. [DOI] [PubMed] [Google Scholar]
- Pompella A, Visikis A, Paolicchi A, DeTata V, Casini AF. The changing faces of glutathione, a cellular protagonist. Biochem Pharmacol. 2003;66:1499–1503. doi: 10.1016/s0006-2952(03)00504-5. [DOI] [PubMed] [Google Scholar]
- Rapp M, Drew D, Daley DO, Nilsson J, Carvalho T, Mel|n K, De Gier JW, Von Heijne G. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 2004;13:937–945. doi: 10.1110/ps.03553804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeor JF, Connolly GC, Dumont ME, Ballatori N. ATP-dependent transport of reduced glutathione on YCF1, the yeast orthologue of mammalian multidrug resistance associated protein. J Biol Chem. 1998;273:33449–33454. doi: 10.1074/jbc.273.50.33449. [DOI] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kumagai H, Tochikura T. Isolation, genetic mapping, and characterization of Escherichia coli K-12 mutants lacking γ-glutamyltranspeptidase. J Bacteriol. 1987;169:3926–3931. doi: 10.1128/jb.169.9.3926-3931.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous γ-glutamyl peptide as an amino acid source, for which γ-glutamyltranspeptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kamatani S, Kim ES, Kumagai H. Amino-peptidases A, B, and N and dipeptidase D are the four cysteinylglycinases of Escherichia coli K-12. J Bacteriol. 2001;183:1489–1490. doi: 10.1128/JB.183.4.1489-1490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Koyanagi T, Izuka S, Onishi A, Kumagai H. The yliA, -B, -C, and -D genes of Escherichia coli K-12 encode a novel glutathione importer with an ATP-binding cassette. J Bacteriol. 2005;187:5861–5867. doi: 10.1128/JB.187.17.5861-5867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe K. Overview of bacterial expression systems for heterologous protein production: From molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- Yuan L, Kaplowitz N. Glutathione in liver diseases and hepatotoxicity. Mol Aspects Med. 2009;30:29–41. doi: 10.1016/j.mam.2008.08.003. [DOI] [PubMed] [Google Scholar]