Abstract
Gene fusions, yielding the formation of multidomain proteins, are evolutionary events that can be utilized as phylogenetic markers. Here we describe a fusion gene comprising the α and β subunits of succinyl-coA synthetase, an enzyme of the TCA cycle, in Pezizomycotina fungi. This fusion is present in all Pezizomycotina with complete genome sequences and absent from all other organisms. Phylogenetic analysis of the α and β subunits of succinyl-CoA synthetase suggests that both subunits were duplicated and retained in Pezizomycotina while one copy was lost from other fungi. One of the duplicated copies was then fused in Pezizomycotina. Our results suggest that the fusion of the α and β subunits of succinyl-CoA synthetase can be used as a molecular marker for membership in the Pezizomycotina subphylum. If a species has the fusion it can be reliably classified as Pezizomycotina, while the absence of the fusion is suggestive that the species is not a member of this subphylum.
Keywords: fused genes, fungi phylogeny, evolutionary marker
Introduction
Amongst eukaryotes, Fungi is the kingdom with most sequenced genomes. The sequenced genomes cover fungal species broadly and include agriculturally, medically, and industrially significant species with very diverse genomes. The fungal kingdom is divided in Chytridiomycota, Neocallimastigomycota, Glomeromycota, Ascomycota, Basidiomycota and Blastocladiomycota, and Microsporidia (Hibbett et al., 2007). Ascomycota, which diverged from Basidiomycota in the region of 741–1195 million years ago (Scannel et al., 2007), is the largest phylum of the Fungi kingdom and is again divided into Saccharomycotina, Taphrinomycotina, and Pezizomycotina (Lumbsch and Huhndorf, 2007). Pezizomycotina is the largest subphylum of Ascomycota and contains more than 33,000 described species (Spatafora et al., 2006).
Pezizomycotina, often referred to as filamentous fungi, grow primarily in a filamentous form and degrade biomass to free sugars that are used as source of energy and carbon (Arvas et al., 2007). Most species exhibit a dominant hyphal growth form, with almost all of the sexually reproducing forms possessing ascomata (Spatafora et al., 2006). Filamentous ascomycetes have been shown to be a monophyletic group distinguished from Taphrinomycotina and Saccharomycotina on the basis of ribosomal DNA sequence analysis (Alexopoulos et al., 1996; Spatafora et al., 2006).
In recent years, comparative genomics rather than gross morphology has been used to analyze specific sub groups of fungi. Notably, analysis of the phylum Saccharomycotina has revealed major events of evolution such as recent whole genome duplication and subsequent gene loss or subspecialization (Arvas et al., 2007; Dietrich et al., 2004). In Pezizomycotina, on the other hand, only a subset of protein families related to plant biomass degradation and secondary metabolism revealed signs of recent expansion, likely because of a repeat-induced point mutation mechanism for the active removal of duplicated sequences that has been identified in varying levels in Pezizomycotina species (Arvas et al., 2007). Gene duplication significantly influences evolution, as duplicated genes may be retained if they provide an advantage, either because of an increased gene dosage effect or because of increased functional diversification, or may alternatively be lost (Wagner, 1998; Li and Graur, 2000; Cornell et al., 2007). Multiple copies of the genes coding for the α and β subunits of succinyl-CoA synthetase have been shown to exist in several species of Aspergillus and in Neosartorya fisheri, placing succinyl-CoA synthetase among the subset of proteins showing gene expansion in Pezizomycotina (Flipphi et al., 2009).
Succinyl-CoA synthetase, also known as succinate-CoA ligase and succinate thiokinase, is an acid-thiol ligase that forms carbon-sulfur bonds. Succinyl-CoA synthetase is an enzyme of the citrate cycle, a major metabolic pathway in which acetyl-CoA derived from the catabolism of sugars and lipids is oxidized to carbon dioxide and energy in the form of GTP, NADH, and FADH2 (Hanson, 2008). This enzyme catalyzes the reversible transformation of succinyl-CoA, orthophosphate, and GDP to succinate, GTP, and the four-carbon metabolite coenzyme A in a three-step substrate-level phosphorylation reaction (Kanehisa and Goto, 2000; Hamblin et al., 2008). Succinyl-CoA synthetase is composed of two subunits, α and β, which generally form dimers in eukaryotes and tetramers in prokaryotes. Nucleotide specificity of succinyl-CoA synthe-tase is thought to be determined by the N-terminal domain of the beta subunit (Hamblin et al., 2008), and the alpha subunit may contain a targeting sequence for the enzyme (Birney and Klein, 1995). While prokaryotes are capable of using both types of nucleotides, eukaryotic succinyl-CoA synthetase enzymes usually occur as isoforms specific for either GTP or ATP (Hamblin et al., 2008). Multiple copies of the succinyl-coA synthetase genes tallies with the finding that duplications are most common in Pezizomycotina when the proteins produce secondary metabolites (Arvas et al., 2007), and with the common belief that natural selection on metabolic phenotypes mainly targets enzymes that control the rate of turnover of molecules through a metabolic pathway, such as succinyl-CoA synthetase (Flipphi et al., 2009).
In Pezizomycotina, one set of the duplicated α and β gene copies for succinyl-CoA synthetase has become fused. Gene fusion, yielding formation of multidomain proteins, is a major driving force in protein evolution and is a rare evolutionary event in comparison to the frequency of point mutations. Consequently, gene fusion events were recently utilized as an evolutionary marker to pinpoint the root of the eukaryotic tree (Nara et al., 2000; Stechmann and Cavalier-Smith, 2002, 2003). Here we examine how the presence of the succinyl-CoA synthetase fusion gene can be used as a simple phylogenetic marker for Pezizo-mycotina, and attempt to trace the origin and nature of this fusion in Fungi.
Materials and Methods
GenBank sequence data
We used the fungi genome BLAST tool from NCBI to search all complete fungal genomes with predicted proteins in GenBank. We started with the sequence of a Pezi-zomycotina succinyl-CoA synthetase α and β fusion protein, and used blastp to identify all other succinyl-CoA synthetase proteins in different fungal genomes.
With the exception of Microsporidia species, we were able to identify at least one α and one β subunit in each fungal genome available in the Genomic BLAST page (Table S1). The searches returned both fused and non-fused genes. For aligning the fused proteins with non-fused α and α copies of the protein, the fusions were artificially split into α′ and β′ subunits. To increase the diversity of fungal species analyzed, we also included the sequences of the α and β succinyl-CoA synthetase subunits of two Neocallimasti-gomycota fungi. As an outgroup we used the α and β subunits of succinyl-CoA synthetase from three mammals, Homo sapiens, Mus musculus and Canis familiaris.
Phylogenetics analyses
An alignment of the α subunits of succinyl-CoA synthetase for 53 organisms, including 25 Pezizomycotina, 14 Saccharomycotina, 2 Taphrinomycotina, 7 Basidiomy-cota, 2 Neocallimastigomycota as well as 3 metazoans to provide an outgroup, was created in ClustalX v.2.0.11 (Larkin et al., 2007) using default settings. A total of 80 sequences were aligned, including the region corresponding to the α subunit in the fusion genes in Pezizomycotina. A separate alignment of β and β′ subunits of the same organisms was created in the same manner. In total, 83 sequences were used in the β subunit alignment, including the region corresponding to the β subunit in the fusion genes in Pezi-zomycotina.
Each set of aligned sequences corresponding to the α and β subunits was used as input to infer phylogenetic trees using maximum likelihood and Bayesian analysis. For the maximum likelihood analysis we first used the program ProtTest (Abascal et al., 2005) to select the amino acid substitution model that best fits the protein alignments. In both datasets, the best model of amino acid substitution was the LG model (Le and Gascuel, 2008), with invariable sites and across site rate variation. We used PhyML 3.0 (Guindon and Gascuel. 2003), as implemented in SeaView (Galtier et al., 1996) to create maximum likelihood trees using the LG model of amino acid substitution (Le and Gascuel, 2008) with optimized number of invariable sites and optimized across site rate variation. Bootstrap support was calculated using the aLRT model (Anisimova and Gascuel, 2006). The Bayesian analysis was performed using MrBayes (Ronquist and Huelsenbeck, 2003) with four Monte Carlo Markov Chains and variable rates of substitution. Each chain was run for 1,000,000 generations and trees were sampled every 1,000 generations. The first 100 trees were excluded as burn-in and we used the remaining 900 trees to calculate bootstrap values. The trees were visualized and prepared for publication using Dendroscope (Huson et al., 2007) and FigTree.
Results
The fusion of the α and β subunits of succinyl-CoA can serve as a phylogenetic marker for Pezizomycotina fungi
We used the sequence of the fused α and α subunits of succinyl-CoA synthetase from Pyrenophora tritici-repentis, a Pezizomycotina, to search all the complete fungal genomes with protein predictions available in the genomic blast webpage form NCBI.
As of August 1st 2009, the genomic blast webpage allowed the search of the protein sequences of 51 fungal genomes: 41 Ascomycota, 7 Basidiomycota and 3 Micros-poridia. The 41 Ascomycota are divided in 25 Pezizo-mycotina, 14 Saccharomycotina, 2 Taphrinomycotina. All genomes analyzed had both the α and α subunits of succinyl-CoA synthetase, with the exception of the three Microsporidial genomes that completely lack both subunits of the protein and were not analyzed further. The genome of the Basidiomycota Postia placenta also seems to be lacking both subunits of the gene; however, an additional search of the genome page for this organism in the Joint Genome Institute (JGI), revealed that the genes are present, and we used the sequences obtained from JGI in the analyses for this organism.
In addition to the sequences obtained from the genomic blast searches we included the sequences of 2 Neocallimastigomycota fungi in the analysis to increase the diversity of the fungal species. The Ascomycota, together with Basidiomycota, form the subkingdom Dikarya, and are more closely related to each other than to Neocalli-mastigomycota or Microsporidia. Finally, we used the sequences of the α and β subunits of succinyl-CoA synthetase from three Metazoans as outgroup.
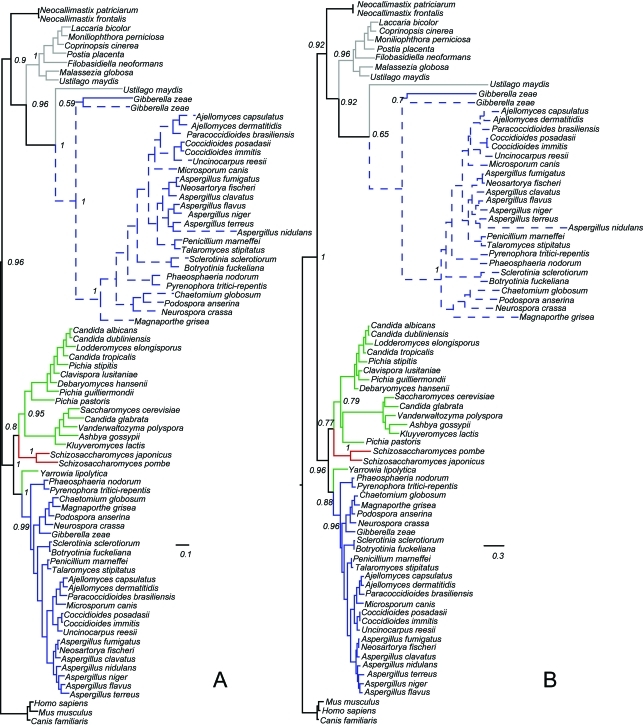
Out of the 25 Pezizomycotina fungi with complete genomes available in NCBI, we immediately detected the fusion gene in 22 genomes. According to the annotation in NCBI the fusion appears to be absent from Aspergillus niger, Coccioides immitis and Gibberella zeae. Interestingly, each of these species has at least two α and two β subunits of succinyl-CoA synthetase. Thus, we further analyzed these genomes to determine if the fusions are really absent or if they are just not annotated.
In Aspergillus niger, there are two subunits α and two subunits β annotated in the genome; a tblastn search of the fusion protein against A. niger genome showed that a subunit β gene immediately follows a subunit α gene in the same strand, indicating that the genes could be fused in this organism. Furthermore, a blastp search of the gene predictions in this organism’s genome webpage in JGI revealed that a fusion gene is present (gene model number jgi|Aspni1|176118). To maintain consistency, we used the proteins in GenBank in the analyses. The proteins involved in the putative fusion protein in A. niger group with the fusion proteins in the other Pezizomycotina in the phylogen-etic trees, further confirming that these two proteins are indeed fused in this genome (Figures 1 and 2, Table S1).
Figure 1.
Phylogenetic trees of the α subunit of succinyl-CoA synthetase in fungi. Pezizomycotina sequences are in blue, Saccharomycotina in green, Taphrinomycotina in red and Basidiomycota in gray. Neocallimastigomycotina and Metazoans (outgroup) are in black. Dashed lines represent fused genes. Bootstrap support values are given in relevant nodes. (a) Mr Bayes tree; (b) PhyML Maximum likelihood tree.
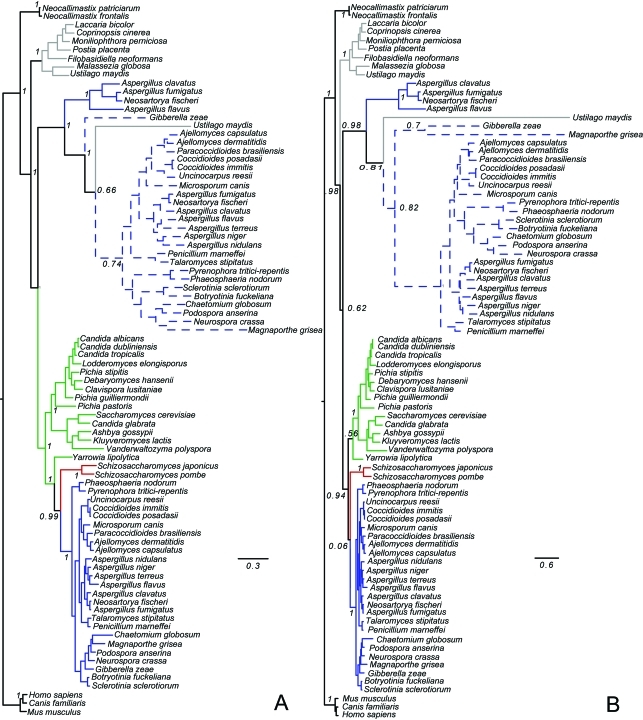
Figure 2.
Phylogenetic tree of the β subunit of succinyl-CoA synthetase in fungi. Pezyzomycotina sequences are in blue, Saccharomycotina in green, Taphrinomycotina in red and Basidiomycota in gray. Neocallimastigomycotina and Metazoans (outgroup) are in black. Dashed lines represent fused genes. Bootstrap support values are given in relevant nodes. (a) Mr Bayes tree; (b) PhyML Maximum likelihood tree.
A similar problem occurs in Coccidioides immitis. A tblastn search revealed that two consecutive genes in the genome hit the fusion gene. In this case, however, the first gene only hits part of subunit α while the second gene hits the remainder of subunit α and the full length of subunit β. We aligned both genes with the fusion genes and determined the boundaries between the α and β subunits assuming the proteins are fused and used these sequences in the analyses. Like in A. niger, the putative fusion proteins groups with the fusion proteins in the other Pezizomycotina in the phylogenetic trees (Figures 1 and 2, Table S1).
Finally, while the fusion is not annotated in Giberella zeae, in this organism a subunit β gene follows a subunit α gene. Both subunits are in the same strand but are 3 kb apart and there seems to be a small intervening gene in the opposite strand between the subunits. A large intron between the subunits could have confounded the gene prediction software, and it is not unknown for a gene to be nested inside an intron of another (Kumar, 2009). It is worth noting that the version of the genome available in GenBank is based on a preliminary whole-genome-shotgun that has been superceded by a new assembly of the genome. Unfortunately, the new assembly doesn’t seem to be available at the time of this writing. Interestingly, these two proteins group with the fusion proteins in the phylogenetic trees (Figures 1 and 2, Table S1), indicating that they could be fused. The alternative explanation would be that the proteins were once fused and that the insertion of a gene in between the α and β subunits broke the fusion into two working copies of the subunits; while plausible, this second explanation seems to be unlikely as the insertion would have to have occurred right at the junction between the two subunits. This organism also has an extra copy of the subunit α that groups with the fusions, suggesting that the fusion gene was duplicated in this organism and the subunit β was eliminated in one of the copies of the fusion.
These results indicate that, even though it might not have been annotated as such, the fusion of the α and β subunits of succinyl-CoA synthetase is present in all 25 Pezi-zomycotina genomes available at GenBank, with the possible exception of that of G. zeae in which the fusion protein seem to at least have once been present. In addition, the fusion gene was not detected in any other fungal genome and a blast search of the entire NR database revealed that the fusion is only present in Pezizomycotina fungi.
The apparent presence of the fusion gene in most Pezizomycotina and its absence in all other organisms suggest that the fused succinyl-CoA synthetase gene can serve as a phylogenetic marker for Pezizomycotina. If a species has the fusion it can be reliably classified as Pezizomy-cotina; if a species does not have the fusion it most likely does not belong to this subphylum.
Evolution of the α and β subunits of succinyl-CoA synthetase in fungi
We used the sequences of the succinyl-CoA synthe-tase of 53 species to build phylogenetic trees for both the α and β subunits of the protein. The data set contained 48 Dikarya, 2 Neocallimastigomycota and 3 metazoans. The Dikarya are divided in 41 Ascomycota (25 Pezizomy-cotina, 14 Saccharomycotina, 2 Taphrinomycotina) and 7 Basidiomycota (Table S1). The sequences of the fusion genes in the 25 Pezizomycotina were split into their α and β constituents.
All sequences were aligned in ClustalW using default parameters and trees were built using MrBayes and PhyML. The resulting trees are shown in Figures 1 and 2. In all trees the fused genes form a monophyletic group (with strong bootstrap support) and the non-fused copies in Pezi-zomycotina form another monophyletic group (with strong bootstrap support).
The phylogenetic trees of the α subunits (Figure 1) suggest that this subunit duplicated before the divergence of Neocallimastigomycota from Dikarya. Saccharomyco-tina and Taphrinomycotina lost the copy that would later become fused in Pezizomycotina, while Neocallimasti-gomycota and Basidiomycota lost the other copy. The α subunit present in Neocallimastigomycota and Basidio-mycota groups with the fused copy in Pezizomycotina, while the α subunit present in Saccharomycotina and Taphrinomycotina groups with the unfused copies of the subunit in Pezizomycotina. G. zeae has an extra copy of subunit α that groups with the fused genes, indicating that it probably resulted from a recent duplication of the fused gene followed by the loss of the β subunit. This loss should not have a large phenotypic effect, as another copy of the fusion gene would be present in the genome.
The trees of the β subunit (Figure 2) indicate a different evolutionary story for this subunit. The β subunit duplicated in Dikarya after their divergence from Neocallimas-tigomycota, and after the divergence of basidiomycetes from ascomycetes, but before the divergence of Pezizomy-cotina from Saccharomycotina and Taphrinomycotina. The copy that would become fused in Pezizomycotina was lost from Saccharomycotina and Taphrinomycotina.
A second unfused copy of the β subunit is present in three Aspergillus species (A. clavatus, A. flavus and A. fumigatus) and in Neosartorya fischeri (Figure 2), all members of Trichocomaceae, and groups with the fusion proteins. This suggests that the β subunit duplicated a second time before becoming fused in Pezizomycotina and was lost from most Pezizomycotina with the exception of some Trichocomaceae.
Interestingly, one Basidiomycota, Ustilago maydis, also has two copies of each subunit of succinyl-CoA syn-thetase. One of the α and one of the β subunits group with the fusion gene in Pezizomycotina, which would suggest that this gene could have been the result of a lateral gene transfer between a Pezizomycotina and U. maydis. This would not be the first gene shown to have been transferred from Pezizomycotina to U. maydis (Shen et al., 2009). We checked the location of these genes in the genome of U. maydis and found that they are consecutive but in opposite strands, and therefore cannot be not fused in this species. A closer look at the trees reveals that the sequence of the U. maydis subunits that group with the fusions has a long branch, and its grouping with the fusions could be explained by long branch attraction. The presence of two copies of each of the subunits in the genome of U. maydis might have left one copy of each subunit free to accumulate mutations, explaining why they would be in such a long branch. However, it is also possible that the fusion gene was horizontally transferred to U. maydis and a genome rearrangement broke the gene in two parts and reversed one of the subunits in relation to the other, although this explanation is less likely considering that such rearrangement would need to have happened in such a way as to generate working versions of the genes.
With the exception of U. maydis, the Basidiomycota form a monophyletic group in the trees. The Neocalli-mastigomycota, Taphrinomycotina and Pezizomycotina also form monophyletic groups. The Saccharomycotina only form a monophyletic group in the maximum likelihood tree of the β subunit (Figure 2b); in all other trees Yarrowia lipolytica groups with the unfused copy of the proteins in Pezizomycotina and not with Saccharomy-cotina. This unexpected placement of Yarrowia lipolytica has been seen in other analyses as well (Arvas et al., 2007, Cornell et al., 2007) and has been attributed to Y. lipolytica ORFs bearing more similarity to other fungi than to Saccharomycotina (Arvas et al., 2007).
Discussion
Our results show that the sub-phylum Pezizomy-cotina is characterized by the presence of a fusion of the α and β subunits of succinyl-CoA synthetase that can be used as a phylogenetic marker to determine if an organism belongs in Pezizomycotina. If a species has the fusion it can be reliably classified as Pezizomycotina, while the absence of the fusion is suggestive that the species is not a member of this subphylum.
Phylogenetic analyses show that the two subunits of succinyl-CoA synthetase duplicated at different times in fungal evolution and then became fused in an ancestor of Pezizomycotina. The gene for the α subunit of succinyl-CoA synthetase duplicated in an ancestor of the Neocalli-mastigomycota and Dikarya. One copy of the gene was then lost in all fungi with the exception of Pezizomycotina. The gene for the subunit β of succinyl-CoA synthetase most likely duplicated in an ancestor of the Ascomycota after the divergence of this group from Basidiomycota. One copy of the gene was then lost from Saccharomycotina and Taphrinomycotina, and kept in Pezizomycotina.
In Pezizomycotina, one copy of the α subunit fused to a copy of the β subunit, generating a fusion protein while retaining the unfused copies as well. As this fusion gene involves two subunits of the same enzyme it is possible that it provides a selective advantage by leading to greater catalytic activity or more efficient co-regulation of the expression of the two succinyl-CoA subunits.
Even though the presence of this fused gene could lead to a more efficient enzyme, either by greater catalytic activity or by more efficient co-regulation, Pezizomy-cotina genomes maintained two copies (one fused one unfused) of each subunit of succinyl-CoA synthetase. This might indicate that this enzyme is needed in large quantities in Pezizomycotina or, more interestingly, it is possible that one of the copies of succinyl-CoA synthetase in Pezizomycotina has acquired an as yet unreported new function, as suggested by Flipphi et al. (2009) for other duplicated genes in the primary carbon metabolism of Aspergillus species.
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Science Foundation under Award No. 0920697. AMK was funded with a grant of the HHMI to Pomona College’s Biology Department. The authors thank Daniel Martinez (Pomona College) for help generating and interpreting the phylogenetic trees.
Footnotes
Associate Editor: Célia Maria de Almeida Soares
References
- Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Alexopoulos CJ, Mims CW, Blackwell M. Introductory Mycology. 4th edition. John Wiley & Sons; New York: 1996. p. 880. [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood ratio test for branchs: A fast, accurate and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Arvas M, Kivioja T, Mitchell A, Saloheimo M, Ussery D, Penttila M, Oliver S. Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccha-romycotina. BMC Genomics. 2007;8:e325. doi: 10.1186/1471-2164-8-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney MA, Klein C. Cloning and expression of the α subunit of succinyl-CoA synthetase from Dictyostelium discoideum. Arch Biochem Biophys. 1995;319:93–101. doi: 10.1006/abbi.1995.1270. [DOI] [PubMed] [Google Scholar]
- Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG. Comparative genome analysis across a kingdom of eukaryotic organisms: Specialization and diversification in the fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. The Ashyba gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Flipphi M, Sun J, Robellet X, Karaffa L, Fekete E, Zeng A, Kubicek C. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol. 2009;46:S19–S44. doi: 10.1016/j.fgb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SeaView and Phy-lo_win, two graphic tools for sequence alignment and molecular phylogeny. Comp Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Hamblin K, Standley D, Rogers M, Stechmann A, Roger A, Maytum R, van der Giezen M. Localization and nucleotide specificity of Blastocystis succinyl-CoA syn-thetase. Mol Microbiol. 2008;68:1395–1405. doi: 10.1111/j.1365-2958.2008.06228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J. The Chemistry of Fungi. Royal Society of Chemistry; Publishing, Cambridge: 2008. p. 244. [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Huson D, Richter D, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:e460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. An overview of nested genes in eukaryotic genomes. Eukaryotic Cell. 2009;8:1321–1329. doi: 10.1128/EC.00143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X v. 2.0. Bioin-formatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino-acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Li WH, Graur D. Fundamentals of Molecular Evolution. 2. Sinauer Associates Inc; Sunderland: 2000. p. 439. [Google Scholar]
- Lumbsch HT, Huhndorf SM. Outline of Ascomycota. Myconet. 2007;13:1–58. [Google Scholar]
- Nara T, Hshimoto T, Aoki T. Evolutionary implications of the mosaic pyrimidine-biosynthetic pathway in euka-ryotes. Gene. 2000;257:209–222. doi: 10.1016/s0378-1119(00)00411-x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Scannell D, Butler G, Wolfe K. Yeast genome evolution – The origin of the species. Yeast. 2007;24(11):929. doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lang B, Burger G. Diversity and dispersal of a ubiquitous protein family: Acyl-CoA dehydrogenases. Nucleic Acids Res. 2009;37:5619–5631. doi: 10.1093/nar/gkp566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora J, Sung G, Johnson D, Hesse C, O’Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, et al. A five-gene phylogeny of Pezizomycotina. Mycologia. 2006;98:1018–1028. doi: 10.3852/mycologia.98.6.1018. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the euka-ryote tree pinpointed. Curr Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Wagner A. The fate of duplicated genes: Loss or new function? Bioessays. 1998;20:785–788. doi: 10.1002/(SICI)1521-1878(199810)20:10<785::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Fungi genome BLAST tool. http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=fungi(August 2009)
- FigTree Software. http://tree.bio.ed.ac.uk/software/figtree/(July 2010)
- Postia placenta genome page at Joint Genome Institute (JGI) http://genomeportal.jgi-psf.org/Pospl1/Pospl1.home.html(June 2010)
- Aspergillus niger genome page at Joint Genome Institute (JGI) http://genome.jgi-psf.org/Aspni5/Aspni5.home.html(June 2010)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.