As an important mechanism of gene regulation, the microRNA pathway itself requires fine regulation. This work shows that an Importin β protein modulates the activity of microRNAs by influencing their loading into the ARGONAUTE1 complex.
Abstract
As key components in the eukaryotic gene regulatory network, microRNAs (miRNAs) themselves are regulated at the level of both metabolism and activity. To identify factors that modulate miRNA activity, we used an Arabidopsis thaliana transgenic line expressing an artificial miRNA that causes trichome clustering and performed a screen for mutants with compromised miRNA activity (cma mutants) or enhanced miRNA activity (ema mutants). From this screen, we identified two novel mutant alleles of SERRATE, which is known to be required for miRNA biogenesis and dozens of other cma and ema mutants. In this study, we analyzed ema1. SAD2/EMA1 encodes an Importin β protein. The ema1 mutation had no effects on the accumulation of miRNAs and ARGONAUTE1 (AGO1) or on their cytoplasmic and nuclear distributions. Intriguingly, we found that the miRNA effector complexes purified from ema1 contained a larger amount of miRNAs and displayed elevated mRNA cleavage activities, indicating that EMA1 modulates miRNA activity by influencing the loading of miRNAs into AGO1 complexes. These results implicate EMA1 as a negative regulator of the miRNA pathway and reveal a novel layer of miRNA activity modulation.
INTRODUCTION
Small silencing RNAs play key roles in gene regulation in eukaryotes. Three major types of small silencing RNAs have been discovered in Arabidopsis thaliana. These include microRNAs (miRNAs), trans-acting small interfering RNAs (ta-siRNAs), and heterochromatic small interfering RNAs (hc-siRNAs). The vast majority of miRNAs and ta-siRNAs repress target gene expression at the posttranscriptional level via mRNA cleavage and translational inhibition, whereas hc-siRNAs mediate DNA methylation at homologous loci, often resulting in transcriptional gene silencing (Baulcombe, 2004; Voinnet, 2009).
Most Arabidopsis miRNA loci are encoded by independent transcriptional units in intergenic regions that are transcribed by RNA polymerase II (Xie et al., 2005, 2010). The primary transcript of a miRNA (pri-miRNA) is likely stabilized by DAWDLE (a nuclear RNA binding protein) (Yu et al., 2008) and processed by Dicer-Like1 (DCL1) into a stem-loop structured miRNA precursor (pre-miRNA), which is further excised into a miRNA/miRNA* (miRNA* is the strand that pairs with miRNA) duplex with 2-nucleotide overhangs at the 3′-termini (Park et al., 2002; Kurihara and Watanabe, 2004; Park et al., 2005). Both processing steps are thought to take place in nuclear dicing bodies containing DCL1, Hyponastic Leaves1 (HYL1), and Serrate (SE) (Fang and Spector, 2007; Song et al., 2007). Biochemical studies have shown that HYL1 and SE promote the efficiency and accuracy of DCL1-catalyzed miRNA processing (Dong et al., 2008). The 3′-termini of the miRNA/miRNA* duplex are methylated by Hua Enhancer1 to provide protection from polyuridylation and subsequent degradation (Li et al., 2005; Yu et al., 2005). The miRNA/miRNA* duplex is exported from the nucleus to the cytoplasm at least partly by HASTY, the Arabidopsis homolog of Exportin 5 (Park et al., 2005), where the miRNA strand of the duplex is selectively loaded onto an Argonaute (AGO) protein to form miRNA-induced silencing complexes (miRISCs). Among the 10 AGO family members in Arabidopsis, AGO1 predominates in the miRNA pathway, as evidenced by both genetic and biochemical analyses (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005; Mi et al., 2008). miRISCs are guided by miRNAs to their target transcripts through base pairing to mediate target mRNA cleavage or translational repression (Voinnet, 2009). A genetic study has shown that KATANIN and the mRNA decapping factor Varicose are involved in miRNA-mediated translational repression but not in miRNA-directed mRNA cleavage (Brodersen et al., 2008). The association of a number of miRNAs and AGO1 with polysomes also supports the notion that miRNA-mediated translation repression is a general mechanism of miRNA action in plants (Lanet et al., 2009). Recently, a class of DCL3-dependent long miRNAs was identified in rice (Oryza sativa). These long miRNAs are associated with AGO4 and mediate DNA methylation at their target genes, revealing a novel mode of plant miRNA action (Wu et al., 2010).
As an important mechanism of gene regulation, the miRNA pathway itself is subject to fine regulation. The regulation could be imposed at the level of miRNA processing, assembly of miRISC, miRISC action, and miRNA turnover. In Arabidopsis, Cyclophilin 40 (CyP40) promotes miRNA activity, probably by promoting the activity of AGO1 (Smith et al., 2009), whereas an F-box protein, F-BOX WITH WD-40 2, negatively regulates miRNA activity by reducing the AGO1 protein level (Earley et al., 2010). A recent biochemical study demonstrated that the loading of miRNAs into AGO1 is facilitated by HSP90 (Iki et al., 2010), consistent with the genetic study that showed the synergistic interaction between HSP90 and AGO1 (Smith et al., 2009). It has also been shown that miRNAs can be degraded by small RNA degrading nucleases to maintain proper levels; plants with reduced levels of small RNA degrading nucleases overaccumulate miRNAs and display pleiotropic developmental defects (Ramachandran and Chen, 2008). It is noteworthy that the two key enzymes in the miRNA pathway are subjected to miRNA-mediated feedback regulation: DCL1, the enzyme that catalyzes miRNA processing, is targeted by miR162 (Xie et al., 2003), whereas AGO1, the main miRNA effector protein, is regulated by miR168 (Vaucheret et al., 2006). Such feedback regulation may maintain the homeostasis of the miRNA pathway for proper functioning (Vaucheret et al., 2006).
Although a decade of study has established a mechanistic framework for the plant miRNA pathway, many questions remain. Is processing of miRNAs from their precursors regulated, and, if so, how? How are miRNAs loaded into effector proteins, and, if so, is their loading regulated? How do miRISCs access their target mRNAs within the cellular environment? As a step to address these questions, we used a transgenic Arabidopsis line, amiR-triOX (Mi et al., 2008), to carry out a forward genetic screen for novel components in the miRNA pathway. From this screen, we obtained dozens of mutants with compromised miRNA activity (cma mutants) or enhanced miRNA activity (ema mutants). Among the cma mutants, we identified two additional se alleles, validating the efficacy of the screen. Among the ema mutants, we cloned the EMA1 locus, which encodes an Importin β. We found that the accumulation of miRNAs and ARGONAUTE1 (AGO1) or their cytoplasmic and nuclear distributions were not changed in the ema1 mutants. Interestingly, larger amounts of miRNAs were detected in the ema1 miRISCs that displayed stronger cleavage activities, indicating that EMA1 negatively regulates miRNA activity by regulating miRNA loading into effector complexes.
RESULTS
A Forward Genetic Screen for Novel Components in the Arabidopsis miRNA Pathway
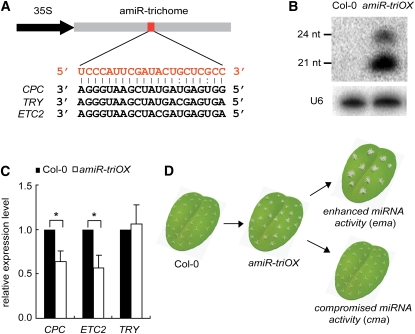
To obtain a more detailed mechanistic picture of the plant miRNA pathway, we performed a forward genetic screen for novel factors that are involved in miRNA-mediated gene regulation in Arabidopsis. We used a transgenic line, amiR-triOX, that overexpresses an artificial miRNA, amiR-trichome (Mi et al., 2008), as the starting material for this screen. amiR-trichome is designed to target three R3 MYB genes, CAPRICE (CPC), TRIPTYCHON (TRY), and ENHANCER of TRIPTYCHON AND CAPRICE2 (ETC2) (Schwab et al., 2006). These genes suppress trichome initiation and differentiation in a redundant manner, and mutations in these genes cause an increase in trichome density and/or trichome clustering and a reduction in the number of root hairs (Ishida et al., 2008). The overexpression of amiR-trichome in the transgenic plants resulted in clustered trichomes on leaf blades (Schwab et al., 2006; Mi et al., 2008), which provides a visual readout for miRNA activity in the transgenic Arabidopsis (Figures 1A and 1B). amiR-triOX had an ~40 to 50% reduction in the expression of CPC and ETC2 but no change in TRY expression, which resulted in a moderate degree of trichome clustering (Figure 1C). Thus, using this screen system, we could obtain two types of mutants, cma mutants with normal trichomes or less-clustered trichomes and ema mutants with more-clustered trichomes (Figure 1D).
Figure 1.
A Genetic Screen for Components in the Arabidopsis miRNA Pathway.
(A) An amiR-trichome designed to target the three indicated MYB genes.
(B) Detection of amiR-trichome expression in the transgenic line amiR-triOX by small RNA gel blot. U6 was also probed and used as the loading control. nt, nucleotide.
(C) Detection of the target genes of amiR-trichome in Col-0 and amiR-triOX by quantitative RT-PCR. Error bars indicate sd (n = 6), and asterisks indicate a significant difference between the indicated samples (t test, P value < 0.05).
(D) Schematic representation of the strategy for screening for ema or cma mutants. See Supplemental Table 1 online for a summary of isolated mutants.
Thus far, 36 cma and 14 ema mutants were obtained from the screen (see Supplemental Table 1 online). Among the cma mutants, we identified two novel alleles of SE (which we named cma1-1/se-6 and cma1-2/se-7), validating the efficacy of the screen (see Supplemental Table 2 online). se-6 carries a G-to-A transition at position 627, while se-7 carries a G-to-A transition at position 1897; both mutations cause splicing defects (see Supplemental Table 2 online). Consistent with the established role of SE in pri-miRNA processing, se-7 had reduced levels of miRNAs (see Supplemental Figure 1A online) and elevated accumulation levels of miRNA target genes (see Supplemental Figure 1B online).
In this study, we analyzed one of the ema mutants, ema1-1.
Characterization of ema1-1
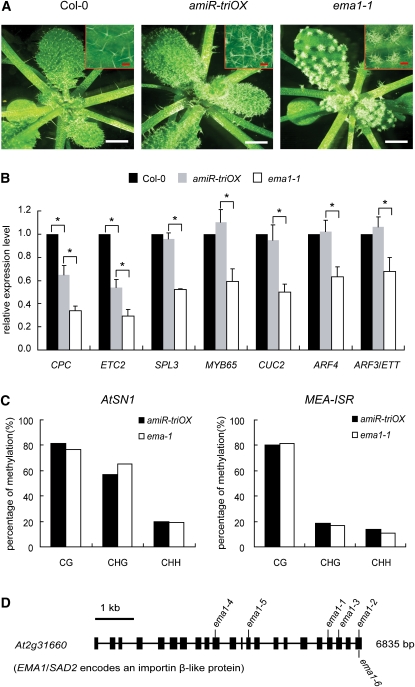
As a result of amiR-trichome–mediated downregulation of CPC and ETC2 (Figure 1C), the amiR-triOX line displayed a moderate degree of trichome clustering on the leaf blades (Figure 2A). In the ema1-1 mutant, the clustering of trichomes was significantly enhanced (Figure 2A). No other obvious developmental phenotypes were observed in ema1-1 (Figure 2A). The enhanced clustering of trichomes was consistent with the decreased accumulation of CPC and ETC2 as measured by quantitative RT-PCR (Figure 2B). It was possible that CPC and ETC2 were methylated by the 24-nucleotide variant of amiR-trichome (Figure 1B), and the reduced levels of these two genes were attributed to increased DNA methylation in ema1-1. To test this possibility, we used bisulfite sequencing to examine the cytosine methylation levels at the targets sites within these two genes in amiR-triOX and ema1-1. Negligible amounts of cytosine methylation were detected at both genes, and the methylation levels remained the same in ema1-1 (see Supplemental Table 3 online). Thus, we concluded that the decreased levels of CPC and ETC2 resulted from enhanced activity of amiR-trichome in ema1-1.To examine whether the activities of endogenous miRNAs were also elevated in ema1-1, we measured the levels of several endogenous miRNA targets. These included SPL3 (a target of miR156), MYB65 (a target of miR159), and CUC2 (a target of miR164). In ema1-1, the levels of all three mRNAs decreased by ~40 to 50%, compared with those in Columbia-0 (Col-0) and amiR-triOX (Figure 2B). This indicated that the ema1-1 mutation has a general impact on the miRNA pathway.
Figure 2.
Isolation of ema1-1.
(A) The more-clustered trichome phenotype of ema1-1. Photographs of 3-week-old seedlings were taken.
(B) Detection of the target genes of amiR-trichome and endogenous miRNAs in Col-0, amiR-triOX, and ema1-1 by quantitative RT-PCR. Error bars indicate sd (n = 5), and asterisks indicate a significant difference between the indicated samples (t test, P value < 0.05).
(C) The effect of ema1-1 on DNA methylation at AtSN1 and MEA-ISR loci. Cytosine methylation at AtSN1 and MEA-ISR in amiR-triOX and ema1-1 were determined by bisulfite sequencing. More than 20 clones were sequenced for each line.
(D) Gene structure of At2g31660. Thick lines indicate exons, and thin lines indicate introns. The six ema1 alleles identified in this study are shown. See Supplemental Figure 2 online for positional cloning of ema1-1.
Next, we examined the effects of ema1-1 on the ta-siRNA and hc-siRNA pathways. We found that ema1-1 had decreased levels of ARF4 and ARF3/ETT, two target genes of ta-siARF (Figure 2B), indicating that the ta-siRNA activity was affected by the ema1-1 mutation. We used bisulfite sequencing to measure the cytosine methylation levels at two representative loci, AtSN1 and MEA-ISR, whose methylation is known to be directed by hc-siRNAs (Zilberman et al., 2003). In ema1-1, the methylation levels at these two loci were comparable to those in Col-0 (Figure 2C), suggesting that ema1-1 has no effect on hc-siRNA activity.
EMA1 Encodes an Importin β Protein
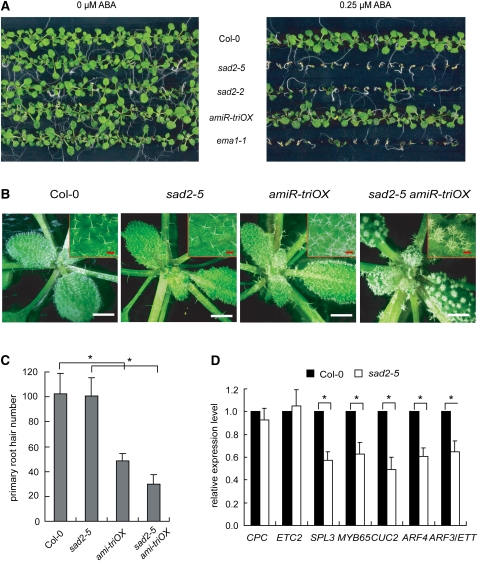
Map-based cloning revealed that ema1-1 carries a G-to-A transition in At2g31660 (Figure 2D; see Supplemental Figure 2A and Supplemental Table 4 online). This mutation causes a splicing defect, which leads to a frame shift and reduced accumulation of At2g31660 transcripts (see Supplemental Figure 2B online). At2g31660 encodes an Importin β-domain protein likely to be involved in nuclear transport. Allelic analyses identified five additional alleles of ema1 among the ema mutants; they were designated as ema1-2~6 (Figure 2D; see Supplemental Table 4 online). At2g31660 was first isolated from a genetic screen for mutants hypersensitive to abscisic acid (ABA) and was named SAD2 (Verslues et al., 2006). SAD2 was later shown to be required for the UV-B response by mediating MYB4 nuclear trafficking (Zhao et al., 2007). Like the sad2 mutants (Verslues et al., 2006), ema1-1 exhibited ABA hypersensitivity in seedling growth (Figure 3A). However, this ABA phenotype likely reflects the requirement of SAD2/EMA1 for nuclear import of a negative regulator of ABA sensitivity as proposed previously (Verslues et al., 2006) and unlikely reflects its role in the miRNA pathway, as a number of studies have demonstrated that the mutants defective in miRNA function are hypersensitive to ABA (Lu and Fedoroff, 2000; Zhang et al., 2008; Earley et al., 2010).
Figure 3.
Characterization of SAD2/EMA1 Mutants.
(A) ABA sensitivity assay for Col-0, sad2-5, sad2-2, amiR-triOX, and ema1-1. Seeds of these lines were planted on Murashige and Skoog medium with 0 or 0.25 μM ABA. The photographs were taken at 16 d after sowing.
(B) Photographs of 3-week-old Col-0, sad2-5, amiR-triOX, and sad2-5 amiR-triOX seedlings. White bars = 2 mm; red bars = 0.25 mm.
(C) Primary root hair numbers of 3-d-old seedlings of Col-0, sad2-5, amiR-triOX, and sad2-5 amiR-triOX. Error bars indicate sd (n = 10), and asterisks indicate a significant difference between the indicated samples (t test, P value < 0.05).
(D) Detection of the target genes of amiR-trichome and endogenous miRNAs in Col-0 and sad2-5 by quantitative RT-PCR. Error bars indicate sd (n = 4), and asterisks indicate a significant difference between the indicated samples (t test, P value < 0.05).
It is noteworthy that this Importin β protein has been shown to be involved in trichome initiation by regulating GLABRA3 (GL3) (Gao et al., 2008; Yoshida et al., 2009). Mutations in the gene that encodes this protein cause a reduction in trichome number but have no effect on root hair number (Gao et al., 2008; Yoshida et al., 2009). As shown in Figures 3B and 3C and Supplemental Figure 3 online, in a null allele of this Importin β, sad2-5, the trichome number on leaf blades was slightly reduced, but the root hair number remained essentially the same as Col-0, confirming the results of previous studies (Gao et al., 2008; Yoshida et al., 2009). It should be emphasized that sad2-5 had no clustered trichomes (Figure 3B). Thus, the highly clustered trichomes in ema1 mutants are unlikely due to the misregulation of GL3 but are rather caused by increased activity of amiR-trichome by mutations in Importin β. To unambiguously show that the enhanced miRNA activity in ema1-1 was due to the mutation of At2g31660, we crossed the amiR-triOX line with sad2-5. The progeny homozygous for both amiR-triOX and sad2-5 showed highly clustered trichomes (Figure 3B) and a reduced number of root hairs (Figure 3C). We also examined the expression of target mRNAs of amiR-trichome and endogenous miRNAs and ta-siRNAs and found that all examined targets of endogenous miRNAs and ta-siRNAs were reduced in sad2-5 (Figure 3D).
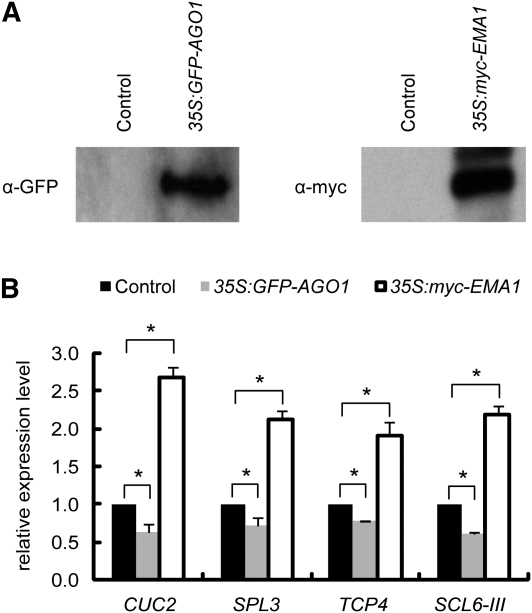
These genetic data indicate that SAD2/EMA1 negatively regulates miRNA activity. Thus, we next tested whether overexpression of SAD2/EMA1 in plant cells could lead to decreased miRNA activity. We transfected Arabidopsis protoplasts with a DNA construct that expresses myc-EMA1 under the 35S promoter. As controls, an empty vector and 35S:GFP-AGO1 (GFP for green fluorescent protein) were also transfected. Overexpression of GFP-AGO1 in cells led to a mild decrease in the accumulation of miRNA targets, compared with that in cells transfected with an empty vector (Figures 4A and 4B). Consistent with its function as a negative regulator of the miRNA activity, overexpression of myc-EMA1 resulted in an approximately two- to threefold increase in the accumulation of miRNA targets (Figures 4A and 4B).
Figure 4.
Overexpression of SAD2/EMA1 Reduces miRNA Activity.
(A) Detection of GFP-AGO1 and myc-EMA1 in Arabidopsis protoplasts transfected with 35S:GFP-AGO1 and 35S:myc-EMA1 constructs by immunoblot analyses.
(B) Detection of the expression levels of several miRNA target genes in Arabidopsis protoplasts transfected with 35S:GFP-AGO1 and 35S:myc-EMA1 by quantitative RT-PCR. Error bars indicate sd (n = 3), and asterisks indicate a significant difference between the indicated samples (t test, P value < 0.05).
Taken together, these results demonstrate that EMA1 encodes an Importin β protein and is a negative regulator of the miRNA pathway.
Loss of SAD2/EMA1 Partially Rescues the hyl1 But Not the ago1 Mutant Phenotype
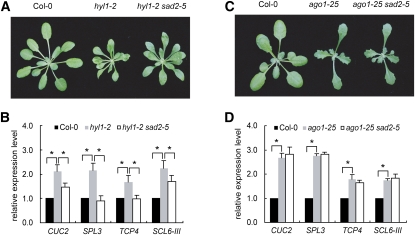
To provide further evidence for the involvement of SAD2/EMA1 in the miRNA pathway, we examined the genetic interaction between SAD2/EMA1 and some of the known miRNA pathway components. We crossed sad2-5 to hyl1-2, a null allele of HYL1 (Vazquez et al., 2004). We found that sad2-5 partially suppressed the developmental phenotypes caused by hyl1-2 (Figure 5A). Consistent with this phenotypic observation, hyl1-2 overaccumulated several miRNA targets, including CUC2, SPL3, TCP4, and SCL6-III, as previously reported (Han et al., 2004), but such overaccumulation was greatly compromised in the hyl1-2 sad2-5 double mutant, with two of the target genes (SPL3 and TCP4) being restored to wild-type levels (Figure 5B). These data indicate a genetic interaction between SAD2/EMA1 and HYL1.
Figure 5.
Loss of SAD2/EMA1 Partially Rescues the hyl1 but Not the ago1 Mutant Phenotype.
(A) Phenotypes of hyl1-2 and hyl1-2 sad2-5 mutants. Photographs were taken of 4-week-old plants.
(B) Detection of the expression levels of several miRNA target genes in Col-0, hyl1-2, and hyl1-2 sad2-5 by quantitative RT-PCR. Error bars indicate sd (n = 3), and asterisks represent a significant difference between the indicated samples (t test, P value < 0.05).
(C) Phenotypes of ago1-25 and ago1-25 sad2-5 mutants. Photographs were taken of 3-week-old plants.
(D) Detection of the expression levels of several miRNA target genes in Col-0, ago1-25, and ago1-25 sad2-5 by quantitative RT-PCR. Error bars indicate sd (n = 3), and asterisks indicate a significant difference between the indicated samples (t test, P value < 0.05).
Next, we crossed sad2-5 to ago1-25 (Morel et al., 2002), a hypomorphic allele of AGO1, which encodes the core protein of the miRNA effector complex. We observed no difference in developmental phenotype between ago1-25 single and ago1-25 sad2-5 double mutants (Figure 5C). In the ago1-25 single mutant, the expression of several miRNA target genes was increased by approximately one- to twofold compared with Col-0 (Figure 5D). Intriguingly, the expression levels of these genes in the ago1-25 sad2-5 double mutant were not reduced and were comparable to those in ago1-25 (Figure 5D). This suggests that SAD2/EMA1 functions through AGO1 to modulate miRNA activity.
Accumulation of miRNAs and AGO1 Is Unaffected by Mutations in SAD2/EMA1
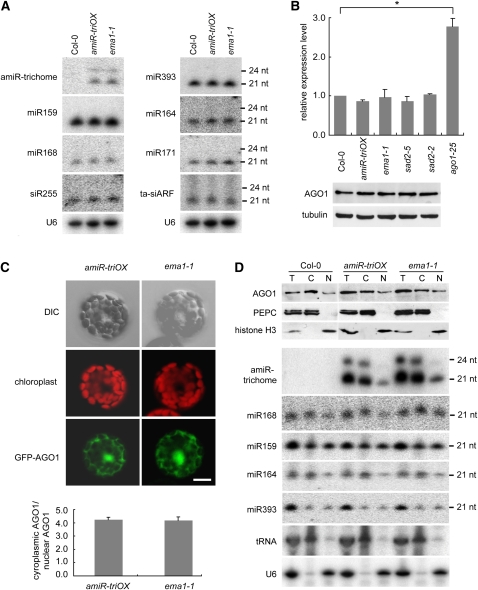
Next, we sought to investigate the mechanism through which SAD2/EMA1 regulates miRNA activity. It was possible that the enhanced miRNA activity in ema1-1 resulted from increased accumulation of miRNAs and/or their effector protein AGO1. Thus, we examined the accumulation of miRNAs in ema1-1 by small RNA gel blot. As shown in Figure 6A, the accumulation levels of amiR-trichome and all examined endogenous miRNAs were unchanged in ema1-1, suggesting that SAD2/EMA1 is not involved in miRNA biogenesis and metabolism. We also detected similar amounts of AGO1 in Col-0, amiR-triOX, and ema1-1, at both the mRNA and protein levels (Figure 6B). The same results were obtained for sad2-5 and sad2-2 (Figure 6B). These data indicate that SAD2/EMA1 modulates miRNA activity not through affecting the biogenesis or stability of miRNAs or AGO1.
Figure 6.
Accumulation and Nuclear and Cytoplasmic Distributions of AGO1 and miRNAs Are Not Perturbed in ema1-1.
(A) Small RNA gel blot analysis of miRNA accumulation in Col-0, amiR-triOX, and ema1-1. U6 was probed and used as a loading control.
(B) Detection of AGO1 mRNA and protein in the indicated plants. Top panel: relative AGO1 transcript level in the indicated plants, as measured by quantitative RT-PCR. Error bars indicate sd (n = 3), and the asterisk indicates a significant difference between the indicated samples (t test, P value < 0.05). Bottom panel: AGO1 protein levels in the indicated plants, as measured by immunoblots. Tubulin was also probed and served as the loading control.
(C) Transient expression of GFP-AGO1 under its native promoter in Arabidopsis wild-type (Col-0) and ema1-1 protoplasts (top panels). The fluorescence intensities of cytoplasmic and nuclear GFP-AGO1 in more than 50 protoplasts were quantified by Image J software, and the mean values are shown (bottom panel). Error bars indicate sd. Bar = 10 μm.
(D) Immunoblot and small RNA gel blot analyses showing the nuclear (N) and cytoplasmic (C) distributions of AGO1 protein and miRNAs in Col-0, amiR-triOX, and ema1-1. PEPC and tRNA were used as protein and RNA markers for the cytoplasmic fraction, and histone H3 and U6 were used as nuclear markers. Note that 2.5 times more protein was loaded for the nuclear extract (N) than for the total (T) and cytoplasmic (C) extracts for the immunoblots. nt, nucleotide.
Cytoplasmic and Nuclear Allocations of miRNAs and AGO1 Are Unaltered by Mutations in SAD2/EMA1
After their maturation in the nucleus, plant miRNAs are exported into the cytoplasm, where they interact with AGO1 to direct target mRNA cleavage (Voinnet, 2009; Xie et al., 2010). It has been shown that miRNAs and AGO1 are present in both the cytoplasm and the nucleus (Park et al., 2005; Fang and Spector, 2007). This raised the possibility that miRNA activity could be modulated at the step of allocating miRNA and AGO1 between the cytoplasm and nucleus. This hypothesis was particularly attractive considering that SAD2/EMA1 is an Importin β protein, whose homolog in humans (Importin 8 [Imp8]) interacts with Ago2 and is required for nuclear import of Ago2 (Weinmann et al., 2009). We thus investigated whether the cytoplasmic and nuclear distribution of miRNAs and AGO1 were altered in ema1-1.
Consistent with previous observations (Fang and Spector, 2007), GFP-AGO1 transiently expressed from either the 35S or AGO1 promoter localized in both the cytoplasm and the nucleus of wild-type Col-0 cells (Figure 6C; see Supplemental Figure 4 online). However, the localization pattern remained essentially unchanged in ema1-1 cells, suggesting that EMA1 has no effect on the cytoplasmic and nuclear distribution of AGO1. To confirm this result, we prepared the cytoplasmic and nuclear fractions of Col-0, amiR-triOX, and ema1-1 leaves and examined the AGO1 protein level in the fractions by immunoblot analysis. As a quality control of the fractionation procedure, a cytosolic protein (phosphoenolpyruvate carboxylase [PEPC]) and a nuclear protein (histone H3) were also probed by immunoblot. As shown in Figure 6D, PEPC was only detected in the cytoplasmic fractions, while histone H3 was only detected in the nuclear fractions, indicating that there was minimal contamination, if any, between the cytoplasmic and nuclear fractions. Consistent with the subcellular localization of GFP-AGO1 (Figure 6C), endogenous AGO1 was detected in both the cytoplasmic and nuclear fractions, and the ratio of AGO1 protein in the cytoplasm and that in the nucleus of ema1-1 was comparable to those of Col-0 and amiR-triOX.
Similarly, we prepared RNAs from the cytoplasmic and nuclear extracts and used RNA gel blot analysis to probe miRNAs, tRNA (a cytoplasmic RNA), and U6 (a nuclear RNA). As expected, the tRNA gave strong signals in the cytoplasmic fractions and was barely detectable in the nuclear fractions, whereas U6 was predominantly found in the nuclear fractions (Figure 6D). Consistent with published results (Park et al., 2005), all examined miRNAs could be detected in both the cytoplasmic and nuclear fractions (Figure 6D). The allocation of miRNAs between the cytoplasm and the nucleus in ema1-1 was similar to that in Col-0 and amiR-triOX (Figure 6D), indicating that SAD2/EMA1 does not play an obvious role in the cytoplasmic and nuclear allocation of miRNAs.
AGO1 Complexes in ema1-1 Contain a Larger Amount of miRNAs and ta-siRNAs and Have Increased Cleavage Activity
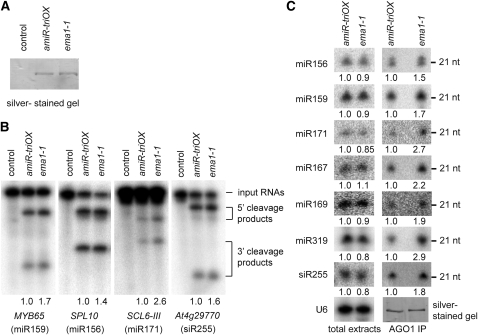
We next asked whether the enhanced miRNA activity in ema1-1 was attributed to increased enzymatic activity of RISCs. To address this question, we purified AGO1 complexes from amiR-triOX and ema1-1 plants by immunoprecipitation using an antibody that specifically recognizes AGO1 (Qi et al., 2005) and measured their cleavage activities by incubation with synthetic target mRNAs (MYB65, a target of miR159; SPL10, a target of miR156; SCL6-III, a target of miR171; and At4g29770, a target of the ta-siRNA, siR255). In agreement with the results that showed that AGO1 expression level was not altered in ema1-1 (Figure 6B), the same amount of AGO1 was purified from amiR-triOX and ema1-1 plants (Figure 7A). However, intriguingly, all three tested miRNA targets were more efficiently cleaved by the AGO1 complexes prepared from ema1-1, as shown by the in vitro cleavage assay (Figure 7B). Quantification of the signals from the cleavage products indicated that the cleavage activities of miRISCs in ema1-1 were elevated by 0.4- to 1.6-fold (Figure 7B). This indicates that the AGO1 complexes formed in ema1-1 have stronger cleavage activity for miRNA targets.
Figure 7.
miRISCs in ema1-1 Are Loaded with a Larger Amount of miRNAs and Have Increased Cleavage Activity.
(A) A silver-stained gel showing purified AGO1 complexes from amiR-triOX and ema1-1. No antibody was added in the control purification.
(B) miRISC activity assays using AGO1 complexes purified from amiR-triOX and ema1-1. The signals for the 3′ cleavage products were quantified and normalized to the input RNAs, and the relative values are shown below the gels.
(C) Detection of miRNAs and ta-siRNAs in total extracts or AGO1 complexes prepared from amiR-triOX and ema1-1. U6 RNA was probed and used as a loading control for total extracts. A silver-stained gel shows that equal amounts of AGO1 complexes were used for small RNA gel blot analysis. The miRNA signals were normalized to U6 RNA or AGO1 protein, and the relative values are shown below the gels. nt, nucleotide.
To this end, we reasoned that AGO1 complexes in ema1-1 could be loaded more efficiently with miRNAs and ta-siRNAs. Although the six examined miRNAs (miR156, miR159, miR171, miR167, miR169, and miR319) and siR255 were detected at similar levels in total extracts from amiR-triOX and ema1-1 plants (Figure 7C), AGO1 complexes purified from ema1-1 contained 0.5 to 1.9 times more miRNAs or ta-siRNAs than did those from amiR-triOX (Figure 7C).
Considered together, these data indicate that SAD2/EMA1 negatively regulates miRNA activity by influencing the loading of miRNAs into AGO1.
DISCUSSION
In the last decade, about a dozen genes have been shown to be required for miRNA biogenesis or functioning (Voinnet, 2009; Xie et al., 2010). Most of these genes were identified through their homology with known RNA interference components in animals (Park et al., 2002; Han et al., 2004; Vaucheret et al., 2004; Park et al., 2005), a candidate-gene approach (Ramachandran and Chen, 2008), mutants with developmental phenotypes resembling those of mutants deficient in miRNA biogenesis (Park et al., 2002; Lobbes et al., 2006; Yu et al., 2008) or with elevated levels of miRNA target genes (Smith et al., 2009), and forward genetic screening using a GFP sensor targeted by endogenous miRNA (Brodersen et al., 2008). In this study, we set up a forward genetic screen for mutants with altered miRNA activity, taking advantage of a transgenic line expressing an amiRNA that causes trichome clustering (Figure 1). This system has several unique features: (1) the visible phenotype allows high-throughput mutant screening, (2) the degree of trichome clustering is sensitive to the alteration of miRNA function, making it possible to isolate genes with a mild impact on the miRNA pathway (cma mutants), and (3) perhaps more advantageously, this screen allows the identification of suppressors of the miRNA pathway (ema mutants). Indeed, using this screen system, we have obtained dozens of cma or ema mutants (see Supplemental Table 1 online). Characterization of these mutants shall yield a more detailed picture of the plant miRNA pathway.
In this work, we identified EMA1, which encodes an Importin β protein. We found that SAD2/EMA1 functions as a negative regulator of the Arabidopsis miRNA pathway. This was in contrast with the function of its homolog in humans, Imp8, which is required for miRNA-guided gene silencing (Weinmann et al., 2009). Imp8 interacts with human Ago proteins, is involved in loading of Ago complexes onto mRNA targets, and is not involved in miRNA loading into Ago complexes (Weinmann et al., 2009). In this study, we show that AGO1 complexes in the ema1-1 mutant were loaded with increased amounts of miRNAs (Figure 7C), indicating that SAD2/EMA1 negatively regulates the loading of miRNAs into AGO1 complexes. Thus, the functions of Importin β proteins in the plant and human miRNA pathways are mechanistically different. It remains to be investigated whether such differences are in accordance with the different modes of miRNA action in plants and humans.
How does SAD2/EMA1 modulate miRNA loading into the AGO1 complex? We envision that SAD2/EMA1 might have a direct role in miRNA loading by regulating the accessibility and/or binding affinity of AGO1 to miRNAs. This would require a physical interaction between SAD2/EMA1 and AGO1. However, we were not able to detect the interaction between SAD2/EMA1 and AGO1 in the coimmunoprecipitation experiments (see Supplemental Figure 5 online). This suggests that the interaction is either transient or very weak, if SAD2/EMA1 interacts with AGO1. It is also possible that SAD2/EMA1 could sequester some amount of miRNAs to prevent their loading into the AGO1 complex. Little is known about how miRISCs are assembled in plants. Given that the majority of miRNAs accumulate in the cytoplasm (Park et al., 2005), it is widely believed that plant miRISCs are formed in the cytoplasm (Xie et al., 2010). SAD2/EMA1 could also modulate miRNA loading through affecting the cytoplasmic and nuclear allocations of some factors involved in miRNA loading. Genetic analyses have shown that SQN/CyP40 promotes the activity of AGO1, possibly through functioning with HSP90 (Smith et al., 2009). Using a cell-free system, it was recently found that HSP90 interacts with AGO1 and facilitates RISC loading (Iki et al., 2010). It will be of great interest to examine whether the cytoplasmic and nuclear distributions of these proteins are regulated by SAD2/EMA1. Besides its role in miRNA loading, we cannot exclude the possibility that SAD2/EMA1 can also negatively regulate the recruitment of miRISCs to their target mRNAs. Future investigation is warranted to clarify the precise actions of SAD2/EMA1 in miRISC loading and functioning.
We showed that SAD2/EMA1 had a moderate effect on the accumulation of miRNA targets. In SAD2/EMA1 mutants, the examined miRNA targets were decreased by less than twofold (Figure 2B). This was not unexpected, with the knowledge that very few miRNA target genes were significantly upregulated in mutants that are deficient in miRNA biogenesis or functioning (Lobbes et al., 2006; Ronemus et al., 2006; Laubinger et al., 2010). This could be attributed to a feedback regulation of target gene expression or a functional redundancy between SAD2/EMA1 and other Importin β–like proteins in Arabidopsis (Bollman et al., 2003), although we favor the idea that SAD2/EMA1 regulation of miRISC loading is a fine-tuning step of the miRNA pathway.
METHODS
Plant Materials, Growth and ABA Treatment
The transgenic Arabidopsis thaliana (Col-0 ecotype) line overexpressing amiR-trichome, designated here as amiR-triOX, was generated previously (Mi et al., 2008). The mutants sad2-5 and sad2-2 are T-DNA insertion lines described previously (Verslues et al., 2006; Zhao et al., 2007) and provided by Y. Guo. hyl1-2 was described previously (Vazquez et al., 2004), and ago1-25 is a hypomorphic allele (Morel et al., 2002). Both mutants were provided by H. Vaucheret. All plants were grown in a 16-h-light/8-h-dark photoperiod or in a10-h-light/14-h-dark photoperiod for protoplast isolation. For the ABA sensitivity assay, wild-type and mutant seeds were surface sterilized and plated on Murashige and Skoog medium with 0 or 0.25 μM ABA.
Mutagenesis and Screening
The amiR-triOX line was used as the parental line for mutagenesis. About 13,000 amiR-triOX seeds were treated with 0.4% ethyl methanesulfonate in 100 mM KPO4 buffer, pH 7.5, for 8 h at room temperature with gentle agitation, washed 20 times with water, and sowed in soil. M2 seeds were collected from 1400 pools of plants, and each pool contained nine plants. Approximately 500 seeds from each pool (~700,000 seeds in total) were grown in soil for mutant screening. Seedlings with a normal distribution of trichomes, less-clustered trichomes, or more-clustered trichomes were selected for further analysis. The obtained mutants were maintained by backcrossing to the amiR-triOX plants.
Map-Based Cloning of ema1-1
To map ema1, ema1-1 ami-triOX in Col-0 ecotype was crossed with Landsberg erecta. The F2 plants for positional cloning were selected with 25 μg/mL hygromycin B on plates. Crude mapping was performed on 30 F2 plants that had more-clustered trichomes, and fine mapping was performed on 909 F2 plants. The ema1-1 mutation was narrowed to the region between F16D14 and F20M17. The markers used to map ema1-1 were designed according to the Monsanto Arabidopsis polymorphism and Landsberg sequence collections (Jander et al., 2002), and the primers are listed in Supplemental Table 5 online.
Quantitative RT-PCR
Total RNA was extracted with the Trizol reagent (Invitrogen) from 18-d-old soil-grown plants. Total RNA was treated with RNase-free DNase I (Promega) to remove DNA and reverse-transcribed by M-MLV reverse transcriptase (Promega) using oligo(dT). Quantitative PCR was performed with SYBR Premix EX Taq (TAKARA). Actin mRNA was detected in parallel and used for data normalization. The primers used for PCR are listed in Supplemental Table 5 online.
Small RNA Gel Blotting
Small RNA gel blot analysis with enriched small RNAs from total extract or RNAs prepared from purified AGO1 complexes was performed as described (Qi et al., 2005). 32P-end–labeled oligonucleotides complementary to miRNA sequences were used as probes. The sequences of the probes are listed in Supplemental Table 5 online.
Bisulfite Sequencing
Genomic DNAs were isolated from 14-d-old seedlings grown on plates using the DNeasy plant mini kit (Qiagen). Bisulfite treatment and PCR amplification were performed as described (Wu et al., 2010). The PCR products were cloned into the pGEM-T easy vector (Promega), and individual clones were sequenced. Sequencing data were analyzed using Kismeth software (Gruntman et al., 2008). The primers used for bisulfite sequencing are listed in Supplemental Table 5 online.
Construction of Vectors Expressing AGO1 and EMA1
The AGO1 fragment in pEmycAGO1 (Mi et al., 2008) was moved into the destination vector pMDC43 (Curtis and Grossniklaus, 2003) by LR recombination to obtain a construct expressing AGO1 that is N-terminally fused to GFP under the 35S promoter. This construct was named 35S:GFP-AGO1. To generate the construct that expressed GFP-AGO1 under its native promoter, an ~1.6-kb DNA fragment containing the putative AGO1 promoter was obtained by PCR using AGO1Pro-F and AGO1Pro-R primers (see Supplemental Table 5 online). This fragment was inserted between the HindIII and KpnI sites to replace the 35S promoter, resulting in ProAGO1:GFP-AGO1. To construct the vector expressing 3xflag-tagged AGO1, the AGO1 coding sequence was amplified from pEmycAGO1 and inserted between the SalI and SpeI sites in a modified pCambia1307 vector with the 3xflag coding sequence. This vector was named 35S:3xflag-AGO1.
EMA1 was RT-PCR amplified using primers EMA1-F and EMA1-R and then cloned into pENTR/D-TOPO (Invitrogen) to generate pESAD2. Then, the EMA1 fragment was transferred from pESAD2 into the pEarleyGate203 vector (Earley et al., 2006) through LR recombination (Invitrogen), resulting in a construct that expressed N-terminally myc-tagged EMA1 under the 35S promoter. This vector was named 35S:myc-EMA1.
Protoplast Transformation
Protoplast preparation and transfection were performed as described (Yoo et al., 2007). GFP fluorescence was observed after 10 to 15 h of recovery using a Zeiss LSM 510 META confocal microscope.
Nuclear-Cytoplasmic Fractionation
Three-week-old plants (0.5 g) were harvested and ground to a fine powder in liquid nitrogen and mixed with 2 mL/g of lysis buffer (20 mM Tris-HCl, pH 7.5, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 250 mM Suc, and 5 mM DTT) supplemented with protease inhibitor cocktail (Roche). The homogenate was filtered through a double layer of Miracloth. The flow-through was spun at 1500g for 10 min, and the supernatant, consisting of the cytoplasmic fraction, was centrifuged at 10,000g for 10 min at 4°C and collected. The pellet was washed four times with 5 mL of nuclear resuspension buffer NRBT (20 mM Tris-HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100) and then resuspended with 500 μL of NRB2 (20 mM Tris-HCl, pH 7.5, 0.25 M Suc, 10 mM MgCl2, 0.5% Triton X-100, and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail (Roche) and carefully overlaid on top of 500 μL NRB3 (20 mM Tris-HCl, pH 7.5, 1.7 M Suc, 10 mM MgCl2, 0.5% Triton X-100, and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail (Roche). These were centrifuged at 16,000g for 45 min at 4°C. The final nuclear pellet was resuspended in 400 μL lysis buffer. As quality controls for the fractionation, PEPC protein and tRNA were detected and used as cytoplasmic markers, and histone H3 and U6 RNA were probed and used as nuclear markers.
Immunoblot Analysis
Total, cytoplasmic, and nuclear protein extracts were loaded on an 8% SDS-polyacrylamide gel for protein separation. After transfer to polyvinylidene fluoride fluoropolymer membranes, proteins were detected using antibodies against AGO1 (Qi et al., 2005), PEPC (Sigma-Aldrich), Histone H3 (Sigma-Aldrich), GFP (Roche), myc (Roche), and flag (Sigma-Aldrich).
Immunopurification
Immunopurification of the AGO1 complex was performed as previously described (Qi and Mi, 2010). The quality and quantity of purified AGO1 were examined by SDS-PAGE followed by silver staining. Immunoprecipitation of myc-EMA1 and flag-AGO1 was performed similarly using commercial antibodies.
miRISC Activity Assay
The miRISC activity assay was performed essentially as described (Qi et al., 2005; Qi and Mi, 2010) using in vitro–transcribed miRNA target transcripts. The PCR primers used for generating in vitro transcription templates are listed in Supplemental Table 5 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: SAD2/EMA1 (At2g31660), AGO1 (At1g48410), CPC (At2g46410), ETC2 (At2g30420), and TRY (At5g53200).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Characterization of se-7.
Supplemental Figure 2. Positional Cloning of ema1-1.
Supplemental Figure 3. Statistics of Trichomes on Leaf Blades of Col-0 and sad2-5.
Supplemental Figure 4. Localization of GFP-AGO1 in Col-0 and ema1-1 Cells.
Supplemental Figure 5. No Interaction Was Detected between SAD2/EMA1 and AGO1.
Supplemental Table 1. Summary of Isolated Mutants.
Supplemental Table 2. se Alleles.
Supplemental Table 3. DNA Methylation at CPC and ETC2.
Supplemental Table 4. ema1 Alleles.
Supplemental Table 5. Primers and Probes.
Acknowledgments
We thank M. Carmell for critical reading of the manuscript and Y. Guo for sad2 mutants and helpful discussions. We thank H. Vaucheret for hyl1-2 and ago1-25 mutants. We also thank Y. Zhang and his lab members for instructions on genetic screen and positional cloning. This work was supported by Transgenic Plant Breeding Program of China (2009ZX08009-026B and 2009ZX08009-009B) and National Basic Research Program of China (2011CB812601 and 2011CB100703). Y.Q. is supported by the Chinese Ministry of Science and Technology.
AUTHOR CONTRIBUTIONS
W.W., R.Y., and Y.Q. designed the research. W.W., R.Y., Y.X., X.F., C.L., and H.S. performed the research. W.W., R.Y., X.Z., and Y.Q. analyzed the data. Y.Q. wrote the article.
References
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman K.M., Aukerman M.J., Park M.Y., Hunter C., Berardini T.Z., Poethig R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Han M.H., Fedoroff N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K., Smith M., Weber R., Gregory B., Poethig R. (2010). An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Gong X., Cao W., Zhao J., Fu L., Wang X., Schumaker K.S., Guo Y. (2008). SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1 and GL2 expression. J. Integr. Plant Biol. 50: 906–917 [DOI] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. (2008). Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Goud S., Song L., Fedoroff N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. (2010). In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Zeller G., Henz S.R., Buechel S., Sachsenberg T., Wang J.W., Rätsch G., Weigel D. (2010). Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc. Natl. Acad. Sci. USA 107: 17466–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D., Rallapalli G., Schmidt D.D., Martin C., Clarke J. (2006). SERRATE: A new player on the plant microRNA scene. EMBO Rep. 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J.B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qi Y., Mi S. (2010). Purification of Arabidopsis argonaute complexes and associated small RNAs. Methods Mol. Biol. 592: 243–254 [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Chen X. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M., Vaughn M.W., Martienssen R.A. (2006). MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R., Willmann M.R., Wu G., Berardini T.Z., Möller B., Weijers D., Poethig R.S. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. USA 104: 5437–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A.C., Bartel D.P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crété P., Vaucheret H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Guo Y., Dong C.H., Ma W., Zhu J.K. (2006). Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. 47: 776–787 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Weinmann L., Höck J., Ivacevic T., Ohrt T., Mütze J., Schwille P., Kremmer E., Benes V., Urlaub H., Meister G. (2009). Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Kasschau K.D., Carrington J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Xie Z., Khanna K., Ruan S. (2010). Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21: 790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.F., Yuan L.J., Shao Y., Du W., Yan D.W., Lu Y.T. (2008). The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 31: 562–574 [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhang W., Zhao Y., Gong X., Guo L., Zhu G., Wang X., Gong Z., Schumaker K.S., Guo Y. (2007). SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]