CBSX1 affects lignin deposition by regulating the H2O2 level in anthers and by modulating plant growth via regulation of photosynthesis. CBSX1 activates thioredoxins (Trxs) and further enhances the enzymatic activity of Trxs in the presence of AMP. CBSX1 is a redox regulator modulating Trxs for development and maintaining homeostasis under conditions that are threatening to the cell.
Abstract
Plant thioredoxins (Trxs) participate in two redox systems found in different cellular compartments: the NADP-Trx system (NTS) in the cytosol and mitochondria and the ferredoxin-Trx system (FTS) in the chloroplast, where they function as redox regulators by regulating the activity of various target enzymes. The identities of the master regulators that maintain cellular homeostasis and modulate timed development through redox regulating systems have remained completely unknown. Here, we show that proteins consisting of a single cystathionine β-synthase (CBS) domain pair stabilize cellular redox homeostasis and modulate plant development via regulation of Trx systems by sensing changes in adenosine-containing ligands. We identified two CBS domain–containing proteins in Arabidopsis thaliana, CBSX1 and CBSX2, which are localized to the chloroplast, where they activate all four Trxs in the FTS. CBSX3 was found to regulate mitochondrial Trx members in the NTS. CBSX1 directly regulates Trxs and thereby controls H2O2 levels and regulates lignin polymerization in the anther endothecium. It also affects plant growth by regulating Calvin cycle enzymes, such as malate dehydrogenase, via homeostatic regulation of Trxs. Based on our findings, we suggest that the CBSX proteins (or a CBS pair) are ubiquitous redox regulators that regulate Trxs in the FTS and NTS to modulate development and maintain homeostasis under conditions that are threatening to the cell.
INTRODUCTION
The cystathionine β-synthase (CBS) domain–containing proteins (CDCPs) comprise a large superfamily of proteins that have evolutionarily conserved CBS domains, and they are ubiquitous in all three domains of life: archaea, bacteria, and eukaryotes. The CBS domain was first discovered in the genome of an archaebacterium, Methanococcus jannaschii, as a highly conserved domain of ~60 amino acids in a variety of proteins (Bateman, 1997). Computational genome analysis has subsequently revealed that this domain is also present in eubacterial and eukaryotic proteins with known specific functions (Ignoul and Eggermont, 2005). CBS domains usually occur in tandem repeats. Crystallographic studies have demonstrated that two CBS domains associate to form a CBS pair with a symmetrical structure that takes the form of two (β1)-α1-β2-β3-α2 units in an antiparallel arrangement (Ignoul and Eggermont, 2005). Functional analyses of various CBS domain–containing proteins have shown the physiological importance of this CBS pair by linking mutations in the CBS domain of enzymes and other proteins to a number of hereditary diseases in humans, including cystathionine-β-synthase in homocystinuria (Shan et al., 2001), inosine-5′-monophosphate dehydrogenase in retinitis pigmentosa (Kennan et al., 2002), AMP-activated protein kinase in familial hypertrophic cardiomyopathy (Blair et al., 2001; Gollob et al., 2001; Arad et al., 2002), and chloride channels in myotonia congenital (Pusch, 2002). Based on these studies, as well as structural analyses (Zhang et al., 1999; Mindell et al., 2001), it is now known that the two CBS domains forming a CBS pair bind adenosine-containing ligands, such as AMP, ATP, or S-adenosylmethionine, and that point mutations within this domain impair the function of those disease-related CDCPs.
In contrast with the other kingdoms of life, the role of CBS domains in plants is still obscure. Based on a whole-genome analysis of Arabidopsis thaliana and rice (Oryza sativa), Kushwaha et al. (2009) reported a total of 34 CDCPs (encoded by 33 genes) in Arabidopsis and 59 CDCPs (encoded by 37 genes) in rice. The authors classified these proteins into two major groups, namely, proteins containing only a single CBS pair and those with two CBS pairs, and then further classified them into subgroups based on additional structural domains. No defined function(s) has yet been determined for the CBS domain, but it has been suggested to have a role in the regulation of many enzymes, thereby contributing to the maintenance of the intracellular redox balance. Based on a comprehensive analysis of expression patterns using existing transcriptome profiles and the Massively Parallel Signature Sequencing (MPSS; http://mpss.udel.edu/at/mpss_index.php) database, Kushwaha et al. (2009) suggested that a few CDCPs may play an important role in stress response/tolerance and development in plants. However, the precise function of CBS domain(s) and CDCPs in plants still remains to be elucidated.
Six different types of thioredoxins (Trxs) have been identified in Arabidopsis: the f-, m-, x-, and y-types in the chloroplast, the o-type in mitochondria, and the h-type in the cytosol, mitochondria, endoplasmic reticulum, and extracellular space (Johnson et al., 1987; Marcus et al., 1991; Rivera-Madrid et al., 1995; Gelhaye et al., 2004; Juárez-Díaz et al., 2006). Trxs are members of two redox systems found in different cell compartments (i.e., the NADP-Trx system [NTS] in the cytosol and mitochondria and the ferredoxin-Trx system [FTS] in the chloroplast), where they function as redox regulators of various target enzymes. In the chloroplast FTS, light energy activates ferredoxin (Fdx), and the activated Fdx reduces oxidized Trxs in a reaction catalyzed by ferredoxin/thioredoxin reductase (FTR). Enzymes that function as an antioxidant, such as peroxiredoxin (Prx) and glutathione peroxidase, are directly regulated by Trxs (Buchanan et al., 2002). The thiol-peroxidases Prx and glutathione peroxidase reduce H2O2 via the thiol group and are essential for regulating the H2O2 level in the cell. After reducing H2O2, these now oxidized thiol-peroxidases need to be reactivated via reduction by Trxs (Meyer et al., 2008). Photosynthesis-related enzymes, such as those involved in five of the control points of the Calvin-Benson cycle, are also known to be regulated by Trxs (Montrichard et al., 2009). However, to date, there has been no report of regulators/effectors of the redox system being indispensable for maintaining cellular homeostasis and/or survival.
In most flowering plants, anthers are dehisced as the endothecial secondary wall is thickened to provide an outwardly bending force for opening the stomium (Bonner and Dickinson, 1989). The main component of secondary wall thickening in the anther is lignin, and the polymerization of lignin in the anther endothecium is associated with the level of H2O2 (Kawasaki et al., 2006). Villarreal et al. (2009) reported that the ectopic overexpression of carbonic anhydrase 2 (CA2) impairs the production of reactive oxygen species (ROS) by the mitochondrial respiratory chain, which in turn affects the H2O2-dependent polymerization of lignin that occurs during anther dehiscence (Villarreal et al., 2009). Although it is well known that the H2O2 level in the anther endothecium is a major determinant of anther dehiscence through its effect on lignin polymerization, information on the regulatory mechanism of H2O2 level in the anther is very limited.
None of the plant CDCPs has been functionally characterized. In this study, we determined the crystal structure at 1.9-Å resolution of homodimeric CBSX2 (the name coined by Kawasaki et al. [2006]), one of the six Arabidopsis CDCPs that contains only a single CBS pair, and compared it with the structure of CDCPs from different species. The structure of the plant protein showed a unique oligomeric assembly, although the folding pattern of each monomer unit is quite similar, possibly due to its function. Overexpression of CBSX1 activated Trxs and H2O2 scavengers, leading to lignin deficiency due to an insufficiency of ROS and thereby causing defective secondary wall thickening in the anther endothecium and, ultimately, sterility through anther indehiscence. CBSX1 and CBSX2 were found to interact with (as well as regulate) all types of Trx in the chloroplast. Moreover, the o-type Trx in mitochondria was also activated by a predicted mitochondrial CBSX member, CBSX3. Based on our findings and those of previous reports, we suggest that the CBSX proteins are ubiquitous redox regulators that control the enzymatic activity of Trxs in FTS and NTS and, as such, play a key role in development and homeostasis in plants.
RESULTS
CBSX1 Is Primarily Expressed in the Cotyledon and Anther
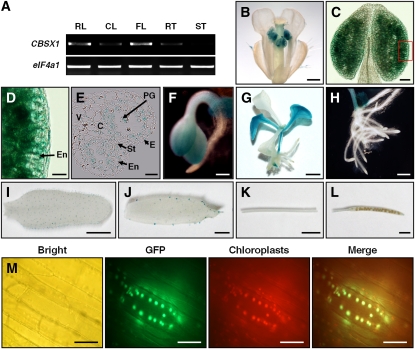
CBSX1 comprises seven introns and eight exons, with an open reading frame of 711 nucleotides that encodes 236 amino acids (see Supplemental Figure 1A online). To identify the spatial expression pattern of CBSX1, we constructed transgenic plants expressing the β-glucuronidase (GUS) reporter gene under the control of a 2-kb fragment of the putative CBSX1 promoter. We obtained a total of 57 T2 lines from proCBSX1:GUS transgenic plants that survived selection on kanamycin. Of these, six individual lines of transgenic plants were selected and examined further for their expression of GUS. ProCBSX1 drove strong GUS expression in both cotyledon and floral tissues, but expression was especially strong in the anthers (Figures 1B to 1G; see Supplemental Figures 2A to 2H online). Histochemical examination of transverse sections of the GUS-expressing anther revealed that CBSX1 was broadly expressed in most anther tissues, such as the epidermal cell, endothecium layer, tapetum, and pollen grain (Figure 1E), whereas in rosette and cauline leaves, GUS expression was detected only in trichomes (Figures 1I and 1J). This spatial expression pattern of the CBSX1 was verified by RT-PCR analysis (Figure 1A). To confirm our experimental results on spatial expression, we compared our data to data in the MPSS database (http://mpss.udel.edu/at/mpss_index.php) and Genevestigator (https://www.genevestigator.com/gv/index.jsp) subroutine. This comparison revealed that CBSX1 was dominantly expressed in the inflorescence (MPSS) and in the seedling and stamen (Genevestigator). These results are in good agreement with our spatial expression data using the GUS fusion constructs. Therefore, based on these spatial and developmental expression patterns, we focused on anther development to identify the function of CBSX1.
Figure 1.
Expression Patterns and Subcellular Localization of CBSX1.
(A) RT-PCR analysis of CBSX1 expression. CL, cauline leaf; FL, flower; RL, rosette leaf; RT, root; ST, stem.
(B) to (L) Histochemical localization of GUS activity in ProCBSX1:GUS transgenic plant. From the 57 T2 transgenic lines, six individual lines were selected and examined for their GUS expression. C, connective tissue; E, epidermis; En, endothecium; PG, pollen grain; St, stomium; V, vascular region.
(B) Inflorescence cluster. Bar = 500 μm.
(C) Close-up view of anther. Bar = 50 μm.
(D) Magnified image of small box in (C). Bar = 20 μm.
(E) Transverse section of anther. Bar = 20 μm.
(F) Cotyledon of GUS-tagged line. Bar = 200 μm.
(G) Seedling stage of GUS-tagged line. Bar = 0.25 cm.
(H) Root of GUS-tagged line. Bar = 500 μm.
(I) Rosette leaf of GUS-tagged line. Bar = 0.5 cm.
(J) Cauline leaf of GUS-tagged line. Bar = 0.2 cm.
(K) Stem of GUS-tagged line. Bar = 0.5 cm.
(L) Silique of GUS-tagged line. Bar = 0.2 cm.
(M) Subcellular localization of CBSX1:smGFP protein. CBSX1:smGFP localized to chloroplasts. Bright, bright-field image; GFP, smGFP fluorescence; Chloroplasts, autofluorescence of chloroplasts; Merge, merged smGFP and chloroplast autofluorescence. The hypocotyl epidermal tissues of Pro35S:CBSX1:smGFP transgenic Arabidopsis seedlings (T2 generation) were observed under a fluorescence microscope. Bars = 20 μm.
CBSX1 Is Localized in the Chloroplast
To determine the subcellular localization of CBSX1 protein, we fused CBSX1 with the soluble modified green fluorescent protein (smGFP) (David and Vierstra, 1996) gene under control of the 35S promoter and then transformed this construct into Arabidopsis. The GFP signal was detected in the chloroplast (Figure 1M; see Supplemental Figure 2I online), which was the expected result based on its gene ontology cellular component category in The Arabidopsis Information Resource (TAIR) database and the prediction by the ChloroP program (http://www.cbs.dtu.dk/services/ChloroP/). It is known that some proteins show tightly defined subcellular localizations, whereas others show low specificity in targeting and are characterized by complicated accumulation patterns (Millar et al., 2009). Therefore, to reconfirm the chloroplast localization of CBSX1 protein, we searched the Plant Proteome Database (http://ppdb.tc.cornell.edu/) and Subcellular Localization of Proteins in Arabidopsis database (http://suba.plantenergy.uwa.edu.au/) and found that chloroplast localization of CBSX1 had already been submitted to those databases.
Structural Features of CBSX Proteins
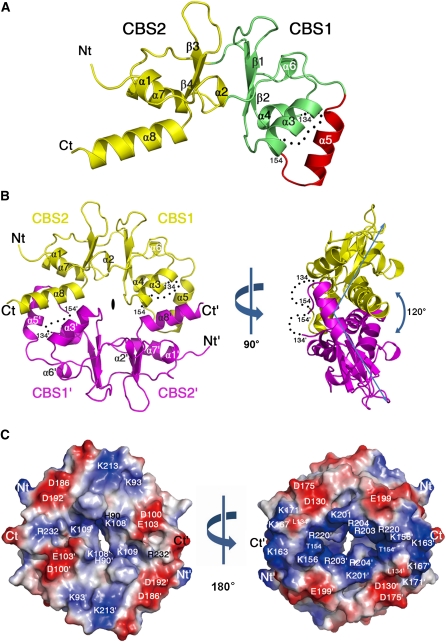
CBSX1 and CBSX2 have close sequence similarity (see Supplemental Figure 1B online). CBSX1 and CBSX2 cDNA sequences, excluding the sequence encoding the signal peptide, were cloned, overexpressed, and purified. The recombinant CBSX1 and CBSX2 formed a homodimer in solution during the purification step, as evidenced by size exclusion chromatography, which revealed a molecule that was double the size (44 kD) of the expected monomer (18.2 kD) of CBSX1 (see Supplemental Figure 3 online). Analysis of the crystal structure of CBSX2 also confirmed that it forms a homodimer consisting of two identical subunits (Figures 2A and 2B). Each subunit consists of two conserved CBS domains that form a pair facing each other; these are linked by two central β-strands (β1-β2 and β3-β4) and flanked by eight α-helices (Figure 2A). The second CBS domain in each subunit contained a characteristic additional α-helix (α5) followed by an invisible region (residues 135 to 153) in the CBSX2 structure. The presence of helix α5 and C-terminal helix α8 is the main characteristic of known dimeric CBS domain proteins, and both helices are critical for the dimer assembly of CBSX2 (Figure 2B).
Figure 2.
Three-Dimensional Structure of CBSX2.
(A) Ribbon diagram showing the monomeric subunit structure of CBSX2. The first CBS domain (CBS1), the second CBS domain (CBS2), and an additional α-helical segment (α5) are colored green, yellow, and red, respectively. The secondary structural elements are sequentially labeled, and invisible residues (from 135 to 153) are indicated as black dots. The N and C termini of CBSX2 are labeled Nt and Ct, respectively.
(B) The overall structure of dimeric CBSX2 viewed along the twofold molecular symmetry axis (left). The two subunits are colored yellow and magenta. Prime (') is added to all labels of one subunit for clarity. Right: 90° rotation along the vertical axis, as indicated.
(C) Electrostatic potential surface of CBSX2 as viewed in (B), left. Positive and negative electrostatic potential are shown in blue and red, respectively. Right: 180° rotation along the vertical axis, as indicated.
Depending on the orientation of the twofold axis, dimeric CBS domain proteins are structurally classified into two groups: parallel or antiparallel assemblies (see Supplemental Figure 4 online). The ribbon pattern of CBSX2 indicates that it is an antiparallel dimer, such that CBS1 interacts with CBS2' and CBS2 interacts with CBS1' on its central twofold axis. It is also characterized by a unique ~120° bend at the side of the molecule (Figure 2B). This feature contrasts with all other parallel and antiparallel CBS domain proteins, which are characterized by an ~180° flat structure at the side (Ragunathan et al., 2008; Tuominen et al., 2010). The orientation and molecular symmetry of CBSX2 may determine the interacting surface for its ligands, which affect its function. A positively charged pocket formed by each CBS pair subunit on the twofold molecular axis was also identified, and this may be a binding site for adenosine-containing ligands, such as AMP, ADP, ATP, NADP+, NADPH, or S-adenosylmethionine, as reported by Ignoul and Eggermont (2005) (Figure 2C).
CBSX1 Activates All Four Trxs in the Chloroplast and Further Augments Activity in the Presence of AMP
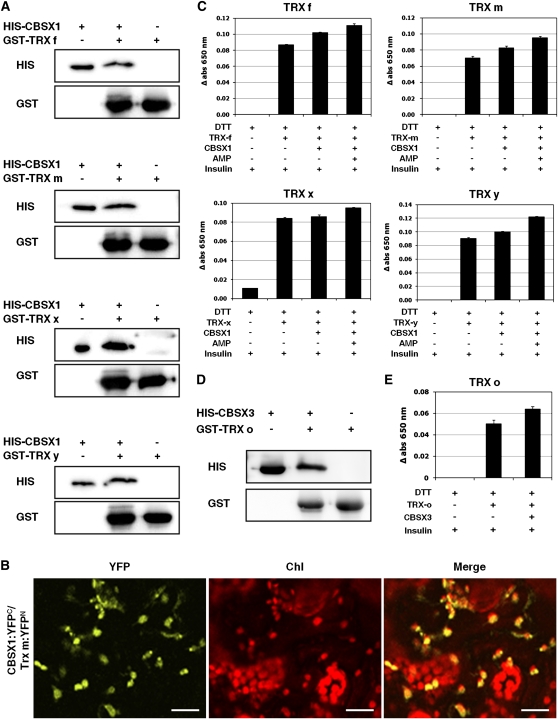
To characterize the precise function of CBSX1 in chloroplasts, we first identified interacting proteins by means of a yeast two-hybrid screen. Of the 96 positive clones sequenced, 29 clones were considered to be strong genuine interactors following confirmation by the binary test and the removal of redundant clones (see Supplemental Table 1 online). Of these 29 interactors, CBSX1 interacted with several chloroplast redox regulators, including Trx f, Trx m, Trx x, and Trx y. The results of our in vitro pull-down analysis using recombinant proteins confirmed that the CBSX1 protein interacts with Trx f, Trx m, Trx x, and Trx y proteins (Figure 3A). To confirm their interaction in living plant cells, we performed bimolecular fluorescence complementation (BiFC) analysis. CBSX1 was fused with sequences encoding the yellow fluorescent protein (YFP) N-terminal fragment and Trx m with sequences encoding the YFP C-terminal fragment (Hu et al., 2002; Walter et al., 2004). Both constructs were then introduced into tobacco leaves by Agrobacterium tumefaciens–mediated infiltration. YFP fluorescence was strongly detected in the chloroplast, thereby confirming the interaction of CBSX1 and Trx m in this plant organelle (Figure 3B).
Figure 3.
In Vitro Interaction of CBSX1 with Trxs and Modulation of Trx Activity by CBSX1 with or without AMP.
(A) Pull-down assay with chloroplast Trx f, m, x, and y proteins and CBSX1.
(B) Interactions of CBSX1 and Trx m in the chloroplast by the BiFC assay. Merge indicates an overlay of the YFP and chlorophyll autofluorescence image. Bars = 20 μm.
(C) Assay for the reducing activity of Trx f, m, x, and y using insulin protein and measured by absorbance at 650 nm. The error bars were calculated based on three independent experiments. The values are means ± sd (n = 3).
(D) Pull-down assay with mitochondrial Trx o protein and CBSX3.
(E) Assay for the reducing activity of Trx o using insulin protein and measured by absorbance at 650 nm. The error bars were calculated based on three independent experiments. The values are means ± sd (n = 3).
FTR reduces Trx enzymes that subsequently act as electron donors to Prx enzymes to eliminate H2O2 (Rouhier et al., 2008). Therefore, we hypothesized that CBSX1 positively regulates Trx to reduce target proteins, such as Prx, and consequently reduces H2O2 to water. To test this hypothesis, we measured changes in the activity of purified recombinant Trx f, m, x, and y proteins in vitro in the presence of chloroplast members of the CBSX family (i.e., CBSX1 or CBSX2) using insulin as a substrate. Insulin consists of two polypeptides held together by disulfide bonds that can be reduced in a Cys thiol-disulfide exchange reaction by Trx, releasing the individual polypeptides. In this reaction, a white precipitate of free insulin β-chain is formed when the reduction reaction initiates (Holmgren, 1979). The rate of precipitate formation was measured by spectrophotometer at 650 nm. As expected, both CBSX1 (Figure 3C) and CBSX2 (see Supplemental Figure 5C online) increased the activities of all four Trxs. The activity of mitochondrial Trx o also increased following the addition of a predicted mitochondrial member of the CBSX family, CBSX3 (Figures 3D and 3E). It is evident that CBSXs activate Trxs in vitro using the artificial reducer DTT (Figures 3C and 3E). To confirm the activation of Trx by CBSX1, we examined Trx activity in the presence of the natural reducer FTR (the cyanobacterium Synechocystis FTR, kindly provided by P. Schürmann) and NADPH. In this trial, CBSX1 was much more effective in increasing the activity of Trx m than DTT. Following the addition of CBSX1, Trx m activity was twofold higher. Moreover, the addition of AMP augmented Trx activity by ~25% compared with that following CBSX1 addition without AMP (see Supplemental Figure 5A online). However, Trx f, Trx x, and Trx y were not efficiently reduced by Synechocystis FTR. Taken together, these results suggest that CBSX1 functions as a redox regulator wherever it is located.
Because many CBS domain proteins bind adenosine-containing ligands (Ignoul and Eggermont, 2005), we tested the effect of adenosine-containing ligands on the activity of Trx in the presence of CBSX1. The ability of CBSX1 (Figure 3C) and CBSX2 (see Supplemental Figure 5C online) to increase the activity of all four Trxs was augmented by AMP binding, but ADP and ATP did not show this enhancing effect (see Supplemental Figure 5B online). Our structural analysis revealed that several basic residues from each subunit, namely, Lys-201, Arg-203, Arg-204, and Arg-220, form a ligand binding pocket (Figure 2C, right), and previous studies have demonstrated the presence of adenosine-containing ligands in similar pockets in the structures of several CBS domain proteins (Xiao et al., 2007; King et al., 2008; Tuominen et al., 2010). As noted, CBSX2 is an antiparallel dimer and thus most likely accommodates two adenosine-containing ligands simultaneously in one pocket (Figure 2C). This structure explains how two smaller AMPs can occupy this pocket, whereas the presence of additional β- and γ-phosphate groups in the bulkier ADP and ATP moieties, respectively, causes electrostatic repulsion in the pocket. This modulation of Trx activity by CBSX proteins in vitro led us to an exploration of their physiological role in in vivo experiments using the CBSX1 gene.
Overexpression of CBSX1 Reveals Severe Sterility Caused by Anther Indehiscence
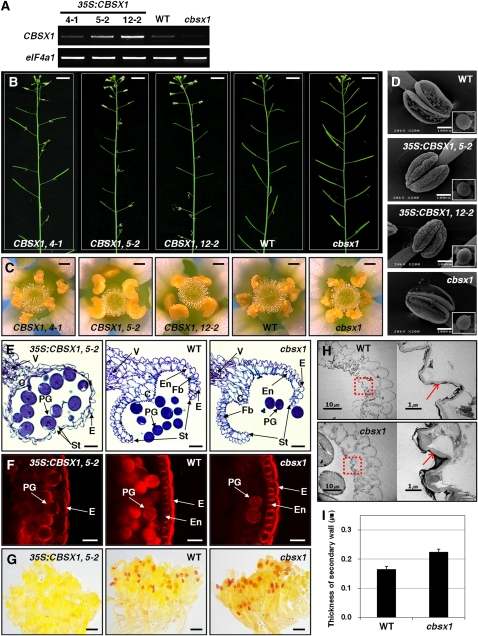
To investigate the physiological function of CBSX1, we obtained one T-DNA insertional mutant line from GABI-Kat (http://www.GABI-Kat.de) and constructed overexpression transgenic lines. The absence of an amplification product by RT-PCR using CBSX1-specific primers indicated its knockout in the cbsx1 line (Figure 4A). Fifteen different transgenic lines overexpressing CBSX1 were developed, of which three independent lines (4-1, 5-2, and 12-2) were chosen for further analysis (Figure 4A). Of these three lines, two (5-2 and 12-2) showed a higher expression of CBSX1 than the wild type, and the third (4-1) showed similar CBSX1 expression to the wild type.
Figure 4.
Phenotypic Analysis of 35S:CBSX1 Overexpressors and cbsx1 Mutant Plants.
(A) Relative amount of CBSX1 transcript in wild-type (WT), 35S:CBSX1, and cbsx1 plants.
(B) Male-sterility phenotype of 35S:CBSX1 plants. Bars = 1 cm.
(C) Anther indehiscence phenotype of 35S:CBSX1plants. Bars = 0.2 mm.
(D) Scanning electron microscopy image of anthers and pollen grains from open flowers. Bars = 100 μm.
(E) Transverse section of wild-type, 35S:CBSX1, and cbsx1 mutant anthers. C, connective tissue; E, epidermis; En, endothecium; Fb, fibrous bands; PG, pollen grain; St, stomium; V, vascular region. Bars = 20 μm.
(F) Histochemical staining of lignin components. Anthers were stained with EtBr and visualized with a fluorescence microscope. Abbreviations as in (E). Bars = 20 μm.
(G) Phloroglucinol staining of lignin components in a bud cluster. Bars = 0.2 cm.
(H) TEM image of secondary cell walls of wild-type and cbsx1 anthers. Right: red-dotted squares magnified ×10. Red arrows indicate the secondary wall of the endothecium. Bars = 10 μm in left panels and 1 μm in right panels.
(I) Secondary cell wall thickness of wild-type and cbsx1 anthers. Error bars indicate the sd of the average of 10 independent measurements.
The two 35S:CBSX1 lines (5-2 and 12-2) were severely sterile, as evidenced by their failure to produce seeds and their shorter siliques relative to the wild type (Figure 4B). Because the phenotype of the 4-1 line was not dissimilar to that of the wild type (Figures 4B and 4C), we used the 5-2 and 12-2 lines for further analyses. There was a relation between CBSX1 expression and sterility: the higher the expression of the transgene, the greater the severity of the sterility. Sterility was more severe in the 12-2 line than in the 5-2 line, and, although a few siliques did successfully develop in both lines, fewer than 10 seeds were ever set. In comparison, the cbsx1 and 4-1 lines showed normal fertility (Figure 4B), indicating that the observed sterility in the overexpressing transgenic lines was caused by the increased expression of CBSX1. Examination of the anthers of wild-type and transgenic plants under the dissecting microscope revealed that anthers of 35S:CBSX1 lines were not dehisced at flowering, whereas the other floral organs were very similar to those of the wild type (Figure 4C; see Supplemental Figure 6A online). Subsequent examination of anther indehiscence by scanning electron microscopy also revealed that anthers of the 35S:CBSX1 lines were indehiscent (Figure 4D) but that the presence of a very narrow crevice that formed in the stomium regions of these 35:CBSX1 anthers allowed these plants to set a very small amount of seeds in these lines (see Supplemental Figure 7A online). As shown in Figure 4D, the anthers of cbsx1 showed no phenotypic differences with the anthers of the wild type. The 35S:CBSX1 lines also had the typical characteristics of well-known fertilization defective mutants, such as increased numbers of flower buds in the inflorescence (see Supplemental Figures 6B and 6C online) and delayed apical senescence (see Supplemental Figure 6D online).
Sterility in plants is often caused by disorders of the male and/or female reproductive system. The scanning electron microscopy analysis was unable to distinguish any morphological differences between the pollens (insets in Figure 4D) and stigmas (see Supplemental Figure 7B online) of the wild type and transgenics, suggesting that any differences between the mutant lines and the wild type were functional ones. To test whether overexpression of CBSX1 caused the incompatibility, even though the phenotype was normal, we cross-pollinated wild-type pollen to the transgenic stigma and vice versa. In both cases, seeds were set successfully, leading us to conclude that the sterility of the transgenic plants was solely caused by anther indehiscence.
Failure of Secondary Wall Thickening in the Endothecium Causes Anther Indehiscence in 35S:CBSX1
To address the cause of anther indehiscence, we observed transverse sections of anthers obtained from 35S:CBSX1, wild-type, and cbsx1 plants at flower development stage 13 (Sanders et al., 1999). As shown in Figure 4E, there were no differences between wild-type and cbsx1 anthers. The wild-type and the cbsx1 pollen grains were released normally from the anther as a consequence of stomium cell degeneration through the shearing force of endothecium thickening (Figure 4E). By contrast, in 35S:CBSX1 anthers, although the tapetum, septum, and stomium cells degenerated normally, the anthers did not dehisce beyond flower stage 13; in addition, the secondary walls of the endothecial cell layer were not expanded (thickened) and the endothecium was entirely collapsed (Figure 4E). These results demonstrate that the failure of secondary wall thickening in the endothecium caused anther indehiscence in 35S:CBSX1 lines.
Deficient Deposition of Lignin Is the Primary Reason for the Failure of Secondary Wall Thickening in 35S:CBSX1
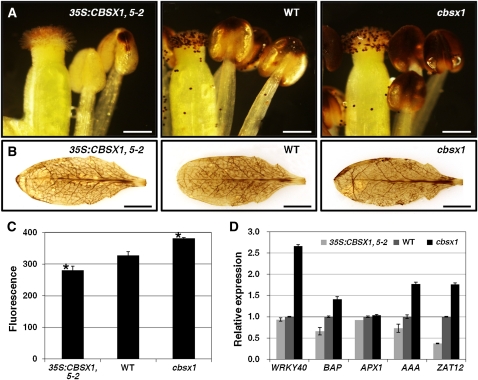
The plant secondary cell wall consists mainly of lignin polymers (Kawasaki et al., 2006). To test our conjecture that the observed failure of secondary wall thickening in the endothecium of 35S:CBSX1 plants was caused by the defective deposition of lignified material, we investigated the amount of lignified material in the secondary wall of ethidium bromide (EtBr)–stained anther endothecium by confocal microscopy. The lignin polymer in lignified cells is stained in red by EtBr. As shown in Figure 4F, the endothecium (spring-shaped) was clearly detectable in wild-type and cbsx1 anthers but was barely visible in 35S:CBSX1 anthers. The large reduction in lignin deposition in 35S:CBSX1 anthers compared with the wild-type and cbsx1 anthers was confirmed histochemically using phloroglucinol-HCl stain (Figure 4G), which targets hydroxycinnamaldehyde residues within the lignin polymer (Newman et al., 2004; Rogers et al., 2005).
There were no conspicuous phenotypic differences in cbsx1 plants, but there did appear to be much more lignin deposition in cbsx1 anthers than in wild-type anthers, as evidenced by the relatively thicker endothecium (based on EtBr staining; Figure 4F) and darker red coloration (phloroglucinol-HCl stain) of the former (Figure 4G). Fewer pollen grains were observed in the cbsx1 anthers than in the wild-type and 35S:CBSX1 anthers (Figure 4F), possibly due to the easier releasing of the pollen grains in the cbsx1 anthers as a result of the excessive anther dehiscence caused by the increased secondary wall thickening of the endothecium relative to that of the wild-type and the 35S:CBSX1 anthers at the same anther developmental stage. To investigate lignin deposition in cbsx1 anthers in more detail, we examined the ultrastructure of the endothecial layer by transmission electron microscopy (TEM). As expected, the secondary wall of the endothecium in the cbsx1 anther was much thicker than that of the wild type (Figures 4H and 4I). In the 35S:CBSX1 anther, the endothelial layer showed an almost collapsed phenotype and the secondary wall was scarcely detected (see Supplemental Figure 7C online). These results confirm that the failure of secondary wall thickening in the 35S:CBSX1 anther endothecium was caused by defective lignin deposition.
Insufficient H2O2 Caused a Lignin Deficiency in the Endothecium in the 35S:CBSX1 Anther, and CBSX1 Regulates the H2O2 Level in the Plant Cell
The indehiscent mutants myb26 and nst1/2 show a loss of secondary wall thickening in the anther endothecium (Yang et al., 2007) similar to that described here for the 35S:CBSX1 lines. The transcription factors MYB26, NST1, and NST2 have also been shown to regulate secondary wall biosynthesis-related genes, such as the cellulose-synthesizing complex genes IRXs (Yang et al., 2007). Therefore, the possibility that these transcription factors are directly regulated by CBSX1 was examined by analyzing their expression patterns in the wild-type, cbsx1, and 35S:CBSX1 plants by real-time PCR. The results demonstrated that MYB26, NST1, and NST2 were not directly regulated by CBSX1 expression level (see Supplemental Figure 8A online).
The chloroplast is the major organelle producing ROS, through the linear electron transfer process as well as by additional electron pathways, and H2O2 contributes to the structural reinforcement of plant cell walls by increasing lignin polymer formation (Boerjan et al., 2003; Ralph et al., 2004). This knowledge and our results led us to hypothesize that CBSX1 plays a role in regulating the chloroplast protein(s) that produces or scavenges H2O2 and/or alters the redox state in the chloroplast. To test this hypothesis, we analyzed H2O2 accumulation in anthers of wild-type, cbsx1, and 35S:CBSX1 plants by the 3,3′-diaminobenzidine (DAB) staining method, which visualizes H2O2 as dark-brown precipitates. Anthers of wild-type and cbsx1 plants showed a dark-brown precipitate, but the 35S:CBSX1 anthers remained unstained (Figure 5A). The darker coloration of the brown spots in cbsx1 anthers in comparison to those of the wild-type anthers was consistent with the histochemical results of lignin staining (Figures 4F and 4G). There was a similar pattern of H2O2 accumulation in rosette leaves, but the H2O2 deficit in 35S:CBSX1 rosette leaves was much less than that in the anthers (Figure 5B).
Figure 5.
Detection of H2O2 and RT-PCR Analysis of ROS-Regulated Genes in 35S:CBSX1 Overexpressors, the Wild Type, and cbsx1 Mutant Plants.
(A) DAB staining of flowers. Bars = 0.5 mm. WT, wild type.
(B) DAB staining of rosette leaves. Bars = 0.5 cm.
(C) Measurement of endogenous H2O2. Data from three separate experiments are shown (mean ± sd; n = 5; statistical significances were determined using one-way analysis of variance followed by a Dunnett test). Asterisk indicates significant difference from the wild type at P < 0.001.
(D) Analysis of ROS-regulated gene expression in 35S:CBSX1, wild-type, and cbsx1 plants. The error bars were calculated based on three independent experiments. The values are means ± sd (n = 3).
[See online article for color version of this figure.]
To confirm the difference in H2O2 accumulation in these leaves, we measured the endogenous concentration of H2O2. As shown in Figure 5C, cbsx1 plants showed the highest level of H2O2 content, but 35S:CBSX1 plants showed absolutely the lowest level. Quantitative RT-PCR was then performed to test whether these differences in H2O2 content affected the expression of several H2O2-regulated marker genes (Gadjev et al., 2006; Adhikari et al., 2011), such as WRKY40, BAP, APX, AAA-type ATPase, and ZAT12. As expected, the expressions of these marker genes were well matched to our experimental H2O2 content measurements (Figure 5D). These results indicate that the defective lignification process in the anther endothecium is caused by insufficient H2O2 and that CBSX1 regulates the level of H2O2 in the plant cell.
Overexpression of CBSX1 Promotes Plant Growth by First Activating Trxs and Then Regulating Their Many Target Proteins
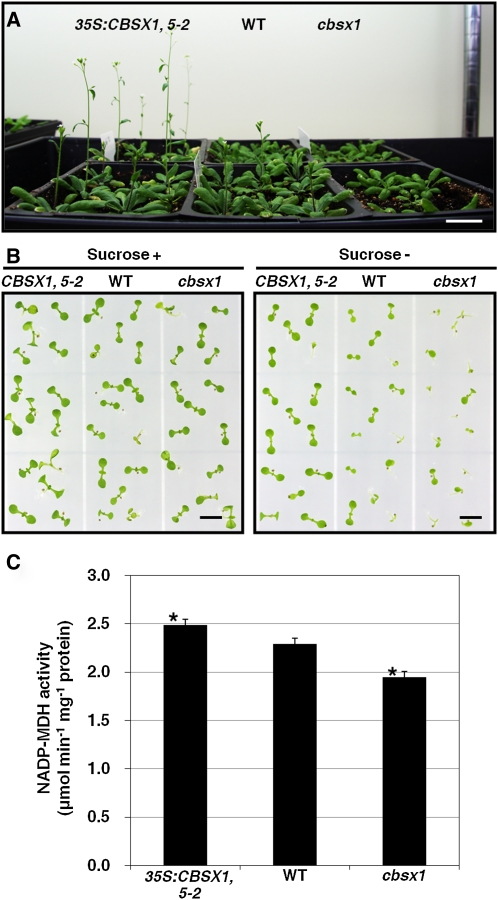
Trxs function as regulators of essential photosynthesis-related enzymes (Montrichard et al., 2009). Therefore, we hypothesized that alterations of CBSX1 expression affecting Trxs and would also influence the photosynthesis-related enzymes regulated by Trxs. To obtain experimental evidence in support of this hypothesis, we investigated the phenotypic differences among wild-type, cbsx1, and 35S:CBSX1 plants. As shown in Figure 6A, the cbsx1 mutant showed slight growth retardation, while 35S:CBSX1 plants grew faster than wild-type plants, under a normal long-day condition. In the early seedling stage, the 35S:CBSX1 plants grew normally on Murashige and Skoog medium lacking Suc, a growth condition that severely affects the early growth of most photosynthesis-related gene mutants. By contrast, the wild-type plants showed slight growth retardation on the Suc-free medium and the cbsx1 plants showed severe growth retardation (Figure 6B). These results clearly demonstrate that CBSX1 directly regulates the Trxs and thereby regulates many of the target proteins of Trxs, including the Calvin cycle enzymes.
Figure 6.
Regulation of Plant Growth by CBSX1.
(A) Morphological comparison of 35S:CBSX1, wild-type (WT), and cbsx1 plants under normal growth conditions. Bar = 2 cm.
(B) Early-stage growth of 35S:CBSX1, wild-type, and cbsx1 plants on medium with or without Suc. Bars = 0.5 cm.
(C) Leaf NADP-MDH activity in 35S:CBSX1, wild-type, and cbsx1 plants. The NADP-MDH activity was measured by monitoring NADPH oxidation to NADP+ at 340 nm; values represent means ± sd (n = 3). Three independent experiments were performed for MDH activity and the one-way analysis of variance with a Dunnett test was used for statistical analysis (asterisk indicates significant difference from the wild type at P < 0.05).
[See online article for color version of this figure.]
Therefore, to confirm the effect of CBSX1 on the Calvin cycle, we measured malate dehydrogenase (MDH) activity in total protein extracts obtained from the wild-type, cbsx1, and 35S:CBSX1 plants. The 35S:CBSX1 plants were found to have the strongest MDH activity and the cbsx1 the weakest (Figure 6C). Based on these results, we suggest that CBSX1 affects photosynthesis-related enzymes through the Trx system.
DISCUSSION
CBSX1 Affects Anther Dehiscence by Regulating the H2O2 Level in the Anther Endothecium through Redox Regulation
Research has identified two major classes of plant male sterility that are due to defective anther dehiscence, which is indispensable for the release of functional pollen grains. One class is due to delayed dehiscence by defects in jasmonic acid biosynthesis/signaling genes, such as coi1 (Feys et al., 1994), dde1/opr3 (Sanders et al., 2000; Stintzi and Browse, 2000), dad1 (Ishiguro et al., 2001), and dde2-2/aos (Park et al., 2002; von Malek et al., 2002). The second is indehiscence by insufficient secondary wall thickening in the anther endothecium. The indehiscence mutants ms35/myb26 (Dawson et al., 1999; Steiner-Lange et al., 2003; Yang et al., 2007) and nst1/nst2 (Mitsuda et al., 2005) are defective in lignified secondary thickening in endothecial cells of the anther wall. Lignin, which is a polymer of three monolignol mononers (p-coumaryl, coniferyl, and sinapyl alcohols) incorporated as phenylpropanoids, is the major component of the secondary wall. It is polymerized by an oxidative coupling between a monolignol molecule and the growing polymer by oxidative enzymes, such as peroxidases that use H2O2 as a substrate (Boerjan et al., 2003; Ralph et al., 2004). Karlsson et al. (2005) reported that H2O2 is required for the lignification of the secondary wall of the tracheary element and that the addition of H2O2 scavengers (ascorbic acid, catalase, and reduced glutathione) decreases the amount of lignin (Karlsson et al., 2005). More recently, Villarreal et al. (2009) reported that the ectopic overexpression of CA2 drastically decreases ROS content and that this reduction possibly prevents H2O2-dependent lignin polymerization in the secondary wall of the anther endothecium (Villarreal et al., 2009). Our 35S:CBSX1 plants had an indehiscent phenotype that was very similar to that of the CA2 overexpression mutant, indicating the involvement of CBSX1 in H2O2-dependent lignin polymerization.
Our study of 35S:CBSX1 plants revealed that the severe sterility of these plants was due to anther indehiscence that resulted from the failure of secondary wall thickening in the anther endothecium caused by defective lignin deposition, which in turn was caused by a decreased H2O2 level (Figures 4 and 5). We also observed anther-dominant expression and chloroplast localization of CBSX1 (Figure 1; see Supplemental Figures 2A to 2I online). We therefore suggest that CBSX1 affects lignin deposition by regulating the level of H2O2 in the anther endothecium. This hypothesis is supported both by the results of the yeast two-hybrid, pull-down, and BiFC experiments, which showed that CBSX1 interacted with all of the Trxs (f, m, x, and y) (Figures 3A and 3B), and by assays showing that the activities of all Trxs increased following the addition of CBSX1 (Figure 3C). These results provide very important clues toward defining the role of CBSX1 in the regulation of the redox system in the anther endothecium with respect to proper dehiscence. H2O2 contributes to the structural reinforcement of plant cell walls by increasing lignin polymerization through the action of peroxidase (Boerjan et al., 2003; Ralph et al., 2004), and H2O2 is reduced by Prxs following their activation by Trxs (Montrichard et al., 2009).
We therefore suggest that CBSX1 directly activates all four chloroplast Trxs and that overexpression of CBSX1 scavenges the H2O2 that is essential for lignin polymerization in the anther endothecium, resulting in severe sterility due to defective secondary wall thickening. Under normal conditions, anthers produce very high amounts of ROS because they are metabolically active due to their vigorous development. As such, CBSX1 needs to be highly expressed to regulate the level of ROS and trigger programmed cell death for the correct and timely development of anther. However, in the endothecium, lignification is inevitably needed for dehiscence to occur during the final stages of anther development, necessitating an increment in ROS content in the endothecium. Anther dehiscence is a series of senescence processes, and an ROS burst occurs during the senescence process. Accordingly, the amount of CBSX1 may not be sufficient to deal with this ROS burst and subsequently allow adequate lignification for anther dehiscence. Alternatively, the expression of CBSX1 may be decreased in the endothecium to allow for the ROS burst and lignification for dehiscence. This inference process emphasizes the importance of accurate redox regulation by CBSX1 in the chloroplast for timely anther development.
The Targets of Trxs Are Regulated by the CBSX1-Induced Activation of Trxs
The FTS in chloroplasts is activated by light energy that is transferred to Trxs via photosynthetic electron flow through thiol-disulfide exchange intermediates according to the following sequence: light→Fdx→FTR→Trxs→target proteins. Therefore, under dark conditions, the regulatory sulfhydryl groups of the enzymes in this system become oxidized; as such, the chloroplast is more oxidative under dark conditions than under light conditions. Antioxidant enzymes (e.g., Prx) and photosynthesis-related enzymes, such as FBPase, NADP-MDH, PRK, SBPase, GAPDH, ATP synthase, and ribulose-1,5-bis-phosphate carboxylase/oxygenase activase, are known to be activated by reduction and inactivated by oxidation. The reduction of these enzymes is subject to regulation by reduced Trx. Trx also functions as an effector of each of the five control points in the Calvin cycle (Marri et al., 2009; Montrichard et al., 2009). That is to say, Trx enhances CO2 fixation and carbohydrate synthesis under light (reducing) conditions. Therefore, any interception of light would directly inhibit the reduction of photosynthetic enzymes by Trxs, thereby reducing the photosynthetic rate and resulting in growth retardation of normal wild-type plants. During the night, Trxs are inactivated and the Calvin cycle is blocked, and the oxidative pentose phosphate pathway provides the ATP and NADPH necessary for basal metabolism. However, the function of Trxs is not devoted to the regulation of day/night alteration. Therefore, mutant plants with the capacity to keep Trxs in the reduced active state will have an increased capacity to activate the photosynthetic enzymes and grow faster than wild-type plants under normal growth conditions because photosynthetic reactions will then be able to occur even under dark conditions. These plants would also grow well on Suc-free media because carbohydrate synthesis would be able to occur even under a restricted supply of the appropriate carbon source.
As expected, the 35S:CBSX1 plants grew more rapidly than the wild-type plants under normal long-day conditions and suffered less growth retardation on Suc-free media (Figures 6A and 6B). The cbsx1 knockout mutant showed more severe growth retardation than the wild-type plants in the seedling stage on Suc-free medium (Figure 6B). In addition, transgenic plants that overexpressed CBSX1 showed a delayed senescence that resembled transgenics overexpressing Trx m (Benitez-Alfonso et al., 2009) (see Supplemental Figure 6D online). These results provide additional evidence that the ectopic overexpression of CBSX1 decreases the level of ROS in the whole-plant system. The finding that 35S:CBSX1 plants had the highest MDH activity (Figure 6C) indicates that CBSX1 can regulate Calvin cycle enzymes via the regulation of Trxs. Slight growth retardation of the cbsx1 mutant under normal growth conditions also supports these MDH results. We therefore suggest that CBSX1 directly controls the Trxs; as such, the targets of Trxs, including Prxs and Calvin cycle enzymes, are regulated through this series of reduction and oxidation processes. However, in terms of H2O2-dependent lignification in most plant tissues, there were no differences between the cbsx1 knockout, wild-type, and 35S:CBSX1 plants, with the only exception being the anther endothecium. We suggest that this lack of any distinct difference is due to only minor decreases in the H2O2 level taking place in the tissues, with the exception of the anther and/or functional redundancy of the six different types of each of the CBSXs and Trxs, even though we still consider that they individually may play different roles in different types of tissues and during different developmental stages.
A CBS Domain–Containing Protein with a Single CBS Pair Forms a Homodimer and Functions as a Master Redox Effector
Protein domains are highly conserved across species and time that have shared functional and structural characteristics. It is commonly inferred that a specific protein domain performs a specific function. However, CBS domains are found in proteins with completely different functions and exert different effects on these respective proteins. The significance of the CBS domain has been identified in studies of human diseases, and a number of hereditary diseases are now known to be caused by point mutations in the CBS domain (Blair et al., 2001; Shan et al., 2001; Kennan et al., 2002; Pusch, 2002). Studies of the crystal structures of CDCPs show that most CBS pairs are at the periphery of the protein complex and fall outside the catalytic center of these proteins. Despite not being part of the catalytic centers, however, mutations in CBS domains abolish the activity of proteins by impairing the binding affinity of metabolites. It is therefore generally considered that CBS domains play a regulatory role for many enzymes and function as sensors of intracellular metabolite ligands, even though their specific functions have not yet been identified.
One of the best approaches to unraveling the precise function of a specific domain of proteins is to identify a protein for which most of the sequences are encoded by only that domain (without any other functional domains or protein portions) and attempt to elucidate its function in a cell. We were able to identify the CBSX proteins and therefore attempted to characterize their function. Based on the TAIR classification and PSORT prediction, CBSX1 and CBSX2 were localized to the chloroplast, CBSX3 to the mitochondria, CBSX4 to the cytosol, and CBSX5 and CBSX6 to the endoplasmic reticulum. The chloroplast CBSX proteins were shown to interact with all four chloroplast Trxs (f, m, x, and y) and with the redox regulators in the chloroplast FTS (Figures 3A and 3B) and to increase their activities (Figure 3C).
We also were able to demonstrate that the CBSX1 protein formed a homodimer (see Supplemental Figure 3 online) and that the effect of CBSX1 on increasing the activity of the various Trxs could be further elevated with the addition of AMP, but not ADP and ATP (Figure 3C; see Supplemental Figure 5B online). Another chloroplast CBSX member, CBSX2, also formed an antiparallel homodimer (Figure 2B) and increased the activities of Trxs following the addition of adenosine-containing ligands (see Supplemental Figure 5C online). A third CBSX member, CBSX3, which was predicted to be localized in the mitochondria, was shown to interact with and activate mitochondrial Trx o (Figures 3D and 3E). Overall, these results suggest that CBSXs are ubiquitous redox effectors that play a role in redox regulation of protein function through regulating the activities of Trxs.
Based on their analysis of microarray data, Kushwaha et al. (2009) reported that CBSXs are differentially expressed under various stress conditions, such as cold, UV, wounding, drought, salt, osmotic stress, and oxidative stress, and that the expression of CBSXs exhibits tissue and developmental stage specificity. Our observations that CBSX1 expression is also induced by treatment with stress-related hormones and various stress, with the exception of cold stress (see Supplemental Figures 8B and 8C online), and that it manifests an anther-dominant pattern (Figures 1A to 1E) led to our hypothesis that CBSXs not only participates in cell homeostasis surveillance and play a defensive role against various threats to the cell by monitoring the changes of redox state or metabolites, but also participate in timed development. In other words, biotic and abiotic stimuli, especially those that evoke bursts in ROS production in the cell, disturb cellular metabolism and also initiate apoptotic cell death processes. These potentially detrimental events induce the expression of CBSXs and may change the proportion of metabolites in subcellular organelles. For example, it is evident that plastid ATP is consumed and that the concentration of ADP increases under such stressful conditions. These increases in ADP catalyze a reaction that converts ADP to ATP and AMP via adenylate kinase (EC 2.7.4.3) (Roberts et al., 1997). These variations in adenosine-containing ligands are sensed by CBSXs, which have already formed a functional homodimer, and activated Trxs, which bind to a specific adenosine-containing ligand, regulating the FTS in the chloroplast and the NTS in the mitochondria and cytosol through direct regulation of the Trxs in the respective systems, thereby maintaining cellular homeostasis. CBSXs also have roles in the regulation of the development of specific tissues, such as the mediation by CBSX1 of anther dehiscence through H2O2-dependent lignin polymerization in the endothecium secondary wall. Although we were unable to characterize all six CBSXs, it appears that those CDCPs having only a single CBS pair without any other domains are evolutionally conserved to help the cell adapt to a critical situation and to regulate the timed development that enables survival.
We suggest that CBSXs are involved in the redox system but that each may have its own specific regulatory role. Our results indicate that CBSX1 and CBSX2 activate all Trxs in the FTS and that this activation is increased by binding with AMP in the chloroplast, thereby enabling these molecules to exert a regulatory effect on both the Calvin cycle and H2O2 levels (Figure 7). However, their role in the cell appears to be slightly different since their spatial expression and amino acid sequence differed from each other. CBSX1 was mainly expressed in nongreen tissues, such as anthers and hypocotyls (Figures 1B to 1H). Another chloroplast CBSX, CBSX2, was found to be highly expressed in green tissues, such as rosette and cauline leaves (data not shown). The CBS domains are highly conserved, but the other regions of each protein are quite different (see Supplemental Figure 1B online). These spatial expression and amino acid variations may result in CBSX1 and CBSX2 differentially interacting with their respective downstream partners. This conjecture is well supported by two earlier reports on the spatial expression of Trxs. In the Arabidopsis genome, nine plastidial TRXs have been found and classified these into four subgroups (two f, four m, one x, and two y). These Trxs are known to be expressed differently in green and nongreen tissues, with Trx y1 found in nonphotosynthetic organs, such as roots and seeds, while Trx y2 appears to be expressed in green tissues, especially in leaves (Collin et al., 2004). de Dios Barajas-López et al. (2007) reported that Trx m and Trx f are present together with their corresponding reductase in nongreen tissues, such as roots and flowers. Based on our results and these two reports, we suggest that CBSX1 mainly functions in the development and responses of nongreen tissues and that CBSX2 performs essentially the same functions in green tissues, together with their relevant downstream partner Trxs, which are expressed in the same tissues. However, to a certain extent, it is possible that CBSX1 and CBSX2 are functionally complementary to each other because their spatial expression slightly overlaps and their biochemical functions are basically the same. In a comparable process, CBSX3 also activates mitochondrial Trx o in the NTS (Figure 3E). The other members of the CBSX family, CBSX4, CBSX5, and CBSX6, may also have a similar role as effectors in other subcellular organelles, and this effect may be enhanced by the ability to recognize specific adenosine-containing ligands.
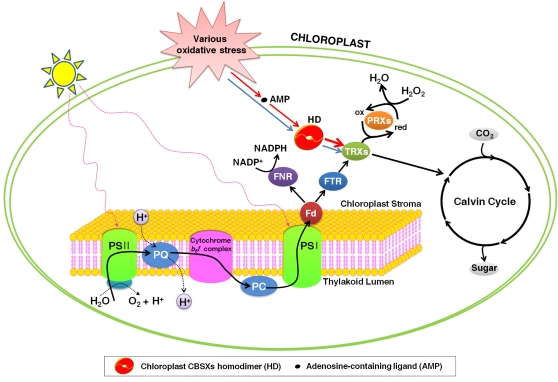
Figure 7.
Schematic Diagram of Chloroplast CBSX Function.
Under various oxidative stress conditions, CBSX1 is overexpressed and forms a functional homodimer, which then activates Trxs in the FTS. These stresses result in changes in the concentration of various adenosine-containing ligands, which are sensed by CBSX1 in the chloroplast. The CBSX1 functional homodimer is augmented by binding to a specific adenosine-containing ligand, resulting in an enhancement of the activation of all Trxs in the FTS. As a result, the cell maintains homeostasis through regulation of the Calvin cycle and H2O2 levels. Fd, ferredoxin; FNR, ferredoxin-NADP+ reductase; ox, oxidized; PC, plastocyanin; PQ, plastoquinone; PSI, photosystem I; PSII, photosystem II; red, reduced.
In conclusion, the significance of this research is that it provides evidence that CBS domains are evolutionarily conserved units that, by means of regulating the redox system, regulate the timely development and also enable the cell to cope with life-threatening conditions. Oxidation reduction is one of the most important factors in the regulation of enzyme activity, and redox regulation by Trx embraces virtually all life processes, such as carbon assimilation, seed germination, transcription, translation, cell division, redox signaling, radical scavenging, and detoxification. Therefore, our initial finding that proteins consisting of only a single CBS pair (without any other protein domains) are able to activate Trxs in both the NTS and FTS redox systems increases our understanding of signaling networks that include metabolite ligands and redox systems, both of which are indispensable for the development and maintenance of cellular homeostasis.
METHODS
Plant Materials
Arabidopsis thaliana (ecotype Columbia-0) plants, 35S:CBSX1 lines, and the T-DNA insertion mutant (cbsx1, AL753455) were grown on a Sunshine mix #5 (Sun Gro) at 22 to 24°C and 50 to 60% relative humidity under long-day conditions (16/8-h light/dark at 120 μmol m−2 s−1) in a controlled environment chamber (Conviron).
RNA Isolation and RT-PCR Analysis
Total RNA was extracted using TRIzol reagent (Gibco BRL). A 5-μg sample of total RNA was reverse transcribed using the Superscript III reverse transcriptase (Invitrogen). For the RT-PCR analysis, the PCR conditions consisted of 26 to 34 cycles of 95°C for 20 s, 54°C for 20 s, and 72°C for 40 s, followed by a 5-min final extension at 72°C. Real-time quantitative PCR analysis was performed in three independently harvested biological replicates on the Light Cycler 480 II real-time PCR system (Roche). Each cDNA sample was amplified in triplicate for CBSX1 and eIF4a1, with the SYBR GreenI as a florescent reporter. The eIF4a1 gene was used as the internal control. The relative abundance of transcripts was measured by PCR efficiency and the threshold cycle (Ct) values were measured according to the geNorm manual (Vandesompele et al., 2002). Coefficient of variation (Cv) was calculated by following formula: Cv = 100 × (sd of Ct/average of Ct).
Construction of Transgenic Lines and Mutant Plants
All binary plasmid constructs were introduced into Arabidopsis plants using Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog (1962) medium containing 50 mg/mL kanamycin and screened for the presence of the transgene by PCR. Homozygous 35S:CBSX1 lines were selected in the T3 generation and used in this study. T-DNA insertion mutant (cbsx1, AL753455) was identified from the GABI-Kat T-DNA Arabidopsis population.
Histochemical and Subcellular Localization Analysis
For GUS detection, plant tissues were stained in a solution of 1 mM 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide cyclohexylammonium salt, 100 mM sodium phosphate buffer, pH 7.0, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 10 mM EDTA, and 0.1% (v/v) Triton X-100. After GUS staining, chlorophyll was removed using 100% ethanol. For detecting lignified cells, we used an EtBr stain (Yang et al., 2007). Fresh tissues were washed (with 1× PBS and 2% [v/v] Tween 20 for 10 min, and then briefly with 1× PBS) and stained with 0.5 ppm EtBr (1 h, room temperature). The tissues were observed with a confocal laser scanning microscope (LSM510 Meta; Carl Zeiss). For detecting lignin deposition, the samples were stained with 2% (w/v) phloroglucinol in 95% ethanol for 2 min and then washed in 9 N HCl for 1 min and mounted in 5 N HCl. The tissues were observed under a light microscope (Olympus model BX-51). For H2O2 detection, the samples were incubated with DAB (Sigma-Aldrich) at room temperature for 2 h. Once brown spots were clearly evident, the leaves were bleached by immersing in 95% ethanol to visualize the spots. The endogenous concentration of H2O2 was measured using the Amplex Red H2O2/peroxidase assay kit (Molecular Probe) as described previously (Shin and Schachtman, 2004). Fluorescence was measured with a fluorescence microplate reader at an excitation of 535 nm and fluorescence emission detection at 590 nm. Transgenic Arabidopsis plants expressing the CBSX1:smGFP protein (pSMGFP, CD3-326; David and Vierstra, 1996) were generated as described for the 35S:AtCBSX1 lines. Hypocotyl epidermal tissues of T2 Arabidopsis were observed under the Olympus BX-51 fluorescence microscope with XF116-2 and XF111-2 filter sets (Omega). For the BiFC assay, CBSX1-YN and Trx m-YC were constructed by cloning the coding regions of CBSX1 and Trx m into the BamHI-XhoI sites of pUC-SPYNE and pUC-SPYCE, respectively (Walter et al., 2004) and were introduced into tobacco leaves by the Agrobacterium-mediated infiltration method. Fluorescence was analyzed with a confocal laser scanning microscope (LSM 510 META; Carl Zeiss). YFP was excited with an argon laser at 514 nm, and the emission wavelength was captured with a 530- to 600-nm band-pass filter.
Scanning Electron Microscopy and TEM Analysis
For scanning electron microscopy, the plant organs were mounted on stubs over double-sided carbon tape and coated with gold particles using a sputter coater (SEM Coating System; Bio-Rad). Specimens were observed with a scanning electron microscope (JEOL5300; Jeol) at an accelerating voltage of 25 kV. For the semithin sections and TEM, the tissues were first immersed in a fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.5 M cacodylate buffer, pH 7.5) and then vacuum infiltrated (25 Pa for 30 min). After fixing overnight at 4°C, the samples were rinsed with 0.5 M cacodylate buffer, postfixed with 1% OsO4 in 0.5 M cacodylate buffer, pH 7.5, overnight at 4°C, and then washed again with 0.5 M cacodylate buffer. The samples were dehydrated at 4°C stepwise through an ethanol series (10% increments, 20 min per step, 10 to ~100%) and then transferred successively to 3:1, 1:1, and 1:3 (v/v) mixtures of ethanol and Spurr’s resin (Ted Pella; 6 h at each step) and finally to 100% Spurr’s resin, and left overnight. Each sample was cured in the flat embedding mold for 2 d at 70°C. Semithin sections (1 μm) were cut on an ultramicrotome model MTX (RMC) using a glass knife and then stained with 1% (w/v) toluidine O. For TEM, ultrathin sections (70 nm thick) were collected in copper grids (200 mesh), stained with 2% (w/v) uranyl acetate and Reynols’s lead citrate, and analyzed by JEM 1010 (Jeol) at 80 kV.
Yeast Two-Hybrid Screening
The yeast two-hybrid Matchmaker system (Clontech) was used for analysis of protein–protein interactions according to the manufacturer's protocols. The open reading frame of CBSX1 cDNA cloned in frame into pGBKT7 vector was used as a bait to screen the Arabidopsis cDNA expression library constructed by Matchmaker library construction system (Clontech) using total RNA extracted from Arabidopsis leaves. The transformed yeast cells were screened on the synthetic dropout medium (lack of Leu, Trp, His, and Ade).
Preparation of Protein Crude Extracts
Leaf protein crude extracts from Arabidopsis (ecotype Columbia-0) plants, 35S:CBSX1 lines, and the T-DNA insertion mutants (cbsx1, AL753455) were prepared as described (Keryer et al., 2004). The Bradford protein assay (Bio-Rad) was used to determine the protein concentration of the clear crude extracts.
Protein Overexpression and Purification
CBSX1 (72-236), CBSX2 (76-232), and CBSX3 (40-206) were purified as described (Jeong et al., 2008). Arabidopsis Trx f (58-179), m (73-187), x (68-183) y (63-173), and o (44-195) protein, except for the N-terminal signal sequence, were purified in a similar manner to the CBSX1 protein, followed by gel filtration chromatography equilibrated with 20 mM Tris-HCl, pH 7.7, 300 mM NaCl, and 1 mM EDTA. CBSX and Trx proteins were concentrated to 12 and 6 mg/mL, respectively. His-tagged (GE Healthcare) CBSXs were isolated by Ni–nitrilotriacetic acid affinity, and glutathione S-transferase (GST)–tagged (GE Healthcare) Trxs were isolated by glutathione affinity, followed by anion exchange chromatography; both were concentrated to 10 mg/mL in a final solution of 50 mM Tris-HCl, pH 8.0, 150 to ~200 mM NaCl, and 1 mM DTT. SynFTR (FTR from Synechocystis sp PCC6803) protein was purified as described (Schürmann, 2002).
Gel Filtration, Pull-Down, and Enzyme Assays
The concentrated CBSX1 protein was injected onto Superose 12 10/300 GL gel filtration columns equilibrated with 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT, and 5% (w/v) glycerol. The elution of protein was monitored by absorbance at 280 nm. BSA (66 kD) and ovalbumin (45 kD) were used as molecular mass markers. Purified His-CBSX1 and His-CBSX2 (each 200 μg) were incubated with purified GST-Trx f, m, x, and y (each 400 μg) in 1 mL of assay buffer (1× PBS and 1 mM DTT) containing 30 μL GST-Sepharose 4B beads (GE Healthcare Bio-Sciences) for 1 h at 4°C; purified His-CBSX3 was also incubated with purified GST-Trx o. The beads were washed at least five times with assay buffer. SDS sample buffer was added, and the beads were boiled for 7 min. The interaction between CBSXs and Trxs was checked by immunoblotting with anti-His and anti-GST antibodies (GE Healthcare). Trx and FTR activity were determined using the insulin-disulfide reduction assay as described (Holmgren, 1977, 1979; Li et al., 1996; Laloi et al., 2001; Reichheld et al., 2007). Trx activity was measured at 24°C and defined as the maximal increase rate in turbidity for 1 h at 650 nm due to insulin precipitation. CBSXs were used at a final concentration of 30 μM in 100 μL of 0.1 M HEPES, pH 7.0, 1 mM EDTA, 20 μM Trx, 5 mM DTT, and 80 μM bovine insulin (Sigma-Aldrich). FTR activity was assayed by measuring the consumption of NADPH as a decrease of absorbance at 340 nm, indicating Trx reduction. The reaction was catalyzed by SynFTR protein, not DTT, as the primary reducer. Absorbance was measured in 100-μL aliquots containing 0.1 M HEPES, pH 7.0, 1 mM EDTA, 20 μM Trx, 30 μM CBSX1, 5 μM SynFTR, 150 μM NADPH, and 80 μM bovine insulin after a 30-min preincubation at 24°C. Adenosine derivatives (1 mM) were used as a cofactor, and the same buffer without CBSXs was used as a reference. The NADP-dependent MDH activity was also assayed by measuring the consumption of NADPH as the decrease of absorbance at 340 nm (Keryer et al., 2004; Tomaz et al., 2010). The assay was performed at 24°C using 0.1 M Tris-HCl, pH 8.0, 1 mM EDTA, 150 μM NADPH, 740 μM oxaloacetate, and fleshly isolated protein extracts (80 mg). The absorbance was monitored at 340 nm and converted to micromoles using the molar extinction coefficient of 6.22 × 103 M−1 cm−1.
CBSX2 Crystallization and Structure Determination
Crystals of CBSX2 were grown by mixing each 1 μL of protein (~12.0 mg/mL in 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5% [w/v] glycerol, and 1 mM DTT) with reservoir buffer [0.1 M MES, pH 6.1, 0.2 M Ca(OAc)2, and 21 to 22% (w/v) polyethylene glycol 8000] at 22°C by the hanging-drop vapor diffusion method. For multiwavelength anomalous dispersion (MAD) phasing, selenomethionine-substituted CBSX2 was expressed using B834(DE3) cells (Novagen) and purified in a similar manner to the wild-type protein. Crystals were transferred to a cryoprotectant solution containing reservoir buffer with 20% (w/v) glycerol and then flash-frozen in a cold nitrogen stream at 100K. Diffraction data were collected on an ADSC Quantum charge-coupled device detector at the 6B beamline of the Pohang Light Source, Korea and the AR-NW12A beamline of the Photon Factory, Tsukuba, Japan (see Supplemental Table 2 online). The data were processed and scaled using the HKL2000 software package (Otwinowski and Minor, 1997). The asymmetric unit in the P6522 space group contains a subunit of dimeric CBSX2. The phases were determined using a four-wavelength MAD data set with SOLVE (Terwilliger and Berendzen, 1999), and model building was performed with ARP/wARP (Cohen et al., 2008) and completed by manual rebuilding with Coot (Emsley et al., 2010). Refinement was performed with PHENIX software (Adams et al., 2010), and the assessment of model geometry and assignment of secondary structural elements were achieved using the PROCHECK program (Laskowski et al., 1993). All figures for the structures were prepared with PyMOL (http://www.pymol.org).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CBSX1 (At4g36910), CBSX2 (At4g34120), CBSX3 (At5g10860), Trx f (At3g02730), Trx m (At4g03520), Trx x (At1g50320), Trx y (At1g76760), Trx o (At2g35010), WRKY40 (At1g80840), BAP (At3g61190), APX1 (At1g07890), AAA-type ATPase (At3g28580), ZAT12 (At5g59820), eIF4a-1 (At3g13920), smGFP (AAB16957), and YFP (ABU49612). The T-DNA insertion line cbsx1 is AL753455 from the GABI-Kat collection. Atomic coordinates and structure factors for CBSX2 have been deposited in the Protein Data Bank with ID code 3SL7.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Structures of CBSX1 and Sequence Alignment between CBSX1 and CBSX2.
Supplemental Figure 2. Expression Patterns and Subcellular Localization of CBSX1.
Supplemental Figure 3. Size Exclusion Chromatography (Superose 12 GL 10/300; Amersham-Pharmacia) Elution Profile of the CBSX1 Homodimer.
Supplemental Figure 4. Ribbon Diagram Comparing the Overall Structure of CBS Domain Proteins.
Supplemental Figure 5. Enhancing the Activities of Trxs and FTR in the Presence of CBSXs and Adenosine-Containing Ligands.
Supplemental Figure 6. Dehiscence and Senescence of Wild-Type and 35:CBSX1 Transgenic Plants.
Supplemental Figure 7. Images of Scanning Electron Microscopy and Transmission Electron Microscopy.
Supplemental Figure 8. Expression Levels of NST1, NST2, and MYB26 and Expression Level of CBSX1 under Various Stress Conditions and Hormone Treatments.
Supplemental Table 1. Proteins Interacting with CBSX1.
Supplemental Table 2. Data Collection and Refinement Statistics.
Acknowledgments
We thank Hong Joo Cho at Korea University for their technical assistance in the TEM studies, Soon-Kap Kim at Korea University for the BiFC assay, and the staff at 6B beamline, Pohang Accelerator Laboratory, Korea and NW12 beamline, Photon Factory, Japan for help with data collection. We also thank Peter Schürmann (Université de Neuchâtel, Switzerland) for the expression plasmid Synechocystis FTR. This research was supported by a grant (2011-0015848 to J.S.S.) from the National Research Foundation of Korea and grants (KRF-2008-313-C00521 to H.K.S. and KRF-2008-359-F00005 to S.H.O.) from the Korea Research Foundation funded by the Ministry of Science and Technology and grants (PJ008198 and PJ008160 to J.S.S.) from the Next-Generation BioGreen21 Program funded by the Rural Development Administration, Republic of Korea. The Seoul Science Fellowship supported K.S.Y.
AUTHOR CONTRIBUTIONS
K.S.Y., S.H.O., B.-C.J., K.W.J, H.K.S., and J.S.S. designed research, analyzed data, and wrote the article. K.S.Y., B.-C.J., M.H.C., S.H., and M.-R.L. performed experiments and analyzed data. H.K.S. and J.S.S. contributed equally to this work.
References
- Adams P.D., et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari N.D., Froehlich J.E., Strand D.D., Buck S.M., Kramer D.M., Larkin R.M. (2011). GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23: 1449–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arad M., Benson D.W., Perez-Atayde A.R., McKenna W.J., Sparks E.A., Kanter R.J., McGarry K., Seidman J.G., Seidman C.E. (2002). Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J. Clin. Invest. 109: 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. (1997). The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22: 12–13 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Cilia M., San Roman A., Thomas C., Maule A., Hearn S., Jackson D. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair E., Redwood C., Ashrafian H., Oliveira M., Broxholme J., Kerr B., Salmon A., Ostman-Smith I., Watkins H. (2001). Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: Evidence for the central role of energy compromise in disease pathogenesis. Hum. Mol. Genet. 10: 1215–1220 [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonner L., Dickinson H. (1989). Anther dehiscence in Lycopersicon esculentum Mill. I. Structural aspects. New Phytol. 113: 97–115 [DOI] [PubMed] [Google Scholar]
- Buchanan B.B., Schürmann P., Wolosiuk R.A., Jacquot J.P. (2002). The ferredoxin/thioredoxin system: from discovery to molecular structures and beyond. Photosynth. Res. 73: 215–222 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohen S.X., Ben Jelloul M., Long F., Vagin A., Knipscheer P., Lebbink J., Sixma T.K., Lamzin V.S., Murshudov G.N., Perrakis A. (2008). ARP/wARP and molecular replacement: The next generation. Acta Crystallogr. D Biol. Crystallogr. 64: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin V., Lamkemeyer P., Miginiac-Maslow M., Hirasawa M., Knaff D.B., Dietz K.J., Issakidis-Bourguet E. (2004). Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol. 136: 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S.J., Vierstra R.D. (1996). Soluble derivatives of green fluorescent protein (GFP) for use in Arabidopsis thaliana. Weeds World 3: 43–48 [Google Scholar]
- Dawson J., Sozen E., Vizir I., Van Waeyenberge S., Wilson Z., Mulligan B. (1999). Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 144: 213–222 [Google Scholar]
- de Dios Barajas-López J., Serrato A.J., Olmedilla A., Chueca A., Sahrawy M. (2007). Localization in roots and flowers of pea chloroplastic thioredoxin f and thioredoxin m proteins reveals new roles in nonphotosynthetic organs. Plant Physiol. 145: 946–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E., et al. (2004). A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc. Natl. Acad. Sci. USA 101: 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollob M.H., Green M.S., Tang A.S., Gollob T., Karibe A., Ali Hassan A.S., Ahmad F., Lozado R., Shah G., Fananapazir L., Bachinski L.L., Roberts R. (2001). Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N. Engl. J. Med. 344: 1823–1831 Erratum. N. Engl. J. Med. 345: 552 [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1977). Bovine thioredoxin system. Purification of thioredoxin reductase from calf liver and thymus and studies of its function in disulfide reduction. J. Biol. Chem. 252: 4600–4606 [PubMed] [Google Scholar]
- Holmgren A. (1979). Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254: 9627–9632 [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Ignoul S., Eggermont J. (2005). CBS domains: Structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 289: C1369–C1378 [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong B.C., Yoo K.S., Jung K.W., Shin J.S., Song H.K. (2008). Purification, crystallization and preliminary X-ray diffraction analysis of a cystathionine beta-synthase domain-containing protein, CDCP2, from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64: 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.C., Cao R.Q., Kung J.E., Buchanan B.B. (1987). Thioredoxin and NADP-thioredoxin reductase from cultured carrot cells. Planta 171: 321–331 [PubMed] [Google Scholar]
- Juárez-Díaz J.A., McClure B., Vázquez-Santana S., Guevara-García A., León-Mejía P., Márquez-Guzmán J., Cruz-García F. (2006). A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNase in vitro. J. Biol. Chem. 281: 3418–3424 [DOI] [PubMed] [Google Scholar]
- Karlsson M., Melzer M., Prokhorenko I., Johansson T., Wingsle G. (2005). Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J. Exp. Bot. 56: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Koita H., Nakatsubo T., Hasegawa K., Wakabayashi K., Takahashi H., Umemura K., Umezawa T., Shimamoto K. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 103: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennan A., et al. (2002). Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(-/-) mice. Hum. Mol. Genet. 11: 547–557 [DOI] [PubMed] [Google Scholar]
- Keryer E., Collin V., Lavergne D., Lemaire S., Issakidis-Bourguet E. (2004). Characterization of Arabidopsis mutants for the variable subunit of ferredoxin:thioredoxin reductase. Photosynth. Res. 79: 265–274 [DOI] [PubMed] [Google Scholar]
- King N.P., Lee T.M., Sawaya M.R., Cascio D., Yeates T.O. (2008). Structures and functional implications of an AMP-binding cystathionine beta-synthase domain protein from a hyperthermophilic archaeon. J. Mol. Biol. 380: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of T L-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Genet. Genomics 204: 383–396 [Google Scholar]
- Kushwaha H.R., Singh A.K., Sopory S.K., Singla-Pareek S.L., Pareek A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 10: 200–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C., Rayapuram N., Chartier Y., Grienenberger J.M., Bonnard G., Meyer Y. (2001). Identification and characterization of a mitochondrial thioredoxin system in plants. Proc. Natl. Acad. Sci. USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R., MacArthur M., Moss D., Thornton J. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291 [Google Scholar]
- Li X., Nield J., Hayman D., Langridge P. (1996). A self-fertile mutant of Phalaris produces an S protein with reduced thioredoxin activity. Plant J. 10: 505–513 [DOI] [PubMed] [Google Scholar]
- Marcus F., Chamberlain S.H., Chu C., Masiarz F.R., Shin S., Yee B.C., Buchanan B.B. (1991). Plant thioredoxin h: An animal-like thioredoxin occurring in multiple cell compartments. Arch. Biochem. Biophys. 287: 195–198 [DOI] [PubMed] [Google Scholar]
- Marri L., Zaffagnini M., Collin V., Issakidis-Bourguet E., Lemaire S.D., Pupillo P., Sparla F., Miginiac-Maslow M., Trost P. (2009). Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2: 259–269 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Siala W., Bashandy T., Riondet C., Vignols F., Reichheld J.P. (2008). Glutaredoxins and thioredoxins in plants. Biochim. Biophys. Acta 1783: 589–600 [DOI] [PubMed] [Google Scholar]
- Millar A.H., Carrie C., Pogson B., Whelan J. (2009). Exploring the function-location nexus: Using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell J.A., Maduke M., Miller C., Grigorieff N. (2001). Projection structure of a ClC-type chloride channel at 6.5 A resolution. Nature 409: 219–223 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrichard F., Alkhalfioui F., Yano H., Vensel W.H., Hurkman W.J., Buchanan B.B. (2009). Thioredoxin targets in plants: The first 30 years. J. Proteomics 72: 452–474 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Newman L.J., Perazza D.E., Juda L., Campbell M.M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37: 239–250 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pusch M. (2002). Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 19: 423–434 [DOI] [PubMed] [Google Scholar]
- Ragunathan P., Kumarevel T., Agari Y., Shinkai A., Kuramitsu S., Yokoyama S., Ponnuraj K. (2008). Crystal structure of ST2348, a CBS domain protein, from hyperthermophilic archaeon Sulfolobus tokodaii. Biochem. Biophys. Res. Commun. 375: 124–128 [DOI] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P., Marita J., Hatfield R., Ralph S., Christensen J. (2004). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 3: 29–60 [Google Scholar]
- Reichheld J.P., Khafif M., Riondet C., Droux M., Bonnard G., Meyer Y. (2007). Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19: 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Madrid R., Mestres D., Marinho P., Jacquot J.P., Decottignies P., Miginiac-Maslow M., Meyer Y. (1995). Evidence for five divergent thioredoxin h sequences in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 92: 5620–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J., Aubert S., Gout E., Bligny R., Douce R. (1997). Cooperation and competition between adenylate kinase, nucleoside diphosphokinase, electron transport, and ATP synthase in plant mitochondria studied by 31P-nuclear magnetic resonance. Plant Physiol. 113: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers L.A., Dubos C., Cullis I.F., Surman C., Poole M., Willment J., Mansfield S.D., Campbell M.M. (2005). Light, the circadian clock, and sugar perception in the control of lignin biosynthesis. J. Exp. Bot. 56: 1651–1663 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Koh C.S., Gelhaye E., Corbier C., Favier F., Didierjean C., Jacquot J.P. (2008). Redox based anti-oxidant systems in plants: biochemical and structural analyses. Biochim. Biophys. Acta 1780: 1249–1260 [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K., Hsu Y.C., Lee P.Y., Truong M.T., Beals T., Goldberg R. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P. (2002). Ferredoxin-dependent thioredoxin reductase: A unique iron-sulfur protein. Methods Enzymol. 347: 403–411 [DOI] [PubMed] [Google Scholar]
- Shan X., Dunbrack R.L., Jr, Christopher S.A., Kruger W.D. (2001). Mutations in the regulatory domain of cystathionine beta synthase can functionally suppress patient-derived mutations in cis. Hum. Mol. Genet. 10: 635–643 [DOI] [PubMed] [Google Scholar]
- Shin R., Schachtman D.P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange S., Unte U.S., Eckstein L., Yang C., Wilson Z.A., Schmelzer E., Dekker K., Saedler H. (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34: 519–528 [DOI] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Berendzen J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaz T., Bagard M., Pracharoenwattana I., Lindén P., Lee C.P., Carroll A.J., Ströher E., Smith S.M., Gardeström P., Millar A.H. (2010). Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 154: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H., Salminen A., Oksanen E., Jämsen J., Heikkilä O., Lehtiö L., Magretova N.N., Goldman A., Baykov A.A., Lahti R. (2010). Crystal structures of the CBS and DRTGG domains of the regulatory region of Clostridiumperfringens pyrophosphatase complexed with the inhibitor, AMP, and activator, diadenosine tetraphosphate. J. Mol. Biol. 398: 400–413 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal F., Martín V., Colaneri A., González-Schain N., Perales M., Martín M., Lombardo C., Braun H.P., Bartoli C., Zabaleta E. (2009). Ectopic expression of mitochondrial gamma carbonic anhydrase 2 causes male sterility by anther indehiscence. Plant Mol. Biol. 70: 471–485 [DOI] [PubMed] [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Xiao B., et al. (2007). Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 449: 496–500 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Yang C., Xu Z., Song J., Conner K., Vizcay Barrena G., Wilson Z.A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Evans G., Rotella F.J., Westbrook E.M., Beno D., Huberman E., Joachimiak A., Collart F.R. (1999). Characteristics and crystal structure of bacterial inosine-5′-monophosphate dehydrogenase. Biochemistry 38: 4691–4700 [DOI] [PubMed] [Google Scholar]