This work examines the role of light in root growth and finds a key role for the actin nucleating complex ARP2/3-SCAR in light-induced promotion of root elongation. Direct perception of light by the root suppresses ubiquitin-mediated SCAR degradation and promotes the formation of longitudinal F-actin arrays, which is critical for sustained root expansion.
Abstract
The ARP2/3 complex, a highly conserved nucleator of F-actin, and its activator, the SCAR complex, are essential for growth in plants and animals. In this article, we present a pathway through which roots of Arabidopsis thaliana directly perceive light to promote their elongation. The ARP2/3-SCAR complex and the maintenance of longitudinally aligned F-actin arrays are crucial components of this pathway. The involvement of the ARP2/3-SCAR complex in light-regulated root growth is supported by our finding that mutants of the SCAR complex subunit BRK1/HSPC300, or other individual subunits of the ARP2/3-SCAR complex, showed a dramatic inhibition of root elongation in the light, which mirrored reduced growth of wild-type roots in the dark. SCAR1 degradation in dark-grown wild-type roots by constitutive photomorphogenic 1 (COP1) E3 ligase and 26S proteasome accompanied the loss of longitudinal F-actin and reduced root growth. Light perceived by the root photoreceptors, cryptochrome and phytochrome, suppressed COP1-mediated SCAR1 degradation. Taken together, our data provide a biochemical explanation for light-induced promotion of root elongation by the ARP2/3-SCAR complex.
INTRODUCTION
It is well established that filamentous actin (F-actin) plays an essential role in root growth (Gilliland et al., 2003; Nishimura et al., 2003; Rahman et al., 2007). The organization and dynamics of F-actin are tightly controlled by an array of regulatory proteins, including the actin related protein 2/3 (ARP2/3) complex and its activator, the SCAR/WAVE complex (called SCAR here for simplicity), which facilitate nucleation of new short F-actin branches from existing filaments (Machesky and Insall, 1998; Mullins and Pollard, 1999). All components of the ARP2/3-SCAR complex are present in Arabidopsis thaliana (Deeks and Hussey, 2005; Szymanski, 2005), and through mutant analyses, this complex has recently been implicated in the control of root elongation (Dyachok et al., 2008). However, the mechanisms of ARP2/3-SCAR complex function in the regulation of root growth are not known.
BRICK1 (BRK1), the plant homolog of the mammalian HSPC300 subunit of the SCAR complex, and SCAR1 were found to be enriched at the periphery of root cells, and the loss of subunits of the ARP2/3-SCAR complex was shown to be correlated with depletion of cortical F-actin in root cells, suggesting a role for the ARP2/3-SCAR pathway in nucleating F-actin and root elongation growth (Dyachok et al., 2008). In addition to work with roots, support for the importance of the ARP2/3-SCAR complex in F-actin–dependent elongation growth comes from studies of the moss Physcomitrella patens. For example, BRK1 and ARPC4, an ARP2/3 complex subunit, are both localized at the expanding tip of the Physcomitrella protonema. Loss of BRK1 causes depletion of the dense actin enrichments at the protonema tip, resulting in a concomitant reduction in growth (Perroud and Quatrano, 2008).
One environmental stimulus that has received attention lately in regard to F-actin–modulated cellular processes is light. For instance, there is accumulating evidence that the movement and positioning of organelles such as chloroplasts in aboveground plant organs in response to light is facilitated by an F-actin–based motility system (reviewed in Wada and Suetsugu, 2004). The molecular components that comprise F-actin–mediated movement of chloroplasts have been uncovered recently, and these include the blue light photoreceptor phototropin (PHOT), the actin binding protein CHUP1, and the two kinesin-like proteins KAC1 and KAC2 (Kadota et al., 2009; Suetsugu et al., 2010). F-actin–mediated positioning of nuclei in Arabidopsis leaf cells has also been shown to require the PHOT blue light receptors (Iwabuchi et al., 2010). Recently, the actin bundling protein THRUMIN1 was discovered to provide an essential link between PHOT receptor activity at the plasma membrane (PM) and F-actin–dependent chloroplast movement. Together with CHUP1, THURMIN1 may be involved in actin remodeling that drives chloroplast movement upon blue light perception by PHOT at the PM (Whippo et al., 2011).
Because of their belowground location, detailed molecular studies on the effect of light on root development have not been as numerous as studies with aboveground organs. However, even in soil-grown plants, light can invoke dramatic changes in root growth as manifested by increased root elongation in deetiolating seedlings. Although not exposed to the same intensity and quality of light as aboveground organs, roots are exposed to light filtered through the soil (Mandoli et al., 1990) or piped light through the vascular cylinder (Mandoli et al., 1984). Many growth responses to light have been documented to occur in roots, including primary root elongation, gravitropism, and phototropism (Sakai et al., 2000; Kiss et al., 2003; Correll and Kiss, 2005; Galen et al., 2007; Tong et al., 2008). Like aboveground organs, roots contain photoreceptors (Somers and Quail, 1995; Tóth et al., 2001; Sakamoto and Briggs, 2002) that may perceive light directly (Somers and Quail, 1995; Kiss et al., 2003; Salisbury et al., 2007). Indeed, photoreceptors, including phytochromes (PHYs) A, B, and D, have been shown to control red light–mediated root elongation (Correll and Kiss, 2005), while PHOT1 and cryptochromes (CRYs) are important for root elongation in response to blue light (Canamero et al., 2006; Galen et al., 2007). Furthermore, PHYs and CRYs both participate in greening of roots under blue light (Usami et al., 2004). The biological significance of light regulation of root growth is not entirely clear. However, there are indications that light could be important for plant fitness when exposed to certain environmental conditions. For example, there is experimental evidence that light through PHOT1 receptors could promote plant drought tolerance by enhancing the efficiency of root growth away from the soil surface (Galen et. al., 2007).
Although examples for light-triggered root growth responses have accumulated over several years, mechanisms by which the light signal perceived by photoreceptors is translated into changes in root growth at the cellular and molecular level are not fully understood. Here, we present cellular, genetic, and biochemical evidence that the rapid growth of roots in the light requires a functional ARP2/3-SCAR complex. We show that light is essential for stabilizing the SCAR complex in the PM, which in turn is necessary for maintaining longitudinally organized F-actin to sustain rapid root growth. This process requires an intact photosensory pathway, including photoreceptors, COP1, and proteasomes, that operate directly in roots independent of aboveground organs.
RESULTS
Growth of Roots of Mutants in the ARP2/3 and SCAR Complex Is Affected Differentially by Light and Darkness
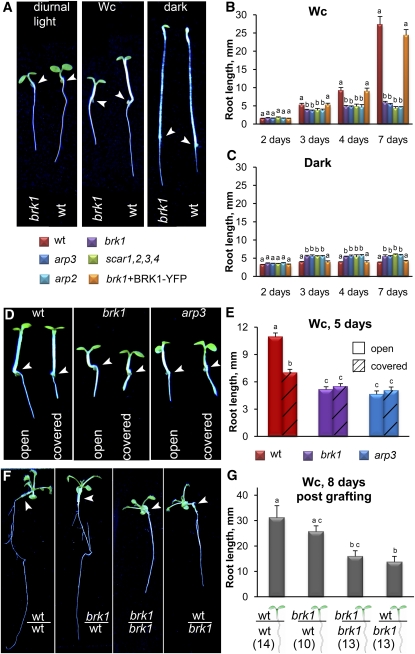
ARP2/3 and SCAR complex subunit mutants grown on sugar-free agar media under diurnal light conditions (i.e., 16-h-light and 8-h-dark cycles) display a short primary root phenotype (Dyachok et al., 2008). To gain deeper insight into the molecular basis of this root growth defect, we compared root growth in wild-type seedlings to four SCAR and ARP2/3 complex subunit mutants (brk1, arp3, and arp2 single mutants and scar1,2,3,4 quadruple mutants) under various light conditions and growth media. We found that on sugar-free agar media, mutant and wild-type seedlings germinated at equal rates and at day 2 had similar root lengths in continuous white light (Wc) and darkness (first data points in Figures 1B and 1C). When kept in Wc for an additional 1 to 5 d, all mutants developed significantly shorter roots compared with wild-type seedlings (Figures 1A and 1B). Surprisingly, root elongation was slightly enhanced in all the mutants compared with the wild type in complete darkness (Figures 1A and 1C). By day 7, all mutants had similar average root length in Wc and in darkness (cf. Figures 1B and 1C). Expression of BRK1-YFP (for yellow fluorescent protein) under the control of the endogenous BRK1 promoter in brk1 (Dyachok et al., 2008) rescued the root growth defects of brk1 (Figures 1B and 1C), confirming that the root phenotypes observed in brk1 mutants were due to a loss of BRK1 function. Thus, comparison of the root phenotypes of mutant and wild-type seedlings grown on sugar-free agar media suggests that in young seedlings, intact SCAR and ARP2/3 complexes promote elongation growth of roots in Wc while inhibiting their elongation in darkness.
Figure 1.
Root Growth of ARP2/3 and SCAR Complex Mutants Is Affected by the Light Environment.
(A) Representative images of 4-d-old brk1 and wild-type (wt) seedlings grown in 16-h-white-light and 8-h-dark cycles, Wc, or continuous darkness. Note that roots of brk1 mutants are inhibited in diurnal light conditions and more so in Wc but not in darkness.
(B) and (C) Root lengths in 2- to 7-d-old wild type, brk1, arp3, arp2, scar1,2,3,4, and brk1 expressing BRK1p:BRK1-YFP (brk1+BRK1-YFP) seedlings in Wc (B) or continuous darkness (C).
(D) and (E) Growth is inhibited in the wild type but not in brk1 or arp3 mutant roots shielded from light. Seeds were placed on the surface of noncovered (open) solid media or inside small openings in black foil covering the media surface (covered). Representative images (D) and root lengths (E) of 5-d-old seedlings grown inside media in horizontally oriented plates in Wc.
(F) and (G) Roots of brk1 mutants are inhibited in seedlings when combined with wild-type or brk1 aerial parts. Aerial parts were swapped between 5-d-old wild-type and brk1 seedlings by grafting. Chimeric seedlings continued to grow for 8 more days in vertically oriented plates in Wc before imaging (F) and measuring root length (G). Individual bars in (G) represent seedlings combining aerial parts and roots with the respective genotypes indicated below. The numbers of successful grafts are indicated in parenthesis.
Arrowheads in (A), (D), and (F) point to the root-hypocotyl junction. Values (means + se) with different letters in (B), (C), (E), and (G) are significantly different as determined by Tukey’s tests at the 99% confidence level (n > 25 seedlings for [B], [C], and [E]).
It has previously been shown that ARP2/3-SCAR complex mutants have expansion defects in leaf epidermal cells (Smith and Oppenheimer, 2005; Szymanski, 2005). Thus, defects in cell expansion could also account for reduced root lengths in mutants. Alternatively, root elongation defects could result from a decreased number of dividing cells in mutants. To distinguish between these possibilities, we evaluated cell elongation and division in brk1 and arp3 mutants under light conditions used in this study. We found that the final length of mature cortical cells in roots of Wc- or dark-grown plants was similar to that of brk1 and arp3 mutants, indicating that root cells expand to the same extent in mutant and wild-type seedlings (see Supplemental Figure 1A online). Although the final cell length of mature cortical cells in mutants and the wild type was similar, we cannot rule out the possibility that the rate of cell expansion in the elongation zone differed between the genotypes. We therefore used confocal microscopy to measure changes in cortical cell length profiles along the root axis of the wild type and mutants (see Supplemental Figure 1B online). We found that cells located near the beginning or in the middle of the elongation zone were shorter in roots of Wc-grown mutants compared with wild-type seedlings. However, cells near the end of the elongation zone had similar lengths in wild-type and mutant roots consistent with measurements made on mature cortical cells (see Supplemental Figures 1A and 1B online). Thus, the slower cell expansion rates could partly explain reduced root growth of Wc-grown brk1 and arp3. We then examined whether frequencies of dividing cells were affected in mutant roots by counting the number of mitotic figures (mitotic spindles and phragmoplasts). We found that the number of mitotic figures was lower in roots of Wc-grown mutants compared with wild-type seedlings. On the contrary, the number of mitotic figures was greater in the mutants compared with wild-type roots in darkness (see Supplemental Figure 1C online). Taken together, the data indicate that the root growth defect of ARP2/3-SCAR complex mutants is due to a combination of altered cell division and expansion.
In addition to short roots, all mutants displayed short hypocotyls compared with the wild type when grown on Suc-free agar media in Wc but not in darkness (see Supplemental Figure 2 online). Because mutants displayed root and hypocotyl phenotypes under our experimental conditions, it is possible that light perceived exclusively by shoots could explain ARP2/3- SCAR–dependent elongation of both hypocotyls and roots. Alternatively, the light signal that facilitates ARP2/3-SCAR–dependent root elongation could be perceived directly by roots. To tease apart the site of light perception, we used two approaches. First, we reasoned that if light stimulates root elongation directly, shielding only the roots of wild-type seedlings but not the shoots from light would be sufficient to inhibit root elongation. We also hypothesized that because seedlings of Wc- and dark-grown ARP2/3-SCAR pathway mutants have similar root lengths (cf. Figures 1B and 1C), differential shielding should have no effect on root elongation in the mutants. To test the effect of limiting light exposure on root elongation, we grew seeds in Wc on horizontal plates on solid media that had either the standard translucent surface or a black foil–covered surface with small openings for seeds, as described by Tong et al. (2008). On plates with a translucent surface, roots of brk1 and arp3 were shorter compared with roots of wild-type seedlings (Figures 1D and 1E). Limiting light exposure by growing roots under a black foil inhibited their elongation dramatically in wild-type but had no effect on root elongation in brk1 or arp3 seedlings. Interestingly, seedlings of the same genotype had similar hypocotyl lengths when grown on covered or translucent surface. Thus, shielding only the roots did not influence hypocotyl elongation (Figure 1D). Altogether, these results indicate that direct exposure of roots to light and an intact ARP2/3-SCAR pathway are both required for root elongation growth.
A second approach to verify whether light activates ARP2/3-SCAR–dependent root elongation directly was to use grafting to combine aerial parts (shoots) of the wild type and roots of mutants and vice versa. We reasoned that if roots perceived light directly, the genetic origin of shoots should not influence root elongation. Indeed, we found that roots of grafts where brk1 seedlings were used as root donors were always shorter compared with roots of grafts where the wild type was used as the root donor, regardless of the source of the shoot (Figures 1F and 1G). Thus, an intact ARP2/3-SCAR pathway in roots but not in aerial parts is required for Wc-dependent root elongation.
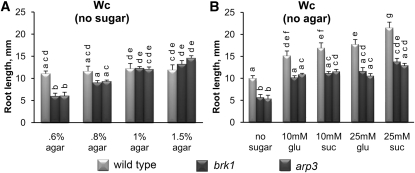
Stunted hypocotyls but not roots were previously reported for SCAR and ARP2/3 complex subunit mutants grown on Suc-supplemented agar media either under diurnal light conditions or in complete darkness (Basu et al., 2004, 2005; Li et al., 2004; Saedler et al., 2004; Zhang et al., 2008; Jörgens et al., 2010). In agreement with these reports, when brk1 and arp3 seedlings were grown on Suc-supplemented agar media, neither Wc- nor dark-grown mutants exhibited short roots (see Supplemental Figure 3 online). Thus, the short root phenotypes observed in the mutants on sugar-free agar media could be due to the altered nutritional status of the seedling. Alternatively, agar in the growth media could be the cause of aberrant root growth in the mutants because increasing the concentration of agar in the media rescued the root growth defects of the mutants even in the absence of sugar (Figure 2A). To dissect the effect of sugars on root elongation, we grew wild-type, brk1, and arp3 seedlings in liquid media supplemented with increasing concentrations of Glc or Suc. Both Glc and Suc promoted root elongation in wild-type and mutant seedlings in a concentration-dependent manner. However, mutant roots remained significantly shorter compared with the wild type (Figure 2B). Because stunted root growth under Wc cannot be rescued by sugars alone, it is unlikely that the defects in root elongation growth of brk1 and arp3 mutants are due to the nutritional status of the seedlings.
Figure 2.
The Short Root Phenotype of brk1 and arp3 Is Light Dependent but Sugar Independent.
Seedlings were grown for 4 d before measuring root length.
(A) Seedlings grown in Wc on sugar-free media supplemented with increasing concentrations of agar.
(B) Seedlings grown in Wc in liquid media supplemented with 10 or 25 mM Glc (glu) or Suc. Values (means + se) with different letters are significantly different by Tukey’s tests at the 99% confidence level (n ≥ 20 plants for each variant).
Disorganized F-Actin Accompanies Root Growth Inhibition in brk1, arp3, and Dark-Grown Wild-Type Seedlings
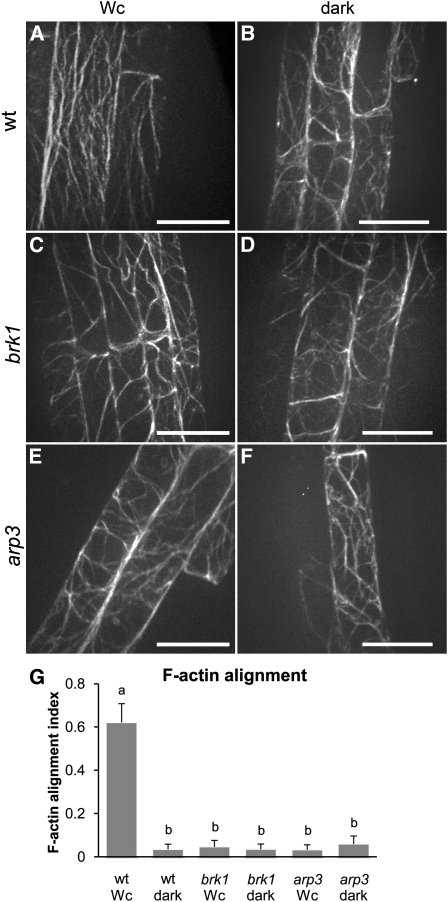
We asked whether a change in the organization of cortical F-actin accompanies inhibited root growth in Wc-grown mutants and dark-grown wild-type seedlings. F-actin was visualized in wild-type, brk1, and arp3 plants expressing an actin binding domain of fimbrin tagged at both ends with green fluorescent protein (GFP-ABD2-GFP; Wang et al., 2008). Expanding cells in roots of wild-type seedlings grown in Wc had prominent arrays of predominantly longitudinal actin filaments (Figure 3A). This longitudinal alignment of F-actin was disturbed in wild-type roots grown in the dark (Figure 3B) and roots of Wc- or dark-grown brk1 and arp3 mutants (Figures 3C to 3F). The predominantly longitudinal F-actin bundles were replaced with randomly oriented filaments. Changes in F-actin organization were quantified using an algorithm we developed in MATLAB for fast Fourier transform and entropy analysis (see Methods). Cells of Wc- or dark-grown brk1 and arp3 roots and cells of dark-grown wild-type roots showed a dramatic reduction in F-actin alignment compared with Wc-grown wild-type roots (Figure 3G). Loss of F-actin alignment correlated with and could potentially explain short roots in mutant and in dark-grown wild-type seedlings.
Figure 3.
Cortical F-Actin Is Misaligned in brk1and arp3 and in Dark-Grown Wild-Type Seedlings.
(A) to (F) Representative confocal images of F-actin in expanding root cells of living 4-d-old seedlings grown in Wc ([A], [C], and [D]) or continuous darkness ([B], [D], and [F]). Images are projections of 5.5-μm Z-stacks focused on the center of the cells. Bars = 20 μm. wt, wild type.
(G) Average indexes of F-actin alignment (means + se) calculated based on the entropy maps of F-actin images of 50 to 70 individual cells for each genotype and light treatment. A higher degree of organization of filaments results in higher alignment index (maximum value of 1), whereas disorganization results in lower alignment index. Values with different letters are significantly different by Tukey’s tests at the 99% confidence level.
Because the growth defects of Wc-grown brk1 and arp3 roots were rescued to some extent in higher percentages of agar, we asked if higher agar concentrations could rescue the stunted growth of wild-type roots in the dark. We found that growth of wild-type seedlings on 1% agar in the dark caused only a minimal increase in average root length compared with growth on 0.6% agar (see Supplemental Figure 4A online). The average root length of wild-type seedlings grown on 1% agar in the dark was still about half the average length of wild-type seedlings in the light (cf. Supplemental Figure 4A online and Figure 1B). Interestingly, similar to dark-grown roots on 0.6% agar, dark-grown roots on 1% agar lacked longitudinal F-actin arrays; thus, growth of wild-type seedlings on 1% agar did not restore F-actin alignment in the dark (see Supplemental Figure 4B online).
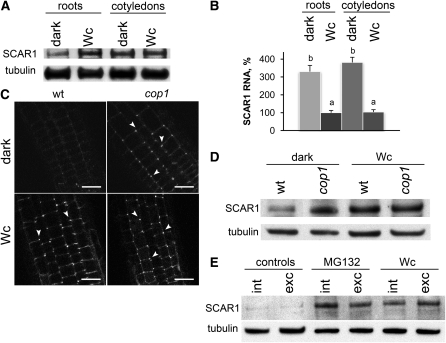
BRK1-YFP Is Mislocalized and SCAR1 Is Depleted in Dark-Grown Roots
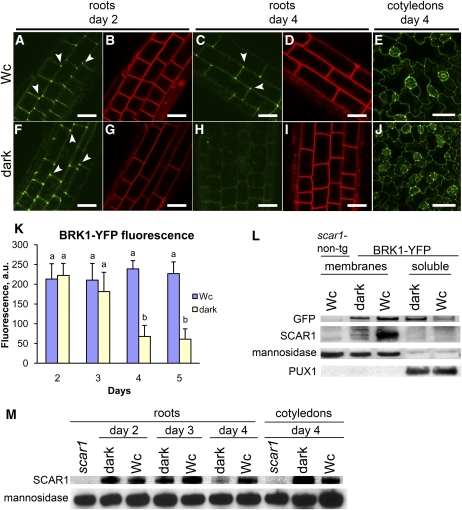
The loss of longitudinally aligned F-actin in roots of dark-grown wild-type plants suggests that the localization of components of the F-actin nucleating ARP2/3-SCAR machinery could also be modified in the dark. We examined the localization of BRK1-YFP under the light conditions used in this study. At day 2, BRK1-YFP fluorescence was found at the periphery of expanding root cells both in Wc and in darkness and colocalized with the styryl dye FM4-64, which incorporated into the PM (Figures 4A, 4B, 4F, and 4G). Bright foci of higher BRK1-YFP fluorescence were consistently observed at the cell corners (Figures 4A and 4F). At day 4, elongating Wc-grown roots retained similar patterns of intracellular BRK1-YFP localization at the cell periphery (Figures 4C and 4D), whereas the signal was dramatically depleted in dark-grown roots (Figures 4H and 4I). We quantified the reduction in BRK1-YFP–associated fluorescence by measuring fluorescence intensity at the cell corners of Wc- and dark-grown roots. Average fluorescence intensity remained stable in Wc-grown roots at all time points but declined dramatically in roots of dark-grown seedlings between days 2 and 5 (Figure 4K). Interestingly, while most of the BRK1-YFP signal disappeared from the roots of dark-grown seedlings at day 4, their cotyledons retained BRK1-YFP fluorescence surrounding the cell periphery in a pattern similar to Wc-grown cotyledons (Figures 4E and 4J). Thus, unlike in roots, BRK1-YFP localization to the cell periphery of cotyledons is not light dependent.
Figure 4.
BRK1-YFP Relocalizes to the Cytoplasm, and SCAR1 Protein Levels Decline in Dark-Grown Roots.
(A) to (J) Representative confocal images of BRK1p:BRK1-YFP (green panels) in expanding cells of roots and cotyledons of seedlings grown in Wc ([A] to [E]) or continuous darkness ([F] to [J]). Roots were counterstained with FM4-64 to mark the outline of the cell (red panels). Arrowheads point to cell corners showing local enrichments of fluorescence. Note that BRK1-YFP fluorescence dissipates in roots but not in cotyledons of dark grown seedlings. Bars = 20 μm.
(K) Average intensities of fluorescence associated with cell corners in the apical 200-μm zone of roots. Values (means + se) with different letters are significantly different by Tukey’s tests at the 99% confidence level (n ≥ 50 cells for each variant). a.u., arbitrary units.
(L) Biochemical characterization of BRK1-YFP and SCAR1 in 4-d-old seedlings grown in Wc or in darkness. Proteins were extracted from roots of nontransgenic scar1 (negative controls) or transgenic brk1 plants expressing BRK1p:BRK1-YFP (BRK1-YFP). Extracts were ultracentrifuged to separate microsomal membranes from supernatants (soluble). Immunoblots were probed subsequently with anti-GFP to detect BRK1-YFP, with anti-SCAR1, with anti-α-1,2-Mannosidase I (membrane protein marker), and with anti-PUX1 (cytoplasmic protein marker). Note that BRK1-YFP is associated with the insoluble membrane pellet in light but is partially solubilized in darkness. SCAR1 is associated with the membrane pellet in light but is degraded in darkness.
(M) SCAR1 protein levels decline in roots of wild-type seedlings in darkness. Proteins were extracted from roots of 2-, 3-, and 4-d-old seedlings and from cotyledons of 4-d-old seedlings grown in Wc or in darkness. Proteins were also extracted from roots and cotyledons of 4-d-old scar1 (negative controls) grown in Wc. Immunoblots were probed with the indicated antibodies.
Dark- and Wc-grown roots had similar expression of BRK1-YFP at the gene and protein level (see Supplemental Figure 5 online). Thus, the absence of the BRK1-YFP fluorescence in dark-grown root cells could be due to the dissociation of BRK1-YFP from the PM. We used immunoblotting to determine whether BRK1-YFP can be detected in the nonmembranous, soluble fractions of root extracts from 4-d-old seedlings. In roots of Wc-grown seedlings, the majority of BRK1-YFP was found in the membrane fraction with the membrane marker protein α-1,2-Mannosidase I (Preuss et al., 2004) (Figure 4L). In roots of dark-grown plants, however, a substantial proportion of BRK1-YFP was found in the soluble fraction with the cytoplasmic marker protein PUX1 (Rancour et al., 2004), while a smaller proportion of BRK1-YFP remained associated with the membrane fraction (Figure 4L). Thus, it appears that a portion of BRK1-YFP moves from the PM to the cytoplasm in roots grown in darkness, which is consistent with the reduction of cell periphery–associated BRK1-YFP fluorescence.
BRK1 localization to the PM requires intact SCAR proteins (Dyachok et al., 2008). To investigate if the release of BRK1-YFP from the PM in darkness is accompanied by changes in SCAR1 localization and/or levels, we probed root extracts with anti-SCAR1 antibodies. Whereas a majority of SCAR1 protein was always found in the microsomal membrane fraction, its levels were dramatically reduced in roots of dark-grown compared with Wc-grown seedlings (Figure 4L).
Analyses of SCAR1 dynamics in wild-type seedlings indicated that SCAR1 levels remained stable in roots of Wc-grown seedlings but declined dramatically in roots of dark-grown seedlings between days 2 and 4 (Figure 4M). In contrast with the reduction of SCAR1 levels in roots, cotyledons of 4-d-old dark- and Wc-grown plants had similar amounts of SCAR1 (Figure 4M). Thus, light deprivation appears to gradually reduce the levels of SCAR1 and the amounts of cell periphery–associated BRK1 in roots but not in cotyledons of Arabidopsis. The high levels of SCAR1 and cell periphery–associated BRK1 in roots is correlated with their faster growth in the light.
Degradation of SCAR1 Protein in Dark-Grown Roots Is 26S Proteasome and COP1 Dependent but Is Independent of Aboveground Organs
To understand the biochemical basis for the reduction of SCAR1 in dark-grown roots, we compared SCAR1 gene expression and protein levels in 3.5-d-old wild-type seedlings grown in Wc or in darkness. Roots of dark-grown seedlings had dramatically reduced SCAR1 protein levels compared with roots and cotyledons of Wc-grown seedlings and to cotyledons of dark-grown seedlings (Figure 5A). Quantitative RT-PCR (qRT-PCR) analyses of total RNA extracted from the same plant material revealed significantly increased levels of SCAR1 mRNA in dark-grown seedlings, both in roots and in cotyledons, compared with roots and cotyledons of Wc-grown seedlings (Figure 5B). Thus, expression of the SCAR1 gene is enhanced in dark-grown compared with Wc-grown seedlings. These results suggest that SCAR1 levels are likely regulated posttranslationally in Wc- and dark-grown seedlings.
Figure 5.
Posttranslational Degradation of SCAR1 in Roots in Darkness Requires 26S Proteasomes and COP1 and Is Independent of Aerial Parts.
(A) and (B) Comparison of SCAR1 protein (A) and RNA levels (B) in seedlings grown in darkness or in Wc. Proteins and RNA were extracted from roots and cotyledons of 3.5-d-old seedlings. Immunoblots were probed with antibodies to SCAR1 and tubulin (A). Real-time qRT-PCR analyses of SCAR1. Histograms represent quantification of the SCAR1 gene transcripts normalized relative to the constitutive control EIF4A-2 (B). Values are means + se. The value for roots of Wc-grown seedlings is set to 100% as reference. Values with different letters in (B) are significantly different by Tukey’s tests at the 99% confidence level (n = 3 biological replicates for each treatment).
(C) BRK1-YFP fluorescence dissipates in the wild type but not in cop1 roots in darkness. Representative confocal images of BRK1p:BRK1-YFP in root tips of 4-d-old wild-type (wt) and cop1 seedlings grown in continuous darkness or in Wc. Arrowheads point to cell corners with local enrichments of BRK1-YFP fluorescence (n ≥ 30 plants for each treatment). Bars = 30 μm.
(D) Dark-induced degradation of SCAR1 is inhibited in cop1 roots. Proteins were extracted from roots of 4-d-old wild-type and cop1 seedlings grown in darkness or in Wc. Immunoblots were probed in succession with antibodies to SCAR1 and tubulin.
(E) Both 26S proteasome antagonist MG132 and light inhibit dark-induced degradation of SCAR1 proteins in roots of intact seedlings and isolated roots. Intact 4-d-old dark-grown seedlings, or roots excised from dark-grown seedlings, were either treated in darkness with DMSO alone (controls) or with 50 μM MG132 dissolved in DMSO, or were transferred to Wc. Proteins were extracted from roots of intact seedlings (int) or from excised roots (exc) after 3 h. Immunoblots were probed with antibodies to SCAR1 and tubulin.
The proteasome system is the major mechanism for regulated protein degradation in eukaryotic organisms (Ciechanover, 1994) and is responsible for the degradation of transcription factors required for photomorphogenesis in dark and light (Jiao et al., 2007; Vierstra, 2009). In the dark, plants use a negatively acting E3 ligase, COP1, which targets positively acting transcription factors (e.g., HY5, HFR1, and LAF1) for proteasome-mediated degradation to repress photomorphogenesis in darkness (Jiao et al., 2007). If COP1 and proteasomes are involved in the degradation of SCAR1 in dark-grown roots, their suppression should theoretically enhance SCAR1 levels. In support of this notion, it was found that both cell periphery–associated fluorescence of BRK1-YFP and levels of SCAR1 protein increased in roots of dark-grown cop1 mutants compared with dark-grown wild type (Figures 5C and 5D). Furthermore, SCAR1 protein levels were elevated in roots of dark-grown seedlings (either excised or intact roots) treated with the 26S proteasome inhibitor MG132 or after their exposure to Wc (Figure 5E). Thus, posttranslational degradation of SCAR1 in dark-grown roots requires the activity of 26S proteasomes and COP1. Because treatment of isolated roots with MG132 or exposing them to Wc resulted in increased SCAR1 levels (Figure 5E), the 26S proteasome–dependent SCAR1 degradation operates directly in roots, consistent with our growth assays (Figures 1D to 1G).
SCAR1 and COP1 Interact
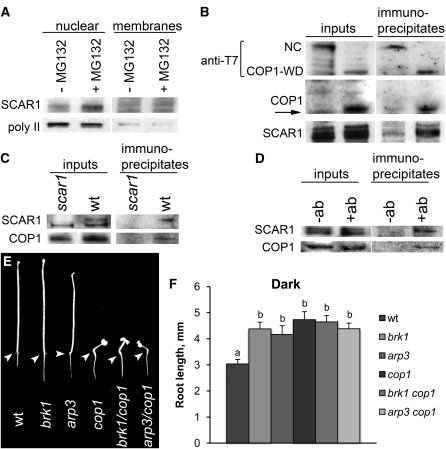
COP1 is predominantly nuclear in darkness, where it interacts with substrate proteins, targeting them for degradation (Jiao et al., 2007). If SCAR1 is a COP1 substrate, we would expect it to localize to the nucleus prior to dark-induced degradation in roots. To test whether SCAR1 is targeted to the nucleus prior to its degradation, we treated roots with MG132 to suppress SCAR1 degradation in darkness. Root extracts were then separated into nuclear and membrane fractions. SCAR1 was found in the nuclear fraction with the nuclear marker protein, the α-subunit of RNA polymerase II (Figure 6A), indicating that dark-induced SCAR1 degradation occurs in the nucleus.
Figure 6.
SCAR1 and COP1 Interact Biochemically and Genetically.
(A) The 26S proteasome antagonist MG132 increases SCAR1 levels in nuclear fractions. Four-day-old dark-grown wild-type seedlings were either treated in darkness with DMSO alone (-MG132) or with 50 μM MG132 dissolved in DMSO (+MG132). Nuclear and microsomal membrane fractions were isolated from root extracts after 3 h of treatment. Immunoblots were probed with antibodies to SCAR1 or RNA polymerase II (nuclear marker protein).
(B) SCAR1 and COP1 interact in vitro. SCAR1 was cotranscribed and translated with T7-tagged WD domain of COP1 (COP1-WD; amino acids 216 to 675) or T7-tagged negative control protein (NC). Input lanes show total samples before antibody incubation. Immunoprecipitate lanes show proteins bound to T7 beads. Immunoblots were probed with antibodies to T7 to detect T7-tagged NC and COP1-WD, to COP1, or to SCAR1. Arrow indicates an antigen band of a lesser molecular weight observed in all anti-COP1 probed samples.
(C) and (D) SCAR1 and COP1 interact in root extracts. Proteins were extracted from 3-d-old dark-grown seedlings. Extracts of scar1 and wild-type (wt) seedlings were incubated with anti-SCAR1 antibodies and A/G protein agarose (C). Extracts of wild-type seedlings were incubated with A/G protein agarose alone (-ab) or with anti-COP1 antibodies and A/G protein agarose (+ab) (D). Input lanes show total protein extracts before antibody incubation. Immunoprecipitate lanes show proteins bound to A/G protein agarose. Immunoblots were probed with antibodies to SCAR1 and COP1.
(E) and (F) Representative images (E) and root length measurements (F) of dark-grown 5-d-old single arp3, brk1, and cop1, and arp3 cop1 and brk1 cop1 double mutant seedlings. Arrowheads in (E) point to the root-hypocotyl junction. Values (means + se) with different letters in (F) are significantly different by Tukey’s tests at the 99% confidence level (n ≥ 30 plants for each variant).
COP1 binds proteins targeted for degradation via its WD domain (COP1-WD) (Torii et al., 1998; Wang et al., 2001). If SCAR1 is a substrate of COP1, direct interaction between SCAR1 and COP1-WD would be expected. To examine the physical interaction between SCAR1 and COP1-WD, we cotranslated SCAR1 in vitro together with T7-tagged COP1-WD or with a T7-tagged negative control protein and immunoprecipitated T7-tagged proteins using anti-T7 antibodies. SCAR1 coprecipitated with T7-tagged COP1-WD, but not with T7-tagged negative controls (Figure 6B), indicating direct interaction between SCAR1 and COP1-WD. Interaction between SCAR1 and COP1 was further confirmed using root extracts immunoprecipitated with anti-SCAR1 or anti-COP1 antibodies (Figures 6C and 6D).
We also generated brk1 cop1 and arp3 cop1 double mutants to evaluate genetic interactions between COP1, and BRK1 and ARP3. The root lengths of 5-d-old dark-grown double brk1 cop1 and arp3 cop1 mutants were very similar to those of single cop1, brk1, and arp3 mutants, indicating that COP1 functions in a genetic pathway with BRK1 and ARP3 in regulating Wc-dependent root growth (Figures 6E and 6F). Taken together, our biochemical and genetic analyses support an interaction between COP1 and SCAR/ARP2/3 pathways.
Monochromatic Light Promotes Maintenance of SCAR Complex, F-Actin, and Root Growth via Photoreceptors
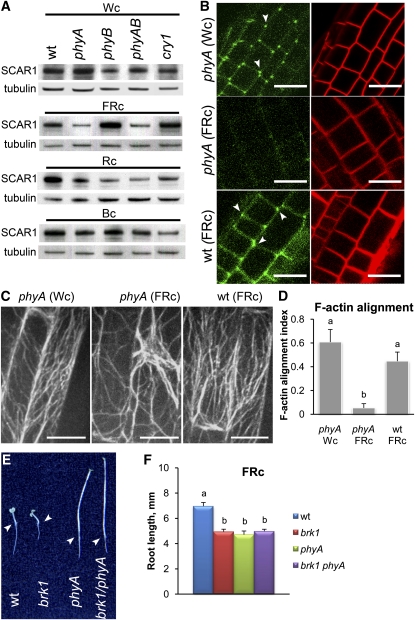
To determine if the promotion of SCAR1 levels and ARP2/3-SCAR–dependent root elongation observed under Wc are regulated by a particular light wavelength(s) and/or fluence rate, we analyzed growth of 4-d-old wild-type, brk1, and arp3 seedlings in continuous monochromatic red (Rc), far-red (FRc), and blue (Bc) light. Both brk1 and arp3 displayed shorter roots compared with wild-type seedlings under increasing fluence rates of Rc, FRc, or Bc light conditions (see Supplemental Figure 6 online). Thus, Rc, FRc, and Bc light appear to all contribute to ARP2/3-SCAR–mediated root growth. Light responses are mediated through photoreceptors (Schäfer and Nagy, 2006). When grown under FRc, seedlings carrying mutations in the gene encoding the FRc light receptor phytochrome A (phyA) were morphologically very similar to wild-type seedlings grown in darkness, indicating that PHYA is the major photoreceptor responsible for perceiving FRc (Schäfer and Nagy, 2006). We therefore decided to use FRc-grown phyA mutants to test whether photoreceptors are involved in regulating ARP2/3- SCAR–dependent root growth. Roots of FRc-grown phyA seedlings exhibited a dramatic decline in SCAR1 levels, while roots of FRc-grown seedlings carrying mutations in genes encoding the Rc light receptor phytochrome B (phyB) or Bc light receptor cryptochrome 1 (cry1) maintained SCAR1 levels comparable to those of the wild type (Figure 7A). In addition, roots of FRc-grown phyA seedlings lacked the cell periphery–associated BRK1-YFP fluorescence pattern observed in FRc-grown wild-type seedlings, indicating the degradation of SCAR complex in roots of phyA in FRc (Figure 7B). To test the effect of PHYA on F-actin organization, roots of FRc-grown phyA seedlings expressing GFP-ABD2-GFP were imaged. F-actin was randomly organized in these roots, in contrast with the predominantly longitudinal alignment of F-actin observed in roots of the wild type grown under the same FRc light (Figures 7C and 7D). Loss of ordered F-actin thus parallels SCAR complex degradation in FRc-grown phyA roots. We further reasoned that if PHYA is part of the machinery responsible for the promotion of root growth in FRc via ARP2/3- SCAR–dependent F-actin polymerization, then root phenotypes of seedlings carrying both brk1 and phyA mutations would not be more severe than those of individual phyA mutants. Indeed, we found that the morphologies of FRc-grown double brk1 phyA and single phyA mutants were very similar, indicating that the PHYA acts in a common pathway with BRK1 (Figures 7E and 7F). Thus, our study of FRc-grown phyA suggests that PHYA is required for FRc light to promote SCAR complex, F-actin organization, and root elongation growth in wild-type roots.
Figure 7.
Stability of SCAR1 and Maintenance of Aligned F-Actin Arrays in Roots Requires Photoreceptors.
(A) SCAR1 protein levels decline in roots of phyA seedlings grown in FRc light. Proteins were extracted from roots of 4-d-old wild-type (wt), phyA, phyB, phyAB, and cry1 seedlings grown in Wc (18 μmol m−2 s−1), FRc (1 μmol m−2 s−1), Rc (42 μmol m−2 s−1), or Bc (10 μmol m−2 s−1) light. Immunoblots were probed with antibodies to SCAR1 and tubulin.
(B) BRK1-YFP fluorescence dissipates in roots of phyA mutant seedlings in FRc. Confocal images of BRK1p:BRK1-YFP (green panels) in root tips of 4-d-old wild-type and phyA seedlings grown in Wc or in FRc. Roots were counterstained with FM4-64 to mark the outline of the cell (red panels). Arrowheads point to cell corners showing local enrichments of fluorescence.
(C) and (D) F-actin is misaligned in roots of phyA mutants in FRc. Representative confocal images (C) and alignment index (D) of F-actin in expanding root cells of 4-d-old wild-type and phyA seedlings grown in Wc or in FRc. Images are projections of 5.5-μm Z-stacks focused on the center of the cells. Indexes of F-actin alignment (means + se) were calculated for 50 to 70 cells for each genotype and light treatment. Values with different letters in (D) are significantly different by Tukey’s tests at the 99% confidence level. Bars = 20 μm.
(E) and (F) Genetic interactions between scar1 and phyA. Representative images (E) and root length measurements (F) of 4-d-old wild-type, brk1 and phyA single, and brk1 phyA double mutants grown in 1 μmol m−2 s−1 FRc. Arrowheads in (E) point to the root-hypocotyl junction. Values (means + se) with different letters in (F) are significantly different by Tukey’s tests at the 99% confidence level (n ≥ 30 plants for each variant).
To dissect photoreceptors involved in the regulation of SCAR1 by Rc and Bc monochromatic light, we analyzed SCAR1 in root extracts of phyB and cry1 mutants. Levels of SCAR1 proteins were reduced in phyB seedlings grown under Rc, and in cry1 under Bc, compared with corresponding wild-type seedlings grown under the same light conditions (Figure 7A). Thus, PHYB and CRY1 appear to be involved in suppressing SCAR1 degradation by Rc and Bc accordingly. Altogether, our data indicate that light perceived by photoreceptors suppresses dark-induced SCAR complex degradation and promotes ARP2/3-SCAR–dependent F-actin organization and root growth.
DISCUSSION
When grown in the dark, roots of Arabidopsis seedlings are significantly shorter than roots of seedlings exposed to light. However, the underlying molecular mechanisms by which light promotes root growth remain poorly understood. Here, we report on a signaling pathway through which light could regulate root development. This pathway involves direct sensing of light by root photoreceptors and the subsequent transmission of the light signal to the actin cytoskeleton. A link between light and actin in mediating root growth is supported by two key observations. First, mutants disrupted in genes encoding various components of the actin nucleating ARP2/3 and SCAR complex displayed shorter roots in Wc that were reminiscent of the short roots of wild-type seedlings grown in the dark (Figure 1). Second, actin organization in the short roots of SCAR complex mutants exposed to light resembled that of wild-type roots grown in the dark (Figure 3). These observations indicate that light is essential for roots of wild-type seedlings to maintain normal actin organization to sustain rapid elongation growth.
How might the light signal sensed by the seedling be relayed to the actin cytoskeleton? On the basis of the similar root phenotypes of ARP2/3 and SCAR complex mutants and dark-grown wild-type seedlings, we hypothesized that ARP2/3-SCAR proteins link light perception to actin reorganization in roots. The mechanism by which this is accomplished is through the tight control of the localization and stability of the SCAR complex when exposed to various light regimes. In the absence of light, the SCAR1 protein in roots was degraded, and this degradation was associated with BRK1-YFP relocalizing from the PM to the cytoplasm (Figure 8). We previously demonstrated that BRK1 localization to the PM requires SCARs (Dyachok et al., 2008). Because BRK1 physically binds to three of the four SCARs (Frank et al., 2004; Uhrig et al., 2007; Zhang et al., 2005), BRK1-YFP dissociation from the PM most likely reflects the degradation of multiple SCAR proteins in the dark. The degradation of SCARs in roots deprived of light could then result in a concomitant rearrangement of the actin cytoskeleton leading to an inhibition of root elongation. Indeed, we found that loss of longitudinal F-actin arrays accompanied growth inhibition in roots of brk1 and arp3 mutants in the light and also in dark-grown wild-type plants (Figure 3). Loss of F-actin alignment also accompanied growth impairments in trichomes and epidermal pavement cells of Arabidopsis ARP2/3-SCAR pathway mutants (Mathur et al., 2003a, 2003b; Brembu et al., 2004). The importance of light in SCAR complex function could be preserved beyond Arabidopsis, as light is also a potent activator of SCAR and ARP2/3-dependent actin polymerization in other systems, including phototaxis in Dictyostelium (Pollitt and Insall, 2009) and light-activated polar cell growth in Physcomitrella (Perroud and Quatrano, 2006, 2008).
Figure 8.
Model for Regulation of SCAR Complex by Light.
(A) Upon perception of red, far-red, or blue light stimuli by photoreceptors, the degradation of SCAR is suppressed. The complex containing SCAR and BRK1 proteins is recruited to the PM, where it promotes ARP2/3-dependent longitudinal F-actin arrays and root elongation growth.
(B) In the absence of light, SCAR and BRK1 proteins dissociate from the PM. The SCAR protein is recruited to the nucleus and is degraded there via COP1 and 26S proteasomes.
It is well established that cell expansion in plants relies on the abundance of ARP2/3-SCAR–dependent F-actin (Le et al., 2003; Li et al., 2003; Deeks et al., 2004; Dyachok et al., 2008; Perroud and Quatrano 2008). In this study, we demonstrate that in addition to abundance, light-dependent ARP2/3-SCAR maintenance of F-actin organization into longitudinal arrays is important for the elongation growth of roots (Figure 3). Why is the longitudinal alignment of F-actin critical for root elongation growth? It is possible that longitudinally oriented F-actin promotes cell expansion by stretching the polysaccharide matrix of the cell wall (Thimann et al., 1992) and/or by promoting the delivery of endocytic vesicles to expanding ends of root cells, similar to their function in endocytosis in mammalian and yeast cells (Kaksonen et al., 2006). Longitudinal arrays of F-actin could also promote myosin-driven transport of building material for cell expansion (Peremyslov et al., 2010). The role of longitudinal arrays of ARP2/3-polymerized F-actin in cell expansion is consistent with the short cells in the beginning and middle parts of the elongation zone, indicating slow expansion rates in Wc-grown ARP2/3-SCAR mutant roots (see Supplemental Figure 1 online). Alternatively, cortical F-actin might influence cell division by directing endocytic trafficking of pectins and other compounds required for new cell wall formation (Baluska et al., 2002) or through distributing compounds regulating cell division, including auxin (Dhonukshe et al., 2008).
In this article, we provide experimental evidence that the degradation of SCAR1 in dark-grown roots is facilitated by the 26S proteasome and COP1 E3 ligase pathway similar to proteasome-mediated degradation of animal SCARs (Rogers et al., 2003; Mitsushima et al., 2006). We also found that COP1 and SCAR1 coprecipitated in vivo, and SCAR1 coprecipitated with the WD domain of COP1 in vitro (Figure 6). Thus, SCAR1 could be a direct target of the COP1- and proteasome-mediated degradation pathway similar to proteasome-mediated degradation of the mammalian SCAR-related N-WASP protein (Suetsugu et al., 2002). Alternatively, the degradation of SCAR1 could be a consequence of downregulation of other components of the SCAR complex, such as PIR1, NAP1, BRK1, and ABILs, which have been proposed to protect plant SCARs from proteasome-mediated degradation, as demonstrated in animals (Blagg et al., 2003; Kunda et al., 2003; Rogers et al., 2003).
The presence of SCAR in the nucleus of dark-grown roots as shown by our in vitro assays (Figure 6) indicates that there must be a mechanism for localizing it there for proteolysis. Both SCAR and SCAR-related N-WASP can accumulate in the nucleus in mammalian cells (Westphal et al., 2000; Suetsugu and Takenawa, 2003). However, unlike N-WASP, neither mammalian nor plant SCARs appear to have a nuclear localization signal; thus, the mechanism of SCAR targeting to the nucleus is currently unknown. It is tempting to speculate that SCAR proteins in dark-grown root cells might bind to yet unidentified proteins with a nuclear localization signal or enter the nucleus with other SCAR complex components via complex formation. For example, the Abi1 subunit of the mammalian SCAR complex is known to undergo nucleocytoplasmic shuttling via its SCAR/WAVE binding domain (Echarri et al., 2004). It would be interesting to determine if other components of the SCAR complex localize to the nucleus and if constitutively targeting SCAR to the PM impacts light-dependent root elongation. Moreover, nuclear ARPs, including ARP2 and ARP3 components of the ARP2/3 complex, are proposed to function in regulating gene expression in animals and in plants; thus, SCARs could also have yet unidentified nuclear-specific functions (Yoo et al., 2007; Fenn et al., 2011; Meagher et al., 2011).
Most published work on the SCAR and ARP2/3 complex class of mutants has focused on phenotypes associated with leaf development, such as abnormal trichome morphology, reduced lobe formation, and altered intercellular adhesion in epidermal pavement cells (Le et al., 2003, 2006; Li et al., 2003; Mathur et al., 2003a, 2003b; Basu et al., 2004, 2005; Brembu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004; Li et al., 2004; Saedler et al., 2004; Zimmermann et al., 2004; Zhang et al., 2005; Djakovic et al., 2006). It was only recently that obvious primary root growth phenotypes have been revealed in these mutants (Dyachok et al., 2008). It is perhaps due to a peculiar interplay between agar and Suc, two standard ingredients of Arabidopsis growth media, which have circumvented a better appreciation of the importance of the ARP2/3 or SCAR/WAVE complex in root development. It is noteworthy that short roots are mirrored by short hypocotyls in young mutant seedlings grown in the absence of sugars, but not by other experimental conditions (e.g., in mutants grown on an agar- and Suc-supplemented media; Basu et al., 2004, 2005; Li et al., 2004; Saedler et al., 2004; Zhang et al., 2008; Jörgens et al., 2010). Thus, it is possible that light regulates ARP2/3-SCAR–dependent growth in roots via mechanisms that are different from those in hypocotyls. The organ-specific nature of ARP2/3- SCAR–dependent growth can be further inferred from our observations of cotyledons. In contrast with roots and hypocotyls, we found no evidence that light regulates the SCAR complex in cotyledons. BRK1 and SCAR1 complex subunits are present in the PM of cotyledon cells regardless of the light environment (Figure 4). This is consistent with the fact that expansion of cotyledons is not affected by mutations in any of the components of the ARP2/3-SCAR complex. These data suggest that the ARP2/3-SCAR complex may function in an organ-specific manner possibly via association with organ-specific factors.
We found that the decline in root growth of wild-type seedlings in darkness is associated with loss of SCAR complex subunits, consistent with the SCAR complex promoting light- but not dark-dependent root growth. Moreover, because root length in darkness at day 3 is greater in SCAR and ARP2/3 complex subunit mutants than it is in the wild type (Figure 1), the SCAR and ARP2/3 complex could actually be inhibitory to early root growth in the dark. This observation is seemingly contradicted by the evidence that the SCAR1 protein is degraded in darkness. It is important to note, however, that the SCAR1 degradation and BRK1-YFP dissociation from the PM do not occur until day 4 (Figure 4). Modifications of the SCAR complex in the absence of light stimuli could trigger both inhibition of root growth at day 3 and degradation of the SCAR protein at a later stage. Such transient activities of proteins targeted for degradation are known. One example is in the SCAR-related N-WASP that activates ARP2/3 complex in developing neurons. Rapid phosphorylation of N-WASP under the conditions that trigger cell extension promotes both initial activation of ARP2/3 complex and subsequent degradation of N-WASP concomitant with the decline in cell extension (Suetsugu et al., 2002). Opposite effects of the ARP2/3-SCAR pathway on early root growth imply alternative pathways activated downstream of the complexes in the light and in the dark. Existence of a parallel pathway controlling plant growth in the absence of light is also consistent with the subtlety of growth alterations in mutants grown under diurnal light conditions (Figure 1A; Szymanski, 2005; Dyachok et al., 2008).
It is generally accepted that roots of plants are exposed to light in soil, but whether roots can perceive light directly is largely debated (Galen et al., 2007). It has been demonstrated that the root is the major site of the perception of UV-B light that inhibits root growth (Tong et al., 2008; Leasure et al., 2009). Here, we demonstrated that the light signal, which triggers root elongation growth, is also sensed primarily by the root. Three observations support this hypothesis. First, shading only the root of wild-type seedlings was sufficient to inhibit their growth to almost the same extent as whole seedlings grown in the dark (Figure 1E). Second, roots of chimeric seedlings wherein brk1 is the root donor were always shorter than roots of chimeric seedlings with wild-type roots regardless of the genotype of the shoot (Figure 1G). Third, exposure to light inhibited dark-induced degradation of SCAR1 similarly in excised and intact roots (Figure 5E).
Although the mechanism of UV-B sensing in roots is photoreceptor independent (Tong et al., 2008), our findings show that roots transmit light to a growth response via photoreceptors, similar to light signaling in aerial parts (Schäfer and Nagy, 2006). For example, PHYA is critical for sensing of FRc light by roots, since root phenotypes of FRc-grown phyA mutants are very similar to those of the wild type grown in the dark, including degradation of SCAR1, dissociation of BRK1-YFP from the PM, and loss of longitudinally aligned F-actin (Figure 7). Thus, both photoreceptor-dependent and -independent mechanisms of light signaling might be present in roots.
In summary, this study provides additional genetic and biochemical evidence that roots possess the molecular machinery for perceiving light signals. An important component of the light signal transduction machinery in roots is the light-activated recruitment of the SCAR complex to the PM where it activates the ARP2/3 complex, which in turn promotes the formation of longitudinal F-actin arrays and elongation growth (Figure 8).
METHODS
Plant Material
Mutants used in this study were in the Columbia background and included brk1-1 (Djakovic et al., 2006; Le et al., 2006), arp3 (Li et al., 2003), arp2 (Li et al., 2003), scar1,2,3,4 quadruple mutant (Dyachok et al., 2008), cop1-6 (McNellis et al., 1994), cry1 (Lin et al., 1996), phyA-211, phyB-9, and phyA phyB double mutant (Reed et al., 1994). BRK1p:BRK1-YFP (Dyachok et al., 2008) was introduced into brk1 mutants, and 35Sp:GFP-ABD2-GFP (Wang et al., 2008) was introduced into Columbia wild type, brk1, arp3, and phyA mutants via Agrobacterium tumefaciens–mediated transformation using the floral dip method (Clough and Bent, 1998). BRK1p:BRK1-YFP was introduced into cop1 and phyA mutants via crossing to transgenic brk1 expressing BRK1p:BRK1-YFP. Seeds used in this study were obtained from plants grown at 80 μmol m−2 s−1 intensity of fluorescent light on an 18-h-light/6-h-dark cycle at 22°C.
Growth Assays
Surface-sterilized seeds were dried on Whatman filter paper for 7 to 10 d before use. For root growth assays, seeds were sown on half-strength Murashige and Skoog medium without Suc, solidified with 0.6% agar (Sigma-Aldrich). After stratification at 4°C for 4 d, seeds were exposed to 2 h of 60 μmol m−2 s−1 Wc at room temperature to induce germination before placing them in darkness at 21°C for 22 h. Vertically oriented plates with seedlings were then either maintained in darkness or transferred to 18 μmol m−2 s−1 Wc at 21°C for up to an additional 7 d. Daily increments of root growth were determined in light-grown seedlings by marking the positions of the root tips on the back of the plate at 24-h intervals. Roots were measured daily for the individual dark-grown seedlings.
To assay the effect of shielding from light, seedlings were grown horizontally on a black foil–covered Petri dish containing half-strength Murashige and Skoog medium with 0.45% gelzan (Caisson Laboratories) for 5 d (Tong et al., 2008). Holes were generated in black foil using 21-gauge needles to allow seeds to contact the growth medium.
For grafting studies, 5-d-old root and shoot donor seedlings were grafted using a transverse-cut butt alignment and a silicon tubing splint (0.3 mm i.d., HelixMark*/VWR) (Turnbull, 2010). Plates were kept at 27°C for 2 d after grafting and then at 21°C for 6 d. Plates were kept in a horizontal position for 3 d after grafting and then restored to a vertical position.
For monochromatic light experiments, plates with seeds were exposed to 3000 μmol FRc before transferring to growth chambers equipped with monochromatic red, far-red, and blue light emitting diodes (Model E30LED; Perceival Scientific) for an additional 3 d (Shen et al., 2005). Plants were imaged using a Nikon DXM 1200C camera controlled by the Nikon ACT1-C software.
Root and cell lengths were measured using ImageJ software version 1.42o (http://rsb.info.nih.gov/ij/). For measuring lengths of expanding and mature cortical cells, seedlings were mounted in 5 μg/mL FM4-64 (Molecular Probes/Invitrogen) in water and imaged using a Leica TCS SP2 AOBS confocal laser scanning microscope equipped with a ×20 HC Plan-Apo objective (numerical aperture 0.7). Lengths of six to eight mature cortical cells near the beginning of root hair zones were measured in each root. Length of expanding cells was measured in files of cells between the meristematic and root hair zone.
Mitotic figures (mitotic spindles and phragmoplasts) were counted in division zones (~200 μm from the root tip) of fixed roots mounted in 100 nM 4’,6-diamidino-2-phenylindole (Invitrogen) in PBS and imaged using the confocal microscope.
Confocal Microscopy
Seedlings expressing BRK1p:BRK1-YFP were mounted in 5 μg/mL FM4-64 in water on slides and examined for YFP and FM4-64 fluorescence using a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems) equipped with a ×63 HCX Plan-Apo water immersion objective (numerical aperture 1.20). Individual images were acquired using Leica LCS confocal software. Fluorescence of GFP-ABD2-GFP was visualized using a Perkin-Elmer UltraView ERS spinning disk confocal microscope equipped with a ×63 Zeiss C-Apochromat water immersion objective (numerical aperture 1.20) and Hamamatsu electron-multiplying–charge-coupled device digital camera. Z-series of 50 to 55 images were acquired at a 0.1-μm interval, and projections were assembled using Volocity 4.3.1 software (Improvision).
For the analysis of F-actin alignment, colored images from the confocal microscope were first converted into gray-scale images followed by the Canny edge detection process to eliminate background noise and covert the images into a gradient mix (Canny, 1986). Fast Fourier transformation was then applied to generate a two-dimensional power spectrum of each gradient matrix. Entropy values for all spectrum elements of each image were calculated based on their 9 × 9 neighborhood elements with a 50% power spectrum threshold. The alignment index of F-actin in an image was then calculated based on the two-dimensional entropy matrix, defined as the ratio between the number of pixels with zero entropy value and the total number of pixels in the matrix. The MATLAB scripts for the aforementioned processing steps are part of a larger image analysis software package that is available upon request.
RNA and Protein Analyses
To prepare total RNA and proteins, ~0.5 g of roots or cotyledons were extracted with Trizol reagent (Invitrogen). After phase separation, RNA was precipitated from upper (aqueous) phase, and proteins were precipitated from lower (organic) phase, according to the manufacturer’s protocol. RNA was purified using the RNeasy Micro Kit (Qiagen), and genomic DNA was removed using the RQ1 RNase-free DNase (Promega). Single-strand cDNA was synthesized using the SUPERSCRIPT III first-strand synthesis system (Invitrogen). For qRT-PCR, cDNA was amplified using Power Sybr Green (Applied Biosystems), according to the manufacturer’s instructions. qRT- PCR was performed with an ABI PRISM 7900 HT sequence detection system (Applied Biosystems), and data were collected and analyzed using the SDS 2.2.1 software (Applied Biosystems). Arabidopsis thaliana SCAR1 cDNA was amplified with two primer pairs: 5′-CTTAAAGAACATTCAGCGGTTG-3′ and 5′-CCCTTCGCTTTCAGTCTCAT-3′, and 5′-AATGTGCTGCTACATCAGAGC-3′ and 5′-GCCACTACTTATTTCCGAAGC-3′. These primer pairs gave similar estimates of expression levels of SCAR1 for each sample. The first primer pair was then selected to present the expression data for SCAR1. EIF4A-2 cDNA was amplified with the 5′-GCTTGACTATGCCCTTCTCC-3′ and 5′-CATGAACCTTGACACCTTGG-3′ primer pair. Expression of EIF4A-2 (At1g54270) was similar in all variants and under all light conditions tested. EIF4A-2 was then selected to normalize the expression data for SCAR1 gene.
Microsomal fractions were prepared from ~0.5 g of roots or cotyledons of Wc- or dark-grown seedlings as described (Dyachok et al., 2008). Nuclear fractions were prepared from ~1.5 g of roots from 4-d-old dark-grown seedlings treated with or without MG132 (50 μM) for 3 h in darkness as described (Liu et al., 2001). Gel electrophoresis and immunoblotting of protein extracts were performed as previously described (Djakovic et al., 2006). SCAR1 was visualized using anti-SCAR1 antibodies (Frank et al., 2004) diluted to 0.1 μg/mL and horseradish peroxidase–conjugated goat anti-chicken IgY antibody (GenWay) diluted 1:20,000. Anti-α-1,2-Mannosidase I (Preuss et al., 2004) and anti-PUX1 (Rancour et al., 2004) were used to detect membrane and cytoplasmic proteins, respectively. Both antibodies were a gift from David Rancour (University of Wisconsin). For the detection of BRK1-YFP, mouse anti-GFP (Zymed/Invitrogen) was used at 0.05 μg/mL followed by alkaline phosphatase–conjugated goat anti-mouse IgG (Promega) diluted 1:10,000. Anti-RNA polymerase II antibody (1.BB.61; Santa Cruz Biotechnology) was used to detect nuclear proteins. Anti-α-tubulin antibody (AbD Serotec) was used to ensure equal loading of total proteins. Blots were developed using ECL Plus Western Blotting Detection Reagents (GE Healthcare/Amersham) and imaged using BioMax XAR film (Perkin-Elmer). Blots were stripped between labeling with different antibodies using Restore Plus Stripping Buffer (Pierce/ThermoScientific) according to the manufacturer’s protocol.
In Vitro Transcription-Translation and in Vivo Binding Experiments
To produce T7-tagged COP1-WD, sequence amplified from COP1 cDNA with primers 5′- GAATTCTCTGTAAAGTTGCGGATGCTC-3′ and 5′-TCACGCAGCGAGTACCAGGTCGAC-3′ was cloned in frame with T7 tag into pET28a vector (Novagen). Full-length SCAR1 cDNAs in pBluescript SK+ and T7-NC (the N-terminal half of At3g05330) were described previously (Frank et al., 2004). Pairs of DNA plasmids (SCAR1 plus the T7-fusion construct) were cotranscribed and cotranslated in vitro using the TNT T7 wheat germ extract system (Promega) according to the manufacturer’s protocols for the nonradioactive reaction. T7-tagged proteins were precipitated using anti-T7 Tag antibody agarose beads (Novagen) as described previously (Frank et al., 2004). Aliquots of total samples (1/100) and immunoprecipitates (1/2) were analyzed via gel electrophoresis and immunoblotting. For the detection of T7-tagged proteins, horseradish peroxidase–conjugated anti-T7 tag antibody (EMD Chemicals) was used diluted 1:10,000. For the detection of COP1-WD, anti-COP1 antibody (L20; Santa Cruz Biotechnology) was used at 0.1 μg/mL followed by horseradish peroxidase–conjugated donkey anti-goat antibody (Santa Cruz Biotechnology) diluted 1:20,000.
To analyze binding between COP1 and SCAR1 proteins in root extracts, ~3 g of 3-d-old dark-grown seedlings were ground in liquid nitrogen and homogenized in 3 mL extraction buffer (TBS with 1 mM DTT, 1/100 plant protease inhibitor cocktail) containing 0.2% Tween 20 using an Omni TH homogenizer on ice. Extracts were incubated at room temperature for 1 h with end-over-end rotation in darkness, diluted 10-fold with no detergent extraction buffer, passed twice over eight layers of Miracloth, and centrifuged twice at 3000g for 10 min. Cleared extracts were incubated with 0.5 μg anti-SCAR1 antibody and 20 μL protein A/G PLUS agarose beads (Santa Cruz Biotechnology). Alternatively, extracts were incubated with 10 μL protein A/G PLUS agarose beads in the presence or in the absence of 0.25 μg anti-COP1 antibody. Antibody incubations were performed at room temperature for 2 h with end-over-end rotation in darkness. Immunoprecipitates were collected by centrifugation at 1700g for 10 min, washed twice with TBS, resuspended in SDS loading buffer, and analyzed via gel electrophoresis and immunoblotting as described above.
BRK1p:GUS Construct and Fixed Material Staining
The BRK1p:β-glucuronidase (GUS) construct was created by amplifying 1.4-kb sequence upstream of the BRK1 coding region (At2g22640) from Columbia genomic DNA with primers 5′-AACTGCAGAGCAATGGAACAGCTTCTCG-3′ and 5′-GCTCTAGATATTAAACCCCAAAACTCTCTCTTC-3′ and cloning the PCR product upstream of UidA (GUS) into T-DNA binary vector pDW137 (Blázquez et. al., 1997). BRK1p:GUS was introduced into Columbia wild type via Agrobacterium-mediated transformation using the floral dip method (Clough and Bent, 1998).
Histochemical staining for GUS activity was performed with 125 μg/mL 5-bromo-4-chloro-3-indolyl β-d-glucuronide cyclohexylamine salt (Gold Biotechnology) as described previously (Meister et al., 2002). The samples were washed with 0.24 n HCl in 20% methanol at 57°C for 15 min and then 60% ethanol at room temperature for 15 min, then rehydrated in 40, 20, 10 ethanol at room temperature for 5 min each, followed by 5% ethanol in 25% glycerol at room temperature for 15 min. The samples were mounted in 50% glycerol, imaged using a Nikon Microphot-FX microscope equipped with Nikon Digital Sight DS-Ri1 Camera, run by Nikon NIS-Elements F3.0 software.
Accession Numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: ARP2 (At3g27000), ARP3 (At1g13180), BRK1 (At2g22640), SCAR1 (At2g34150), SCAR2 (At2g38440), SCAR3 (At1g29170), SCAR4 (At5g01730), COP1 (At2g32950), CRY1 (At4g08920), PHYA (At1g09570), PHYB (At2g18790), and EIF4A-2 (At1g54270).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cell Expansion and Division Is Altered in brk1 and arp3 Compared with Wild-Type Roots.
Supplemental Figure 2. Hypocotyls Are Inhibited in Continuous White Light–Grown but Not in Dark-Grown brk1 and arp3 Mutants Compared with the Wild Type.
Supplemental Figure 3. Roots Are Not Inhibited in brk1 and arp3 Mutants Grown on Media Combining Agar and Suc.
Supplemental Figure 4. Root Length and F-Actin Organization in Roots of 4-d-Old Seedlings Grown in the Dark on Media Supplemented with Low (0.6%) or High (1%) Agar.
Supplemental Figure 5. BRK1 Is Expressed in Light- and Dark-Grown Plants.
Supplemental Figure 6. Root Length of brk1 and arp3 Seedlings Is Inhibited in Red, Far Red, and Blue Light Compared with Wild-Type Seedlings.
Acknowledgments
We thank Li Quan for design of qRT-PCR primers, David Rancour for the antibodies, Hui Shen for helpful discussions, and Laurie Smith, Amanda Wright, Richard Nelson, Carolyn Rasmussen, and Phillip Harries for critical review of the manuscript. This work was supported by the Samuel Roberts Noble Foundation (J.D. and E.B.B.), the Oklahoma Center for the Advancement of Science and Technology (Grant OCAST PSB10-004 to E.B.B.), and the National Science Foundation (Grants IOS-0822811 and IOS-0849287 to E.H.). The confocal microscopes used in this study were supported by National Science Foundation Major Research Instrumentation Grants DBI-0722635 and DBI-0400580 to E.B.B.
AUTHOR CONTRIBUTIONS
J.D. conceived and designed the project. J.D., L.Z., and E.H. conducted light experiments. F.L. and J.H. developed the algorithm for F-actin quantification. J.D., E.H., and E.B.B. analyzed and interpreted the data. J.D. wrote the article with the help of E.H. and E.B.B.
References
- Baluska F., Hlavacka A., Samaj J., Palme K., Robinson D.G., Matoh T., McCurdy D.W., Menzel D., Volkmann D. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol. 130: 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D., El-Assal Sel.-D., Le J., Mallery E.L., Szymanski D.B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131: 4345–4355 [DOI] [PubMed] [Google Scholar]
- Basu D., Le J., El-Essal Sel.-D., Huang S., Zhang C., Mallery E.L., Koliantz G., Staiger C.J., Szymanski D.B. (2005). DISTORTED3/SCAR2 is a putative arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17: 502–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagg S.L., Stewart M., Sambles C., Insall R.H. (2003). PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13: 1480–1487 [DOI] [PubMed] [Google Scholar]
- Blázquez M.A., Soowal L.N., Lee I., Weigel D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Brembu T., Winge P., Seem M., Bones A.M. (2004). NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16: 2335–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canamero R.C., Bakrim N., Bouly J.P., Garay A., Dudkin E.E., Habricot Y., Ahmad M. (2006). Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224: 995–1003 [DOI] [PubMed] [Google Scholar]
- Canny J.F. (1986). A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8: 679–698 [PubMed] [Google Scholar]
- Ciechanover A. (1994). The ubiquitin-proteasome proteolytic pathway. Cell 79: 13–21 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Correll M.J., Kiss J.Z. (2005). The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol. 46: 317–323 [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Hussey P.J. (2005). Arp2/3 and SCAR: Plants move to the fore. Nat. Rev. Mol. Cell Biol. 6: 954–964 [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Kaloriti D., Davies B., Malhó R., Hussey P.J. (2004). Arabidopsis NAP1 is essential for Arp2/3-dependent trichome morphogenesis. Curr. Biol. 14: 1410–1414 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 105: 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic S., Dyachok J., Burke M., Frank M., Smith L.G. (2006). BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133: 1091–1100 [DOI] [PubMed] [Google Scholar]
- Dyachok J., Shao R.M., Vaughn K., Bowling A., Facette M., Djakovic S., Clark L., Smith L.G. (2008). Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol. Plant 1: 990–1006 [DOI] [PubMed] [Google Scholar]
- Echarri A., Lai M.J., Robinson M.R., Pendergast A.M. (2004). Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol. Cell. Biol. 24: 4979–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal Sel.-D., Le J., Basu D., Mallery E.L., Szymanski D.B. (2004). Arabidopsis GNARLED encodes a NAP125 homolog that positively regulates ARP2/3. Curr. Biol. 14: 1405–1409 [DOI] [PubMed] [Google Scholar]
- Fenn S., Breitsprecher D., Gerhold C.B., Witte G., Faix J., Hopfner K.P. (2011). Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J. 30: 2153–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M., Egile C., Dyachok J., Djakovic S., Nolasco M., Li R., Smith L.G. (2004). Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc. Natl. Acad. Sci. USA 101: 16379–16384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen C., Rabenold J.J., Liscum E. (2007). Functional ecology of a blue light photoreceptor: effects of phototropin-1 on root growth enhance drought tolerance in Arabidopsis thaliana. New Phytol. 173: 91–99 [DOI] [PubMed] [Google Scholar]
- Gilliland L.U., Pawloski L.C., Kandasamy M.K., Meagher R.B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33: 319–328 [DOI] [PubMed] [Google Scholar]
- Iwabuchi K., Minamino R., Takagi S. (2010). Actin reorganization underlies phototropin-dependent positioning of nuclei in Arabidopsis leaf cells. Plant Physiol. 152: 1309–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörgens C.I., Grünewald N., Hülskamp M., Uhrig J.F. (2010). A role for ABIL3 in plant cell morphogenesis. Plant J. 62: 925–935 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kadota A., Yamada N., Suetsugu N., Hirose M., Saito C., Shoda K., Ichikawa S., Kagawa T., Nakano A., Wada M. (2009). Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 13106–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Toret C.P., Drubin D.G. (2006). Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7: 404–414 [DOI] [PubMed] [Google Scholar]
- Kiss J.Z., Mullen J.L., Correll M.J., Hangarter R.P. (2003). Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 131: 1411–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., Baum B. (2003). Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13: 1867–1875 [DOI] [PubMed] [Google Scholar]
- Le J., El-Assal Sel.-D., Basu D., Saad M.E., Szymanski D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Le J., Mallery E.L., Zhang C., Brankle S., Szymanski D.B. (2006). Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr. Biol. 16: 895–901 [DOI] [PubMed] [Google Scholar]
- Leasure C.D., Tong H.Y., Yuen G.G., Hou X.W., Sun X.F., He Z.H. (2009). ROOT UV-B SENSITIVE2 acts with ROOT UV-B SENSITIVE1 in a root ultraviolet B-sensing pathway. Plant Physiol. 150: 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Blanchoin L., Yang Z., Lord E.M. (2003). The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 132: 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sorefan K., Hemmann G., Bevan M.W. (2004). Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol. 136: 3616–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Ahmad M., Cashmore A.R. (1996). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10: 893–902 [DOI] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J., Wagner D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M., Insall R.H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8: 1347–1356 [DOI] [PubMed] [Google Scholar]
- Mandoli D.F., Ford G.A., Waldron L.J., Nemson J.A., Briggs W.R. (1990). Some spectral properties of several soil types: Implications for photomorphogenesis. Plant Cell Environ. 13: 287–294 [Google Scholar]
- Mandoli D.F., Tepperman J., Huala E., Briggs W.R. (1984). Photobiology of diagravitropic maize roots. Plant Physiol. 75: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Kernebeck B., Hülskamp M. (2003b). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Kirik V., Kernebeck B., Srinivas B.P., Hülskamp M. (2003a). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130: 3137–3146 [DOI] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.B., Kandasamy M.K., King L. (2011). Actin functions in the cytoplasmic and nuclear compartments. In The Plant Cytoskeleton, Liu B., (New York; Springer; ), pp. 3–32 [Google Scholar]
- Meister R.J., Kotow L.M., Gasser C.S. (2002). SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Mitsushima M., Sezaki T., Akahane R., Ueda K., Suetsugu S., Takenawa T., Kioka N. (2006). Protein kinase A-dependent increase in WAVE2 expression induced by the focal adhesion protein vinexin. Genes Cells 11: 281–292 [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Pollard T.D. (1999). Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 9: 244–249 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Yokota E., Wada T., Shimmen T., Okada K. (2003). An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polymerization, reveals its distinctive function in root development. Plant Cell Physiol. 44: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Dolja V.V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud P.F., Quatrano R.S. (2006). The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil. Cytoskeleton 63: 162–171 [DOI] [PubMed] [Google Scholar]
- Perroud P.F., Quatrano R.S. (2008). BRICK1 is required for apical cell growth in filaments of the moss Physcomitrella patens but not for gametophore morphology. Plant Cell 20: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt A.Y., Insall R.H. (2009). Loss of Dictyostelium HSPC300 causes a scar-like phenotype and loss of SCAR protein. BMC Cell Biol. 10: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M.L., Serna J., Falbel T.G., Bednarek S.Y., Nielsen E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16: 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Bannigan A., Sulaman W., Pechter P., Blancaflor E.B., Baskin T.I. (2007). Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J. 50: 514–528 [DOI] [PubMed] [Google Scholar]
- Rancour D.M., Park S., Knight S.D., Bednarek S.Y. (2004). Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of arabidopsis CDC48. J. Biol. Chem. 279: 54264–54274 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Wiedemann U., Stuurman N., Vale R.D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R., Zimmermann I., Mutondo M., Hülskamp M. (2004). The Arabidopsis KLUNKER gene controls cell shape changes and encodes the AtSRA1 homolog. Plant Mol. Biol. 56: 775–782 [DOI] [PubMed] [Google Scholar]
- Sakai T., Wada T., Ishiguro S., Okada K. (2000). RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Briggs W.R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury F.J., Hall A., Grierson C.S., Halliday K.J. (2007). Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 50: 429–438 [DOI] [PubMed] [Google Scholar]
- Schäfer E., Nagy F., (2006). Photomorphogenesis in Plants and Bacteria, 3rd ed: Function and Signal Transduction Mechanisms. (Dordrecht, The Netherlands: Springer; ). [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Oppenheimer D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Somers D.E., Quail P.H. (1995). Phytochrome-mediated light regulation of PHYA- and PHYB-GUS transgenes in Arabidopsis thaliana seedlings. Plant Physiol. 107: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S., Hattori M., Miki H., Tezuka T., Yamamoto T., Mikoshiba K., Takenawa T. (2002). Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev. Cell 3: 645–658 [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Takenawa T. (2003). Translocation of N-WASP by nuclear localization and export signals into the nucleus modulates expression of HSP90. J. Biol. Chem. 278: 42515–42523 [DOI] [PubMed] [Google Scholar]
- Suetsugu N., Yamada N., Kagawa T., Yonekura H., Uyeda T.Q.P., Kadota A., Wada M. (2010). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 8860–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D.B. (2005). Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8: 103–112 [DOI] [PubMed] [Google Scholar]
- Thimann K.V., Reese K., Nachmias V.T. (1992). Actin and the elongation of plant cells. Protoplasma 171: 153–166 [Google Scholar]
- Tong H.Y., Leasure C.D., Hou X.W., Yuen G., Briggs W., He Z.H. (2008). Role of root UV-B sensing in Arabidopsis early seedling development. Proc. Natl. Acad. Sci. USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., McNellis T.W., Deng X.W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17: 5577–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R., Kevei E., Hall A., Millar A.J., Nagy F., Kozma-Bognár L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C.G. (2010). Grafting as a research tool. Methods Mol. Biol. 655: 11–26 [DOI] [PubMed] [Google Scholar]
- Uhrig J.F., Mutondo M., Zimmermann I., Deeks M.J., Machesky L.M., Thomas P., Uhrig S., Rambke C., Hussey P.J., Hülskamp M. (2007). The role of Arabidopsis SCAR genes in ARP2-ARP3-dependent cell morphogenesis. Development 134: 967–977 [DOI] [PubMed] [Google Scholar]
- Usami T., Mochizuki N., Kondo M., Nishimura M., Nagatani A. (2004). Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol. 45: 1798–1808 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wada M., Suetsugu N. (2004). Plant organelle positioning. Curr. Opin. Plant Biol. 7: 626–631 [DOI] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang Y.-S., Yoo C.-M., Blancaflor E.B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Westphal R.S., Soderling S.H., Alto N.M., Langeberg L.K., Scott J.D. (2000). Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 19: 4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo C.W., Khurana P., Davis P.A., DeBlasio S.L., DeSloover D., Staiger C.J., Hangarter R.P. (2011). THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr. Biol. 21: 59–64 [DOI] [PubMed] [Google Scholar]
- Yoo Y., Wu X., Guan J.L. (2007). A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J. Biol. Chem. 282: 7616–7623 [DOI] [PubMed] [Google Scholar]
- Zhang C., Mallery E.L., Schlueter J., Huang S., Fan Y., Brankle S., Staiger C.J., Szymanski D.B. (2008). Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20: 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dyachok J., Krishnakumar S., Smith L.G., Oppenheimer D.G. (2005). IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17: 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I., Saedler R., Mutondo M., Hulskamp M. (2004). The Arabidopsis GNARLED gene encodes the NAP125 homolog and controls several actin-based cell shape changes. Mol. Genet. Genomics 272: 290–296 [DOI] [PubMed] [Google Scholar]