This work demonstrates that the NPH3 protein of Arabidopsis represents a core component of a CULLIN3-based E3 ubiquitin ligase that targets the phototropin1 (phot1) photoreceptor for blue light–stimulated mono/multi- and polyubiquitination. In addition, it was shown that phot1 ubiquitination by this E3 complex is necessary for normal phototropic responsiveness.
Abstract
Plant phototropism is an adaptive response to changes in light direction, quantity, and quality that results in optimization of photosynthetic light harvesting, as well as water and nutrient acquisition. Though several components of the phototropic signal response pathway have been identified in recent years, including the blue light (BL) receptors phototropin1 (phot1) and phot2, much remains unknown. Here, we show that the phot1-interacting protein NONPHOTOTROPIC HYPOCOTYL3 (NPH3) functions as a substrate adapter in a CULLIN3-based E3 ubiquitin ligase, CRL3NPH3. Under low-intensity BL, CRL3NPH3 mediates the mono/multiubiquitination of phot1, likely marking it for clathrin-dependent internalization from the plasma membrane. In high-intensity BL, phot1 is both mono/multi- and polyubiquitinated by CRL3NPH3, with the latter event targeting phot1 for 26S proteasome-mediated degradation. Polyubiquitination and subsequent degradation of phot1 under high-intensity BL likely represent means of receptor desensitization, while mono/multiubiquitination-stimulated internalization of phot1 may be coupled to BL-induced relocalization of hormone (auxin) transporters.
INTRODUCTION
Plants use sunlight not only for photosynthesis but also as a temporal and spatial cue for regulation of growth and development (Chen et al., 2004). A variety of adaptive responses have evolved such that plants can use light directionality, quantity, and quality to optimize their success. One such response is phototropism, or the bending of plant organs toward (stems and leaves) or away from (roots) a directional blue light (BL) source (Holland et al., 2009). The fitness benefits conferred to a plant by the phototropic response include maximization of photosynthetic light capture in aerial organs and water acquisition via roots (Pedmale et al., 2010).
Several key components of the phototropic signal response system have been identified and at least partially characterized. While the BL-activated Ser/Thr protein kinases, phototropin (phot1 and phot2; for a discussion of the nomenclature of the phototropins see, Briggs et al., 2001), are arguably the most notable proteins identified to date, the NONPHOTOTROPIC HYPOCOTYL3 (NPH3) protein is clearly the most enigmatic (Holland et al., 2009). The phototropins function as phototropic photoreceptors, with phot1 acting as the primary receptor under low-light intensities and both phot1 and phot2 functioning as redundant receptors under moderate to high light intensities (Sakai et al., 2001). By contrast, NPH3 appears necessary for phototropism under all BL conditions (Inada et al., 2004), though its biochemical function has remained elusive (Pedmale and Liscum, 2007).
NPH3 and ROOT PHOTOTROPISM2 (RPT2) represent the founding members of a novel plant-specific family of proteins (Motchoulski and Liscum, 1999; Sakai et al., 2000), designated the NRL (for NPH3/RPT2-Like) family (Holland et al., 2009). Several regions of sequence and predicted structural conservation define members of the NRL family, with three domains being most notable: (1) an N-terminal BTB (broad complex, tramtrack, bric à brac) domain, (2) a centrally located NPH3 domain (Pfam, PF03000), and (3) a C-terminal coiled-coil domain (Pedmale et al., 2010). The coiled-coil region of NPH3 has been shown to function as part of a phot1-interacting domain (Motchoulski and Liscum, 1999), but neither the BTB nor the NPH3 domain have been ascribed a biochemical function for any NRL family member. However, the BTB domains of NPH3 and RPT2 can mediate heterodimerization of these two proteins in yeast (Inada et al., 2004).
In recent years, a common functional role for the wide assortment of BTB domain–containing proteins (hereafter referred to as BTB proteins) has begun to emerge, namely, that BTB proteins act as both a CULLIN3 (CUL3) binding and substrate adapter protein in CUL3-based E3 ubiquitin ligases (Willems et al., 2004). CUL-based E3 complexes, also called CRLs for CULLIN-RING-ligases, catalyze the final step in a sequential three-enzyme process that results in the ubiquitination of a target protein (Hershko and Ciechanover, 1998). Though first described in fungal (Geyer et al., 2003) and animal cells (Pintard et al., 2003; Xu et al., 2003), CRL3s have been observed in plant cells as well (Dieterle et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005; Christians et al., 2009).
Whereas proteolysis is the most commonly recognized outcome of CRL-dependent protein ubiquitination (Hershko and Ciechanover, 1998), protein ubiquitination also regulates a number of proteasome-independent cellular processes, including DNA repair and transcription, membrane protein endocytosis, and subcellular protein trafficking (Miranda and Sorkin, 2007; Chen and Sun, 2009). A number of ubiquitination patterns are also observed from one substrate to the next: a single ubiquitin (Ub) moiety can be ligated to a single Lys residue within the substrate protein (monoubiquitination), single Ub molecules can be attached to multiple Lys residues (multiubiquitination), and/or polyUb chains can be added at one or more Lys residues (polyubiquitination) (Komander, 2009).
The most well characterized ubiquitination events are those using Lys-48 and Lys-63–linked polyUb chains, with the former being the most common proteasomal-targeting signal (Komander, 2009). Though mono/multiubiquitination has also been associated with protein turnover (Boutet et al., 2007), it is typically coupled to nonproteasomal functions. For example, in animal cells, activated plasma membrane receptor-ligand complexes are often mono/multiubiquitinated, internalized via clathrin-dependent endocytosis, and shuttled to the lysosome for destruction (Tanaka et al., 2008).
In this study, we show that the plasma membrane–associated phototropic receptor phot1 is ubiquitinated in response to BL activation. Ubiquitination of phot1 is dependent upon both the phot1-interacting proteins NPH3 and CUL3, suggesting that this posttranslational modification occurs via action of a CRL3NPH3 complex. Under high-light conditions, phot1 is both mono/multiubiquitinated and polyubiquitinated, with the latter stimulating the 26S proteasome–dependent degradation of phot1 likely as a means of sensor desensitization. By contrast, under low intensities of BL, phot1 appears to be exclusively mono/multiubiquitinated, likely targeting the receptor for internalization. This latter hypothesis is certainly intriguing in light of recent findings that phot1 undergoes BL-induced clathrin-associated endocytosis (Kaiserli et al., 2009).
RESULTS
NPH3 Is a CUL3-Interacting Protein
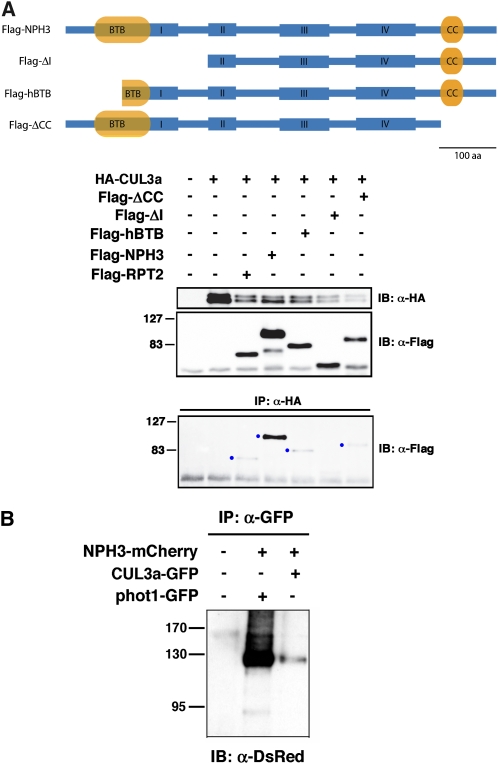
In silico comparisons of the predicted tertiary structure of the NPH3 BTB domain with that of a number of plant BTB proteins known to interact with CUL3 (Dieterle et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005; Christians et al., 2009) suggest that structural differences should not preclude NPH3-CUL3 interaction. Though previous studies failed to detect interaction between CUL3 and members of the NRL family by yeast two-hybrid or in vitro assays (Dieterle et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005), we found that an N-terminal NPH3 polypeptide containing the complete BTB domain could be coimmunopurified with Arabidopsis thaliana CULLIN3a (CUL3a) when coexpressed in insect cells (see Supplemental Figure 1A online). Figure 1A shows that NPH3 can also be coimmunoprecipitated with CUL3a when both are coexpressed in mammalian COS-1 cells, confirming their potential for in vivo interaction. We found that full-length NPH3 interacts more strongly with CUL3a in COS-1 cells than any truncated version of NPH3 retaining the BTB domain (Figure 1A). However, the finding that the Flag-ΔI polypeptide (lacking the BTB domain altogether) cannot interact with CUL3a (Figure 1A, lane 6) is consistent with the requirement of the BTB domain for CUL3 interaction. Moreover, the observed interaction between CUL3a and the Flag-hBTB polypeptide, which retains all of the predicted CUL3 binding sites (Stogios et al., 2005), demonstrates that amino acids within the C-terminal half of NPH3’s BTB domain are sufficient to promote at least weak NPH3-CUL3 interaction (Figure 1A, lane 5). Interestingly, we found that NPH3 could also interact with human CUL3 in COS-1 cells and that this interaction exhibited a similar apparent requirement for both N- and C-terminal portions of NPH3 as that of the NPH3-CUL3a interaction (see Supplemental Figure 1B online). Thus, it appears that either portions within the C-terminal two-thirds of NPH3 are important for the structural integrity and CUL3 binding capacity of the BTB domain or that extra-BTB portions of NPH3 also play a direct role in CUL3 interaction. However, the latter scenario seems unlikely based on what is known about other BTB-CUL3 interactions (Stogios et al., 2005). Unlike NPH3, full-length RPT2 interacted more weakly with CUL3a in COS-1 cells (Figure 1A, lane 3). These latter findings are consistent with the weaker conditional phenotypes of the rpt2 mutants as well as RPT2’s apparent functional dependency on NPH3 (Sakai et al., 2000; Inada et al., 2004).
Figure 1.
NPH3 Interacts with CUL3 in Vivo.
(A) NPH3 interacts with CUL3 in mammalian cells. Flag-tagged NPH3 and RPT2 polypeptides coexpressed with HA-CUL3a in COS-1 cells are shown on top. Constructs expressed the following NPH3 proteins: Flag-NPH3, full-length NPH3 protein; Flag-ΔI, NPH3 from Pro-248 to terminal Ser-746; Flag-hBTB, NPH3 from Pro-100 to terminal Ser-746; Flag-ΔCC, NPH3 from Met-1 to Trp-650. Immunoblots of total cellular proteins probed with anti-HA and anti-Flag antibodies are shown in the middle two blots as controls for expression of the various constructs. The bottom blot in uses immunoblotting (IB) with anti-Flag antibodies to detect Flag-tagged proteins interacting with anti-HA antibody immunoprecipitated (IP) HA-CUL3a. Blue dots denote the presence of a CUL3-interacting Flag-tagged protein. aa, amino acids.
(B) Full-length NPH3 interacts with CUL3 in N. benthamiana cells. Two days after infiltration, microsomal proteins were isolated from plants exhibiting robust fusion protein expression (Figure 2), GFP-CUL3a and phot1-GFP were immunoprecipitated (IP) with anti-GFP antibodies and then NPH3-mCherry detected by immunoblot (IB) analysis with anti-DsRed antibodies.
The findings that NPH3 and CUL3 can interact in nonplant cells prompted us to employ a Nicotiana benthamiana transient coexpression system to ask if this interaction also occurs in plant cells. We first determined that transiently overexpressed NPH3 (with an mCherry tag) is associated with the plasma membrane in N. benthamiana leaf epidermal cells (Figures 2A and 2E), as occurs with native and transiently expressed NPH3 in other plant species (Motchoulski and Liscum, 1999; Lariguet et al., 2006; Inoue et al., 2008). Transiently expressed phot1, tagged with either green fluorescent protein (GFP) or mCherry, is also associated with the plasma membrane (Figures 2F and 2I), as expected (Sakamoto and Briggs, 2002). When coexpressed, NPH3 and phot1 colocalize to the plasma membrane (Figure 2G). Moreover, NPH3 and phot1 are coimmunoprecipitated from microsomal fractions prepared from these transfected leaves (Figure 1B). The proper subcellular localization and physical interaction of NPH3 and phot1 in transfected N. benthamiana leaves suggests that native functions of these two proteins, including possible interaction and function with CUL3, can be examined in this system.
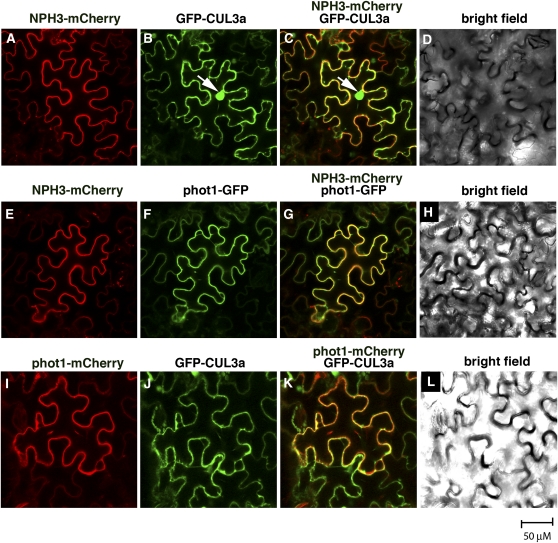
Figure 2.
Transiently Expressed NPH3, phot1, and Arabidopsis CUL3a Colocalize to the Cell Periphery in N. benthamiana Leaf Epidermal Cells.
(A) to (D) Localization of NPH3-mCherry ([A] and [C]) and GFP-CUL3a ([B] and [C]) signals to the periphery in tobacco cells. Signal overlap in (C) is colored yellow. (D) shows a bright-field image of the same cells shown in (A) to (C). White arrow in (B) and (C) indicates the nucleus.
(E) to (H) Localization of NPH3-mCherry ([E] and [G]) and phot1-GFP ([F] and [G]) signals to the cell periphery. Signal overlap in (G) is colored yellow. (H) shows a bright-field image of the same cells shown in (E) to (G).
(I) to (L) Localization of phot1-mCherry ([I] and [K]) and GFP-CUL3a ([J] and [K]) signals to the cell periphery. Signal overlap in (K) is colored yellow. (L) shows a bright-field image of the same cells shown in (I) and (J).
When CUL3a was transiently expressed in N. benthamiana leaves, we found that it localized to the nucleus and the plasma membrane (Figures 2B and 2J; see Supplemental Figure 2 online). These patterns of localization are consistent with observations in mammalian cells, where CUL3 has been found to localize to the nucleus, cytoplasm, Golgi, and plasma membrane (Singer et al., 1999; Meyer-Schaller et al., 2009). Non-nuclear CUL3a colocalizes with phot1 and NPH3 (Figures 2C and 2K), indicating the potential for in planta interaction between CUL3 and NPH3. In fact, NPH3-mCherry coimmunoprecipitates with GFP-CUL3a in microsomal fractions (Figure 1B) prepared from the N. benthamiana leaves coexpressing these proteins (Figures 2A to 2C), confirming not only that CUL3 can localize to the plasma membrane but also that CUL3 and NPH3 can interact in planta.
Normal Phototropism Requires Functional CUL3
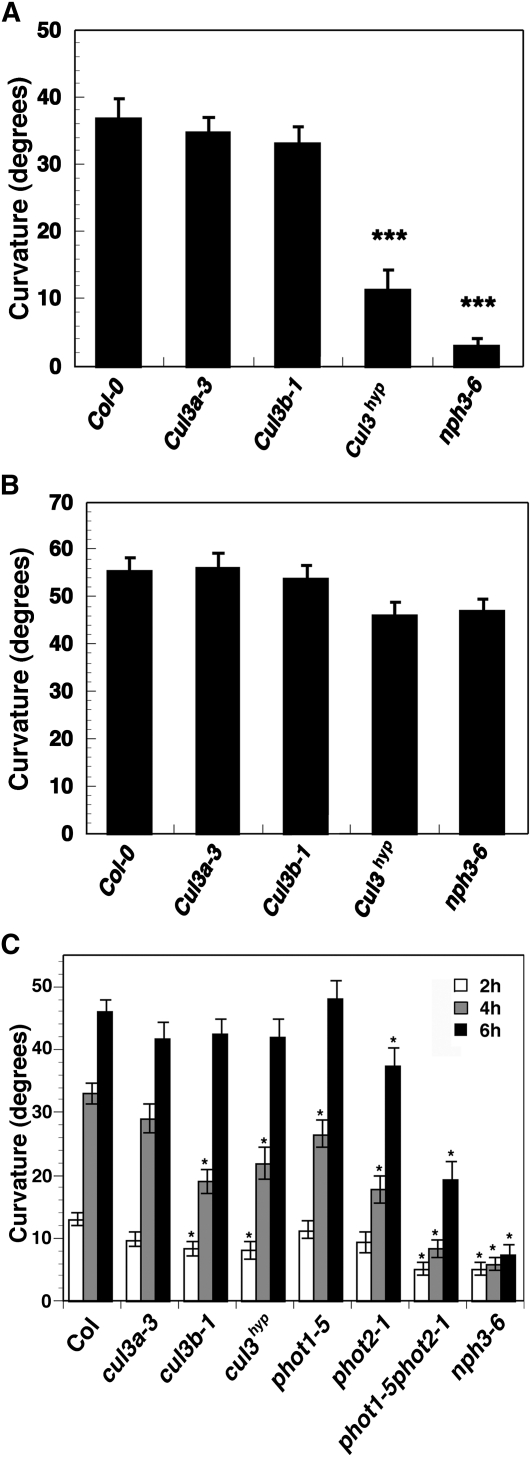
If NPH3 represents a bona fide CUL3-interacting protein, one might expect mutations in CUL3 to result in altered NPH3-dependent responses, such as phototropism. Single null loss-of-function mutations in either of the CUL3 genes of Arabidopsis, CUL3A or CUL3B, result in relatively subtle phenotypes, but embryonic lethality is observed when both genes are knocked out, indicating that the encoded proteins are essential and functionally redundant (Figueroa et al., 2005). Fortunately, a nonlethal hypomorphic double mutant (designated cul3hyp), carrying a knockout allele of cul3b and a partial loss-of-function allele of cul3a, exists; this double mutant facilitates functional studies of postembryonic processes where functional redundancy occurs (Thomann et al., 2009). Figure 3A shows that wild-type, cul3a, and cul3b seedlings exposed to low-intensity BL (0.1 μmol m−2 s−1) exhibited little difference in their hypocotyl phototropic responsiveness. However, cul3hyp double mutants grown under the same conditions exhibited significantly reduced phototropism (Figure 3A). The fact that cu+l3hyp remains more responsive than the nph3-6 null mutant (Figure 3A) should not be surprising since the former retains some CUL3 function (Thomann et al., 2009). However, it would appear that the altered phototropic response of the cul3hyp seedlings under low-light conditions does not result from a general defect in hypocotyl growth or tropic responsiveness (e.g., auxin responsiveness; Harper et al., 2000) since all genotypes exhibited similar robust gravitropic curvatures (Figure 3B).
Figure 3.
CUL3 Function Is Necessary for Normal Hypocotyl Phototropism, but Not Gravitropism, in Arabidopsis.
(A) Hypocotyl phototropism in seedlings exposed to 4 h of low-intensity unilateral BL (0.1 μmol m−2 s−1). Col, the wild type. Data represent the mean ± se of at least 70 seedlings for each genotype. Mean responses differing significantly (P < 0.001; Students t test) from that of the wild type are indicated by asterisks.
(B) Hypocotyl gravitropism in etiolated seedlings. Seedlings were germinated and grown on agar-solidified 0.5× MS medium in vertically oriented plates for 3 d in darkness and then the plates were rotated on-edge 90° to induce a gravitropic response, as described previously (Stowe-Evans et al., 1998). Col, the wild type. Data represent the mean ± se of at least 70 seedlings for each genotype. Student’s t test revealed that no genotype exhibited a response significantly different from that of the wild type (P > 0.05).
(C) Hypocotyl phototropism in seedlings exposed to high-intensity unilateral BL (120 μmol m−2 s−1) for 2 (white bars), 4 (gray bars), or 6 h (black bars). Data represent the mean ± se of at least 53 seedlings for each genotype. Col, the wild type. Mean responses differing significantly (P < 0.01; Student’s t test) from that of the wild type are indicated by asterisks.
When the phototropic response was examined under high-intensity BL (120 μmol m−2 s−1), we found that in addition to the cul3hyp double mutant, the cul3b single mutant also exhibited a significantly diminished response (Figure 3C). Intriguingly, the cul3 mutant lines exhibit a temporal-dependent phenotype where more pronounced phototropic deficiency is observed early in the response (Figure 3C). This suggests that either there is a stronger requirement for CUL3 function during initial stages of the phototropic response, or there is a cumulative effect of retained CUL3 function over time, such that even the reduced levels of CUL3 present in the cul3hyp double mutant are sufficient to establish a nearly normal level of responsiveness by 6 h of exposure to high light.
BL Stimulates Ubiquitination of phot1 in Planta
When taken together with the knowledge that BTB proteins function as substrate adapters for CRL3s (Willems et al., 2004), the findings presented here that NPH3 and CUL3 can interact in vivo (Figure 1) and that CUL3 is required for normal phototropism (Figure 3) suggest that ubiquitination of a substrate protein by a CRL3NPH3 complex likely represents an important component of phototropic signaling. One obvious protein to examine for in planta ubiquitination is phot1, since it is known to interact with NPH3 (Motchoulski and Liscum, 1999; Inada et al., 2004; Lariguet et al., 2006).
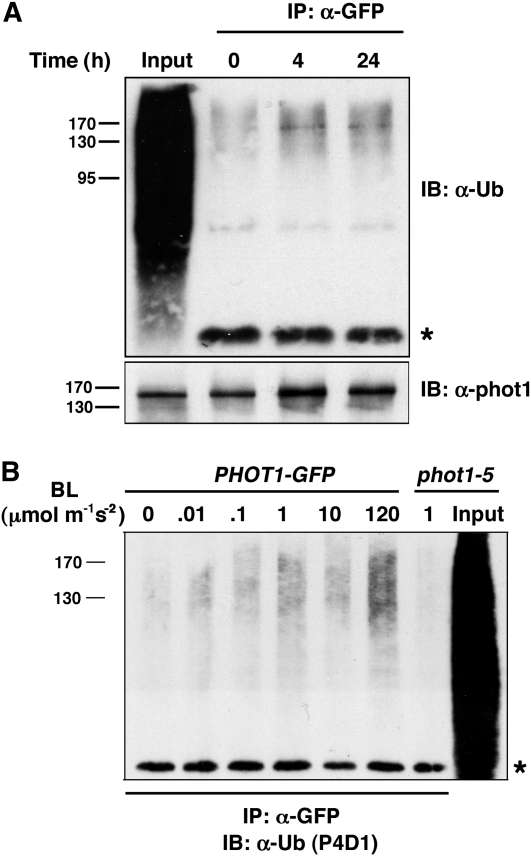
Because a large number of proteins are likely to be ubiquitinated in a cell at any given time (Figure 4A, first lane; also see Stone et al., 2003), we used a transgenic line expressing a functional and immunoprecipitatable phot1-GFP fusion protein (Lariguet et al., 2006) to assess phot1 ubiquitination status specifically. As shown in Figure 4A, immunoblot analysis with an anti-Ub antibody (U5379; Sigma-Aldrich) detects a protein with similar mobility as immunoprecipitable phot1-GFP. Interestingly, a stronger immunoreactive signal was observed in samples from seedlings irradiated with low intensity BL (0.1 μmol m−2 s−1) than in those kept in darkness (Figure 4A, compare lanes 3 and 4 to lane 2), suggesting that not only is phot1 apparently ubiquitinated in planta but that BL influences this posttranslational modification. Because phot1 functions in phototropism over a wide range of light intensities (Sakai et al., 2001), we next determined if phot1 ubiquitination observed under low intensity BL (Figure 4A) also occurs at higher BL intensities. Figure 4B shows that phot1 is ubiquitinated over the same range of BL intensities for which it is active in promoting phototropism (Sakai et al., 2001), with the response being strongest at the highest intensity (120 μmol m−2 s−1).
Figure 4.
BL Stimulates the Ubiquitination of phot1 in Planta.
(A) Phot1 is ubiquitinated in planta. Total cellular proteins were prepared from Arabidopsis phot1-5PHOT1-GFP seedlings that had been mock irradiated (0 h BL treatment) or exposed to low-intensity unilateral BL (0.1 μmol m−2 s−1; 4 or 24 h). Proteins were immunoprecipitated (IP) with anti-GFP antibodies (lanes 2 to 5). Total proteins not subjected to immunoprecipitation are included as a positive control for total cellular ubiquitination (Input, lane 1). Ubiquitinated proteins were detected in the top panel by immunoblot (IB) analysis with U5379 anti-Ub antibodies (Sigma-Aldrich). Asterisk denotes the IgG heavy chain (~50 kD) of the α-GFP antibodies and represents a loading control for IP samples. Phot1-GFP protein was detected in the bottom panel with anti-phot1 antibodies.
(B) BL stimulates ubiquitination of phot1 over a wide range of intensities. Total cellular proteins were prepared from Arabidopsis phot1-5PHOT1-GFP seedlings that had been mock irradiated (0 min) or exposed to 2 h of unilateral BL at the indicated fluence rates, as well as from phot1-5 seedlings exposed to 2 h of BL at a moderate intensity (1.0 μmol m−2 s−1) BL as a control. With the exception of an aliquot from the phot1-5 sample that was retained as a positive control for total cellular ubiquitination (Input, lane 8), all samples were subjected to immunoprecipitation (IP) with anti-GFP antibodies (lanes 1 to 7). Ubiquitinated proteins were detected by immunoblot (IB) analysis with P4D1 anti-Ub antibodies. Asterisk denotes the IgG heavy chain (~50 kD) of the α-GFP antibodies and represents a loading control for IP samples.
High-Intensity BL Stimulates Both Mono/Multi- and Polyubiquitination of phot1, with PolyUb-phot1 Being Targeted for Proteasome-Dependent Degradation
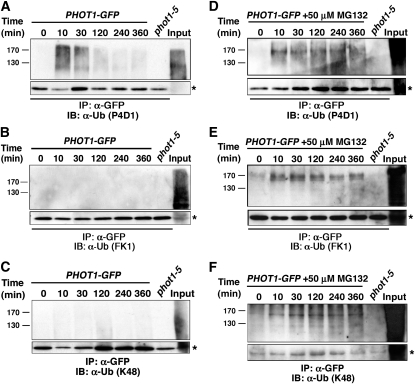
When the temporal dependency of phot1 ubiquitination under high-intensity BL (120 μmol m−2 s−1) was examined, we observed a peak immunoreactive signal between 10 and 30 min after the start of irradiation, which declined and was largely undetectable after 4 h or more of irradiation (Figure 5A). It is worth noting that in Figure 5A, the ubiquitinated phot1 (phot1-Ub) is detected by an antibody (P4D1) that recognizes both mono/multi- and polyubiquitinated proteins. By contrast, when a polyubiquitin-specific antibody (FK1) is used, no such signal is observed in phot1-GFP immunoprecipitates (Figure 5B, lanes 1 to 6), though a large number of polyubiquitinated proteins are recognized in total protein extracts used for immunoprecipitations (Figure 5B, lane 8). Similar results were obtained with anti-K48 antibodies that specifically recognize K48-linked polyUb chains (Figure 5C), suggesting that phot1 is mono/multiubiquitinated rather than polyubiquitinated under high light conditions.
Figure 5.
High-Intensity BL Stimulates Both Mono/Multi- and Polyubiquitination of phot1.
(A) When the 26S proteasome is functional, ubiquitinated phot1 is detected by an antibody that recognizes both mono/multi- and polyubiquitinated proteins. Total cellular proteins were prepared from phot1-5PHOT1-GFP seedlings that had been mock irradiated (0 min) or exposed to high-intensity unilateral BL (120 μmol m−2 s−1) for the indicated times. Total cellular proteins were also prepared from phot1-5 seedlings exposed to 120 min of high intensity BL as a negative control (lane 7). With the exception of an aliquot from the phot1-5PHOT1-GFP sample that was retained as a positive control for total cellular ubiquitination (Input, lane 8), all samples were subjected to immunoprecipitation (IP) with anti-GFP antibodies (lanes 1 to 7). Ubiquitinated proteins were detected by immunoblot (IB) analysis with P4D1 anti-Ub antibodies. Bottom panel shows an extracted portion of the same blot containing IgG heavy chain (~50 kD) from the α-GFP antibody (asterisk) as a loading control for IP samples.
(B) When the 26S proteasome is functional, no phot1 is detected by an antibody that recognizes only polyubiquitinated proteins. Replication of experiment in (A) except that ubiquitinated proteins were detected with the FK1 polyubiquitin-specific anti-Ub antibodies. Bottom panel represents a loading control as described in (A).
(C) When the 26S proteasome is functional, no phot1 is detected by an antibody that recognizes Lys-48–linked polyubiquitin chains. Replication of experiment in (A) except that ubiquitinated proteins were detected with anti-K48-Ub antibodies. Bottom panel represents a loading control as described in (A).
(D) When the 26S proteasome is inhibited, persistent phot1-Ub is detected. Replication of experiment in (A) except that seedlings were treated with 50 μM MG132 for 2 h prior to being mock irradiated or exposed to high-intensity unilateral BL. Bottom panel represents a loading control as described in (A).
(E) When the 26S proteasome is inhibited, persistent phot1 is detected by an antibody that recognizes only polyubiquitinated proteins. Replication of experiment in (B) except that seedlings were treated with 50 μM MG132 for 2 h prior to being mock irradiated or exposed to high-intensity unilateral BL. Bottom panel represents a loading control as described in (A).
(F) When the 26S proteasome is inhibited persistent phot1 is detected by an antibody that recognizes Lys-48–linked polyubiquitin chains. Replication of experiment in (C) except that seedlings were treated with 50 μM MG132 for 2 h prior to being mock irradiated or exposed to high-intensity unilateral BL. Bottom panel represents a loading control as described in (A).
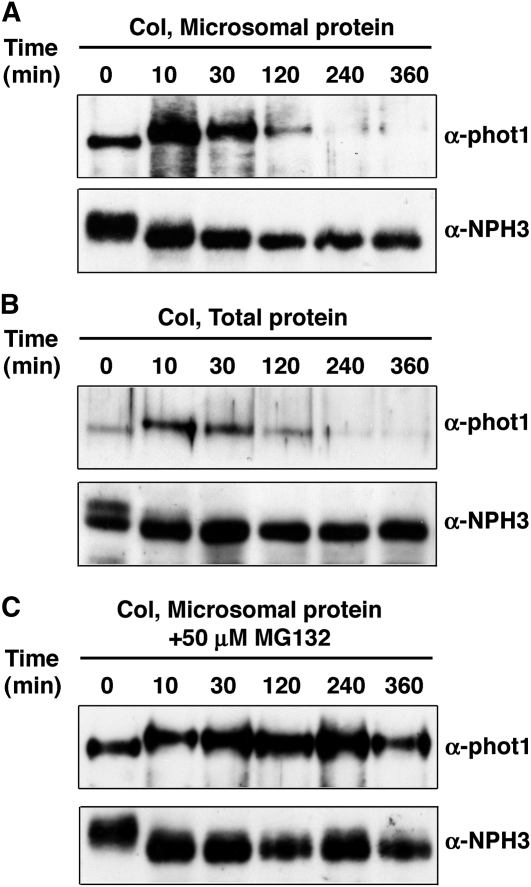
Previous studies by Sakamoto and Briggs (2002) showed that the abundance of phot1 protein declines after prolonged exposure to moderate intensity BL (20 μmol m−2 s−1), suggesting that the observed decrease in phot1-Ub signal under high-intensity BL (Figure 5A) might reflect a downregulation in steady state levels of phot1 itself. Immunoblot analysis with anti-phot1 antibodies shows that both membrane-associated and total cellular phot1 protein abundance does indeed decline in response to irradiation of seedlings with high-intensity BL and with a similar time course as that observed for the decrease in phot1-Ub signal (cf. Figures 6A and 6B to Figure 5A). The observed changes in phot1 abundance appear specific since NPH3 levels remain relatively constant (Figures 6A and B, bottom panels). BL-induced reduction in phot1 protein abundance is blocked by pretreatment of plants with the 26S proteasome–specific inhibitor MG132 (Figure 6C), indicating that phot1 is degraded by the 26S proteasome and that ubiquitination likely targets phot1 for destruction under high BL conditions.
Figure 6.
High-Intensity BL Stimulates 26S Proteasome–Dependent Degradation of phot1.
(A) Membrane-associated phot1 protein levels decrease in response to high-intensity BL. Microsomal proteins were prepared from wild-type (Col) Arabidopsis seedlings that had been mock irradiated (0 min) or exposed to high-intensity unilateral BL (120 μmol m−2 s−1) for the indicated times. Top panel, phot1 was detected by immunoblot (IB) analysis with anti-phot1 antibodies; bottom panel, NPH3 was detected with anti-NPH3 antibodies as a stable loading control.
(B) Total cellular phot1 protein levels decrease in response to high-intensity BL. Wild-type (Col) seedlings were treated and phot1 and NPH3 detected as in (A).
(C) The proteasome inhibitor MG132 stabilizes phot1 protein levels in plants exposed to high-intensity BL. Microsomal proteins were prepared from wild-type (Col) seedlings treated with 50 μM MG132 for 2 h prior to being mock irradiated (0 min) or exposed to high-intensity unilateral BL (120 μmol m−2 s−1) for the indicated times. Phot1 and NPH3 were detected by immunoblot (IB) analysis as described in (A).
Though not unknown, it is rather atypical for mono/multiubiquitinated proteins to be targeted for proteasome-dependent turnover (Boutet et al., 2007). Instead, polyUb chains (most commonly Lys-48 linked) represent the predominant molecular tag targeting proteins for degradation by the 26S proteasome (Komander, 2009). As such, we wondered whether our inability to detect polyubiquitinated phot1 (Figures 5B and 5C) might simply have been precluded by the degradation of phot1 under high-intensity BL, rather than lack of polyubiquitination per se. When phot1 protein levels are stabilized by treating seedlings with MG132 prior to exposure to high intensity BL (Figures 6A and 6B), phot1-Ub is detected over the entire time course by the P4D1, FK1, and K48 anti-Ub antibodies (Figures 5D to 5F), indicating that phot1 is both mono/multi- and polyubiquitinated under high BL conditions and that the latter modification likely targets phot1 for 26S proteasomal-dependent degradation.
A Functional CRL3NPH3 Complex Is Required for Both Mono/Multi- and Polyubiquitination, as Well as Subsequent Degradation of phot1
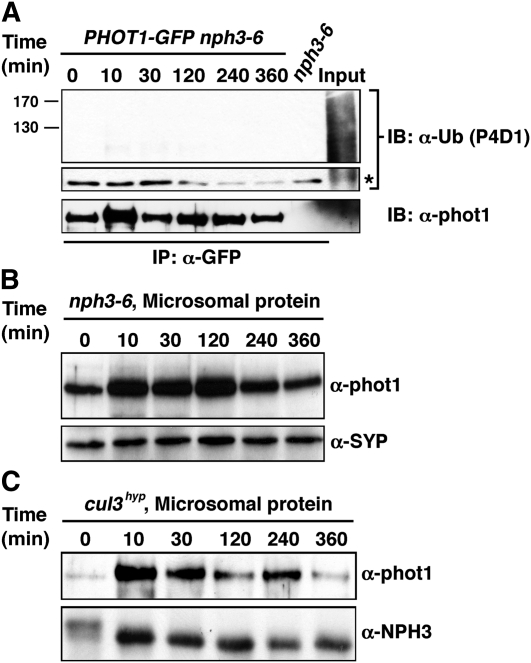
Results presented thus far demonstrate that NPH3 and CUL3 can interact in planta (Figures 1 and 2); the functions of both NPH3 and CUL3 are required, at least genetically, for normal phototropic responsiveness (Figure 3; also see Liscum and Briggs, 1995, 1996; Motchoulski and Liscum, 1999), and phot1 is ubiquitinated in response to BL (Figures 4 and 5). When taken together with the knowledge that NPH3 also interacts with phot1 (Motchoulski and Liscum, 1999; Lariguet et al., 2006; Inoue et al., 2008), these results suggest that the regulatory functions of NPH3 and CUL3 on phototropism may occur via a CRL3NPH3 complex-dependent ubiquitination of phot1. In this context, it is compelling to note that phot1 ubiquitination is not detected in the nph3-6-null mutant under high BL conditions (Figure 7A). Moreover, high-BL-induced phot1 protein degradation is dramatically retarded in both the nph3 single and cul3hyp double mutant backgrounds (Figures 7B and 7C).
Figure 7.
CRL3NPH3 Function Is Required for Ubiquitination and Degradation of phot1.
(A) High-intensity BL-dependent ubiquitination of phot1 is absent in the nph3-null mutant background. Total cellular proteins were prepared from Arabidopsis phot1-5nph3-6PHOT1-GFP seedlings that had been mock irradiated (0 min) or exposed to high-intensity unilateral BL (120 μmol m−2 s−1) for the indicated times (lanes 1 to 6). Total cellular proteins were also prepared from mock irradiated nph3-6 seedlings as a negative control (lane 7). With the exception of aliquots from the phot1-5PHOT1-GFP samples that were retained as positive controls for total cellular ubiquitination (Input, lane 8), all samples were subjected to immunoprecipitation (IP) with anti-GFP antibodies (lanes 1 to 7). In the top two panels, ubiquitinated proteins were detected by immunoblot (IB) analysis with P4D1 anti-Ub antibodies. Middle panel shows an extracted portion of the same blot from the top panel containing IgG heavy chain (~50 kD) from the α-GFP antibody (asterisk) as a loading control for IP samples. Bottom panel shows a replicate blot probed with anti-phot1 antibodies to demonstrate that although ubiquitinated phot1 is not detected by the P4D1 antibody (top panel), phot1-GFP is in fact immunoprecipitated effectively from phot1-5nph3-6PHOT1-GFP seedlings grown under high BL conditions.
(B) BL fails to stimulate phot1 degradation in the nph3-null mutant background. Microsomal proteins were prepared from nph3-6 seedlings, and phot1 was detected by immunoblot (IB) analysis with anti-phot1 antibodies (top panel). Bottom panel shows a replicate blot probed with an anti-syntaxin antibody (α-SYP) as a plasma membrane–specific protein loading control.
(C) BL-induced degradation of phot1 is retarded in the cul3hyp mutant background. Microsomal proteins were prepared from cul3hyp seedlings and phot1 detected in the top panel as described in (B). Bottom panel shows a replicate blot probed with an anti-NPH3 antibody as a plasma membrane–specific protein loading control.
Low-Intensity BL Stimulates Only Mono/Multiubiquitination of phot1
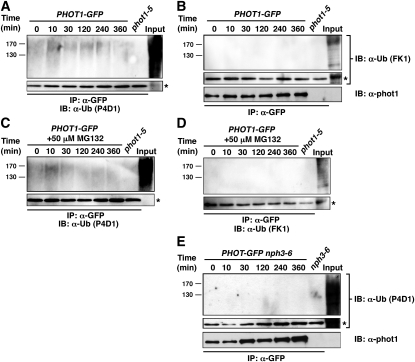
While it now seems clear that high intensity BL stimulates both the mono/multi- and polyubiquitination of phot1 (Figure 5), with the latter targeting the phot1 protein for 26S proteasomal degradation (Figure 6), most studies on phot1-dependent phototropism have been done under low-intensity BL conditions (Motchoulski and Liscum, 1999; Harper et al., 2000; Sakai et al., 2000, 2001; Blakeslee et al., 2004; Esmon et al., 2006; Larigeut et al., 2006; Pedmale and Liscum, 2007; Kaiserli et al., 2009). Therefore, we set out to examine the ubiquitination of phot1 occurring in response to low BL (Figure 4) in more detail. Though the signal is not as intense as observed after irradiation with high-intensity BL (Figure 5A), phot1-Ub is detected with the P4D1 antibody in immunoprecipitated samples from PHOT1-GFP seedlings exposed to as little as 10 min of low-intensity BL (0.1 μmol m−2 s−1) (Figures 8A and 8C). In contrast with what is observed in high BL, it would appear that phot1 is exclusively mono/multiubiquitinated under low-intensity BL conditions, as no immunoprecipitated phot1 is detected by the polyubiquitination-specific FK1 antibody, independent of whether seedlings were pretreated with MG132 or not (Figures 8B and 8D). However, it does appear that the CRL3NPH3 complex may mediate this mono/multiubiquitination response, as it does for both the mono/multi- and polyubiquitination responses under high-intensity BL (Figure 7A), since the P4D1 antibodies fail to detect any immunoprecipitated phot1 in the PHOT1-GFP nph3 background (Figure 8E). Given the apparent differences in phot1 ubiquitination patterns observed in wild-type seedlings exposed to low- versus high-intensity BL, it is interesting to note that low-intensity BL does not appear to stimulate the degradation of phot1 (Sakamoto and Briggs, 2002; Pedmale and Liscum, 2007; Sullivan et al., 2010; see Supplemental Figure 3 online), suggesting that the cellular consequences of phot1 mono/multi- and polyubiquitination are likely to be different.
Figure 8.
Low-Intensity BL Stimulates NPH3-Dependent Mono/Multiubiquitination of phot1.
(A) When the 26S proteasome is functional, ubiquitinated phot1 is detected by an antibody that recognizes both mono/multi- and polyubiquitinated proteins. Total cellular proteins were prepared from phot1-5PHOT1-GFP seedlings that had been mock irradiated (0 min) or exposed to low intensity (0.1 μmol m−2 s−1) unilateral BL for the indicated times. Total cellular proteins were also prepared from phot1-5 seedlings exposed to 120 min of low-intensity BL as a negative control (lane 7). With the exception of an aliquot from the phot1-5PHOT1-GFP sample that was retained as a positive control for total cellular ubiquitination (Input, lane 8), all samples were subjected to immunoprecipitation (IP) with anti-GFP antibodies (lanes 1 to 7). Ubiquitinated proteins were detected by immunoblot (IB) analysis with P4D1 anti-Ub antibodies. Bottom panel shows an extracted portion of the same blot containing IgG heavy chain (~50 kD) from the α-GFP antibody (asterisk) as a loading control for IP samples.
(B) When the 26S proteasome is functional, no phot1 is detected by an antibody that recognizes only polyubiquitinated proteins. Replication of experiment in (A) except that ubiquitinated proteins were detected with the FK1 polyubiquitin-specific anti-Ub antibodies. Middle panel represents a loading control as described in (A). Bottom panel shows a replicate blot probed with anti-phot1 antibodies to demonstrate that although ubiquitinated phot1 is not detected by the FK1 antibody (top panel), phot1-GFP is in fact immunoprecipitated effectively from phot1-5PHOT1-GFP seedlings grown under low BL conditions.
(C) When the 26S proteasome is inhibited, ubiquitinated phot1 is detected by an antibody that recognizes both mono/multi- and polyubiquitinated proteins. Replication of experiment in (A) except that seedlings were treated with 50 μM MG132 for 2 h prior to being mock irradiated or exposed to high-intensity unilateral BL. Bottom panel represents a loading control as described in (A).
(D) When the 26S proteasome is inhibited, no phot1 is detected by an antibody that recognizes only polyubiquitinated proteins. Replication of experiment in (B) except that seedlings were treated with 50 μM MG132 for 2 h prior to being mock irradiated or exposed to high-intensity unilateral BL. Bottom panel represents a loading control as described in (A).
(E) Low-intensity BL-dependent ubiquitination of phot1 is absent in the nph3-null mutant background. Total cellular proteins were prepared from Arabidopsis phot1-5nph3-6PHOT1-GFP seedlings that had been mock irradiated (0 min) or exposed to low-intensity unilateral BL (0.1 μmol m−2 s−1) for the indicated times (lanes 1 to 6). Total cellular proteins were also prepared from mock-irradiated nph3-6 seedlings as a negative control (lane 7). With the exception of aliquots from the phot1-5PHOT1-GFP samples that were retained as positive controls for total cellular ubiquitination (Input, lane 8), all samples were subjected to immunoprecipitation (IP) with anti-GFP antibodies (lanes 1 to 7). In the top two panels, ubiquitinated proteins were detected by immunoblot (IB) analysis with P4D1 anti-Ub antibodies. Middle panel shows an extracted portion of the same blot from the top panel containing IgG heavy chain (~50 kD) from the α-GFP antibody (asterisk) as a loading control for IP samples. Bottom panel shows a replicate blot probed with anti-phot1 antibodies to demonstrate that although ubiquitinated phot1 is not detected by the P4D1 antibody (top panel), phot1-GFP is in fact immunoprecipitated effectively from phot1-5nph3-6PHOT1-GFP seedlings grown under low BL conditions.
DISCUSSION
Eukaryotic cells frequently use ubiquitination as a posttranslational mechanism to regulate the stability, intracellular localization, and function of proteins in response to environmental signals (Gao and Karin, 2005; Miranda and Sorkin, 2007). Here, we show that BL stimulates the ubiquitination of the phototropic receptor phot1 (Figures 4, 5 and 8) and that this response is likely mediated by CRL3NPH3, a CUL3-based E3 ubiquitin ligase that uses the phot1-interacting BTB protein NPH3 as its substrate adapter (Figures 1, 2, 7 and 8E). Under high intensities of BL (e.g., 120 μmol m−2 s−1), phot1 appears both mono/multi- and polyubiquitinated (Figure 5), with the latter modification targeting phot1 for 26S proteasome–dependent degradation (Figure 6). From a biological perspective, it is worth noting that phot1 degradation has only been observed in seedlings exposed to intensities of BL equivalent to, or in excess of, that necessary for maximal phototropic response (Figure 6; see Supplemental Figure 3 online; Sakai et al., 2001; Sakamoto and Briggs, 2002; Pedmale and Liscum, 2007; Sullivan et al., 2010). This suggests that the stimulation of phot1 polyubiquitination under high BL conditions represents a means to desensitize phototropic signaling through receptor turnover. Based on this simple hypothesis, we predicted that mutants compromised in their CRL3NPH3-dependent phot1 degradation, such as nph3-6 and cul3hyp (Figures 7B and 7C), would exhibit enhanced phototropism in response to irradiation with high-intensity BL. However, significantly reduced phototropic curvatures are observed in nph3-6 and cul3hyp mutants under high BL conditions (Figure 3C). While these findings do not in themselves argue against a connection between phot1 polyubiquitination and receptor desensitization, they do demonstrate that this simple model is not sufficient to explain the observed photophysiology. In this context, we would like to note that phototropism is severely impaired in nph3-6 and cul3hyp mutants in response to irradiation with low-intensity BL (Figure 3A), conditions which promote mono/multiubiquitination (Figures 8A and 8C), but not polyubiquitination (Figures 8B and 8D) or subsequent degradation of phot1 (Figure 8B; Sakamoto and Briggs, 2002; Pedmale and Liscum, 2007). These latter results suggest that mono/multiubiquitination of phot1 positively influences functionality of the receptor relative to phototropism and that the abrogation of this effect in nph3-6 (Figure 8E) and cul3hyp mutants masks any reduction in receptor desensitization that might occur due to the stabilization of phot1 protein levels in these mutants under high BL conditions (Figures 6B and 6C). Of course it also remains formally possible that polyubiquitination itself has a positive influence on phot1 quite apart from its proteasomal targeting activity. Additional studies are necessary to distinguish between these potential mechanisms.
Though we have just begun to characterize the low BL-stimulated mono/multiubiquitination of phot1, it seems clear, based on the mutant phenotypes just discussed, that this modification is important for normal receptor function, at least relative to phototropism. In fungal and animal systems, ligand-activated plasma membrane–associated receptors are often mono/multiubiquitinated to mark them for clathrin-dependent endocytosis as a means to modulate downstream signaling (d’Azzo et al., 2005; Miranda and Sorkin, 2007). Several pieces of independent data suggest that such a mechanism may provide a useful framework to help explain our results positively linking mono/multiubiquitination and phototropic function of phot1: First, BL induces the movement of phot1 from the plasma membrane to cytoplasmic locations (Sakamoto and Briggs, 2002). Moreover, pharmacological and immunoprecipitation studies suggest that BL-induced movement of phot1 is clathrin mediated (Kaiserli et al., 2009; Sullivan et al., 2010). We independently found that phot1 associates with clathrin heavy chain and that this association is enhanced by exposure to BL (see Supplemental Figure 4 online). Second, CRL3NPH3-dependent mono/multiubiquitination of phot1 is stimulated by BL (Figures 4, 5 and 8), implying that photoexcited phot1 (i.e., receptor-ligand complex) is targeted more efficiently for ubiquitination than dark-state phot1 (i.e., free receptor). Lastly, it appears that phot1 activation by BL (i.e., formation of a receptor-ligand complex) is prerequisite to its clathrin-associated endocytosis (Kaiserli et al., 2009). It is interesting to note that while an active protein kinase domain is not necessary for BL-stimulated degradation of phot1 (Sullivan et al., 2010), it is the kinase active isoform of phot1 that undergoes clathrin-associated endocytosis (Kaiserli et al., 2009). These latter findings suggest that internalized phot1 is likely to retain the capacity to catalyze the phosphorylation of substrate proteins. As such, endocytosis of phot1 could represent a mechanism to alter which substrates are potentially phosphorylated in the cell, based solely on phot1 localization. For example, BL-induced mono/multiubiquitin-dependent endocytosis could result in limitations on the amount of phot1 capable of modifying substrates at the inner surface of the plasma membrane. Alternatively, endocytosis of even small fractions of phot1 could lead to the phosphorylation of cytoplasmic proteins in sufficient quantity to fundamentally change intracellular signaling.
Though it has yet to be examined, it is tantalizing to speculate that CRL3NPH3-dependent ubiquitination and subsequent internalization/endocytosis of phot1 is functionally coupled to alterations in transport of the plant hormone auxin, a process necessary for the establishment of phototropic curvatures (Esmon et al., 2006). Several apparent connections between NPH3, phot1, and auxin transport have already been established. For example, changes in auxin transport that normally occur in response to phototropic stimulation do not occur in an aphototropic nph3 deletion mutant of rice (Oryza sativa; Haga et al., 2005). What is more, it has been shown that phot1 function is required for BL-induced changes in the intracellular localization of the auxin efflux transporter PIN-FORMED1 (PIN1) (Blakeslee et al., 2004), a response that likely contributes to the overall changes in auxin transport patterns in phototropically responsive seedlings (Friml, 2010). Recent studies have shown that the intracellular localization/activity of PIN1 can be directly regulated via phosphorylation by the protein kinase PINOID (PID) (Skirpan et al., 2009; Huang et al., 2010; Zhang et al., 2010). Interestingly, PID and phot1 are both members of the AGCVIII subfamily of protein kinases (Robert and Offringa, 2008), suggesting that phot1 kinase activity might also directly regulate auxin transporter localization/activity. While the dynamics of PID intracellular localization have not been reported, it is worth noting that PID, like phot1, can be found associated with both the plasma membrane and endosomes (Michniewicz et al., 2007) and that its function as a regulator of auxin transport is genetically coupled to a subfamily of NRL proteins, namely, NAKED PINS IN YUC MUTANTS1 (NPY1) to NPY5 (Cheng et al., 2008). It has previously been proposed that NRL proteins and AGCVIII kinases represent cooperative modules for the regulation of auxin transport (Robert and Offringa, 2008; Holland et al., 2009), and when taken together with the results presented here, it seems reasonable to further postulate that CRL3NRL-dependent ubiquitination and subsequent internalization of plasma membrane–associated AGCVIII kinases may represent a critical biochemical component of these regulatory modules. Future studies will address not only the detail of how ubiquitination of phot1 regulates phototropic responsivness but also more broadly how AGCVIII kinases and NRL proteins/CRL3NRL complexes impact auxin biology at the biochemical and cellular levels.
METHODS
Plant Materials and Growth Conditions
Three-day-old dark-grown Arabidopsis thaliana seedlings used in this study were all in the Columbia (Col-0) accession. With the exception of the phot1-5nph3-6PHOT1-GFP line (gift from C. Fankhauser), all mutant and transgenic genotypes have been described previously: phot1-5 (Huala et al., 1997), phot1 phot2 (Kinoshita et al., 2001), phot1-5PHOT1-GFP (Sakamoto and Briggs, 2002), nph3-6 (Motchoulski and Liscum, 1999), cul3a-3 (Thomann et al., 2009), cul3b-1 (Thomann et al., 2005), and cul3hyp (Thomann et al., 2009). The PHOT1-GFP transgene is functional and complements the phot1-5-null mutant phenotypes (Sakamoto and Briggs, 2002).
Arabidopsis seedlings were grown either on agar-based medium or in liquid culture (depending upon the experiment) as described below. First, seeds were surface sterilized using 30% (v/v) commercial bleach (20 min) and then washed five times with sterile water. Unless noted otherwise, seeds were next placed either on filter paper mounted on 1% (w/v) agar-solidified half-strength Murashige and Skoog (0.5× MS) medium (Murashige and Skoog, 1962) in Petri plates or in 15 mL of 0.5× MS liquid medium in 125- to 250-mL Erlenmeyer flasks. The seeds were then cold treated (4°C) for 3 d in darkness before treatment with red light for 1 h to induce uniform germination (Stowe-Evans et al., 1998). The plates or flasks were next transferred to complete darkness at 22 ± 2°C, and seedlings were allowed to grow for 3 d prior to any treatment. For growth of seedlings in liquid medium, the flasks were shaken at 50 to 60 rpm throughout their growth and subsequent treatments up to tissue collection. For BL treatments, seedlings growing on agar medium were either mock irradiated or exposed to unilateral BL (Stowe-Evans et al., 1998) of various intensities for different time intervals, whereas seedlings grown in liquid medium were irradiated with BL at specified fluence rate from underneath the flasks with constant shaking (Pedmale and Liscum, 2007). Low-intensity BL (0.1 μmol m−2 s−1) was generated as described previously (Stowe-Evans et al., 1998), whereas moderate to high-intensity BL (10 to 120 μmol m−2 s−1) was generated by filtering light from a Navitar Xenon 560 projector (550-W xenon lamp) through one layer of blue acrylic (Rohm and Haas No. 2424, 3.18-mm thick; Cope Plastics). Intensities (fluence rates) were varied by altering the distance between the light source and plant material. For experiments employing proteasome inhibitor treatments (Lee and Goldberg, 1998), seedlings grown in liquid 0.5× MS medium supplemented with either the 50 μM MG132 (Sigma-Aldrich) or solvent in which the inhibitor was initially dissolved (DMSO) for 2 h prior (Feng et al., 2008) to irradiation or mock irradiation.
Four- to five-week-old greenhouse-grown Nicotiana benthamiana plants were used for transient expression of phot1, NPH3, and CUL3a and were grown from seed in the greenhouse under 16-h-light/8-h-dark conditions. Plants were watered twice weekly for the entire growth period. After infiltration plants were kept under continuous white light (~100 μmol m−2 s−1; Stowe-Evans et al., 1998) under laboratory conditions (22 to 24°C) until analyses were performed.
Gene Constructs
Unless otherwise noted, full-length PHOT1, NPH3, and CUL3a proteins (as described in TAIR8; www.Arabidopsis.org) were expressed in insect and mammalian cells. In insect cells, we expressed only the first 188 amino acids of NPH3 (which includes the entire BTB domain; designated 6xHis-NPH3-BTB). In addition to full-length NPH3, various NPH3 truncations were expressed in mammalian cells (Figure 1A, top). Protein coding sequences were generated by PCR amplification from respective cDNA templates and cloned in frame with the indicated epitope tag for subsequent expression. Myc-phot1 was generated using the pCMV-Myc vector (Clontech); Flag-NPH3 (and all derivatives of NPH3; Figure 1A, top) and Flag-RPT2 with pCMV-tag2A (Stratagene); 6xHis-NPH3-BTB with pAcHLT-A (BD Biosciences); HA-At-CUL3a and HA-Hs-Cul3 with pCRUZ-HA (Santa Cruz Biotech) as described previously (Zhang et al., 2004); and GST-6xHis-At-CUL3a with pAcGHLT-A (BD Biosciences). The mouse Rbx1a construct used in insect cell studies has been described elsewhere (Li et al., 2005). Insect cell expression vectors used the Baculovirus polyhedrin promoter, while mammalian cell expression vectors used a CMV (cytomegalovirus) constitutive promoter.
For transient expression in N. benthamiana, GFP-CUL3a was generated by cloning the full-length coding region of Arabidopsis CUL3A (from cDNA) in frame with GFP using the pMDC43 vector (Curtis and Grossniklaus, 2003). Similarly, phot1-GFP was constructed by cloning the full-length coding region of PHOT1 (from cDNA) in frame using the pMDC84 (Curtis and Grossniklaus, 2003). A modified pMDC84 vector, designated pUP-Red, in which the GFP coding sequence was replaced with mCherry coding sequence (Shaner et al., 2004), was used to generate NPH3-mCherry and phot1-mCherry fusion proteins. The pUP-Red vector was also engineered such that a Flag epitope tag is present on the fusion proteins, C-terminal to mCherry. All of the vectors for transient expression in N. benthamiana use a tandem duplicated cauliflower mosaic virus 35S-derived constitutive promoter to drive fusion protein expression.
Insect and Mammalian Cell Culture and Transfections
The Trichoplusia ni cell line Hi5 (Invitrogen) was used for insect cell expression studies. Arabidopsis NPH3 (6xHis-NPH3-BTB) and CUL3a (GST-6xHis-CUL3a) polypeptides were coexpressed together with mouse Rbx1a in Hi5 cells according to established protocols (Li et al., 2005). The COS-1 African green monkey kidney cell line (ATCC) was used in all mammalian cell expression studies. COS-1 cells were grown in knockout Dulbecco’s modified Eagle’s medium supplemented with 15% ES-cell qualified bovine serum, 2.5 mM glutathione, and 100 μM mercaptoethanol. Transfections were performed with Lipofectamine Plus (Invitrogen) according to the manufacturer’s instructions.
Transient Expression of Proteins in N. benthamiana
For the transient expression of proteins in N. benthamiana, we used an established Agrobacterium tumefaciens–based infiltration method (Voinnet et al., 2003; Caplan et al., 2008). Each of the fusion protein expression vectors to be analyzed was first electroporated into the A. tumefaciens strain GV3101 (PMP90). Next, these A. tumefaciens cultures were grown overnight, pelleted, and resuspended in 10 mM MgCl2, 10 mM MES and induced with 200 μM 4′-hydroxy-3′,5′-dimethoxyacetophenone (acetosyringone). An A. tumefaciens strain expressing the silencing suppressor protein P19 (Voinnet et al., 2003) was also prepared and coinfiltrated with all of the test constructs. Leaves from 4- to 5-week-old N. benthamiana plants were infiltrated (Voinnet et al., 2003; Caplan et al., 2008) with a mixed suspension of A. tumefaciens (P19 strain plus one or more of strains harboring a test construct) in which each strain was adjusted to an optical density of 0.5 at 600 nm prior to mixing. Two days after infiltration, a small section of the infiltrated area was excised and used for confocal microscopy, or the entire leaf was used for protein extraction (see below).
Microscopy
Confocal fluorescence images were acquired on a Zeiss LSM 510 META NLO two-photon point-scanning confocal system mounted on an Axiovert 200M inverted microscope with a ×40/1.2 C-apochromat water immersion objective. A 488-nm laser of the Zeiss LSM 510 META NLO two-photon point-scanning confocal system was used to excite GFP, and the fluorescent signal was visualized with a 500- to 550-nm band-pass filter. mCherry was excited with a 543-nm laser, and the emission signal was visualized with a 600- to 630-nm band-pass filter. Microscopy images were initially collected with the LSM510 operating software, and later the images were processed for brightness/contrast and overlaid with Adobe Photoshop CS3.
Preparation of Total Cellular and Microsomal Membrane Proteins from Arabidopsis and N. benthamiana
Total cellular and microsomal membrane proteins were isolated from Arabidopsis seedlings or N. benthamiana leaves after BL or inhibitor treatments essentially as described previously (Pedmale and Liscum, 2007). In brief, Arabidopsis seedling or tobacco leaf tissue was ground in ice-cold homogenization/resuspension buffer (50 mM HEPES, pH 7.8, 300 mM Suc, 150 mM NaCl2, 10 mM NaAcetate, 5 mM EDTA, and 1× Plant Protease inhibitor cocktail [Sigma]), following by filtration through nylon cloth and centrifugation at 10,000g to remove particulate matter. Resultant total cellular proteins (supernatant of low speed centrifugation) were either retained for further use or subjected to ultracentrifugation at 100,000g for 1.25 h to pellet microsomes. Microsomal membranes were then resuspended in homogenization/resuspension buffer using a Potter-Elvenjem homogenizer. The resuspended microsomal membranes were aliquoted and stored in −70°C until used. Protein concentrations of samples were determined by the Bradford colorimetric method (Bio-Rad). All manipulations performed under dim red light in a 4°C temperature-controlled room.
Antibodies, Immunoprecipitation, and Immunoblot Analysis
The anti-NPH3 (Motchoulski and Liscum, 1999), anti-phot1 (Christie et al., 1998), and anti-SYP (Kalde et al., 2007) antibodies have been described previously. Anti-6xHis (BD Biosciences), anti-Myc (Santa Cruz Biotechnology), anti-Flag (Sigma-Aldrich), Anti-DsRed/mCherry (Clontech), anti-GFP (Invitrogen), agarose-conjugated anti-GFP (MBL International), anti-HA (Convance), various anti-ubiquitin (U5379, Sigma-Aldrich; P4D1, Cell Signaling Technology; FK1, Enzo Life Sciences; K48, Millipore; K63, eBioscience), as well as alkaline phosphatase (AP)-conjugated anti-rabbit IgG and anti-mouse IgG (Promega) antibodies were purchased from commercial sources. The P4D1 anti-Ub antibodies have been shown to detect both mono/multi- and polyubiquitinated proteins, whereas the FK1 anti-Ub antibodies detect only mono/multiubiquitinated proteins (Fujimuro et al., 1994; Haglund et al., 2003; Fujimuro and Yokosawa, 2005). Recently, Barberon et al. (2011) used the P4D1 and FK1 antibodies in studies of differentially ubiquitinated proteins in Arabidopsis, demonstrating the utility of these reagents in plants.
In experiments using insect cells, glutathione affinity chromatography was employed to purify GST-6xHis-CUL3a and its interactions with Rbx1a and NPH3-BTB assessed through copurification and subsequent immunoblot analysis with the anti-6xHis antibody (Li et al., 2005). Prior to immunoprecipitation of expressed proteins from mammalian cells, total lysates from an aliquot of cells were collected (48 h after transfection) in sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 100 mM DTT, and 0.1% bromphenol blue) and subjected to immunoblot analysis to assure relatively equal levels of expression of the various proteins of interest. For immunoprecipitation assays, mammalian cells were lysed in radioimmune precipitation assay buffer (10 mM sodium phosphate, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and Complete protease inhibitor mixture [Sigma-Aldrich]). Cell lysates were precleared with protein A beads and then incubated with 2 μg of appropriate affinity-purified antibody for 2 h at 4°C, and finally incubated for 2 h at 4°C with protein A-agarose beads. Immunoprecipitated proteins were washed three times with radioimmune precipitation assay buffer and then resuspended in SDS-PAGE sample buffer and boiled for 4 min before being subjected to immunoblot analysis. For immunoprecipitation of proteins from Arabidopsis and N. benthamiana, total and microsomal protein fractions prepared in homogenization/resuspension buffer with 1% Triton X-100 were handled as described previously (Lariguet et al., 2006).
Protein samples were separated on SDS-polyacrylamide gels and then transferred to nitrocellulose membranes by electroblotting for subsequent immunodetection as described previously (Pedmale and Liscum, 2007). For immunodetection, membranes were first blocked using 5% (w/v) dry milk in 20 mM Tris-HCl, pH 7.6, 137 mM NaCl, and 0.05% Tween 20 (Tris-buffered saline [TBS]/Tween) for 3 h. This was followed by incubation with appropriate primary antibody for 2 h in TBS/Tween plus 1% dry milk. Membranes were then washed three times with TBS/Tween, followed by incubation with appropriate AP-conjugated secondary antibody in TBS/Tween plus 1% dry milk for 3 h. Finally, membranes were washed four times with TBS/Tween (without dry milk), and primary antibody-reactive protein bands were detected by chemiluminescence using Immobilon AP substrate (Millipore).
Phototropic and Gravitropic Response Assays in Arabidopsis Seedlings
Arabidopsis seedlings were grown on horizontally (for phototropic assays) or vertically oriented (for gravitropic assays) 0.5× MS agar-solidified media in darkness for 3 d and then assessed for phototropic or gravitropic responsiveness as described previously (Pedmale and Liscum, 2007).
Identification of Clathrin Heavy Chain as a phot1-Interacting Protein
Seedlings (phot1-5PHOT1-GFP) were grown on filter paper overlaid on 0.5× MS medium for 3 d in darkness and then either mock irradiated or exposed to BL (10 μmol m−2 s−1) for 30 min. Microsomal membranes were then isolated according to methods previously described (Pedmale and Liscum, 2007). Microsomal pellets were resuspended in extraction buffer with 1% Triton X-100 and incubated on ice for 30 min. Samples were then centrifuged at 100,000g for 30 min at 4°C to pellet nonsolubilized proteins. Resultant soluble microsomal membrane protein (250 μg per sample) was used for immunoprecipitation with the anti-GFP microbeads (Miltenyi Biotech) according to the manufacturer’s protocol. For the wash step, the manufacturer supplied lysis buffer was used, except that it was protease inhibitor treated before use. For elution, the native protein elution protocol was used, using 50 μL of 0.1 M triethylamine, pH 11.8. Only one round of elution was performed. Samples were neutralized with 3 μL of 1 M MES, pH 3.0. Colloidal Coomassie Brilliant Blue–stained one-dimensional gel bands were excised and digested with trypsin, and samples were subjected to tandem time-of-flight analysis (Applied Biosystems 4700 Proteomics analyzer). A peptide mass fingerprinting search was conducted with the National Center for Biotechnology Information database to identify phot1-interacting proteins, one of which turned out to be clathrin heavy chain.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PHOT1 (At3g45780), PHOT2 (At5g58140), NPH3 (At5g64330), RPT2 (At2g30520), Arabidopsis CUL3A (At1g26830), Arabidopsis CUL3B (At1g69670), human CUL3 (NM_003590), and clathrin heavy chain (At3g11130).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. NPH3-CUL3 Interactions in Insect and Animal Cells.
Supplemental Figure 2. Transiently Expressed GFP-CUL3A Appears to Associate with the Plasma Membrane in Plasmolyzed N. benthamiana Leaf Epidermal Cells.
Supplemental Figure 3. Phot1 Protein Is Stable under Low-Intensity Blue Light.
Supplemental Figure 4. Clathrin Heavy Chain Represents a phot1-Interacting Protein Whose Abundance Appears to Increase in Coimmunoprecipitates with phot1 in Response to in Planta Blue Light Exposure.
Supplemental References. References for Supplemental Figures 1 to 4.
Acknowledgments
We thank members of the Liscum laboratory for frequent help and advice. We thank Walter Gassmann for providing the P19-expressing A. tumefaciens strain and Christian Fankhauser for providing the phot1-5nph3-6PHOT1-GFP transgenic Arabidopsis line. We also thank Saikat Bhattacharjee (Gassmann Lab) for helping us establish the N. benthamiana system. We also thank Michael Garcia and Joanne Chory for critical reading of the manuscript and Neil Pratt, Lerxst Zivojinovic, and Dirk Lee for ongoing encouragement. U.V.P. was supported by University of Missouri/Monsanto Life Sciences and University of Missouri Maize Biology Training Program (Deparment of Energy/National Science Foundation/USDA) Fellowships. D.R. was supported by University of Missouri Interdisciplinary Plant Group, Department of Education Graduate Assistance in Areas of National Need, and National Institute of General Medical Sciences (T34-GM008396) Fellowships. We acknowledge support from the National Science Foundation (IOS-0415970 and IOS-0817737 to E.L.) and the National Institutes of Health (1RO1-AT003389 to S.S. and M.H.).
AUTHOR CONTRIBUTIONS
D.R., U.V.P., and E. Liscum designed the experiments, analyzed the data, and wrote the article. D.R. and U.V.P. performed the bulk of the experiments, and J.M. helped them. M.H. helped design human cell expression experiments, and U.V.P. performed them with S.S. X.T. and N.Z. performed the insect cell expression experiments. E. Lechner and P.G. provided seed stocks and critical phenotypic information prior to publication of cul3hyp mutant studies.
References
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee J.J., Bandyopadhyay A., Peer W.A., Makam S.N., Murphy A.S. (2004). Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 134: 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S.C., Disatnik M.H., Chan L.S., Iori K., Rando T.A. (2007). Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130: 349–362 [DOI] [PubMed] [Google Scholar]
- Briggs W.R., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13: 993–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J.L., Mamillapalli P., Burch-Smith T.M., Czymmek K., Dinesh-Kumar S.P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Sun L.J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33: 275–286 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. (2008). NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 21017–21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians M.J., Gingerich D.J., Hansen M., Binder B.M., Kieber J.J., Vierstra R.D. (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M., Reymond P., Powell G.K., Bernasconi P., Raibekas A.A., Liscum E., Briggs W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282: 1698–1701 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Azzo A., Bongiovanni A., Nastasi T. (2005). E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6: 429–441 [DOI] [PubMed] [Google Scholar]
- Dieterle M., Thomann A., Renou J.P., Parmentier Y., Cognat V., Lemonnier G., Müller R., Shen W.H., Kretsch T., Genschik P. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41: 386–399 [DOI] [PubMed] [Google Scholar]
- Esmon C.A., Tinsley A.G., Ljung K., Sandberg G., Hearne L.B., Liscum E. (2006). A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl. Acad. Sci. USA 103: 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L., Shen Y., Feng S., Bostick M., Callis J., Hellmann H., Deng X.-W. (2005). Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. (2010). Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235 [DOI] [PubMed] [Google Scholar]
- Fujimuro M., Sawada H., Yokosawa H. (1994). Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349: 173–180 [DOI] [PubMed] [Google Scholar]
- Fujimuro M., Yokosawa H. (2005). Production of antipolyubiquitin monoclonal antibodies and their use for characterization and isolation of polyubiquitinated proteins. Methods Enzymol. 399: 75–86 [DOI] [PubMed] [Google Scholar]
- Gao M., Karin M. (2005). Regulating the regulators: Control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol. Cell 19: 581–593 [DOI] [PubMed] [Google Scholar]
- Geyer R., Wee S., Anderson S., Yates J., Wolf D.A. (2003). BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12: 783–790 [DOI] [PubMed] [Google Scholar]
- Gingerich D.J., Gagne J.M., Salter D.W., Hellmann H., Estelle M., Ma L., Vierstra R.D. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Haga K., Takano M., Neumann R., Iino M. (2005). The rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell 17: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P.P., Dikic I. (2003). Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Harper R.M., Stowe-Evans E.L., Luesse D.R., Muto H., Tatematsu K., Watahiki M.K., Yamamoto K., Liscum E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Holland J.J., Roberts D., Liscum E. (2009). Understanding phototropism: From Darwin to today. J. Exp. Bot. 60: 1969–1978 [DOI] [PubMed] [Google Scholar]
- Huala E., Oeller P.W., Liscum E., Han I.S., Larsen E., Briggs W.R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278: 2120–2123 [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada S., Ohgishi M., Mayama T., Okada K., Sakai T. (2004). RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S.-i., Kinoshita T., Takemiya A., Doi M., Shimazaki K.-I. (2008). Leaf positioning of Arabidopsis in response to blue light. Mol. Plant 1: 15–26 [DOI] [PubMed] [Google Scholar]
- Kaiserli E., Sullivan S., Jones M.A., Feeney K.A., Christie J.M. (2009). Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell 21: 3226–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde M., Nühse T.S., Findlay K., Peck S.C. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. USA 104: 11850–11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Doi M., Suetsugu N., Kagawa T., Wada M., Shimazaki K. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Komander D. (2009). The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Lariguet P., Schepens I., Hodgson D., Pedmale U.V., Trevisan M., Kami C., de Carbonnel M., Alonso J.M., Ecker J.R., Liscum E., Fankhauser C. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. USA 103: 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. (1998). Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Li T., Pavletich N.P., Schulman B.A., Zheng N. (2005). High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 398: 125–142 [DOI] [PubMed] [Google Scholar]
- Liscum E., Briggs W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Briggs W.R. (1996). Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 112: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schaller N., Chou Y.C., Sumara I., Martin D.D., Kurz T., Katheder N., Hofmann K., Berthiaume L.G., Sicheri F., Peter M. (2009). The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc. Natl. Acad. Sci. USA 106: 12365–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Miranda M., Sorkin A. (2007). Regulation of receptors and transporters by ubiquitination: New insights into surprisingly similar mechanisms. Mol. Interv. 7: 157–167 [DOI] [PubMed] [Google Scholar]
- Motchoulski A., Liscum E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Pedmale U.V., Celaya R.B., Liscum E. (2010). Phototropism: Mechanisms and outcomes. In The Arabidopsis Book 8: e0125, doi/10.1199/tab.0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale U.V., Liscum E. (2007). Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J. Biol. Chem. 282: 19992–20001 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J.H., Willems A., Johnson J.L., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., Peter M. (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316 [DOI] [PubMed] [Google Scholar]
- Robert H.S., Offringa R. (2008). Regulation of auxin transport polarity by AGC kinases. Curr. Opin. Plant Biol. 11: 495–502 [DOI] [PubMed] [Google Scholar]
- Sakai T., Kagawa T., Kasahara M., Swartz T.E., Christie J.M., Briggs W.R., Wada M., Okada K. (2001). Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Wada T., Ishiguro S., Okada K. (2000). RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Briggs W.R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Singer J.D., Gurian-West M., Clurman B., Roberts J.M. (1999). Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13: 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan A., Culler A.H., Gallavotti A., Jackson D., Cohen J.D., McSteen P. (2009). BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol. 50: 652–657 [DOI] [PubMed] [Google Scholar]
- Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Privé G.G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans E.L., Harper R.M., Motchoulski A.V., Liscum E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 118: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S., Kaiserli E., Tseng T.S., Christie J.M. (2010). Subcellular localization and turnover of Arabidopsis phototropin 1. Plant Signal. Behav. 5: 184–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Tanaka N., Saeki Y., Tanaka K., Murakami M., Hirano T., Ishii N., Sugamura K. (2008). c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol. Cell. Biol. 28: 4805–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A., Brukhin V., Dieterle M., Gheyeselinck J., Vantard M., Grossniklaus U., Genschik P. (2005). Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 43: 437–448 [DOI] [PubMed] [Google Scholar]
- Thomann A., Lechner E., Hansen M., Dumbliauskas E., Parmentier Y., Kieber J., Scheres B., Genschik P. (2009). Arabidopsis CULLIN3 genes regulate primary root growth and patterning by ethylene-dependent and -independent mechanisms. PLoS Genet. 5: e1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Willems A.R., Schwab M., Tyers M. (2004). A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695: 133–170 [DOI] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T.H., Vidal M., Elledge S.J., Harper J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321 [DOI] [PubMed] [Google Scholar]
- Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. (2004). Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24: 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]