Mobile signals provided by hormones and morphogens are essential to organize multicellular structures. This article demonstrates that the joint activity of two bHLH transcription factors is required at two separate stages during Arabidopsis gynoecium and fruit development. In both instances, these factors mediate their function by ensuring appropriate distribution of the plant hormone auxin.
Abstract
Structural organization of organs in multicellular organisms occurs through intricate patterning mechanisms that often involve complex interactions between transcription factors in regulatory networks. For example, INDEHISCENT (IND), a basic helix-loop-helix (bHLH) transcription factor, specifies formation of the narrow stripes of valve margin tissue, where Arabidopsis thaliana fruits open on maturity. Another bHLH transcription factor, SPATULA (SPT), is required for reproductive tissue development from carpel margins in the Arabidopsis gynoecium before fertilization. Previous studies have therefore assigned the function of SPT to early gynoecium stages and IND to later fruit stages of reproductive development. Here we report that these two transcription factors interact genetically and via protein–protein contact to mediate both gynoecium development and fruit opening. We show that IND directly and positively regulates the expression of SPT, and that spt mutants have partial defects in valve margin formation. Careful analysis of ind mutant gynoecia revealed slight defects in apical tissue formation, and combining mutations in IND and SPT dramatically enhanced both single-mutant phenotypes. Our data show that SPT and IND at least partially mediate their joint functions in gynoecium and fruit development by controlling auxin distribution and suggest that this occurs through cooperative binding to regulatory sequences in downstream target genes.
INTRODUCTION
The evolution of fruits was of key importance in the evolutionary success of flowering plants (angiosperms). Fruits are female reproductive organs, which contain and nurture the developing seeds and finally mediate their efficient dispersal to optimize the chances of success for future generation. Fruits are derived from carpels, which form a gynoecium in the center of the flower. Many key regulators of carpel development also have roles in leaf development, thereby emphasizing the evolutionary origin of carpels as modified leaves (Scutt et al., 2006; Ferrándiz et al., 2010).
In Arabidopsis thaliana, the gynoecium is topped distally with stigmatic tissue that functions to mediate pollen germination (see Supplemental Figure 1A online). It also marks the beginning of the pollen-guiding transmitting tract, which runs down the center of the gynoecium (Nemhauser et al., 2000; Crawford et al., 2007). The solid and radially symmetric style supports the next segment of the transmitting tract, and an intact style is required for efficient fertilization. The ovary is formed as a longitudinal cylinder with mediolateral symmetry that reflects its origin as two fused leaf-like structures. It is composed of two compartments divided by the septum and adjacent placentae from which the ovules arise. The most basal part of the gynoecium is the gynophore, which connects the gynoecium to the receptacle and the rest of the plant.
The importance of the plant hormone auxin in patterning along the different axes of polarity in the gynoecium is well established (Nemhauser et al., 2000; Balanzá et al., 2006; Østergaard, 2009; Ståldal and Sundberg, 2009). It has been convincingly demonstrated that auxin synthesis at the apex of the Arabidopsis gynoecium is required for apical tissues to develop appropriately, and that transcription factors belonging to the SHORT INTERNODES, STYLISH (STY), and NGATHA families are required for this production. For example, members of these families promote expression of YUCCA (YUC) genes known to mediate auxin synthesis (Cheng et al., 2006; Sohlberg et al., 2006; Alvarez et al., 2009; Trigueros et al., 2009; Eklund et al., 2010), and the gynoecium defect of a sty1/2 double mutant can be rescued by application of exogenous auxin (Ståldal et al., 2008).
Basic Helix-Loop-Helix (bHLH) proteins comprise a large family of transcription factors, which exist in all eukaryotes (Massari and Murre, 2000; Pires and Dolan, 2010). Several members of this family act during gynoecium development or later in fruit development. For example, mutations in the SPATULA (SPT) gene lead to early defects in the development of carpel marginal tissues, such as stigma, style, septum, and transmitting tract (Alvarez and Smyth, 1999; Heisler et al., 2001). Similar phenotypes were observed in multiple combinations of mutations in the bHLH-encoding HECATE (HEC) genes, and protein–protein interactions between SPT and HEC proteins in yeast two-hybrid experiments suggest that these factors may have common downstream targets (Gremski et al., 2007).
After fertilization, the Arabidopsis fruit develops as a pod and differentiates many tissues, such as the valves (seedpod walls) and the central replum (see Supplemental Figure 1B online). Valves and replum tissues are separated by narrow files of highly specialized tissue called valve margins. When this tissue approaches maturity, it is composed of a layer of lignified cells and a layer of small cells, which will secrete cell wall–degrading enzymes to promote cell separation and to allow fruit opening and seed dispersal. INDEHISCENT (IND) bHLH is a key regulator of both the lignified layer and the separation layer in valve margin development, and fruits from ind mutants fail to open on maturity (Liljegren et al., 2004).
Recently, we found that IND functions at least partially through direct regulation of hormonal dynamics. In the wild-type fruits, local depletion of auxin at the valve margin is required for specification of the separation layer where fruit opening takes place. IND mediates the formation of this auxin minimum by directly regulating members of the auxin transport machinery (Sorefan et al., 2009). IND is also required to induce expression of GA4 to promote synthesis of the hormone gibberellin. As a consequence, DELLA proteins are degraded, allowing another bHLH protein, ALCATRAZ (ALC), to promote separation layer specification (Arnaud et al., 2010).
In this paper, we show that two bHLH factors, IND and SPT, previously described to function in very distinct aspects of female reproductive tissue development, in fact are closely connected at several levels to promote gynoecium patterning and seed dispersal. Our data demonstrate that the function of these genes is necessary for normal auxin distribution both in the gynoecium and at late stages of fruit development, and that IND and SPT bind together and regulate genes involved in modulating auxin transport.
RESULTS
IND Directly Regulates SPT Expression
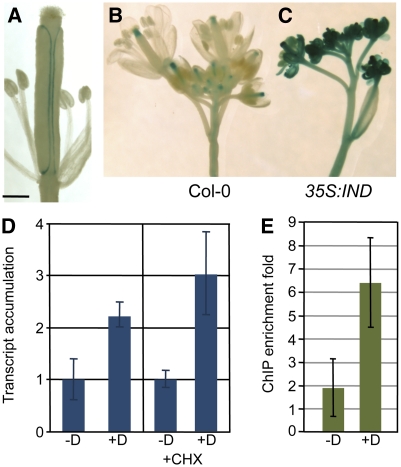
The IND gene encodes a bHLH protein that is expressed at the valve margin during Arabidopsis fruit development and is required for its specification (Liljegren et al., 2004). Although no defect in valve margin formation was previously detected in mutants of another bHLH-encoding gene, SPT, it has been shown that SPT is expressed in this tissue during fruit development, becoming limited to the separation layer, and that this expression depends on a functional IND gene (Figure 1A) (Heisler et al., 2001; Groszmann et al., 2010). In agreement with these observations, we found that ectopic overexpression of the IND gene under control of the 35S promoter led to strong induction of a SPT:β-glucuronidase (GUS) reporter construct in most tissues of the inflorescence (Figures 1B and 1C; see Supplemental Table 1 online). To further analyze this regulation, we used a dexamethasone (Dex)-inducible line (35S:IND:GR) expressing the IND protein fused to the rat glucocorticoid receptor (GR) under control of the 35S promoter (Lloyd et al., 1994; Sorefan et al., 2009). A microarray analysis showed that SPT expression is significantly upregulated when IND activity is induced (2.5-fold, P < 0.01), which was confirmed by quantitative RT-PCR (Q-RT-PCR) analysis (Figure 1D). Moreover, when the plants were treated with cycloheximide (Chx) to block translation, SPT transcript accumulation was still stimulated in response to IND activation (Figure 1D), showing that the regulatory process does not involve de novo protein synthesis and that SPT is an immediate target of IND.
Figure 1.
SPT Expression Is Directly Induced by IND.
(A) Expression of a SPT:GUS reporter construct (−1262 bp) at the valve margin of a stage-16 Arabidopsis fruit (Smyth et al. 1990).
(B) and (C) SPT:GUS (−6253 bp) expression in inflorescences of Col-0 (B) and 35S:IND (C).
(D) Q-RT-PCR analysis of SPT relative transcript accumulation after control treatment (−D) or IND activation by Dex treatment (+D) in a 35S:IND:GR line, in presence or absence of Chxe (+CHX). Values are the average of three biological repeats ± sd. +Dex values are significantly different from their corresponding −Dex-treated control (Student’s t test P value < 0.05).
(E) Fold-enrichment of the SPT promoter region in immunoprecipitated chromatin using an anti-GR antibody on the 35S:IND:GR line after control treatment (−D) or Dex treatment (+D). Values are the average of four biological repeats ± sd. The values are significantly different (Student’s t test P value = 0.01).
Bar in (A) = 0.5 mm.
To test whether IND regulates SPT expression via direct interaction with elements of the SPT gene, we used the 35S:IND:GR line to perform a chromatin immunoprecipitation (ChIP) experiment. Quantitative PCR (Q-PCR) analysis of precipitated DNA showed an enrichment of a SPT promoter fragment in Dex-treated compared with control-treated plants (Figure 1E). Thus, IND directly binds to the SPT promoter or is part of a protein complex that is bound to the promoter. Together these data identify IND as a direct and positive regulator of SPT expression consistent with the overlapping expression pattern of IND and SPT in the valve margin.
SPT Is Necessary but Not Sufficient for the Effect of Ectopic IND Expression
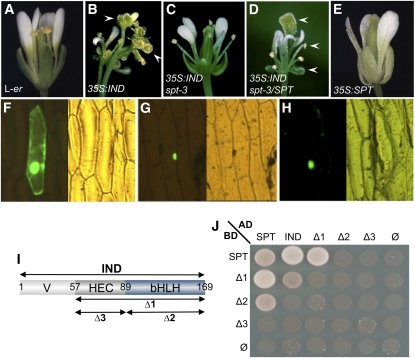
Ectopic expression of IND under the control of a 35S promoter leads to the formation of carpelloid sepals and stamens with stigmatic tissue at their apices and to sterile valveless gynoecia (Figures 2A and 2B; see Supplemental Figure 2 online) (Sorefan et al., 2009) reminiscent of gynoecia of the pinoid (pid) mutant (Bennett et al., 1995; Benjamins et al., 2001). Because SPT is dramatically overexpressed in 35S:IND lines, we wondered whether IND mediates its ectopic effects through activation of SPT. To answer this question, we transformed the spt-2 and spt-3 mutants with the 35S:IND construct and found that the IND-induced defects were partially restored. Although 35S:IND spt flowers develop narrow petals and short gynoecia with enlarged stigmata, ectopic formation of stigmatic tissue was almost never observed (Figure 2C; see Supplemental Figure 2 and Supplemental Table 2 online). To confirm that the phenotypic difference was related to SPT activity, we crossed a sample of these 35S:IND spt-2 and 35S:IND spt-3 lines with Landsberg erecta, thus introducing a wild-type (dominant) allele of SPT. All resulting F1 plants showed more dramatic flower phenotypes, similar to those observed when the wild-type Landsberg erecta was transformed directly with the 35S:IND construct (Figure 2D). These data therefore demonstrate that SPT activity is necessary for the ectopic formation of carpelloid features observed in 35S:IND plants.
Figure 2.
Ectopic IND Function Is SPT-Dependent, and IND and SPT Proteins Interact.
(A) to (E) Flowers of L-er (A), 35S:IND (B), 35S:IND spt-3 (C), 35S:IND spt-3/SPT (D), and 35S:SPT (E). Plants in (C) and (D) derive from the same transformation event. Arrowheads in (B) and (D) indicate ectopic stigma.
(F) to (H) GFP localization in epidermal onion cells transiently transformed with 35S:SPTΔNLS:GFP alone (F), 35S:SPT (G), or 35S:IND (H).
(I) Schematic of IND truncated versions used in (J). Variable N-terminal domain (V), the HEC-conserved domain (HEC), and the bHLH domain (bHLH) are indicated. Amino acid positions are shown. Δ1 construct contains HEC and bHLH domain, Δ2 contains the bHLH domain and Δ3 contains the HEC domain.
(J) Yeast two-hybrid experiment using fusions of SPT and IND (full-length and truncated version) with GAL4 activation and binding domains (AD and BD, respectively). Cells were spotted on selective medium lacking Leu, Trp, His, and adenine.
Interestingly, when ectopically expressing SPT under the 35S promoter, neither floral patterning defects nor ectopic stigma formation was observed (Figure 2E; see Supplemental Figure 2D online) (Penfield et al., 2005; Groszmann et al., 2008; Ichihashi et al., 2010). These data therefore show that the ectopic activities of both IND and SPT are necessary for the development of carpelloid organs.
IND and SPT Proteins Interact
One hypothesis to explain the effect of IND and SPT overexpression is that the transcription factors interact in complexes to regulate the expression of common target genes, leading to the formation of carpeloid and stigmatized tissue. Ectopic expression of SPT by itself would be insufficient to initiate this developmental program (Figure 2E), but 35S:IND alone can do so, because it also induces ectopic expression of its partner, SPT. Protein–protein interactions between IND and SPT would indeed be consistent with studies showing that bHLH transcription factors form homo- and heterodimers, which are crucial for their DNA binding activity (Massari and Murre, 2000; Longo et al., 2008; Pires and Dolan, 2010).
To test whether IND and SPT proteins can interact, we performed in vivo nuclear localization recovery assays by biolistic bombardment of onion cells. A deleted version of the SPT protein, lacking its localization domain and fused to green fluorescent protein (GFP) (35S:SPTΔNLS:GFP), did not accumulate preferentially in any cellular compartment (Figure 2F) (Groszmann et al., 2008). However, on coexpression with a wild-type version of SPT, a specific nuclear signal was detected (Figure 2G), showing that SPT is able to bind the mutated version and bring it to the nucleus. These data therefore demonstrate SPT–SPT homodimerization in plant cells. Coexpression of the 35S:SPTΔNLS:GFP construct with a 35S:IND gave a similar result (Figure 2H), demonstrating that IND protein is localized in the nucleus and can interact in vivo with SPT. Furthermore, these nuclear localization and protein interaction data were confirmed by bimolecular fluorescence complementation (BiFC) in Nicotiana tobaccum (tobacco) cells (see Supplemental Figure 3 online), which also revealed that IND is capable of homodimerization. BiFC with either SPT or IND in combination with unrelated proteins, such as ETTIN (ETT) and BREVIS RADIX (BRX), revealed no fluorescent signal, showing that the SPT–IND and IND–IND interactions are specific (see Supplemental Figure 3 online).
It has been shown by structural analyses that certain bHLH transcription factors interact with partners mainly through their HLH domain (Longo et al., 2008). To test whether this is also true for IND and SPT interaction, we performed a yeast 2-hybrid assay using three deleted versions of IND (Figures 2I and 2J; see Supplemental Figure 4 online). In yeast, the IND bHLH domain was found to be necessary and sufficient for heterodimerization with SPT, at least when SPT was fused to the GAL4 activation domain, because the bHLH region could function alone and its deletion abolished the interaction (Δ2 and Δ3 compared with Δ1 in Figure 2J at left). However, with SPT fused to the GAL4 DNA binding domain, the IND bHLH domain by itself was no longer able to interact (Δ2 in Figure 2J at top) but now seemed to require adjacent sequences present in Δ1. These data indicate that SPT and IND interact through their bHLH domains, and that this interaction may be stabilized by sequence or structure in the region flanking the bHLH domain of IND. This region is called the “HEC domain,” because it shows conservation with the HEC proteins (Heim et al., 2003; Gremski et al., 2007; Pires and Dolan, 2010).
These results show that IND can homodimerize (full-length IND and Δ1 in Figure 2J). Because of interaction-independent activation provided by the N-terminal domain of IND, it was not possible to obtain data using a full-length IND protein fused to the GAL4 DNA binding domain. Even so, yeast interaction assays suggest that all three domains of IND contribute to homodimerization, because partial or total loss of interaction was observed when variously deleting these domains. Because the IND bHLH domain by itself could interact with SPT but could not interact with full-length IND and the other IND truncations, it is likely that the IND–IND interaction through the bHLH domains is weaker than the bHLH interactions in the IND–SPT heterodimerization.
SPT Is Required for Separation Layer Development
SPT is expressed in the valve margins of developing Arabidopsis fruits; moreover, because IND regulates SPT expression and because the two proteins interact, we investigated a possible role for SPT in valve margin specification. For these analyses, we used two T-DNA alleles, spt-11 and spt-12, which are recently described strong mutant alleles in the Columbia (Col-0) background that show morphological defects in the gynoecium similar to the well-described alleles in Landsberg erecta (see Supplemental Figure 5 online) (Alvarez and Smyth, 1999; Ichihashi et al., 2010).
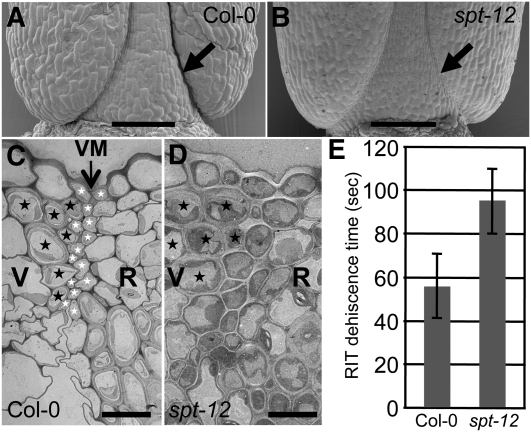
First, we used the shatter-quantification assay known as the Arabidopsis Random Impact Test (Arnaud et al., 2010) to quantify the opening sensitivity of spt fruits. Assaying many mature spt-12 siliques revealed a significant increase in shatter resistance compared with the wild-type control (Figure 3E), although this increase was not as pronounced as observed in ind mutant fruits. As reported earlier, the defect in ind mutant fruits is too severe to quantify dehiscence with this technique (Arnaud et al., 2010). Close analysis by scanning electron microscopy confirmed a defect in valve margin specification, revealing a less defined crease in mature spt-12 mutant fruits compared with the wild type (Figures 3A and 3B). It is possible that the partial indehiscence of spt in these assays is due to reduced fertility, because a reduced number of seeds could affect the fruit dehiscence via mechanical or developmental effects. We therefore characterized the dehiscence of nonfertilized emasculated gynoecia (see Supplemental Figure 6 online). Emasculated gynoecia from spt-11 and spt-12 mutants were still intact 20 d after emasculation, whereas the wild-type gynoecia naturally opened, confirming a direct role of SPT in valve margin formation.
Figure 3.
SPT Is Involved in Valve Margin Specification.
(A) and (B) Scanning electron microscopy of the base of Col-0 and spt-12 fruits at stage 18. Arrows indicate the dehiscence zone.
(C) and (D) Transmission electron micrographs of valve margin (VM) region in Col-0 and spt-12 siliques (stage 17b). Black and white stars indicate cells from the lignified and separation layers of the VM, respectively. Valves (V), VM, and replum (R) regions are indicated.
(E) Dehiscence assessment (Arabidopsis Random Impact Test) of Col-0 and spt-12. Values correspond to the time of shaking required to open 50% of dried siliques. Values are the average of at least three biological repeats (20 mature siliques for each) ± sd. The values are significantly different (Student’s t test P value = 0.02).
Bars in (A) and (B) = 100 μm; bars in (C) and (D) = 10 μm.
To further characterize the role of SPT in valve margin development, we studied the morphology of the valve margin in the spt-12 mutant by transmitting electron microscopy on transverse sections (Figures 3C and 3D). In the wild-type mature fruits, the valve margin differentiates late in development (stage 17) into a layer of lignified cells and a separation layer made of small cells (Figure 3C) (Rajani and Sundaresan, 2001; Liljegren et al., 2004; Arnaud et al., 2010). In spt-12 siliques, the lignified layer was clearly identifiable, but the adjacent cells did not present the characteristics of small separation layer cells (Figure 3D). SPT is thus involved in the specification of the separation layer cells of the valve margins.
IND and SPT Interact to Form Marginal Tissues
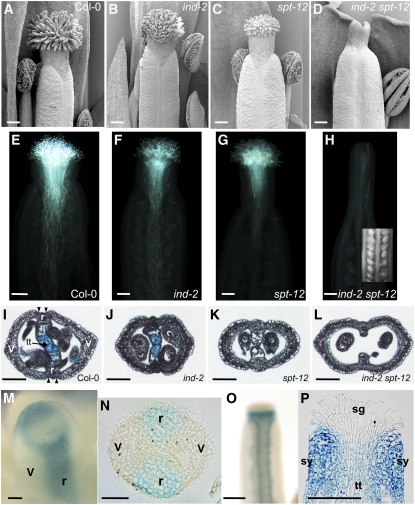
To further understand the function of IND and SPT, we studied the genetic interactions between them. Previously characterized spt single-mutants are defective in the development of marginal tissue structures, such as septum, stigma, and style (Alvarez and Smyth, 1999; Heisler et al., 2001). We observed identical defects in the spt-11 and spt-12 alleles (Figures 4C, 4K5A and 5B; see Supplemental Figure 5 online), including a 30% reduction of stigma hair length (Figures 4C and 5A) and a 20% reduction in style width (Figure 5B) compared with the wild type at Developmental stage 13 (Smyth et al., 1990). In the ovary, transmitting tract cells were completely absent, as seen by Alcian Blue staining (Figure 4K). Formation of unfused styles was only observed occasionally under our growth conditions, reflecting the variability of the spt phenotype also reported by others (see Supplemental Figure 5 online) (Alvarez and Smyth, 2002; Penfield et al., 2005). A reduced amount and growth of pollen tubes in the spt-12 gynoecium compared with the wild type was observed, which reflects the reduction in stigmatic tissue development and lack of transmitting tract (Figures 4E and 4G).
Figure 4.
IND and SPT Promote Formation of Marginal Tissues.
(A) to (D) Scanning electron microscopy images of gynoecia apical tissues at stage 13 in Col-0, ind-2, spt-12, and ind-2 spt-12.
(E) to (H) Pollen-tube growth in Col-0 (E), ind-2 (F), spt-12 (G), and ind-2 spt-12 (H). Inset in (H) is a light microscope image of ovules in an ind-2 spt-12 gynoecium.
(I) to (L) Cross sections of stage-13 ovaries in the wild type (Col-0) (I), ind-2 (J), spt-12 (K), and ind-2 spt-12 (L). Tissues are indicated in the wild-type section (I): v, valve; r, replum; tt, transmitting tract that has been stained with Alcian Blue.
(M) and (N) IND:IND:GUS expression in whole mount (M) and cross section (N) of stage-9 gynoecia. Presumptuous valves (v) and repla (r) are indicated.
(O) and (P) IND:IND:GUS expression in whole mount (O) and longitudinal section (P) of stage-12 gynoecia. Stigma (sg), style (sy), and transmitting tract (tt) are indicated in (P).
Bars in (A) to (H), (I) to (L), (O), and (P) = 100 μm; bars in (M) and (N) = 25 μm.
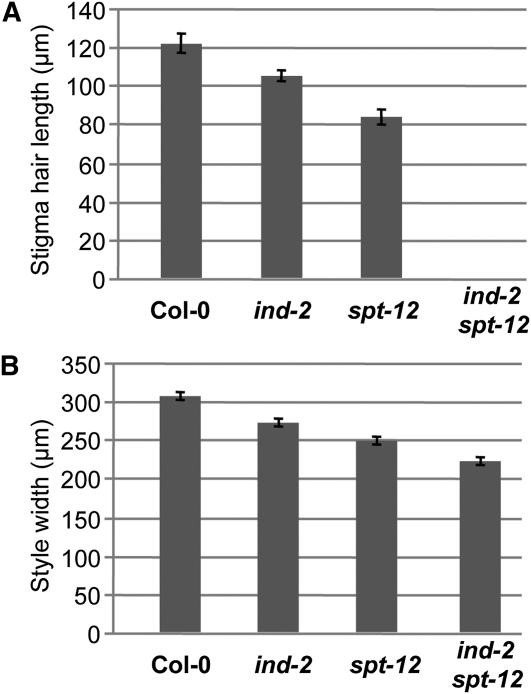
Figure 5.
IND and SPT Regulate Stigma Hair Length and Style Width.
(A) Length of stigma hairs in Col-0, ind-2, spt-12, and ind-2 spt-12.
(B) Width of style in Col-0, ind-2, spt-12, and ind-2 spt-12. Values in (A) and (B) are the average of >28 measurements ± se. All the values in individual panels (A) and (B) are significantly different to each other (Student’s t test P value < 0.01).
Although no effect on gynoecium development has been described for ind mutants, defects in the spt mutant gynoecium were strongly enhanced in the ind spt double mutant (Figures 4A to 4L, 5A, and 5B). ind-2 spt-12 mutant gynoecia were characterized by a complete absence of stigma (Figures 4D and 5A), and styles were split in two to various extents (see Supplemental Figures 7A to 7D online) but maintained style identity with the characteristically wax-crenulated epidermal cells (see Supplemental Figure 7E online). Style width of the ind-2 spt-12 double was more reduced than in spt-12 (29% reduction compared with the wild type) (Figure 5B). Septum tissue was not formed, the internal part of medial tissues being reduced to the inner replum (Figure 4L). Although ovules seemed to develop normally, no pollen germination and pollen tube growth was observed, most likely because of the absence of medial tissues (Figure 4H).
After careful examination of ind-2 gynoecia at stage 13, we identified a slight but significant reduction of stigma hair length and style width (13% decrease in both cases) (Figures 4B, 5A and 5B). Although, the ind-2 mutant has no fertility defects and produces a seed set similar to the wild type, we observed a reduced amount of pollen tubes formed; however, these did not exhibit reduced growth compared with the wild type (Figures 4E and 4F). This effect may be due to the observed reduction in stigmatic tissue length or style width. No effect of the ind-2 mutation was seen on septum and transmitting tract formation (Figure 4J).
A role for IND in septum, style, and stigma development would imply that IND is expressed in these tissues. We checked this using a reporter line, IND:IND:GUS, in which a 3.2-kb fragment of the IND gene containing 2.7 kb of promoter sequence and the 0.5-kb open reading frame was translationally fused to the GUS reporter gene and which is expressed later at the valve margin throughout postfertilization fruit development (Sorefan et al., 2009). At stage 12, when the stigma is fully developed, strong GUS staining was observed in the style and in the valve margins, but not in the stigma (Figure 4O). Longitudinal sections confirmed these observations and showed lower levels of expression in the transmitting tract (Figure 4P). We then checked the expression of the construct at stage 9 of gynoecium development, before the septum and stigma are fully formed. A ring of GUS expression was present at the top of the gynoecium, where the stigma would soon arise (Figure 4M); in the replum; and in the developing septum, just when it arises from the fusion of the septum ridges and well before the transmitting tract differentiates (Alvarez and Smyth, 2002) (Figure 4N). This expression pattern is consistent with the role of IND early in the formation of gynoecium marginal tissues revealed through phenotypic analysis of the ind spt double mutant. It also coincides with SPT expression in marginal tissues at this stage, which has been thoroughly characterized in previous studies (Heisler et al., 2001; Groszmann et al., 2010). Given this overlap of IND and SPT expression, IND may function in early gynoecium development through formation of a regulatory complex with SPT.
IND and SPT Are Required for Normal Auxin Dynamics Throughout Gynoecium and Fruit Development
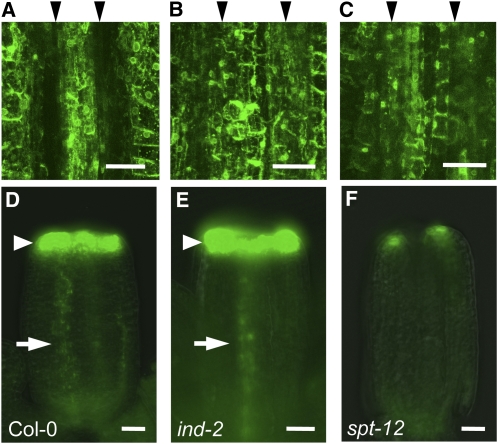
Previously, we showed that IND promotes depletion of auxin from the separation layer and that the resulting auxin minimum is required for fruit opening (Sorefan et al., 2009). Because spt mutants fail to specify separation layer cells, we tested whether SPT has a role in formation of the auxin minimum. To this end, we analyzed the distribution of the auxin signaling reporter DR5:GFP in the spt-12 background. In wild-type fruits at stage 17b, 90% of valve margins showed no DR5:GFP signal (36 out of 40 analyzed valve margins), appearing as a gap between flanking signals in the valve and replum (Figure 6A). None of the ind-2 mutant fruits tested exhibited an auxin minimum at stage 17b consistent with previous analysis (Figure 6B) (Sorefan et al., 2009). For spt-12, a partial effect was observed, with 60% of valve margins (28 out of 46) exhibiting no auxin minimum (Figure 6C), and 40% still having a reduction in the DR5:GFP signal as found in the wild type. Although the penetrance of the spt-12 mutation was weaker than for ind-2, the data suggest that SPT promotes separation layer development through formation of an auxin minimum in this tissue.
Figure 6.
IND and SPT Regulate Auxin Distribution.
(A) to (C) DR5:GFP expression in VM region of stage-17b fruits of Col-0 (A), ind-2 (B), and spt-12 (C). Black arrowheads above the images indicate the position of VM creases, which show a gap in fluorescence in the wild-type but not in the ind-2 and spt-12 fruits.
(D) to (F) DR5:GFP expression in stage-9 gynoecia of Col-0 (D), ind-2 (E), and spt-12 (F). Arrowheads and arrows indicate respectively GFP signals at the top of the gynoecium and in the presumptive replum.
Bars in (A) to (C) = 50 μm; bars in (D) to (F) = 25 μm.
It is well established that modulation of auxin synthesis and transport affects early gynoecium development (Nemhauser et al., 2000; Sohlberg et al., 2006). Given the involvement of IND and SPT in regulating auxin transport in the valve margin, we tested whether IND and SPT also mediate their effects in the earlier development of medial tissues through auxin dynamics. To this end, we characterized the DR5:GFP reporter in ind-2 and spt-12 mutants at stage 9 of gynoecium development, before the septum and stigma tissues are formed and before the mutants exhibit any clear phenotype (Figures 6D to 6F). In the wild type and ind-2, a strong ring of GFP signal was present at the distal end, and a line of weaker signal was observed inside the presumptive replum (Figure 6D). By contrast, in the spt-12 gynoecium, no replum signal could be detected, and the apical signal was reduced in intensity and limited to two spots in the lateral domains above the presumptive valves (Figure 6F). The DR5:GFP signal in the wild-type, ind, and spt correlates with the severity of disruption in medial tissue development observed at later developmental stages for the mutants (Figure 4). These data suggest that SPT-controlled auxin dynamics in early stages of gynoecium development are required for proper formation of medial tissues.
IND and SPT Bind Elements in the PID Promoter and Jointly Regulate PID and WAG2 Gene Expression
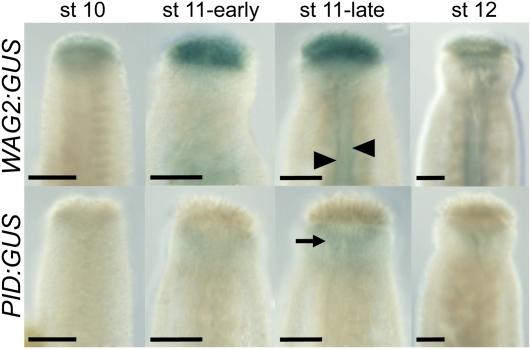
We have established that IND and SPT interact both genetically and through physical protein–protein contact. These data suggest that they function within the same protein complex to regulate common downstream target genes. Two strong candidates for such genes are WAG2 and PID, which we have already demonstrated are direct targets of IND in planta (Sorefan et al., 2009). Our previous work has shown that IND positively regulates the expression of WAG2 but represses expression of PID. To verify whether the expression patterns of these genes are in agreement with them being common targets of IND and SPT during gynoecium development, we analyzed GUS reporter lines for WAG2 and PID during gynoecium development (Figure 7). This analysis showed that WAG2:GUS is expressed in the style and that the dynamics of this expression overlaps with IND and SPT expression and support a role for WAG2 in regulating lateral auxin distribution in the style. Moreover, in gynoecia from late stage 11, WAG2:GUS was also expressed in the two strips of cells where valve margins will form (arrowheads in Figure 7), identical to the expression of IND at this stage of development. Conversely, PID:GUS expression was largely absent from the style in this experiment, except for a weak signal detected in the style at late stage 11 (arrow in Figure 7). These data therefore support the hypothesis that SPT and IND oppositely regulate WAG2 and PID to control distribution of auxin in the style.
Figure 7.
Expression of WAG2 and PID in the Developing Gynoecium.
Histochemical staining of gynoecia from WAG2:GUS (Top) and PID:GUS (Bottom) reporter lines throughout development. Developmental stages are indicated above. Arrowheads in WAG2:GUS (st 11-late) indicate expression in the medial region, and arrow in PID:GUS (st 11-late) points to weak expression in the style.
Bars = 50 μm.
Transcription factors of the bHLH family bind to so-called E-box cis-elements (CANNTG) and predominantly to the G-box form (CACGTG) (Toledo-Ortiz et al., 2003). Interestingly, we identified an E-box variant in the PID promoter (TCTCACGCGTTG) at −136 bp from the predicted transcription start site (TAIR database, www.Arabidopsis.org). An identical E-box variant, although in the opposite orientation, is located in the SPT promoter and confers IND-dependent SPT expression in the valve margin (Groszmann et al., 2010). In a yeast one-hybrid experiment, we found a strong interaction between IND protein and the PID E-box variant (Figure 8B; see Supplemental Figure 8 online). This interaction was specific for IND, because neither SPT nor its closest homolog, ALC, was able to bind. Another 60 bp upstream in the PID promoter, we identified two closely situated canonical G-box elements (GGCACGTGACAACGTCTCACACGTGTC). Yeast one-hybrid interaction assays showed that SPT had a stronger affinity toward this double G-box than the two other bHLH proteins tested here, IND and ALC (Figure 8B; see Supplemental Figure 8 online).
Figure 8.
IND and SPT Cooperate to Regulate Downstream Targets and Bind Separate but Adjacent Elements in the Promoter of PINOID.
(A) Scanning electron microscopy of style and stigmatic tissues in pid-9.
(B) Quantification of yeast one-hybrid interactions between ALC, IND, and SPT proteins and two different elements of PID promoter (E-box variant and Double G-box). ALC was used as a negative control (nonspecific binding of a bHLH protein). Values are averages of five replicates ± sd. Asterisk (*) indicates values that are significantly different from the corresponding ALC control (Student’s t test P value < 0.0001).
(C) and (D) Q-PCR analysis of PID (C) and WAG2 (D) relative to transcript accumulation after control treatment (−D) or IND activation (+D) in 35S:IND:GR (SPT) or 35S:IND:GR spt-12 (spt-12). Values are the average of at least four biological repeats ± se. Asterisk (*) indicates values that are significantly different from their corresponding −Dex-treated control (Student’s t test P value < 0.02).
(E) Schematic of SPT and IND binding to the PID promoter. The double G-box elements are orange and the E-box is green. TATA indicates the position of the transcription initiation TATA box, and the PID coding region is shown in red.
Bar in (A) = 100 μm.
If IND and SPT can bind to closely positioned cis-elements on the same gene and interact both genetically and through protein–protein contact, it is likely that they cooperatively regulate downstream targets. We therefore tested whether the inverse regulation of PID and WAG2 by IND as described earlier (Sorefan et al., 2009) is dependent on SPT activity. In this experiment, we crossed the 35S:IND:GR line to the spt-12 mutant and checked for regulation by comparing untreated and Dex-treated 7-d-old seedlings (Figures 8C and 8D). Repression of PID and induction of WAG2 in Dex-treated 35S:IND:GR seedlings was similar to previous descriptions; however, no significant change was observed between Dex-treated and untreated samples in the 35S:IND:GR spt-12 mutant (Figures 8C and 8D), demonstrating that both PID and WAG2 are cooperatively regulated downstream targets of IND and SPT in plants.
DISCUSSION
IND and SPT Interact to Promote Seed Dispersal
bHLH proteins mediate their effect through homo- and heteromeric interactions (Massari and Murre, 2000; Longo et al., 2008). In this paper, we have revealed that the two bHLH proteins, IND and SPT, interact genetically and through protein–protein contact to control tissue patterning in the Arabidopsis gynoecium and fruit. IND directly promotes SPT expression, and the conversion of floral organs into carpelloid structures covered in stigmatic tissue on IND overexpression is dependent on a functional SPT gene. Because overexpression of SPT alone has no detectable effect, these data suggest that both IND and SPT proteins are required to regulate downstream gene expression.
The observation that IND promotes SPT gene expression and that IND and SPT proteins can interact was surprising, given that their reported functions are in different processes of female reproductive tissue development. However, a recent analysis of the SPT promoter did reveal IND-dependent expression of SPT in the valve margin late in fruit development (Groszmann et al., 2010). Further, expression of an artificially repressive form of SPT resulted in loss of dehiscence (Groszmann et al., 2011). We have now obtained direct evidence for a defect in valve margin development in two strong spt mutant alleles, both by microscopic analyses (scanning electron microscopy and transmission electron microscopy) and by direct quantification of shatter potential. Although it was less pronounced than in ind mutant fruits, most spt fruits did not form the auxin minimum at the valve margins, previously demonstrated to be required for separation layer specification (Sorefan et al., 2009). Together, these data suggest that IND and SPT interact to regulate expression of common downstream targets involved in the timely depletion of auxin from the separation layer.
IND and SPT Interact to Control Gynoecium Development
The effect of spt mutations on earlier gynoecium development is well characterized (Alvarez and Smyth, 1999; 2002; Heisler et al., 2001. spt mutant gynoecia have reduced stigmatic and septum tissues and lack a transmitting tract for pollen-tube guidance. As a consequence, spt mutants have reduced fertility. In contrast with spt mutants, ind mutants have very modest defects in gynoecium development, although a careful analysis of ind gynoecia revealed a slight but significant decrease in stigma length and style width. These reductions may explain the decrease in pollen-tube density observed in the ind-2 mutant used here; however, this has no detectable effect on fertility. In fact, the transmitting tract in ind-2 gynoecia seems to function normally, because the pollen tubes grew down to the very base, as in the wild type. We hypothesize that the full seed set observed in ind-2, despite reduced pollen-tube density, may reflect the fact that the wild type supports growth of an excess number of pollen tubes compared with the available ovules (Crawford et al., 2007).
The combination of ind and spt severely increased the gynoecial defects of spt mutants. The double mutant gynoecia completely lacked medial tissues and were sterile, revealing a function for IND in specifying these tissues. The lack of stigmatic tissue prevented any attachment of pollen and therefore of pollen-tube growth. This synergistic effect suggests that other partners are involved in the IND–SPT regulatory complex, such that in the ind mutant, SPT may interact with other partners to at least partially trigger the developmental process and vice versa for IND in the spt mutant.
IND and SPT Mediate Their Effect Through Regulation of Auxin Dynamics
Accumulation of auxin at the apex of Arabidopsis gynoecia seems to be a requirement for normal development, because mutations in the auxin biosynthetic YUCCA genes or their regulators, STY1 and STY2, lead to severe phenotypes (Sohlberg et al., 2006; Cheng et al., 2006). In agreement with this, we observed a very strong DR5:GFP signal at the apical region, where the style and stigma will form. Interestingly, this signal was dramatically diminished in the spt-12 mutant. This result is consistent with previous reports that apical gynoecium defects in spt mutants are rescued by treatment with the auxin transport inhibitor, NPA (Nemhauser et al., 2000; Ståldal et al., 2008). Nemhauser et al. (2000) suggested that SPT is involved in the transduction pathway downstream of auxin. Alternatively, Dinneny and Yanofsky (2005) proposed that SPT promotes the high auxin level at the apical region by inhibiting polar auxin transport. NPA treatment of the spt mutant would thus mimic SPT function, inhibiting transport of auxin away from the apex and allowing the auxin accumulation necessary for apical tissue development. The data obtained here therefore lend support to the hypothesis of Dinneny and Yanofsky (2005) by suggesting that SPT and IND coregulate auxin transport.
Members of the AGC3 family of protein kinases regulate the direction of auxin transport through phosphorylation at specific sites of the PIN auxin efflux carriers (Michniewicz et al., 2007; Dhonukshe et al., 2010; Huang et al., 2010). Previously, we showed that IND controls the direction of auxin transport through direct regulation of two such AGC protein kinase genes, PID (repression) and WAG2 (activation) (Sorefan et al., 2009). Accordingly, it could be predicted that the perturbed auxin distribution described above in ind and spt mutants is due to misexpression of these kinase-encoding genes. In support of this hypothesis, pid mutants develop gynoecia with enlarged stigmatic tissue, suggesting that PID is a repressor of stigma development (Figure 8A). Moreover, the expression of a WAG2:GUS reporter line overlaps with the expression pattern of IND at the apical region of the gynoecium, whereas PID:GUS expression is largely absent from this tissue, although a very weak PID:GUS signal was observed in the style at late stage 11 (Figure 7). Other factors may be able to slightly overrule the repressive activity of IND and SPT at this stage. The genetic and protein–protein interaction data presented here between IND and SPT, together with the effect of spt mutation on the distribution of DR5:GFP signal, strongly suggest that SPT acts on auxin distribution through the same mechanism as IND. In support of a joint role in regulating auxin transport, we found that the inverse regulation of PID and WAG2 by IND depends on a functional SPT. Moreover, we used yeast one-hybrid to show that both SPT and IND are capable of binding specific sequences located in the PID promoter. SPT was found to bind a typical G-box motif, whereas IND bound a so-called E-box variant. These results are in agreement with a previous prediction based on amino acid signature of the bHLH domain, that SPT would have G-box specificity (Toledo-Ortiz et al., 2003). IND, on the other hand, lacks the amino acid signature of the bHLH domain shown for other bHLH proteins to be in direct contact with DNA (Toledo-Ortiz et al., 2003; Liljegren et al., 2004). IND may therefore use different amino acids for DNA interaction, which could reflect its high preference for the atypical bHLH binding site. An E-box variant identical to the one in the PID promoter is located in the SPT promoter, although in the opposite orientation. This sequence is required for SPT valve margin expression (Groszmann et al., 2010); however, because of self-activation in the yeast one-hybrid assay, we have been unable to verify whether IND binds to this element.
IND and SPT Cooperativity
It was previously suggested that SPT, because of its broad expression pattern throughout plant development, requires protein partners with more specific expression patterns (Gremski et al., 2007). Two observations reported here support this hypothesis: the lack of carpel and fruit defects in 35S:SPT transgenics (Groszmann et al., 2008) and the dramatically stronger binding of IND compared with SPT to their respective cis-elements tested here. At the PID promoter, initial strong binding of IND to the variant E-box may increase the interaction of SPT with the G-boxes, which may otherwise be too weak, leaving SPT unable to regulate PID expression by itself. Similarly, IND may also require the formation of heterocomplexes to regulate transcription of target genes. Based on the genetic and protein interaction data presented here and on previously reported results, it is possible that interaction between different partners on the same promoter could lead to different regulatory outputs and that different downstream targets require different partner combinations.
As mentioned above, SPT may interact with partners other than IND during gynoecium development, because the ind single mutant phenotype in the gynoecium is markedly weaker than the phenotype of spt single mutants. The IND-related HEC proteins are good candidates for such partners, because reduction of HEC1/2/3 activity leads to a phenotype similar to the ind spt mutant: a complete absence of stigma and a strong reduction of septum (Gremski et al., 2007). In support of this hypothesis, it was previously shown that SPT can interact with the HEC proteins in yeast, and their partially overlapping expression pattern suggests that SPT and HEC may interact in planta to regulate a common set of downstream target genes (Gremski et al., 2007). Similarly, IND has been reported to interact with the SPT-related protein ALC in yeast (Liljegren et al., 2004), and like IND, ALC is expressed at the valve margin and is required for separation layer development (Rajani and Sundaresan, 2001). Although we were unable to detect a significant interaction between ALC and elements found in the PID promoter in the yeast one-hybrid assay, it is likely that a complex involving both IND and ALC also regulates common targets. Recently, a shared role for ALC and SPT in both gynoecium and dehiscence zone development has been reported (Groszmann et al., 2011) that has parallels to the IND–SPT interaction found here. Mutations in the ALC gene also enhance the spt mutant phenotype during gynoecium development, and the ALC and SPT proteins are also able to interact in yeast and in planta (Groszmann et al., 2011).
Together, these observations suggest that several combinations of bHLH proteins exist to regulate different stages of reproductive tissue development, and it is possible that specific combinations contribute to different extents. It is also possible that partial redundancy among such transcription-factor combinations provides built-in robustness to buffer against irregularities during reproductive tissue development. Future studies using in vivo and in vitro interaction techniques combined with mutant analyses will shed light on these possibilities by revealing the downstream targets specific for individual combinations.
In conclusion, our data support a model in which SPT and IND mediate aspects of gynoecium and fruit development through cooperative binding and regulation of their target genes (Figure 8E). Our results also suggest that IND and SPT control auxin distribution similarly in the two developmentally distinct scenarios.
METHODS
Plant Materials and Growth Conditions
Plants were grown on soil in glasshouse conditions mimicking long days (16 h light/8 h dark). Developmental stages of flowers and fruits were defined as in Smyth et al. (1990). Mutant lines ind-2 (Liljegren et al., 2004), spt-11, and spt-12 (Ichihashi et al., 2010) and pid-9 (Christensen et al., 2000) were in Col-0 background, and spt-2 and spt-3 were in Landsberg erecta background. Reporter lines of SPT:GUS (Groszmann et al., 2010) were in Landsberg erecta background, and IND:IND:GUS (Sorefan et al., 2009) was in Col-0 background.
Plasmid Construction and Arabidopsis Transformation
The 35S:IND construct was created similarly to 35S:SPT (Groszmann et al., 2008) by amplification of the full-length open reading frame by PCR and insertion into the multiple cloning site of the pART7 vector containing a 5′ 35S promoter element and a 3′ OCS sequence and subsequently in to pMLBART (Gleave, 1992) for further transformation into the Agrobacterium tumefaciens strain AGL1. Arabidopsis thaliana wild-type and mutant plants were transformed with 35S:IND and 35S:SPT by the floral dipping method described by Clough and Bent (1998).
Q-RT-PCR Expression Analysis
Seeds were germinated in 0.5× Murashige and Skoog medium with constant shaking. We then treated 7-d-old seedlings for 4 h with 10 μM Dex, with or without Chx, and with or without 50 μM indole-3-acetic acid (IAA) as described in Sorefan et al. (2009). The addition of auxin is based on our findings that gene expression effects are enhanced in the presence of IAA (Sorefan et al., 2009).
Total RNA was extracted using RNeasy Plant mini kit (Qiagen). cDNA was produced using 2-5 μg of total RNA and a polyT(15) primer. At least three biological and three technical repeats were performed. Q-PCR was performed with SYBR green jumpstart Taq ReadyMix (Sigma-Aldrich) on a Chromo4 Real-Time PCR detector. PCR products were checked by agarose gel electrophoresis. UBQ10 was used as normalization control, because its expression was not affected by the treatment (primers listed in Supplemental Table 3 online).
Unpaired two-sample Student’s t tests were used for statistical analysis. When necessary, data were log-transformed to meet the criteria of equal variances.
Yeast Two-Hybrid
The IND and SPT coding regions were PCR amplified and cloned in pGAD424 and pGBT9 vectors (Clontech). The yeast two-hybrid experiment was performed according to the manufacturer’s instructions. Interactions were assessed via growth on selective yeast media and by quantifying β-galactosidase activity by liquid culture assay using ONPG as substrate (three technical repeats), following the guidelines of the manufacturer.
Yeast One-Hybrid
Sense and antisense oligonucleotides containing a triplication of either the wild-type E-box variant or a mutated version from the PID promoter were designed and included three nucleotides of flanking sequence on both sides. Similarly, sense and antisense oligonucleotides were designed to create a construct containing three copies of the PID double G-box with three flanking nucleotides. The sequences of the oligonucleotides are listed in Supplemental Table 3 online.
Sense and antisense nucleotides were annealed and ligated into the pLacZi plasmid (Clontech). For the double G-box and oligonucleotides, G1F and R and G2F and R were annealed, respectively. Then a three-point ligation was performed to clone them into the pLacZi plasmid. The construct therefore contains three times the double G-box. The three resulting plasmids and an empty pLacZi were transformed into the yeast strain YM4271 for stable integration into the yeast chromosome according to the manufacturer’s manual.
Full-length constructs of IND, SPT, and ALC in pGAD424 were transformed into the resulting lines, and interactions were assessed by quantifying β-galactosidase activity by liquid culture assay using ONPG as substrate (five repeats) and by colony-lift filter assays according the guidelines by the manufacturer.
Alcian Blue Staining
Tissue was fixed in 3.7% formaldehyde, 5% acetic acid, and 50% ethanol and subsequently dehydrated through an ethanol series. The tissue was cleared with Histoclear (National Diagnostics) and embedded in paraffin. An RM 2255 rotary microtome (Leica) was used to make 8-μm transverse stem sections. After deparaffinization with Histoclear, sections were stained with an Alcian Blue 8GX solution (0.05% Alcian Blue 8GX 0.1 M acetate buffer, pH 5.0). Sections were examined under light microscopy.
Scanning Electron Microscopy
Fruits were fixed for ~4 h at 25°C in 3.7% formaldehyde, 5% glacial acetic acid, and 50% ethanol. After critical point drying, tissue was coated with gold and examined in a Philips XL30 FEG microscope using an acceleration voltage of 3 kV.
Transmission Electron Microscopy
Stage-17b fruits were fixed in 2.5% vol/vol glutaraldehyde/0.05 M Na cacodylate (pH 7.2), vacuum-infiltrated, and left overnight at room temperature. Samples were postfixed in 1% osmium tetroxide/0.05 M Na cacodylate for 1 h, briefly washed with water; and dehydrated in ethanol series. Samples were then infiltrated in London Resin White resin (London Resin) and sectioned for transmission electron microscopy imaging with an FEI Technai G2 20 Twin Transmission Electron Microscope.
ChIP
35S:IND:GR seeds were grown for 7 d in 0.5% Glc (w/v) 0.5× Murashige and Skoog medium with constant shaking. Seedlings were treated with 50 μM IAA and 10 μM Dex, and ChIP was performed as described below.
The ChIP experiments were performed as previously described (Sorefan et al., 2009). Q-PCR was performed using SYBR Green JumpStart Taq ReadyMix in a Bio-Rad Chromo4 Q-PCR machine, and the primers used were SPT473 and SPT776 for SPT and Mu-likeF and Mu-likeR for the Mu-like transposon (see Supplemental Table 3 online). The values correspond to the ratios between pull-down DNA with and without the GR antibody, both initially normalized by Mu-like transposon. We used 304 bp of the SPT promoter region upstream of the transcription start site. As described in Sorefan et al. (2009), NRT2.1 served as a negative control not affected by the Dex treatment.
β-Glucuronidase (GUS) staining
GUS assays were performed as described in Arnaud et al. (2010). Plants were fixed in 90% acetone on ice for 20 min, then rinsed with a buffer containing 0.5 mM of K-ferrocyanide (Sigma-Aldrich), 0.5 mM of K-ferricyanide (Sigma-Aldrich), and 0.2% Triton X-100 in 50 mM of sodium phosphate buffer, pH 7.2. Samples were then incubated for 24 to 48 h at 37°C in the buffer containing 2 mM of 5-bromo-4-chloro-3-indolyl p-d-glucuronide (Melford).
Stigma Length and Style Width Quantification
Pictures of stage-13 gynoecia immediately before pollination were taken using a Leica MZ16 dissecting microscope with the replum facing the lens. The width of the style and the length of one representative stigma hair per gynoecium were measured using the Fiji program package (ImageJ software).
BiFC
Open reading frames of full-length IND, SPT, ETT, and BRX were cloned into vectors pYFPN43 and pYFPC43 (http://www.ibmcp.upv.es/FerrandoLabVectors.php), and BiFC was performed as previously described (Scacchi et al. 2009).
Confocal Microscopy
Confocal microscopy was performed using a Leica SP laser-scanning microscope equipped with an Argon krypton laser (Leica) as described in Sorefan et al. (2009).
Onion Single-Cell Layer Biolistics
Assessment of nuclear localization in onion epidermal cells that had been biolistically transformed with GFP reporter constructs was done as described in Brewer et al. (2004).
Assessment of Dehiscence Using an Arabidopsis Random Impact Test
Silique samples at stage 18 or older were selected randomly from Arabidopsis wild-type and mutant plants. After equilibrating to 50% relative humidity, the fruits were subjected to the shatter-resistance assay as previously described (Arnaud et al., 2010).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: IND (AT4G00120), SPT (AT4G36930), PID (AT2G34650), WAG2 (AT3G14370), ALC (AT5G67110), ETT (AT2G33860), BRX (AT1G31880), UBQ10 (AT4G05320), and NRT2.1 (AT1G08090).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphology of the Arabidopsis Fruit.
Supplemental Figure 2. SPT Is Necessary but Not Sufficient for the Effect of Ectopic IND Expression.
Supplemental Figure 3. IND and SPT Interact in Plant Cells.
Supplemental Figure 4. Quantification of IND-SPT Interactions in Yeast.
Supplemental Figure 5. spt-11 and spt-12 Mutant Alleles.
Supplemental Figure 6. SPT Promotes Dehiscence.
Supplemental Figure 7. A Range of Marginal Tissue Defects in ind-2 spt-12 Double Mutant Gynoecia.
Supplemental Figure 8. IND and SPT Bind Separate Elements That Occur Nearby in the Promoter of PINOID.
Supplemental Table 1. Ectopic Expression of INDEHISCENT Activates SPATULA Expression.
Supplemental Table 2. Transformation Data for 35S:IND Transformed into the Wild-Type and spatula Mutant Plants.
Supplemental Table 3. Oligonucleotide Sequences.
Acknowledgments
We thank Steve Penfield for spt-11 and spt-12 mutant seeds, Michael Groszmann for the IND genomic clone, 35S:SPT plants, and for useful discussions, Charlie Scutt for critical comments on the manuscript, Nicolas Arnaud for useful discussions, and Martin F. Yanofsky and Kristina Gremski for sharing unpublished observations and useful discussions. We are grateful to Andrew Davis, Sue Bunnewell, and Kim Findlay at the John Innes Centre for assistance with photography, transmission electron microscopy, and scanning electron microscopy and to Dian Guan for constructing the double G-box vector for the yeast one-hybrid experiment. This work was funded by grants from the Biotechnological and Biological Science Research Council (BB/D018005/1 to L.Ø.), from the Australian Research Council (A19927094 to D.R.S.), from the Spanish Government (BIO2009-09920 to C.F.), and by a Marie Curie Fellowship (MEST-CT-2005-019727 to S.F.).
AUTHOR CONTRIBUTIONS
T.G., D.R.S., and L.Ø. designed the research. T.G., T.P., P.S., S.F., M.O., K.S., T.A.W., V.B., and D.R.S. performed the research. T.G., T.P., P.S., S.F., E.K., M.O., K.S., T.A.W., V.B., C.F., D.R.S., and L.Ø. analyzed the data, and T.G., D.R.S., and L.Ø. wrote the article.
References
- Alvarez J., Smyth D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Alvarez J., Smyth D.R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163: 17–41 [Google Scholar]
- Alvarez J.P., Goldshmidt A., Efroni I., Bowman J.L., Eshed Y. (2009). The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 21: 1373–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T.A., Lawrenson T., Sablowski R., Østergaard L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24: 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzá V., Navarrete M., Trigueros M., Ferrándiz C. (2006). Patterning the female side of Arabidopsis: The importance of hormones. J. Exp. Bot. 57: 3457–3469 [DOI] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykaas P.J., Offringa R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Bennett S.R.M., Alvarez J., Bossinger G., Smyth D.R. (1995). Morphogenesis of pinoid mutants of Arabidopsis thaliana. Plant J. 8: 505–520 [Google Scholar]
- Brewer P.B., Howles P.A., Dorian K., Griffith M.E., Ishida T., Kaplan-Levy R.N., Kilinc A., Smyth D.R. (2004). PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131: 4035–4045 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford B.C., Ditta G., Yanofsky M.F. (2007). The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr. Biol. 17: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Yanofsky M.F. (2005). Drawing lines and borders: How the dehiscent fruit of Arabidopsis is patterned. Bioessays 27: 42–49 [DOI] [PubMed] [Google Scholar]
- Eklund D.M., Ståldal V., Valsecchi I., Cierlik I., Eriksson C., Hiratsu K., Ohme-Takagi M., Sundström J.F., Thelander M., Ezcurra I., Sundberg E. (2010). The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22: 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Fourquin C., Prunet N., Scutt C.P., Sundberg E., Trehin C., Vialette-Guiraud A.C.M. (2010). Carpel Development. Adv. Bot. Res. 55: 1–73 [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gremski K., Ditta G., Yanofsky M.F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601 [DOI] [PubMed] [Google Scholar]
- Groszmann M., Paicu T., Smyth D.R. (2008). Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 55: 40–52 [DOI] [PubMed] [Google Scholar]
- Groszmann M., Bylstra Y., Lampugnani E.R., Smyth D.R. (2010). Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 61: 1495–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M., Paicu T., Alvarez J.P., Swain S.M., Smyth D.R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 10.1111/j.1365–313X [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Atkinson A., Bylstra Y.H., Walsh R., Smyth D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Horiguchi G., Gleissberg S., Tsukaya H. (2010). The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51: 252–261 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Roeder A.H., Kempin S.A., Gremski K., Østergaard L., Guimil S., Reyes D.K., Yanofsky M.F. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Schena M., Walbot V., Davis R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Longo A., Guanga G.P., Rose R.B. (2008). Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: Heterodimer selectivity and DNA recognition. Biochemistry 47: 218–229 [DOI] [PubMed] [Google Scholar]
- Massari M.E., Murre C. (2000). Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Feldman L.J., Zambryski P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888 [DOI] [PubMed] [Google Scholar]
- Østergaard L. (2009). Don't 'leaf' now. The making of a fruit. Curr. Opin. Plant Biol. 12: 36–41 [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani S., Sundaresan V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11: 1914–1922 [DOI] [PubMed] [Google Scholar]
- Scacchi E., Osmont K.S., Beuchat J., Salinas P., Navarrete-Gómez M., Trigueros M., Ferrándiz C., Hardtke C.S. (2009). Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136: 2059–2067 [DOI] [PubMed] [Google Scholar]
- Scutt C.P., Vinauger-Douard M., Fourquin C., Finet C., Dumas C. (2006). An evolutionary perspective on the regulation of carpel development. J. Exp. Bot. 57: 2143–2152 [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg J.J., Myrenås M., Kuusk S., Lagercrantz U., Kowalczyk M., Sandberg G., Sundberg E. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 47: 112–123 [DOI] [PubMed] [Google Scholar]
- Sorefan K., Girin T., Liljegren S.J., Ljung K., Robles P., Galván-Ampudia C.S., Offringa R., Friml J., Yanofsky M.F., Østergaard L. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586 [DOI] [PubMed] [Google Scholar]
- Ståldal V., Sohlberg J.J., Eklund D.M., Ljung K., Sundberg E. (2008). Auxin can act independently of CRC, LUG, SEU, SPT and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytol. 180: 798–808 [DOI] [PubMed] [Google Scholar]
- Ståldal V., Sundberg E. (2009). The role of auxin in style development and apical-basal patterning of the Arabidopsis thaliana gynoecium. Plant Signal. Behav. 4: 83–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigueros M., Navarrete-Gómez M., Sato S., Christensen S.K., Pelaz S., Weigel D., Yanofsky M.F., Ferrándiz C. (2009). The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 21: 1394–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]