This work examines termination of floral stem cell maintenance, finding mechanisms that regulate when stem cells cease to function as stem cells, and finding that AGAMOUS directly represses WUSCHEL (WUS) expression by binding to the WUS locus and recruiting Polycomb Group factors that methylate histone H3 Lys-27 to repress transcription.
Abstract
Floral stem cells produce a defined number of floral organs before ceasing to be maintained as stem cells. Therefore, floral stem cells offer an ideal model to study the temporal control of stem cell maintenance within a developmental context. AGAMOUS (AG), a MADS domain transcription factor essential for the termination of floral stem cell fate, has long been thought to repress the stem cell maintenance gene WUSCHEL (WUS) indirectly. Here, we uncover a role of Polycomb Group (PcG) genes in the temporally precise repression of WUS expression and termination of floral stem cell fate. We show that AG directly represses WUS expression by binding to the WUS locus and recruiting, directly or indirectly, PcG that methylates histone H3 Lys-27 at WUS. We also show that PcG acts downstream of AG and probably in parallel with the known AG target KNUCKLES to terminate floral stem cell fate. Our studies identify core components of the network governing the temporal program of floral stem cells.
INTRODUCTION
Stem cells possess the potential to generate all or some differentiated cell types during development in a multicellular organism. Certain types of stem cells are active throughout the life of an organism, but others, such as the embryonic stem (ES) cells in animals and the floral stem cells in plants, are precisely terminated, meaning that they cease to function and be maintained as stem cells, in a process that is coordinated with other developmental events. Much is known about the factors that confer stemness in both plants and animals (Mayer et al., 1998; Nichols et al., 1998; Mitsui et al., 2003; Masui et al., 2007), but little is known about how stem cell maintenance is precisely terminated within a developmental context.
Several types of stem cells contribute to the generation of the body plan of a plant. The shoot apical meristem (SAM) harbors stem cells that produce the entire aboveground structures of a plant in an indeterminate manner; these stem cells are active throughout plant development. A floral meristem harbors stem cells that give rise to all organs found in a flower, including sepals, petals, stamens, and carpels. In contrast with the stem cells in the SAM, floral stem cells are determinate insofar as they generate a precise number of floral organs and then cease to be stem cells. The termination of floral stem cell fate is coincident with the development of carpel primordia, the final organs to be made from the floral meristem. However, the termination of floral stem cell maintenance is not simply the differentiation of stem cells into carpel cells because we isolated mutants that uncouple carpel identity specification and the termination of floral stem cell maintenance (Ji et al., 2011; this study). Instead, a mechanism independent of, but coordinated with, organ identity specification is responsible for the precise termination of floral stem cell fate. As such, floral stem cells provide a good model for studying the temporal program of stem cells.
The termination of floral stem cell maintenance involves two key transcription factors, AGAMOUS (AG), a MADS domain protein, and WUSCHEL (WUS), a homeodomain protein. WUS is expressed in a few cells known as the organizing center (OC) underneath the floral stem cells and the OC signals to the overlying cells to maintain their stem cell identity (Laux et al., 1996; Mayer et al., 1998). By stage 6 of flower development (stages according to Smyth et al., 1990) when the primordia for the final floral organs (carpels) arise, WUS expression is shut off, which results in the termination of floral stem cell maintenance. The temporally regulated repression of WUS expression requires AG (Laux et al., 1996; Lenhard et al., 2001; Lohmann et al., 2001), which also serves as a key factor in specifying the identities of stamens and carpels (Bowman et al., 1989). In an ag loss-of-function mutant, stamens are transformed into petals and carpels are replaced by an internal flower to result in a flowers-within-flower phenotype (Bowman et al., 1989). AG expression commences at stage 3 in a domain that encompasses that of WUS (Drews et al., 1991; Mayer et al., 1998), yet WUS expression is not shut off until stage 6. Consequently, AG has been considered to be an indirect regulator of WUS. In fact, AG is known to activate the expression of another transcription factor gene KNUCKLES (KNU) at stage 6 in a region that encompasses the WUS-expressing OC in the floral meristem; KNU is in turn necessary for the repression of WUS expression (Sun et al., 2009). However, the floral determinacy defects of the knu-1 mutant are much weaker than those of ag null mutants (Bowman et al., 1989; Payne et al., 2004). Although this could be due to the knu-1 allele not being a null allele, an alternative explanation is that, in addition to activating KNU expression, AG also represses WUS expression through unknown mechanisms.
Polycomb Group (PcG) proteins were first identified as repressors of homeotic genes in Drosophila melanogaster (Lewis, 1978; Jürgens, 1985). PcG proteins in Drosophila associate in various functionally distinct subcomplexes (reviewed in Müller and Verrijzer, 2009), including Polycomb Repressive Complex2 (PRC2), which trimethylates histone H3 Lys-27 (H3K27) at specific target genes, PRC1, which recognizes the H3K27me3 mark to promote the compaction of chromatin for transcriptional repression, and Pho-Repressive Complex, which is responsible for the recruitment of PcG proteins to genes containing Polycomb response elements. Homologs of all Drosophila PRC2 subunit genes are found in Arabidopsis thaliana (reviewed in Pien and Grossniklaus, 2007; Köhler and Villar, 2008; Schatlowski et al., 2008; Hennig and Derkacheva, 2009; Zheng and Chen, 2011). In particular, CURLY LEAF (CLF) and its paralogs SWINGER (SWN) and MEDEA are homologs of Drosophila E(z), the H3K27 methyltransferase. MEDEA acts predominantly in the seed, while CLF and SWN are broadly expressed and act redundantly during vegetative and reproductive development (Goodrich et al., 1997; Grossniklaus et al., 1998; Chanvivattana et al., 2004). TERMINAL FLOWER2/LIKE HETEROCHROMATIN PROTEIN1 (TFL2/LHP1) is considered a functional counterpart of PRC1 because it recognizes, and colocalizes with, H3K27me3 throughout the genome (Turck et al., 2007; Zhang et al., 2007a). TFL2/LHP1 as the functional PRC1 counterpart is further supported by the similar phenotypes exhibited by tfl2 mutants and mutants in PRC2 genes (Kotake et al., 2003; Mylne et al., 2006; Sung et al., 2006). No Pho-Repressive Complex homologs or Polycomb response elements have been identified, and how PcG is recruited to specific targets in plants remains largely enigmatic. One recent study showed that a noncoding RNA recruits PRC2 to the FLC gene (Heo and Sung, 2011).
In this study, we show that PcG is required for AG-mediated repression of WUS expression and termination of floral stem cell maintenance. We show that, in addition to indirectly repressing WUS expression through the activation of KNU expression at stage 6 (Sun et al., 2009), AG directly represses WUS expression before stage 6 by binding to the WUS locus and recruiting PcG to WUS. Our studies establish a direct link between WUS and AG, which has long been known to repress WUS expression but is thought to do so indirectly, and establishes a core network of floral stem cell regulators. Furthermore, mutations in two AG binding sites abrogate the termination of WUS expression in flower development and reveal the unexpected existence of a group of cells with the unique ability to express WUS in mature flowers. This suggests that the earlier WUS-expressing OC remains distinct from neighboring cells in mature flowers.
RESULTS
CLF Is Required for the Temporally Regulated Termination of Floral Stem Cell Maintenance
To identify players that regulate the temporal program of floral stem cells, we performed an ethyl methanesulfonate mutagenesis in the ag-10 background. While ag null alleles are defective in both floral stem cell fate termination and floral organ identity specification, as reflected by reproductive-to-perianth organ transformation, as well as a flowers-within-flower phenotype (Bowman et al., 1989), the weak ag-10 allele is only mildly defective in floral stem cell fate termination and is normal in floral organ identity specification (Ji et al., 2011). ag-10 flowers generate a full complement of floral organs like the wild type (Figures 1A and 1B), but one to a few siliques on an ag-10 plant are short and bulged with additional floral organs inside (Figure 1I), reflecting a mild defect in stem cell fate termination. In the ag-10 mutagenesis screen, mutations that enhanced the mild stem cell defects were isolated based on the presence of bulged siliques throughout the plant.
Figure 1.
Phenotypes of ag and clf Single and Double Mutants.
(A) A wild-type (Ler) flower.
(B) An ag-10 flower with a slightly enlarged gynoecium.
(C) A clf-47 flower.
(D) An ag-10 clf-47 flower with a much more enlarged gynoecium compared with ag-10.
(E) and (F) Longitudinal sections through stage 11 flowers of ag-10 clf-47 (E) and ag-10 (F) genotypes. In (E), the floral meristem continued to generate organs (arrow) inside the carpels.
(G) A clf-2 flower.
(H) An ag-10 clf-2 flower with similar phenotypes to those of ag-10 clf-47.
(I) Siliques from plants of the indicated genotypes. Most siliques on an ag-10 plant were long and thin (represented by the two on the left); one to a few siliques were short and bulged (represented by the one on the right). Most siliques from ag-10 clf-47 or ag-10 clf-2 plants were short and bulged.
(J) Siliques from F1 plants of the cross between ag-10 clf-47 and ag-10 clf-2. The siliques were similar in morphology to those of ag-10 clf-47 or ag-10 clf-2.
Bars = 1 mm in (A) to (C) and (G) to (I), 0.75 mm in (D), 0.5 mm in (J), and 100 μm in (E) and (F).
One such mutant displayed mostly short and bulged siliques (Figures 1D and 1I). Longitudinal sections of stage 7 and older flowers revealed a dome-shaped meristem between the two carpels in this mutant (Figures 1E,2C, and 2D) but not in the majority of ag-10 flowers (Figures 1F, 2A, and 2B). In later-staged flowers of this mutant, additional organs were generated inside the primary gynoecia (Figure 1E). Therefore, this recessive mutation enhanced the ag-10 floral determinacy defect phenotype. Genetic mapping revealed a G-to-A mutation in CLF, which resulted in the conversion of amino acid 794 in the SET domain from Arg (R) to His (H) (see Supplemental Figure 1 online). The Arg-794 residue is highly conserved within the SET domain, which is itself conserved in E(z) homologs and is responsible for the H3K27 methyltransferase activity (see Supplemental Figure 1B online). The enhancer mutation was named clf-47. The ag-10 clf-47 double mutant resembled well-characterized clf single mutants in that it was dwarfed, exhibited early flowering, and had small and curled leaves (Goodrich et al., 1997). The clf-47 single mutant did not exhibit obvious defects in floral stem cells (Figures 1C and 1I).
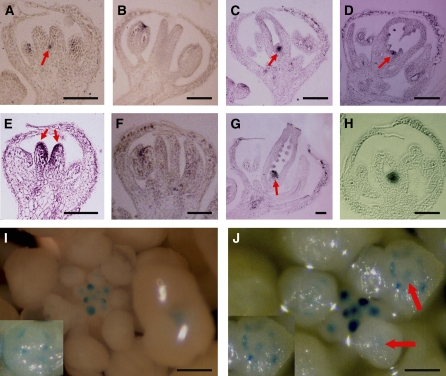
Figure 2.
Expression Patterns of WUS and STM in ag-10 and ag-10 clf-47 Flowers.
(A) to (D) In situ hybridization with a WUS antisense probe. The arrows indicate WUS signals.
(A) and (B) WUS expression was detected in a stage 7 (A) but not a stage 9 (B) ag-10 flower.
(C) and (D) WUS expression was detected in ag-10 clf-47 flowers at stage 9 (C) and stage 11 (D).
(E) to (G) In situ hybridization using an STM antisense probe. The arrows indicate STM signals.
(E) and (F) STM expression was detected at stage 7 (E) but not stage 9 (F) in ag-10 flowers.
(G) STM expression was detected in a stage 12 ag-10 clf-47 flower.
(H) A longitudinal section of a stages 8 or 9 ag-10 clf-47 pWUS:GUS flower showing GUS staining inside the carpels.
(I) and (J) GUS staining in pWUS:GUS (I) and ag-10 clf-47 pWUS:GUS (J) inflorescences. Arrows in (J) indicate GUS signals in the center of stages 8 and 9 flowers. The insets are stages 8 and 9 flowers. The ring-like GUS signals were from anthers. The GUS signals inside the ring in (J) were from the floral meristem.
Bars = 50 μm in (A) to (H) and 250 μm in (I) and (J).
To confirm that the floral phenotype of ag-10 clf-47 was caused by a mutation in CLF, we introduced another clf allele, clf-2 (Figure 1G; Goodrich et al., 1997) into ag-10. Like ag-10 clf-47, the ag-10 clf-2 double mutant exhibited short and bulged siliques with ectopic floral organs inside (Figures 1H and 1I). In addition, an allelic test was conducted by crossing ag-10 clf-47 to ag-10 clf-2. The resulting F1 plants had both enhanced floral determinacy defects in flowers and vegetative phenotypes characteristic of clf mutants (Figure 1J). Collectively, these results show that CLF is required for floral meristem determinacy.
CLF Is Necessary for the Temporally Controlled Repression of WUS Expression
To investigate the molecular basis of the floral determinacy defects of the ag-10 clf-47 double mutant, we first performed in situ hybridization to determine the temporal and spatial expression patterns of WUS, which promotes stem cell identity, and STM, which is required for the acquisition and/or maintenance of meristematic fate (Long et al., 1996), in floral meristems. In the wild type, WUS expression is shut off at stage 6 when carpel primordia are formed (Mayer et al., 1998). In the ag-10 single mutant, stage 7 was the latest stage when WUS expression was observed (Figure 2A); only one out of 10 stage 7 flowers examined expressed WUS. In the ag-10 clf-47 double mutant, nine out of 10 stage 7 flowers examined showed WUS expression, and the expression persisted in much older flowers (Figures 2C and 2D). Expression in such late-staged flowers was not observed in the ag-10 single mutant (Figure 2B). Therefore, CLF was required for the temporally precise repression of WUS expression in the flower. STM expression also persisted much longer in the ag-10 clf-47 double mutant than in the ag-10 single mutant. In the ag-10 single mutant, stage 7 was the latest stage when STM expression was observed (Figures 2E and 2F); only two out of 10 ag-10 stage 7 flowers examined showed STM expression. Seven out of nine ag-10 clf-47 flowers had STM expression, which persisted in much older flowers (Figure 2G). As STM is a marker for meristematic cells, the results suggested that cells retained meristematic activity until a much later stage in ag-10 clf-47 flowers.
Although our results indicated that CLF is required for WUS repression in flower development, it was not clear whether CLF is involved in the initial repression of WUS, which requires AG, or in a later step after the initial repression to maintain the repressed state. To monitor WUS expression in all floral stages in an inflorescence, we introduced a pWUS:GUS (for β-glucuronidase) reporter that recapitulates endogenous WUS expression patterns (Bäurle and Laux, 2005) into ag-10 clf-47. In the wild type, this reporter was active in floral meristems between stages 1 and 6 (Figure 2I). In ag-10 clf-47, continuous GUS expression was observed from early stages until stages 8 and 9 (Figure 2J). In flowers of stages 8 and 9 when GUS signals were visible in anthers, GUS expression was observed in the center of the flowers in ag-10 clf-47 but not in the wild type (Figures 2I and 2J, insets). Longitudinal sections of ag-10 clf-47 pWUS:GUS flowers confirmed prolonged GUS expression in late-staged floral meristems (Figure 2H). Importantly, we did not observe any reduction in GUS expression at stage 6 in ag-10 clf-47. This suggests that CLF is required for the initial repression of WUS.
To determine whether the prolonged WUS or STM expression in ag-10 clf-47 floral meristems underlies the floral determinacy defects, we crossed the loss-of-function wus-1 mutation (Laux et al., 1996) and the partial loss-of-function stm-2 mutation (Clark et al., 1996) into ag-10 clf-47. The wus-1 mutation results in premature termination of the floral meristem such that the flower terminates in a central stamen (Figure 3A). wus-1 was epistatic to ag-10 clf-47 as ag-10 clf-47 wus-1 triple mutant flowers also terminated precociously (Figure 3B; see Supplemental Table 1 online). stm-2 flowers show premature termination of the floral meristem such that they have a reduced number of floral organs (Figure 3C). stm-2 was also epistatic to ag-10 clf-47 for floral meristem determinacy (Figure 3D; see Supplemental Table 1 online). These results indicated that the floral determinacy defects of ag-10 clf-47 were due to prolonged WUS and STM expression.
Figure 3.
Phenotypes of clf-47 and ag-10 clf-47 in Combination with Mutations in Other Floral Meristem Regulators.
(A) A wus-1 flower that lacked a full complement of floral organs.
(B) An ag-10 clf-47 wus-1 flower, which was similar to wus-1 with respect to floral meristem determinacy.
(C) An stm-2 flower that lacked a full complement of floral organs.
(D) An ag-10 clf-47 stm-2 flower, which was similar to stm-2 flowers in terms of floral meristem determinacy.
(E) An ag-1 flower with a flowers-within-flower phenotype.
(F) An ag-1 clf-47 flower, which was morphologically identical to ag-1 flowers.
(G) A sup-1 flower with more stamens than the wild type.
(H) An ag-10 clf-47 sup-1 flower, which developed numerous stamens from an indeterminate floral meristem.
(I) A knu-1 flower.
(J) An ag-10 knu-1 flower with an enlarged gynoecium.
(K) An ag-10 clf-47 flower with an enlarged gynoecium.
(L) An ag-10 clf-47 knu-1 flower with an internal flower replacing the gynoecium.
(M) Siliques from knu-1, ag-10 knu-1, and ag-10 clf-47 plants. The knu-1 silique on the left was a representative silique from young knu-1 plants, while the one on the right was a representative silique from old knu-1 plants.
Bars = 1 mm in (A) to (L) and 2.5 mm in (M).
CLF and AG Confer Floral Meristem Determinacy in the Same Genetic Pathway
To determine the genetic relationship between AG and CLF in the regulation of floral stem cells, we introduced the clf-47 mutation into the ag null mutant ag-1 background (Bowman et al., 1989). The floral phenotypes of the ag-1 clf-47 double mutant were identical to those of the ag-1 single mutant (Figures 3E and 3F), indicating that CLF and AG act in the same pathway in conferring floral meristem determinacy. Consistent with this finding, both ag-1 and ag-10 clf-47 interacted synergistically with sup-1, a mutation in SUPERMAN (SUP), a gene that acts in parallel with AG in the regulation of floral stem cells (Bowman et al., 1992). The sup-1 single mutant flowers exhibit an increased number of stamens and carpels, but the floral meristem eventually terminates in a few carpels (Figure 3G). The clf-47 sup-1 double mutant flowers had more stamens and carpels than sup-1 flowers. The ag-10 clf-47 sup-1 triple mutant exhibited a dramatic enhancement of the floral determinacy defects of both ag-10 clf-47 and sup-1 in that they developed numerous stamens following a spiral phyllotaxy from an indeterminate floral meristem (Figure 3H). Similarly, combining ag-1 with sup-1 resulted in a drastic enhancement of floral meristem activity (Bowman et al., 1992).
A number of genes are known to promote floral determinacy by maintaining AG expression in the center of the floral meristem (Schultz et al., 1991; Alvarez and Smyth, 1999; Carles et al., 2005; Prunet et al., 2008; Das et al., 2009; Maier et al., 2009). To determine whether CLF acts similarly, we performed in situ hybridization to examine AG expression in wild-type, ag-10, and ag-10 clf-47 flowers. As in the wild type, AG transcripts were present in the inner two whorls of ag-10 and ag-10 clf-47 floral meristems (see Supplemental Figures 2A to 2F online). Because in situ hybridization is not a quantitative measure of gene expression, we performed immunoblotting to determine the levels of AG protein in wild-type and ag-10 clf-47 inflorescences and leaves. Consistent with AG being a known PcG target (Goodrich et al., 1997), AG protein levels were elevated in ag-10 clf-47 compared with the wild type in both inflorescences and leaves (see Supplemental Figure 2G online). The fact that clf-47 resulted in compromised floral determinacy without causing a decrease in AG expression suggests that CLF is required for AG-mediated floral stem cell regulation. This, together with the genetic studies showing that CLF and AG act in the same pathway, suggests that CLF acts downstream of AG in terminating floral stem cell maintenance.
Loss of Function in TFL2/LHP1 Also Enhances ag-10
We investigated whether the function of CLF in floral meristem determinacy reflects a similar role for PcG. We crossed tfl2-2 (Larsson et al., 1998), a mutation in the PRC1 component TFL2/LHP1, into ag-10 to determine whether tfl2-2 also enhances ag-10. Since the tfl2-2 allele is in the Columbia (Col) background, we first crossed it to a line in which the ag-10 mutation in the Landsberg erecta (Ler) background was introgressed into Col by five backcrosses. Unlike ag-10 in Ler, ag-10Col did not exhibit any floral determinacy defects (all siliques on a plant were long and thin as in the wild type), indicating that there was a genetic suppressor/modifier in Col. In a tfl2-2 plant, the rosette leaves are curled and the inflorescence terminates in a few disorganized flowers (Figure 4A; Larsson et al., 1998). The gynoecia in these flowers are thin (Figure 4A), suggesting that the tfl2-2 mutant itself did not have any floral determinacy defects. The tfl2-2 mutation enhanced ag-10Col in that some gynoecia of ag-10Col tfl2-2 plants consisted of more than two carpels that were partially fused. However, the phenotypes of ag-10Col tfl2-2 were much weaker than those of ag-10 clf-47. To determine whether this was due to a suppressor in Col, we crossed tfl2-2 to ag-10 in Ler. In the F2 population, all plants with tfl2-2 vegetative phenotypes were genotyped for ag-10. Among 110 ag-10 tfl2-2 plants, 32 had severe floral determinacy defects that were similar to, or even more severe than, those of ag-10 clf-47 plants. The flowers of these ag-10 tfl2-2 plants had bulged or unfused gynoecia with internal floral organs (Figures 4B and 4D). The remaining ag-10 tfl2-2 plants had normal siliques (Figure 4C). Since none of the ag-10 tfl2-2/TFL2 or ag-10 TFL2/TFL2 plants had floral determinacy defects, tfl2-2 was responsible for the loss of floral determinacy in some ag-10 tfl2-2 plants. The segregation of this phenotype among ag-10 tfl2-2 plants was consistent with the existence of a single dominant suppressor or several recessive suppressors of ag-10 in the Col ecotype.
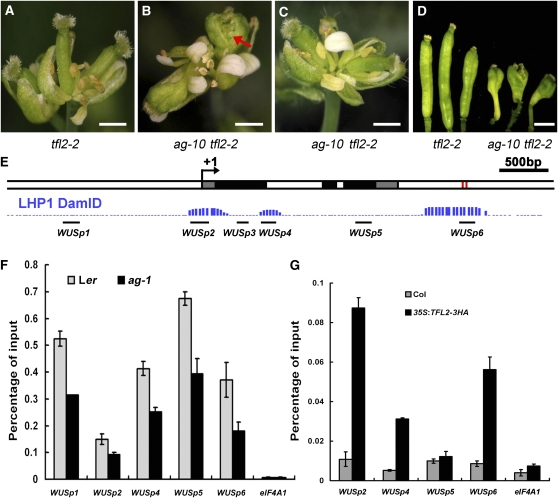
Figure 4.
TFL2/LHP1 Acts in Floral Stem Cell Termination, and WUS Is a PcG Target.
(A) A terminal inflorescence composed of several fused flowers in tfl2-2. Note that the gynoecia were thin.
(B) A representative inflorescence of ag-10 tfl2-2 plants with floral determinacy defects. The flowers had bulged gynoecia with ectopic floral organs inside (arrow).
(C) A representative inflorescence of ag-10 tfl2-2 plants without floral determinacy defects. The gynoecia were thin.
(D) Siliques from plants of the indicated genotypes. The ag-10 tfl2-2 plants were from the F2 population of the cross between ag-10 and tfl2-2. Only siliques from the ag-10 tfl2-2 plants with floral determinacy defects are shown. Bars = 1 mm in (A) to (D).
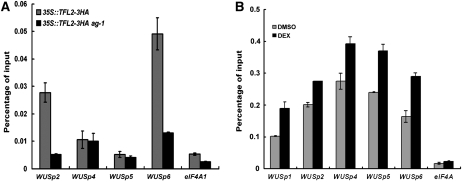
(E) A diagram of the WUS genomic region with “+1” being the transcription start site. Gray, black, and white rectangles represent 5′ or 3′ untranslated regions, coding regions, and introns or intergenic regions, respectively. The two red rectangles represent the two CArG boxes. The three regions of TFL2/LHP1 occupancy at WUS as determined by genome-wide profiling of TFL2/LHP1 binding sites are shown in blue (LHP1 DmID; Zhang et al., 2007a). The regions interrogated for AG, H3K27me3, or TFL2/LHP1 enrichment at WUS in this study are shown as black bars.
(F) ChIP with anti-H3K27me3 antibodies to determine the levels of H3K27me3 at WUS in wild-type (Ler) and ag-1 inflorescences containing stage 8 and younger flowers.
(G) ChIP with anti-HA antibodies in Col (a negative control) and 35S:TFL2-3HA to examine TFL2/LHP1 occupancy at WUS. For (F) and (G), the regions examined are diagramed in (E). eIF4A1 served as a negative control. Error bars represent sd, which were calculated from three technical repeats. Three biological replicates gave similar results.
WUS Is a Target of PcG in Flowers and Seedlings
Genome-wide profiling of H3K27me3 with 10-d-old Arabidopsis seedlings identified ~4000 genes, including WUS, as potential targets of PcG (Zhang et al., 2007b). H3K27me3 was found to be enriched throughout the WUS genomic region, including the entire intergenic region between WUS and its upstream gene and up to 1.5 kb downstream of the 3′ end of the transcript (Zhang et al., 2007b). To confirm that the H3K27me3 mark at WUS was PcG dependent, we performed chromatin immunoprecipitation (ChIP) to examine H3K27me3 levels in wild-type seedlings and in the clf-28 swn-7 double mutant, which germinates into abnormal seedlings that develop into callus-like tissues (Chanvivattana et al., 2004). H3K27me3 was enriched at the known PcG target AG in the wild type, but this enrichment was eliminated in clf-28 swn-7, consistent with previous reports (see Supplemental Figure 3 online; Schubert et al., 2006). Similarly, the levels of H3K27me3 at the WUS locus were drastically reduced in clf-28 swn-7 (see Supplemental Figure 3 online). To determine whether WUS was a target of PcG in flowers, we performed ChIP to examine H3K27me3 levels at WUS in wild-type inflorescences. As in seedlings, H3K27me3 was enriched throughout the WUS genomic region in inflorescences (Figure 4F).
Previous studies suggest that PRC1 binds H3K27me3 through TFL2/LHP1 to effect transcriptional inhibition at PcG targets (Turck et al., 2007; Zhang et al., 2007a; Xu and Shen, 2008). Intriguingly, although H3K27me3 was found throughout the WUS locus, genome-wide profiling of TFL2/LHP1 occupancy in 10-d-old seedlings revealed that three distinct regions at WUS were bound by TFL2/LHP1 (Zhang et al., 2007a). These were WUSp2 at the transcription start site, WUSp4 in the first intron, and WUSp6 located ~800 bp downstream of the coding region (Figure 4E). We determined whether TFL2/LHP1 was associated with the WUS locus at these regions in inflorescences by ChIP with anti-HA antibodies in 35S:TFL2-3HA (Liu et al., 2009). We found that TFL2/LHP1 was indeed enriched at these three specific regions but not at another region (WUSp5) tested at the WUS locus (Figure 4G). The enrichment of both H3K27me3 and TLF2/LHP1 at WUS in inflorescences suggests that WUS is a target of PcG in flowers.
AG Binds the WUS Locus in Vivo and Directly Represses WUS Expression
The restricted distribution of TFL2/LHP1 relative to that of H3K27me3 at WUS indicates that the H3K27me3 mark alone is not sufficient for TFL2/LHP1 recruitment to targets. Given that AG is a major factor in repressing WUS expression, we hypothesized that AG contributes to the recruitment of TFL2/LHP1 to WUS. This hypothesis would only be possible if AG binds the WUS locus in vivo. We decided to test this hypothesis despite previous assumptions that AG represses WUS expression indirectly. AG occupancy at multiple positions along the WUS locus was examined by ChIP using anti-AG antibodies in wild-type inflorescences. The ag-1 null mutant, from which no AG protein was detectable by immunoblotting with the antibodies, served as a negative control. Among six sites spanning the WUS locus from −1367 to +2702 (+1 being the transcription start site) examined, enrichment of AG at two specific sites at WUS was found (Figure 5A). Intriguingly, the AG binding sites overlapped with two of the three TFL2/LHP1 binding sites, WUSp2 and WUSp6 (Figures 4G and 5A).
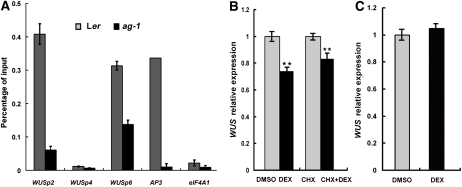
Figure 5.
AG Binds the WUS locus and Represses WUS Expression Directly.
(A) ChIP using anti-AG antibodies to determine AG occupancy at WUS. The null allele ag-1 and the eIF4A1 locus both served as negative controls. AP3, a known direct target of AG (Gómez-Mena et al., 2005), served as a positive control.
(B) Real-time RT-PCR to determine WUS transcript levels in 35S:AG-GR ag-1 inflorescences containing stage 8 and younger flowers. Inflorescences were treated with DMSO, DEX, CHX, or CHX plus DEX. Two hours later, the inflorescences were dissected to remove old flowers and harvested for RNA extraction and RT-PCR. Four biological replicates were performed for the DMSO/DEX experiment, and five were performed for the CHX/DEX experiment. Error bars represent sd, which were calculated from these biological repeats. The calculated P values for both experiments were 0.011.
(C) Real-time RT-PCR to measure WUS transcript levels in 35S:AG-GR ag-1 clf-47 inflorescences. Chemical treatments and RNA isolation were as in (B).
The in vivo binding of AG to WUS prompted us to determine whether AG directly represses WUS expression. We took advantage of the established 35S:AG-GR ag-1 line in which the functional AG-glucocorticoid receptor (GR) fusion protein could be activated by dexamethasone (DEX) (Gómez-Mena et al., 2005; Sun et al., 2009). We treated 35S:AG-GR ag-1 inflorescences with either DMSO (control) or DEX, and at 2 h after treatment, collected inflorescences containing stage 8 and younger flowers (to enrich for tissues with WUS expression) and examined WUS expression by quantitative real-time RT-PCR. A small but consistent and statistically significant decrease in WUS expression was observed upon DEX treatment (Figure 5B). Next, to determine whether this repression was direct, we performed the DMSO and DEX treatments in the presence of the protein synthesis inhibitor cycloheximide (CHX). WUS expression was also reduced in CHX/DEX-treated samples relative to the CHX-treated control samples (Figure 5B). This indicates that AG was able to directly repress WUS expression. We were aware that a previous study using 35S:AG-GR ag-1 inflorescences containing flower buds of stages 1 to 10 showed that AG induction activated WUS expression at 8 h postinduction (Ito et al., 2004). To reconcile these results, we monitored WUS expression in a postinduction time course (0 to 24 h). Consistent with the previous study as well as our results above, we found a reduction in WUS expression in early time points but an increase in WUS expression at 12 and 24 h (see Supplemental Figure 4A online). This increase in WUS expression in later time points probably reflected a role of AG in the activation of WUS expression in anthers and carpels. The expected changes in the expression of APETALA1 and SPOROCYTELESS, two known AG targets (Ito et al., 2004), showed that AG activity was being effectively activated by our treatments (see Supplemental Figures 4B and 4C online).
An AG Binding Site Is Necessary for WUS Repression
MADS domain–containing proteins, including AG, bind a distinct DNA motif called the CArG box with CC(A/T)6GG as the consensus sequence (Huang et al., 1993; Shiraishi et al., 1993). AG has been shown to bind to the consensus sequence and variants with one to a few nucleotide changes (Huang et al., 1993; Shiraishi et al., 1993; Riechmann et al., 1996; Sun et al., 2009). We inspected the WUSp2 and WUSp6 regions (Figure 4E) that were bound by AG and TFL2/LHP1 in vivo for potential CArG boxes with no more than a single nucleotide difference from the consensus sequence. While we did not identify any CArG boxes in the WUSp2 region, two tandem sequences resembling CArG boxes were found in the WUSp6 region (Figure 6A) in Col. A single nucleotide change in each CArG box was found in the Ler sequence, but at least one CArG box remained intact in Ler (Figure 6A).
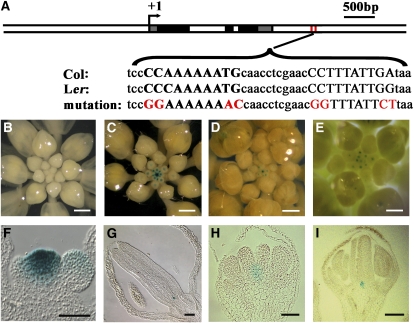
Figure 6.
Two CArG Boxes within the AG and TFL2/LHP1 Binding Sites at WUS Are Required for the Repression of WUS Expression throughout Flower Development.
(A) A diagram of the WUS genomic region as in Figure 4E. The sequences of the region containing the two CArG boxes (capital letters) from Col and Ler as well as the mutated versions are shown. A typical CArG box is CC(A/T)6GG, but slight variants also serve as functional CArG boxes.
(B) A representative inflorescence of WUS1.6:GUS:WUS3′wt transgenic plants showing no GUS staining.
(C) A representative inflorescence of WUS1.6:GUS:WUS3′mut transgenic plants showing strong GUS staining in the inflorescence meristem and floral meristems.
(D) An inflorescence of a WUS3.2:GUS:WUS3′wt transgenic plant showing GUS staining in the inflorescence meristem and young floral meristems.
(E) An inflorescence of a WUS3.2:GUS:WUS3′mut transgenic plant with GUS signals in apparently older flowers than in (D).
(F) and (G) Longitudinal sections of an inflorescence (F) or a stage 14 flower (G) of WUS1.6:GUS:WUS3′mut transgenic plants. In (F), the inflorescence meristem (center) is flanked by a stage 1 and a stage 2 floral primordia. GUS signals were present in the inflorescence meristem. In (G), GUS signals were present at the base of the gynoecium.
(H) A longitudinal section of a stage 7 WUS3.2:GUS:WUS3′wt flower. This was the latest stage when GUS expression could be detected in this genotype.
(I) A longitudinal section of a stage 12 flower from WUS3.2:GUS:WUS3′mut transgenic plants. GUS expression was detected at the base of the gynoecium.
Bars = 250 μm in (B), (D), and (E), 400 μm in (C), and 50 μm in (F) to (I).
We employed a reporter gene approach to determine whether the CArG boxes are necessary for the repression of WUS in flower development. We generated WUS1.6:GUS:WUS3′wt, in which GUS replaced the WUS coding sequence in the WUS genomic region (−1658 to +2943; position 1 being the transcription start site as in Bäurle and Laux, 2005), and WUS1.6:GUS:WUS3′mut, in which the two CArG boxes were mutated (Figure 6A). None of the 43 transgenic plants carrying the wild-type transgene showed any GUS staining in inflorescences (Figure 6B), consistent with previous findings (Bäurle and Laux, 2005). By contrast, all 34 transgenic plants containing the mutant transgene showed strong GUS signals in both the inflorescence meristem and floral meristems (Figure 6C). Longitudinal sections revealed GUS signals in the inflorescence meristem and young floral meristems not only in the rib zone in which WUS is expressed but also in the central zone containing the stem cells (Figure 6F; see Supplemental Figures 5A and 5B online). The expanded GUS-positive domain was not due to the spread of the GUS product to neighboring cells since in situ hybridization using a GUS antisense probe also detected the presence of GUS mRNA in the central zone of inflorescence and floral meristems (see Supplemental Figures 5C and 5D online). More importantly, GUS signals were present in a small number of cells throughout flower development, including in late-staged flowers with well-developed gynoecia (Figure 6G). These results indicate that the CArG boxes are necessary for the repression of WUS expression both spatially and temporally. Since AG is not expressed in the inflorescence meristem, the expression of the mutant reporter transgene in the inflorescence meristem suggests that AG cannot be the only MADS domain protein repressing WUS expression through the two CArG boxes.
We also sought to evaluate the role of the CArG boxes in the context of the 3.2-kb WUS promoter, which was found to be sufficient to drive reporter gene expression in patterns reminiscent of WUS (Bäurle and Laux, 2005). We generated WUS3.2:GUS:WUS3′wt and WUS3.2:GUS:WUS3′mut plants containing a 3.2-kb promoter region. Indeed, plants carrying the wild-type transgene showed GUS expression in the inflorescence meristem and young floral meristems as previously reported (Figure 6D; Bäurle and Laux, 2005). Plants carrying the mutant transgene showed GUS expression not only in the inflorescence meristem and young floral meristems but also in later-staged flowers than those carrying the wild-type transgene (cf. Figures 6D and 6E). This was confirmed by examination of GUS signals in sections of flowers of various stages. While the latest stage when GUS signals were visible for WUS3.2:GUS:WUS3′wt flowers was stage 7 (Figure 6H), GUS staining was evident in a small number of cells at the base of the gynoecia in stage 12 WUS3.2:GUS:WUS3′mut flowers (Figure 6I).
AG Recruits PcG to WUS to Repress WUS Expression
Having shown that AG binds to the WUS locus in vivo and that the binding sites are crucial for the termination of WUS expression in flower development, we proceeded to test the hypothesis that AG is required for the recruitment of PcG to WUS. We first examined whether the ag-1 mutation led to a change in H3K27me3 levels and TFL2 occupancy at WUS. ChIP with inflorescences containing flower buds of stages 1 to 8 (older flowers were removed to enrich for meristematic cells) showed that H3K27me3 levels throughout the WUS locus were reduced in ag-1 compared with Ler (Figure 4F). ChIP was performed to examine TFL2/LHP1 occupancy at WUS in 35S:TFL2-3HA versus 35S:TFL2-3HA ag-1 inflorescences containing stage 8 and younger flowers. TFL2/LHP1 occupancy at WUS was drastically decreased in ag-1 at WUSp2 and WUSp6, regions bound by AG, but not at WUSp4, a region not bound by AG (Figure 7A). These results were consistent with, but not sufficient to support, the conclusion that AG recruits PcG to WUS because the reduction in H3K37me3 levels and TFL2/LHP1 occupancy could be a consequence of prolonged WUS expression in ag-1. To confirm a role of AG in PcG recruitment to WUS, we took advantage of the 35S:AG-GR ag-1 system and examined the levels of H3K27me3 at WUS upon AG induction. H3K27me3 levels were increased throughout the WUS locus at 2 h after AG induction (Figure 7B), suggesting that AG plays an active role in recruiting PcG to WUS.
Figure 7.
AG Recruits PcG to WUS.
(A) ChIP using anti-HA antibodies to determine TFL2/LHP1 occupancy at WUS in 35S:TFL2-3HA and 35S:TFL2-3HA ag-1 inflorescences.
(B) ChIP using anti-H3K27me3 antibodies in DMSO- or DEX-treated 35S:AG-GR ag-1 inflorescences. At 2 h after treatments, inflorescences were dissected to remove stage 9 and older flowers and used for ChIP. In (A) and (B), real-time PCR reactions were performed with immunoprecipitated and total input DNA. Error bars represent sd, which were calculated from three technical repeats. Three biological replicates gave similar results. The regions interrogated are as diagramed in Figure 4E. eIF4A1 served as a negative control.
These findings raised the possibility that AG represses WUS expression through the repressive activities of PcG. We took advantage of the 35S:AG-GR ag-1 system, with which we had shown that WUS transcript levels were detectably reduced at 2 h following DEX treatments, to test this possibility. We first crossed 35S:AG-GR ag-1 with clf-47 to produce 35S:AG-GR ag-1 clf-47 plants. These plants were then treated with DEX or the DMSO control, and WUS transcript levels were assayed by real-time RT-PCR at 2 h following the chemical treatments. A consistent decrease in WUS transcript levels was observed in 35S:AG-GR ag-1 (Figure 5B), but no changes in WUS transcript levels were detected in 35S:AG-GR ag-1 clf-47 (Figure 5C), indicating that AG-mediated repression of WUS expression requires PcG.
AG Has a Direct and an Indirect Role in the Repression of WUS Expression
AG is known to indirectly repress WUS expression through the activation of KNU expression (Sun et al., 2009). Our results indicate that AG also has a direct role in the termination of floral stem cell maintenance by repressing WUS expression through the recruitment of PcG. If these were independent functions of AG, we would expect clf and knu mutations to exhibit additive genetic interactions. Thus, we examined the genetic relationship between KNU and CLF. First, we crossed knu-1, which was isolated in the Wassilewskija background (Payne et al., 2004) but was crossed into Landsberg once, into ag-10. The gynoecia of knu-1 plants were initially short and thin, but some were bulged in late-staged plants consistent with the development of an ectopic gynoecium inside the primary gynoecia (Figures 3I and 3M; Payne et al., 2004). The ag-10 knu-1 double mutant exhibited bulged gynoecia throughout the plant and in younger plants compared with the knu-1 single mutant (Figures 3J and 3M), suggesting that knu-1 enhanced the ag-10 floral determinacy defects. Next, we generated the ag-10 clf-47 knu-1 triple mutant. Flowers of the triple mutant had much more severe floral determinacy defects than either ag-10 knu-1 or ag-10 clf-47 in that the gynoecia were replaced by a new flower (cf. Figures 3L and 3M). The enhancement of knu-1 by clf-47 suggests that KNU and CLF act in parallel in the control of floral determinacy and is consistent with our model that AG confers floral determinacy through two mechanisms: the direct repression of WUS expression through PcG recruitment to WUS and indirect repression of WUS expression through the activation of KNU expression. However, the fact that knu-1 may not be a null allele and that CLF has a paralog with partially redundant functions complicates the genetic interpretations.
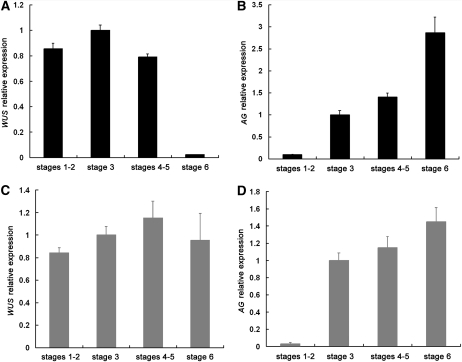
Next, we sought to validate the two independent functions of AG with temporal resolution. The activation of KNU expression occurs at stage 6 in flower development and coincides with the termination of WUS expression (Payne et al., 2004; Sun et al., 2009). However, AG expression commences at stage 3, and our results show that AG can recruit PcG to directly repress WUS expression. This raises the question of when AG starts to repress WUS expression in flower development. Intriguingly, it was noted that in situ hybridization analysis appeared to show that WUS expression was highest at stages 2 and 3 (Lenhard et al., 2001), suggesting that WUS expression began to decline at stages 3 and 4. To quantify WUS expression in flower development and to determine when AG starts to repress WUS expression, we performed laser capture microdissection of floral meristems at various stages followed by real-time RT-PCR in wild-type and ag-1 inflorescences. A circular area identical in size at the center of stages 1 and 2, 3, 4 and 5, or 6 floral meristems was captured from serial sections to ensure that all cells corresponding to a particular meristem zone were collected from a floral meristem (see Supplemental Figure 6 online for an example of the dissected areas). Consistent with prior in situ hybridization results (Drews et al., 1991; Mayer et al., 1998), WUS expression was terminated by stage 6 and AG expression commenced at stage 3 (Figures 8A, 8B, and 8D), suggesting that our laser capture of the floral meristems was precise. In the wild type, WUS expression peaked at stage 3 and decreased by 20% at stages 4 and 5 (Figure 8A). The small decrease in stages 4 and 5 was reproducible in two biological replicates and, more importantly, absent in ag-1 flowers (Figure 8C), suggesting that AG represses WUS expression starting at stages 4 and 5. Taken together, we propose that AG starts to repress WUS expression soon after AG expression begins by recruiting PcG to WUS. This direct effect probably occurs from stages 4 to 6, but the indirect effect through KNU occurs at stage 6, and together the two mechanisms result in the termination of WUS expression (Figure 9).
Figure 8.
Quantitative Measurements of WUS and AG Expression at Various Stages in Flower Development.
Laser capture microdissection was performed to collect cells from the central region of a floral meristem of a defined stage in the wild type ([A] and [B]) and ag-1 ([C] and [D]). Real-time RT-PCR was then performed to examine the levels of WUS ([A] and [C]) and AG ([B] and [D]) transcripts using UBQ5 as the internal control. The levels of expression were shown as relative to those of stage 3, which were set to 1.0. Error bars represent sd, which were calculated from three technical repeats. Two biological replicates gave nearly identical results.
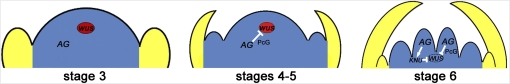
Figure 9.
A Model of the Termination of Floral Stem Cell.
AG terminates floral stem cell maintenance by repressing WUS expression (Lenhard et al., 2001; Lohmann et al., 2001). A previous study (Sun et al., 2009) showed that AG represses WUS expression indirectly by activating KNU, which in turn represses WUS expression directly or indirectly. Data presented in this study show that AG also directly represses WUS expression by recruiting PcG to WUS. Genetic studies are consistent with KNU and PcG acting downstream of AG and in parallel to each other in terminating floral stem cell maintenance.
DISCUSSION
Stem Cell Termination in Plants and Animals Employs a Conserved Mechanism
PcG proteins repress the expression of a multitude of genes in a developmentally regulated manner in both plants and animals. Studies in animal ES cells show that PcG is required for the differentiation of ES cells into other cell types and that key ES cell maintenance genes are targets of PcG (Boyer et al., 2006; Pasini et al., 2007; Shen et al., 2008). Our studies show that the plant PcG is required for the temporally regulated termination of floral stem cell fate by repressing the expression of the stem cell maintenance gene WUS. Therefore, both plants and animals use PcG to regulate stem cell maintenance.
The Fate of the WUS-Expressing OC Cells in the Floral Meristem
The WUS-expressing cells in the SAM and floral meristems constitute the OC that communicates to the overlying cells to specify their stem cell identity. In floral meristems, the OC, as marked by WUS expression, is present from stages 1 to 6. What happens to the OC cells after stage 6? Presumably, the OC cells are incorporated into carpels; they become assimilated and are no different from surrounding carpel cells. An unexpected finding from this study is that the OC cells probably retain their uniqueness even after the cessation of WUS expression. Our data reveal that the expression of the GUS reporter gene continues in a group of cells at the base of the carpels in late-staged WUS1.6:GUS:WUS3′mut and WUS3.2:GUS:WUS3′mut flowers (Figures 6G and 6I), thus revealing the presence of a group of cells at the base of carpels that is molecularly distinguishable from surrounding cells. Since these cells are likely descendants of earlier WUS-expressing OC cells, it is likely that OC cells remain distinct from surrounding carpel cells in late-staged flowers. This, together with the genetic uncoupling of carpel identity specification and stem cell maintenance in ag-10, ag-10 ago10 (Ji et al., 2011), and ag-10 clf-47, reinforces the conclusion that stem cell termination is not simply the differentiation of stem cells or OC cells into carpel cells.
AG Acts in Floral Stem Cell Termination via Two Mechanisms
As a key temporal regulator of floral stem cells, AG has been thought to terminate floral stem cell maintenance by indirectly repressing WUS expression. In this study, we show that AG binds two CArG boxes ~1 kb downstream of the WUS coding region and that these CArG boxes are crucial in the repression of WUS expression. We also show that induction of AG-GR results in the repression of WUS expression in the absence of protein synthesis, suggesting that AG is a direct repressor of WUS. Therefore, we propose that AG achieves the temporally precise repression of WUS expression through two parallel mechanisms: the transcriptional activation of KNU, which in turn acts to repress WUS, and the direct repression of WUS through the recruitment of PcG to WUS (Figure 9). In addition, our laser capture microdissection experiments show that AG starts to repress WUS expression at stages 4 and 5. It is likely AG acts directly on WUS in stages 4 to 6 and exerts its indirect effects on WUS repression through KNU at stage 6 (Figure 9).
A previous study showed that the 5′ regulatory region of WUS between −595 and −99 (+1 being the transcription start site), when provided in four copies, was sufficient to confer the correct spatial and temporal patterns of WUS expression to GUS (Bäurle and Laux, 2005). Our work shows that the CArG boxes in the 3′ region have strong influences on WUS expression and implicates MADS domain proteins in addition to AG that bind the two CArG boxes in vivo. These findings do not contradict each other because transcriptional regulatory regions usually consist of multiple positive and negative elements that exert additive or combinatorial effects on gene expression.
Recruitment of PcG to WUS Requires AG
How PcG is recruited to specific loci is a major outstanding question in both mammals and plants. In this study, we found that the MADS domain transcription factor AG binds two of the three regions that are also bound by TFL2/LHP1 at the WUS locus. The specific reduction in TFL2/LHP1 occupancy at these two regions (but not at the third region) in ag-1 suggests that AG promotes the recruitment of TFL2/LHP1 to WUS. A previous study found that another MADS domain protein, SHORT VEGETATIVE PHASE, promotes the recruitment of TFL2/LHP1 to the SEP3 gene (Liu et al., 2009). Therefore, transcription factors may play a general role in PcG recruitment to targets in plants. How AG recruits TFL2/LHP1 or PRC2 to WUS is currently unknown. Extensive coimmunoprecipitation studies between AG and TFL2/LHP1 or AG and CLF failed to detect any association between the proteins in vivo. However, it cannot be ruled out that AG recruits PcG to WUS through protein–protein interactions because the coimmunoprecipitation experiments were limited in sensitivity because the OC in which the interactions would take place constituted a small portion of the tissues examined. Alternatively, AG may promote the production of noncoding transcripts, which in turn serve to recruit either PRC2 or TFL2/LHP1. Increasing evidence points to the involvement of noncoding RNAs in PcG recruitment to targets in animals and plants (Rinn et al., 2007; Zhao et al., 2008; Khalil et al., 2009; Kanhere et al., 2010; Tsai et al., 2010; Yap et al., 2010; Heo and Sung, 2011).
METHODS
Plant Materials
All mutants or transgenic lines are in the Ler background with the exception of tfl2-2 (Larsson et al., 1998) and 35S:TFL2-3HA (Sun et al., 2009), which are in the Col background; knu-1 (Payne et al., 2004), which was isolated in the Wassilewskija background but was crossed once into Ler; and ag-10col, in which ag-10 in Ler was introgressed into Col through five backcrosses. Arabidopsis thaliana plants were grown at 23°C under continuous light. The clf-47 allele described in this study is an independent isolate (in Landsberg) of the clf-81 mutation described by Schubert et al. (2006) and isolated in the Col-0 background by H. Tsukaya.
Ethyl Methanesulfonate Mutagenesis
ag-10 seeds (1 mL, ~1000 seeds/100 μL) were washed with 0.1% Tween 20 for 15 min, incubated with 0.2% ethyl methanesulfonate for 12 h, and washed three times with 10 mL water each (1 h for each wash on a rotator). ag-10 enhancers were isolated in the M2 generation based the presence of bulged siliques throughout the plant. The mutants were backcrossed at least two times to ag-10 before further studies.
Map-Based Cloning of CLF
ag-10 clf-47 (Ler) was crossed to ag-10col. In the F2 population, plants showing the ag-10 clf-47 phenotypes were selected as the mapping population. Initially, 27 ag-10 clf-47 plants were used for rough mapping, which showed that clf-47 was linked to the marker nga1126 on chromosome 2. For fine mapping, we designed new simple sequence length polymorphic or cleaved-amplified polymorphic sequence markers in this region according to polymorphisms between Ler and Col from the Monsanto Arabidopsis Polymorphism and Ler Sequence database (http://www.Arabidopsis.org/Cereon). clf-47 was mapped to a 150-kb region covered by the BACs T16B14 and F3N11. The CLF gene was identified as a candidate gene and sequenced from the mutant.
Generation of Mutant Combinations
To generate double or triple mutants involving ag-10 or ag-10 clf-47, the ag-10 clf-47 double mutant was crossed to sup-1 (Bowman et al., 1992), stm-2/+ (Clark et al., 1996), ag-1/+ (Bowman et al., 1989), and wus-1/+ (Laux et al., 1996). In the F2 generation, plants resembling clf mutants in vegetative phenotypes were screened for floral phenotypes characteristic of sup-1, stm-2, ag-1, and wus-1 flowers. Then, plants with wild-type and ag-10/ag-10 genotypes at the AG locus were identified by molecular genotyping. To generate ag-10 clf-47 knu-1, ag-10 knu-1, and clf-47 knu-1, ag-10 clf-47 plants were crossed to knu-1. In the F2 population, all three mutations were genotyped to identify plants of the correct genotypes.
For ag-10 genotyping, PCR was performed on genomic DNA using primers JAGp75 and JAGp76 (see Supplemental Table 2 online), and the PCR products were digested by BstXI. The ag-10 mutation abolishes this restriction site. For clf-47 genotyping, PCR products amplified from genomic DNA using primers CLFmuF and CLFmuR (see Supplemental Table 2 online) were subjected to BstuI digestion. The clf-47 mutation abolishes the restriction site. For knu-1 genotyping, PCR was performed on genomic DNA with the primers knu-1genoF and knu-1genoR (see Supplemental Table 2 online), and the PCR products were digested with HpyCH4III. knu-1 abolishes this restriction site.
Plasmid Construction
To construct WUS1.6:GUS:WUS3′wt, PCR was performed with the primers WUSGUSF and WUSGUSR (see Supplemental Table 2 online) using genomic DNA from pWUS:GUS plants (Bäurle and Laux, 2005) as the template. Since both the endogenous WUS locus and the transgene could be amplified, the PCR products were digested with EcoRV to eliminate the WUS genomic DNA, and the remaining PCR products corresponding to the transgene were cloned into pENTR/D-TOPO (Invitrogen). Sequencing was conducted to ensure the integrity of the clone. To construct WUS1.6:GUS:WUS3′mut, site-directed mutagenesis was conducted on the WUS1.6:GUS:WUS3′wt plasmid by 18 cycles of PCR amplification of the entire plasmid with the primers WUSGUSPmF and WUSGUSPmR (see Supplemental Table 2 online) that carry the mutated nucleotides using the Phusion DNA polymerase (Finnzymes). The resulting clones were sequenced to confirm the presence of the introduced mutations and the absence of unwanted mutations. The WUS1.6:GUS:WUS3′wt and WUS1.6:GUS:WUS3′mut plasmids were linearized by MluI digestion, and the inserts were recombined into pEarleyGate303 (Earley et al., 2006) using a Gateway LR Clonase kit (Invitrogen) according to the manufacturer’s instructions.
To construct WUS3.2:GUS:WUS3′wt and WUS3.2:GUS:WUS3′mut, the 3.2-kb WUS promoter was amplified by PCR using primers WUS3.2proF and WUS3.2proR (see Supplemental Table 2 online) and the plasmid pIB39-WUS (HindIII-Bst1107I):GUS (a gift from T. Laux; Bäurle and Laux, 2005) as the template. The PCR product was cloned into pGEM-T-easy (Promega). The 3.2-kb promoter was released by NotI and SmaI digestion and cloned into WUS1.6:GUS:WUS3′wt or WUS1.6:GUS:WUS3′mut to replace the 1.6-kb promoter. The inserts in the WUS3.2:GUS:WUS3′wt and WUS3.2:GUS:WUS3′mut plasmids were then recombined into pEarleyGate303 as described above.
In Situ Hybridization
In situ hybridization was performed as described (Chen et al., 2002). For the WUS probe, the WUS coding region was amplified by RT-PCR and cloned into pGEM-T-easy (Promega). This plasmid was digested with SpeI and transcribed with T7 RNA polymerase to generate the antisense probe. STM probe was prepared as described (Chuck et al., 1996). For generating the GUS probe, the PCR reaction was performed using primers GUST7 and GUSSP6 (see Supplemental Table 2 online) and the WUS1.6:GUS:WUS3′wt plasmid as the template. In vitro transcription was performed with either T7 or SP6 RNA polymerase using the purified PCR product as the template to generate the antisense or sense probe, respectively.
Histochemical Staining
GUS staining was performed as described (Jefferson et al., 1987; Rodrigues-Pousada et al., 1993). Inflorescences were fixed in 90% cold acetone for 15 to 20 min and rinsed with the rinse solution [50 mM NaPO4, pH 7.2, 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6]. The infiltration solution [50 mM NaPO4, pH 7.2, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 2 mM X-Gluc] was added, and the inflorescences were vacuum infiltrated for 10 min followed by incubation at 37°C overnight.
For Toluidine Blue staining, the tissue sections on slides were soaked in 0.1% Toluidine Blue in 0.1% sodium borate briefly and rinsed in water.
RNA Extraction and Gene Expression Analysis
The 35S:AG-GR ag-1 plants (Gómez-Mena et al., 2005; Sun et al., 2009) or 35S:AG-GR ag-1 clf-47 plants (generated in this study) were treated with DMSO, DEX (10 μM), CHX (10 μM), or DEX plus CHX (10 μM for each) in 0.015% Silwet L-77 by applying the solution onto inflorescences. At various time points after the chemical treatments, the inflorescences were dissected under a stereomicroscope to remove flowers of stage 9 or older, and RNA was isolated with TRI reagent (MRC). Contaminating DNA was eliminated with DNaseI (New England Biolabs) treatment, and reverse transcription was performed using M-MLV reverse transcriptase (Promega). Quantitative real-time RT-PCR was conducted in triplicate on the Bio-Rad IQ5 real-time PCR system using the SYBR Green PCR master mix (Applied Biosystems). Four to five biological replicates were conducted, and the results were analyzed with SPSS statistics 17.0 (IBM) using the independent-samples t test.
ChIP
ChIP was performed as described previously (Sun et al., 2009; Zheng et al., 2009) with slight modifications. Inflorescences were ground in liquid nitrogen and cross-linked with 1% formaldehyde in M1 buffer (10 mM phosphate buffer, pH 7.0, 0.1 M NaCl, 10 mM mercaptoethanol, 1 M hexylene glycol, 1× protease inhibitor cocktail [Roche], and 1 mM PMSF) for 10 min. The suspension was filtered through four layers of Miracloth, and the filtrate was centrifuged at 12,000 rpm for 10 min. The pelleted chromatin was washed three times with M2 buffer (M1 buffer plus 10 mM MgCl2 and 0.5% Triton X-100) and once with M3 buffer (10 mM phosphate buffer, pH 7.0, 0.1 M NaCl, 10 mM mercaptoethanol, 1× protease inhibitor cocktail [Roche], and 1 mM PMSF). Chromatin was resuspended in nuclei lysis buffer and sonicated to generate DNA fragments of ~500 bp. The lysate was precleared by incubation with 50 μL protein-A agarose beads/salmon sperm DNA (Millipore) for 1 h and incubated with anti-HA (abcam), anti-H3K27me3 (abcam), or anti-AG antibodies (see below) overnight. The bound DNA fragments were recovered and purified with columns from the Plasmid Extraction kit (Qiagen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on bound and input DNAs. Primers used are listed in Supplemental Table 2 online. AG antibodies were produced in rabbits against an AG-specific peptide at Sigma-Genosys. The antisera were purified using a C-terminal portion of AG protein expressed in Escherichia coli as a fusion to maltose binding protein.
For testing TFL2 occupancy at WUS by ChIP, entire inflorescences of Col (a negative control) and 35S:TFL2-3HA were used. For testing AG binding to WUS by ChIP, entire inflorescences from Ler and ag-1 (a negative control) were used. For ChIP to examine the status of H3K27me3 in Ler and ag-1 or TFL2/LHP1 occupancy at WUS in 35S:TFL2-3HA and 35S:TFL2-3HA ag-1, microdissected inflorescences containing flowers of stage 8 and younger were used.
Laser Capture Microdissection
Laser capture microdissection was performed as described (http://seedgenenetwork.net/arabidopsis#procedure; Cai and Lashbrook, 2006; Hsieh et al., 2011) using the Arcturus laser capture microdissection instrument (Applied Biosystems). In brief, inflorescences from Ler and ag-1 plants were fixed with ethanol/acetic acid, dehydrated, and embedded in paraffin blocks. Ribbons with 8-μm sections were loaded on a slide and deparaffinized. Floral meristems at stages 1 and 2, 3, 4 and 5, and 6 were identified. A circular area covering the center of a meristem was excised from all the serial sections that contained that meristem. This ensured that all cells from the central region of an entire floral meristem were included. Total RNA was extracted with the Arcturus Picopure RNA isolation kit (Applied Biosystems) according to the manufacturer’s protocol. Reverse transcription was conducted with M-MLV reverse transcriptase (Promega). Quantitative RT-PCR was performed in triplicate on the Bio-Rad IQ5 Real-time PCR system usingSYBR Green PCR master mix (Applied Biosystems). Two biological replicates were conducted.
Immunoblotting
One hundred milligrams of leaves or inflorescences from Ler or ag-10 clf-47 plants were ground in liquid nitrogen and homogenized in 2× SDS sample buffer (0.5 M Tris-HCl, pH 6.8, 4.4% [w/v] SDS, 20% [v/v] glycerol, 2% [v/v] 2-mercaptoethanol, and bromophenol blue). The samples were boiled for 10 min, cooled on ice for 5 min, and centrifuged at 16,000g for 5 min at 4°C to precipitate insoluble material. Proteins in the supernatant were resolved in a 12% SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with anti-AG antibodies. Signal development was performed with the ECL+Plus Western Blotting system (GE Healthcare) and by exposure of the membrane to x-ray film (Denville) at a time course of 30 s, 1 min, and 2 min. The time course ensured that the signal detection was within linear range.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AG, AT4G18960; APETALA1, AT1G69120; APETALA3, AT3G54340; CLF, AT2G23380; eIF4A1, AT3G13920; KNU, AT5G14010; SPOROCYTELESS, AT4G27330; UBQ5, AT3G62250; and WUS, AT2G17950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Structure of the CLF Gene and Similarity between CLF and Other Eukaryotic E(z) Homologs within the SET Domain.
Supplemental Figure 2. Effects of the clf-47 Mutation on AG Expression.
Supplemental Figure 3. CLF and SWN Are Responsible for H3K27 Trimethylation at the WUS Locus in Seedlings.
Supplemental Figure 4. A Time-Course Analysis of Gene Expression in Response to AG Induction in 35S:AG-GR ag-1 Inflorescences.
Supplemental Figure 5. GUS Staining and in Situ Hybridization to Examine GUS Expression in WUS1.6:GUS:WUS3′mut Transgenic Lines.
Supplemental Figure 6. Laser Capture Microdissection of Floral Meristems.
Supplemental Table 1. Floral Organ Counts in Flowers of Various Genotypes.
Supplemental Table 2. Oligonucleotides Used in This Study.
Acknowledgments
We thank John Harada for valuable advice and protocols on laser capture microdissection. We thank Toshiro Ito, Thomas Laux, Robert Sablowski, Wenhui Shen, and Hao Yu for sharing materials and Yuanyuan Zhao and Thanh Theresa Dinh for comments on the manuscript. This work was supported by a grant from National Institutes of Health (GM61146) to X.C. R.M. and J.G. were funded by a grant from the UK Biotechnology and Biological Science Research Council. R.E.Y. was supported by a National Science Foundation Integrative Graduate Education and Research Traineeship training grant (DGE0504249). C.L. was supported by the National Natural Science Foundation of China (Grant 90919033) and the Chinese Academy of Sciences (Grant KSCX2-EW-Q-24-02).
AUTHOR CONTRIBUTIONS
X.L., Y.J.K., J.G., and X. Chen designed the research. X.L., Y.J.K., R.M., R.E.Y., C.L., and Y.P. performed research. C.L. and X. Cao contributed new analytic tools. X.L., Y.J.K., R.M., R.E.Y., Y.P., J.G., and X. Chen analyzed data. X. Chen, X.L., and Y.J.K. wrote the article.
References
- Alvarez J., Smyth D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Bäurle I., Laux T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Sakai H., Jack T., Weigel D., Mayer U., Meyerowitz E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114: 599–615 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cai S., Lashbrook C.C. (2006). Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. Plant J. 48: 628–637 [DOI] [PubMed] [Google Scholar]
- Carles C.C., Choffnes-Inada D., Reville K., Lertpiriyapong K., Fletcher J.C. (2005). ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 132: 897–911 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chen X., Liu J., Cheng Y., Jia D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Das P., Ito T., Wellmer F., Vernoux T., Dedieu A., Traas J., Meyerowitz E.M. (2009). Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 136: 1605–1611 [DOI] [PubMed] [Google Scholar]
- Drews G.N., Bowman J.L., Meyerowitz E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Gómez-Mena C., de Folter S., Costa M.M., Angenent G.C., Sablowski R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438 [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J.P., Hoeppner M.A., Gagliano W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Hennig L., Derkacheva M. (2009). Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hsieh T.F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M.J., Hashimoto M., Kirkbride R.C., Harada J.J., Zilberman D., Fischer R.L. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Mizukami Y., Hu Y., Ma H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21: 4769–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., et al. (2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. (1985). A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153–155 [Google Scholar]
- Kanhere A., et al. (2010). Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.M., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Villar C.B. (2008). Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 18: 236–243 [DOI] [PubMed] [Google Scholar]
- Kotake T., Takada S., Nakahigashi K., Ohto M., Goto K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44: 555–564 [DOI] [PubMed] [Google Scholar]
- Larsson A.S., Landberg K., Meeks-Wagner D.R. (1998). The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T., Mayer K.F., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jürgens G., Laux T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Lewis E.B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Maier A.T., Stehling-Sun S., Wollmann H., Demar M., Hong R.L., Haubeiss S., Weigel D., Lohmann J.U. (2009). Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136: 1613–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A.A., Ko M.S., Niwa H. (2007). Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9: 625–635 [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Müller J., Verrijzer P. (2009). Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Mylne J.S., Barrett L., Tessadori F., Mesnage S., Johnson L., Bernatavichute Y.V., Jacobsen S.E., Fransz P., Dean C. (2006). LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. (2007). The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T., Johnson S.D., Koltunow A.M. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131: 3737–3749 [DOI] [PubMed] [Google Scholar]
- Pien S., Grossniklaus U. (2007). Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta 1769: 375–382 [DOI] [PubMed] [Google Scholar]
- Prunet N., Morel P., Thierry A.M., Eshed Y., Bowman J.L., Negrutiu I., Trehin C. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20: 901–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Chang H.Y. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada R.A., De Rycke R., Dedonder A., Van Caeneghem W., Engler G., Van Montagu M., Van Der Straeten D. (1993). The Arabidopsis 1-Aminocyclopropane-1-Carboxylate Synthase Gene 1 is expressed during early development. Plant Cell 5: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatlowski N., Creasey K., Goodrich J., Schubert D. (2008). Keeping plants in shape: Polycomb-group genes and histone methylation. Semin. Cell Dev. Biol. 19: 547–553 [DOI] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E.A., Pickett F.B., Haughn G.W. (1991). The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell 3: 1221–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.J., Fujiwara Y., Kim J., Mao X., Yuan G.C., Orkin S.H. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H., Okada K., Shimura Y. (1993). Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4: 385–398 [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Xu Y., Ng K.H., Ito T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23: 1791–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., He Y., Eshoo T.W., Tamada Y., Johnson L., Nakahigashi K., Goto K., Jacobsen S.E., Amasino R.M. (2006). Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 38: 706–710 [DOI] [PubMed] [Google Scholar]
- Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Shen W.H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yap K.L., Li S., Muñoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007b). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B.J., Khorasanizadeh S., Gaudin V., Jacobsen S.E. (2007a). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14: 869–871 [DOI] [PubMed] [Google Scholar]
- Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Chen X. (2011). Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]