This study evaluates the effect of two actin binding proteins (AIP1 and ADF) on F-actin dynamics in moss. Whereas AIP1 enhances F-actin turnover, ADF is required for it.
Abstract
The remodeling of actin networks is required for a variety of cellular processes in eukaryotes. In plants, several actin binding proteins have been implicated in remodeling cortical actin filaments (F-actin). However, the extent to which these proteins support F-actin dynamics in planta has not been tested. Using reverse genetics, complementation analyses, and cell biological approaches, we assessed the in vivo function of two actin turnover proteins: actin interacting protein1 (AIP1) and actin depolymerizing factor (ADF). We report that AIP1 is a single-copy gene in the moss Physcomitrella patens. AIP1 knockout plants are viable but have reduced expansion of tip-growing cells. AIP1 is diffusely cytosolic and functions in a common genetic pathway with ADF to promote tip growth. Specifically, ADF can partially compensate for loss of AIP1, and AIP1 requires ADF for function. Consistent with a role in actin remodeling, AIP1 knockout lines accumulate F-actin bundles, have fewer dynamic ends, and have reduced severing frequency. Importantly, we demonstrate that AIP1 promotes and ADF is essential for cortical F-actin dynamics.
INTRODUCTION
The actin cytoskeleton is critical for a wide variety of cellular processes, ranging from cell division and morphogenesis to cell polarity and motility (Pollard and Cooper, 2009). In plants, actin is required for the polarized cell expansion termed tip growth. Tip-growing cells are found in all land plants (Hepler et al., 2001; Menand et al., 2007). In mosses, protonemal cells and rhizoids colonize and anchor the plant to the growth surface while acquiring nutrients from the environment. In flowering plants, pollen tubes are necessary for transmission of the male gametophyte to the ovule for sexual reproduction, and root hairs enhance the surface area of the root to promote nutrient uptake (Hepler et al., 2001). Tip growth is therefore a critical process in all land plants because these tissues are required for overall plant fitness and species propagation.
The actin cytoskeleton plays an integral role in promoting tip growth (Hepler et al., 2001). In particular, constant remodeling of the actin filament (F-actin) network is required. Remodeling is a dynamic process whereby F-actin turnover occurs via polymerization, depolymerization, severing, nucleation, as well as large-scale translocation events. Pharmacological studies show that alterations to the equilibrium between F-actin and monomeric actin inhibit the expansion of pollen tubes and root hairs (Gibbon et al., 1999; Miller et al., 1999; Vidali et al., 2001), demonstrating that a fine balance of actin assembly and disassembly is required for tip growth. More recently, time-lapse imaging of the cortical actin cytoskeleton in planta provides further support for rapid remodeling of the actin cytoskeleton (Era et al., 2009; Staiger et al., 2009; Vidali et al., 2009a; Smertenko et al., 2010). Individual actin filaments undergo stochastic dynamics whereby rapid elongation of filaments is interspersed by severing events (Michelot et al., 2007; Staiger et al., 2009) that result in the dramatic reorganization of the actin cytoskeleton on the order of seconds.
The biochemical properties of actin alone are not sufficient to account for the rapid F-actin remodeling observed in vivo. Thus, it is likely that other factors, notably actin binding proteins, are required to regulate actin’s behavior. Indeed genetic studies support the requirement for a number of actin binding proteins in the expansion of tip-growing cells (Blanchoin et al., 2010). These include actin-nucleating proteins, such as formins (Vidali et al., 2009c; Ye et al., 2009; Cheung et al., 2010) and the Arp2/3 complex (Harries et al., 2005; Perroud and Quatrano, 2006; Finka et al., 2008), the filament-stabilizing fimbrins (Wu et al., 2010), actin monomer binding proteins like profilin (Ramachandran et al., 2000; Vidali et al., 2007, 2009b), and actin turnover proteins like actin depolymerizing factor (ADF) (Augustine et al., 2008; Vidali et al., 2009b), villin (Zhang et al., 2010; Zhang et al., 2011), and actin interacting protein1 (AIP1) (Ketelaar et al., 2004). It is tempting to suggest that these actin binding proteins are important for tip growth by promoting the reorganization of F-actin. However, a recent loss-of-function study demonstrated that myosin XI is critical for tip growth but does not alter actin dynamics (Vidali et al., 2010). Therefore, it is imperative to quantify the effects of actin binding protein loss of function on actin dynamics to understand the mechanism by which these proteins function.
The ADF/cofilin family is widely regarded as a major cellular mediator of actin turnover (Bamburg, 1999; Maciver and Hussey, 2002). ADF/cofilins sever actin filaments (Maciver et al., 1991; Andrianantoandro and Pollard, 2006; Pavlov et al., 2007). Severing events can result in rapid disassembly and recycling of actin monomer pools (Hotulainen et al., 2005) or generation of new barbed ends capable of elongation (Ghosh et al., 2004). In budding yeast, severing-defective cofilin alleles are less sensitive to latrunculin A–mediated disassembly of actin patches (Lappalainen and Drubin, 1997), demonstrating that cofilin’s severing activity is necessary for the turnover of these structures. ADF/cofilin proteins are also required for generation of dynamic actin structures, such as lamellipodia (Chan et al., 2000) and comet tails in Listeria monocytogenes (Rosenblatt et al., 1997; Loisel et al., 1999), supporting their role in the remodeling of actin networks.
AIP1, a conserved eukaryotic protein, enhances ADF/cofilin’s actin filament disassembly activity (Ono, 2003). It is well established that AIP1 and ADF/cofilin interact in animals, plants, and fungi based on yeast two-hybrid screens (Rodal et al., 1999; Allwood et al., 2002), genetic interactions (Iida and Yahara, 1999; Rodal et al., 1999), and affinity chromatography (Okada et al., 1999), demonstrating the broad conservation of this interaction. AIP1 has negligible effects on actin filaments. However, in the presence of ADF/cofilin, disassembly occurs at a higher rate than either protein alone (Aizawa et al., 1999; Okada et al., 1999; Rodal et al., 1999). While the mechanism of cooperative disassembly is still unclear, evidence supports a role for AIP1 in capping the barbed end of ADF/cofilin-severed actin filaments (Okada et al., 1999, 2002, 2006; Balcer et al., 2003; Mohri et al., 2004), enhancing actin filament severing by ADF/cofilins (Aizawa et al., 1999; Mohri et al., 2004), or a combination of these mechanisms. AIP1’s activity ultimately results in further fragmentation of ADF/cofilin-severed actin filaments (Aizawa et al., 1999; Okada et al., 1999; Okreglak and Drubin, 2010), showing that AIP1 functions in the turnover of F-actin.
Consistent with its biochemical activities, AIP1 is important for actin-dependent processes in a variety of organisms. In Dictyostelium discoideum, AIP1 knockouts grow slower with defects in cytokinesis, endocytosis, and cell migration (Konzok et al., 1999). AIP1 mutants in Caenorhabditis elegans are severely defective in motility arising from disorganized F-actin in muscles (Ono, 2001). The AIP1 gene in Drosophila melanogaster is essential and required for cell proliferation and the proper assembly of polarized epidermal hair cells (Ren et al., 2007). While AIP1 deletion in budding yeast does not result in any growth defects (Rodal et al., 1999), AIP1 is clearly involved in actin turnover. AIP1 nulls are synthetic lethal with viable cofilin mutant alleles placing them in a common genetic pathway (Iida and Yahara, 1999; Rodal et al., 1999). Furthermore, AIP1 nulls aberrantly accumulate cofilin along actin cables (Iida and Yahara, 1999; Rodal et al., 1999), and these cables are less sensitive to turnover by latrunculin A (Okada et al., 2006). Collectively, these studies highlight a functional link between AIP1 and actin, demonstrating that AIP1 has evolved variable roles and requirements in fungi, animals, and slime molds.
In Arabidopsis thaliana, inducible knockdown of AIP1 reduces the growth of diffusely expanding intercalary cells and tip-growing root hairs (Ketelaar et al., 2004). Under robust AIP1 knockdown conditions, shoot and flower development fails and plants exhibit cell senescence, demonstrating that AIP1 is essential (Ketelaar et al., 2004). Like AIP1 homologs in other eukaryotes, plant AIP1 interacts with ADF to enhance actin filament disassembly in vitro (Allwood et al., 2002). This biochemical activity is manifested in vivo by enhanced actin bundling in AIP1 knockdown plants. In lily (Lilium longiflorum) pollen, AIP1 and ADF colocalize to the subapical actin fringe (Lovy-Wheeler et al., 2006), suggesting that they interact to remodel this dynamic F-actin network. While the biochemical activities and in vivo roles of AIP1 and ADF are consistent with a role in actin remodeling, the extent to which these proteins are required for promoting F-actin turnover has not been established in planta.
The moss Physcomitrella patens is an excellent model system for studying actin binding proteins. The cortical F-actin array in tip-growing protonemal cells is readily accessible for high-resolution imaging and dramatically rearranges as a result of growth, shrinkage, buckling, and translocation of actin filaments (Vidali et al., 2009a). Furthermore, the moss genome has been sequenced (Rensing et al., 2008), and homologous recombination occurs efficiently upon transformation (Schaefer and Zrÿd, 1997), making targeted knockout and allele replacement studies feasible. Finally, moss possesses a single ADF gene (Augustine et al., 2008), precluding the need to dissect isovariant-specific effects on the actin cytoskeleton. In this study, we show that AIP1 promotes tip growth and demonstrate that AIP1 and ADF are critical for promoting cortical actin dynamics in planta.
RESULTS
Moss Has a Single AIP1 Gene
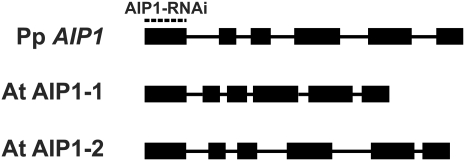
We used the amino acid sequences of the two AIP1 genes (AIP1-1 and AIP1-2) from Arabidopsis in BLAST searches of the P. patens genome (Rensing et al., 2008) to identify AIP1 in P. patens. We found a single AIP1 locus. To determine the full-length open reading frame, we generated cDNA from moss protonemal tissue and amplified the predicted AIP1 coding sequence. The coding region of Pp AIP1 is composed of six exons of similar size to the exons of AIP1-1 and AIP1-2 from Arabidopsis (Figure 1). Furthermore, moss and Arabidopsis AIP1 genes share common exon/intron boundaries, indicating that the gene structure has been conserved through the ~400 million years of evolution separating bryophytes and flowering plants. The moss AIP1 protein sequence is also conserved, sharing 57 to 58% identity and 78% similarity with the two Arabidopsis AIP1 proteins (see Supplemental Table 1 online).
Figure 1.
Schematic of P. patens and Arabidopsis AIP1 Gene Structures.
Exons are thick black boxes; introns are black lines. Intron-exon boundaries were determined by comparing genomic and cDNA sequences. The sequence used for the AIP1 coding sequence-RNAi (AIP1-RNAi) is indicated by a dashed line above the first exon.
AIP1 Is Important for Tip Growth
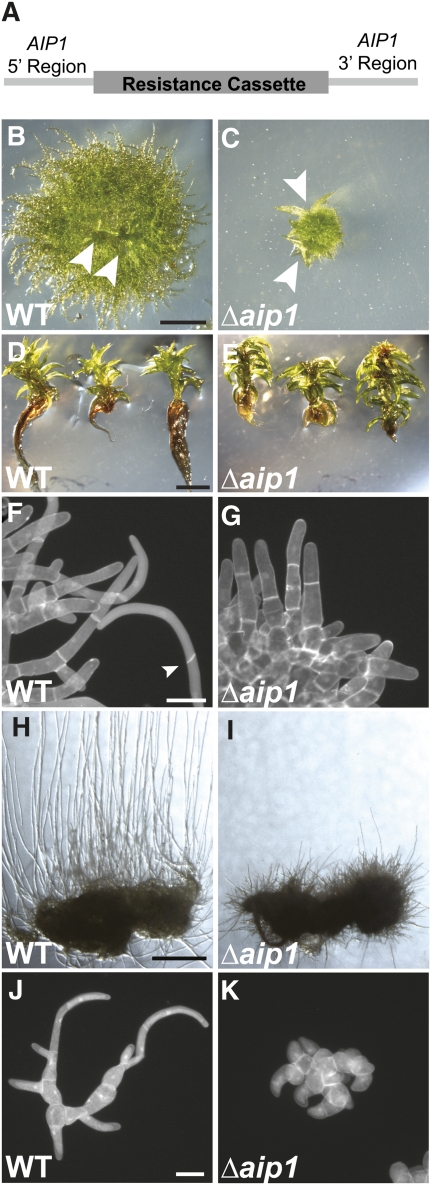
To assess the role of AIP1 in plant growth, we used homologous recombination to remove the moss AIP1 gene. The AIP1 knockout construct contains a resistance cassette flanked by a region of sequence upstream of the AIP1 start codon and a region downstream of the stop codon as targeting arms for homologous recombination (Figure 2A). We transformed this construct into protoplasts from a variety of different moss lines and selected for stable integrants. We confirmed proper targeting of the locus at both 5′ and 3′ ends and stable integration of a single copy of the AIP1 knockout construct using PCR amplification (see Supplemental Figure 1 online). Two independent stable AIP1 knockout (Δaip1) lines were isolated in each moss background for further analysis.
Figure 2.
AIP1 Promotes the Growth and Formation of Tip-Growing Cells.
(A) The AIP1 knockout construct was generated by PCR amplifying a sequence upstream of the start codon (5′ region) and downstream of the stop codon (3′ region) as targeting arms for homologous recombination. The targeting arms flank a resistance cassette to select for AIP1 knockout plants.
(B) and (C) Wild-type (WT) (B) or Δaip1 (C) plants were grown on cellophane-overlain agar plates for 3 weeks. Arrowheads point to emerging gametophores. Bar = 1 mm.
(D) and (E) Wild-type (D) or Δaip1 (E) plants were grown on agar plates for 3 months. Plants were removed from the agar and laid on an agar plate to display the growth of the gametophores and rhizoids. Bar = 2 mm.
(F) and (G) Plants were stained with calcofluor and DAPI to image cell walls and nuclei, respectively, and then visualized by epifluorescence microscopy. Apical cell length in 8-d-old wild type (F) and Δaip1 (G) plants was measured from fluorescence images (Table 1). The arrowhead points to an oblique cell plate of a caulonemal cell. Bar = 50 μm.
(H) and (I) Wild-type (H) and Δaip1 (I) plants were grown on a cellophane-overlaid agar plate for 18 d in the dark. Bar = 2 mm.
(J) and (K) The number of cells per plant was measured in 5-d-old plants stained with calcofluor and DAPI to image cell walls and nuclei, respectively. Wild-type (J) and Δaip1 (L) plants were visualized by epifluorescence microscopy. Bar = 50 μm.
To examine differences in plant growth, we made protoplasts of wild-type and Δaip1 lines and allowed the protoplasts to regenerate into plants. Tip-growing protonemal cells emerge from the protoplast, forming a dense network of filamentous tissue that eventually gives rise to the leaf-like gametophores that expand by diffuse growth. In comparison to the wild type, 3-week-old Δaip1 plants have severe defects in expansion of tip-growing protonemal cells (Figures 2B and 2C). Interestingly, we did not observe differences in the timing of gametophore emergence (Figures 2B and 2C). After 3 months of growth from protoplasts, we found no obvious defects in the development of gametophores in the Δaip1 lines (Figures 2D and 2E). However, we found that the tip-growing rhizoids that emerge from the base of the gametophores are severely stunted (Figures 2D and 2E). These results indicate that AIP1 contributes to tip growth but not diffuse growth in P. patens.
Since the primary defect in Δaip1 plants appears to be growth of tip-growing tissues, we examined this more closely. The tip-growing protonemata are subdivided into two cell types: slow-growing chloronemal cells and faster growing caulonemal cells (Menand et al., 2007). These cell types are readily distinguishable because chloronemal cells have transverse cell plates, whereas caulonemal cells have oblique cell plates with respect to the filament axis. To determine the relative composition of caulonemal and chloronemal cells, we regenerated plants from protoplasts and after 8 d fixed and stained wild-type and Δaip1 plants with calcofluor to image cell walls. While the wild type and Δaip1 have many chloronemal cells (Table 1, Figures 2F and 2G), we observed caulonemal cells in the wild type only (Table 1, Figure 2F). Additionally, we measured the length of apical cells and found that Δaip1 tip cells were shorter than wild-type chloronemal and caulonemal cells (Table 1). These data suggest that AIP1 is important for normal tip cell expansion and for the formation of caulonemal cells.
Table 1.
Analysis of Apical Cell Number and Length in AIP1 Knockout Plants
| No. of Tip Cells |
Average Tip Cell Length (μm) |
|||
| Line | Chloronema | Caulonema | Chloronema | Caulonema |
| Wild type | 310 | 167 | 81.1 ± 27.7 | 120.9 ± 37.9 |
| Wild-type Δaip1 #1 | 374 | 0 | 65.3 ± 19.1* | NA |
| Wild-type Δaip1 #2 | 239 | 0 | 63.7 ± 20.5* | NA |
The number and average length of chloronemal and caulonemal tip cells was determined from 8-d-old plants in ImageJ. Standard deviation is reported after the ± sign. The asterisk indicates that the values are significantly different from wild type with a P value < 0.001. Alpha significance level is set at 0.05. NA, not applicable.
To test whether Δaip1 plants are capable of forming caulonemal cells, we grew the wild type and Δaip1 in the dark for 18 d to induce caulonemal cell production (Cove et al., 1978). Both the wild type and Δaip1 formed caulonemal cells, although there was a striking reduction in the growth of these cells in the Δaip1 lines (Figures 2H and 2I). Collectively, these results suggest that the tip growth phenotype in Δaip1 is due, in part, to the limited formation and expansion of caulonemal cells.
Growth in moss protonemal cells is a combination of directed cell expansion at the cell apex and cell division. Therefore, it is possible that reduced tip growth in Δaip1 results from a delay or defects in cell division. To investigate this, we stained 5-d-old plants with 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor to simultaneously visualize nuclear DNA and cell walls. Δaip1 plants had a similar number of cells per plant compared with the wild type (Figures 2J and 2K; the wild type = 14.5 ± 6.3, n = 59; wild-type Δaip1 #1 = 15.1 ± 5.9, n = 53; wild-type Δaip1 #2 = 14.0 ± 5.9, n = 57; all comparisons not significant). Furthermore, we did not observe any obvious cell division defects, such as binucleate cells or incomplete cell plates, which are associated with mis-segregation of nuclei, suggesting that reduced tip growth is the result of a cell expansion defect rather than aberrant or delayed cell division.
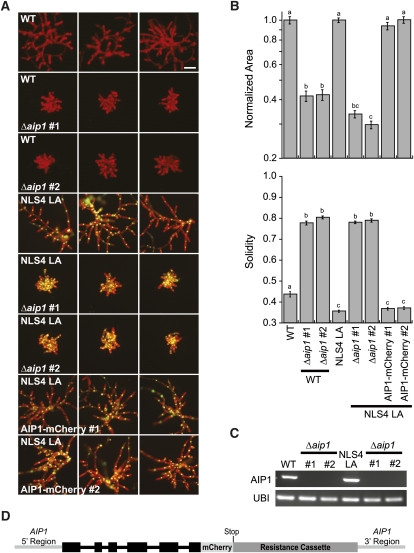
We further examined the growth of protonemal cells by generating wild-type and Δaip1 protoplasts and imaging plants after 1 week of regeneration (Figure 3A). We quantified the growth phenotype by measuring plant area of chlorophyll autofluorescence. The area of Δaip1 plants was reduced by approximately 60% compared with the wild type (Figures 3A and 3B). We also observed that control lines had many branched protonomal filaments, whereas Δaip1 plants appeared as clusters of cells that had severe defects in forming polarized outgrowths. We quantified the morphological differences by measuring plant solidity, a ratio of the total plant area divided by its convex hull area. Plants that expand into their environment and have large spaces between protonemal filaments have lower solidity values than plants that are compact and form few polarized outgrowths. In comparison to the wild type, Δaip1 lines have larger solidity values (Figure 3B), corresponding to their lack of branched outgrowths and stubby appearance.
Figure 3.
Growth and Morphology Are Affected in AIP1 Knockout Plants.
(A) Representative images of 1-week-old plants visualized by epifluorescence stereomicroscopy. Two independent Δaip1 lines are compared with their respective control plants: the wild type (WT) or a line stably coexpressing NLS-GFP-GUS and LA (NLS4 LA). Green nuclei are a result of the NLS-GFP-GUS signal. Two independent lines expressing a C-terminal fusion of AIP1-mCherry at the endogenous locus in the NLS4 LA line are shown. Bar = 100 μm.
(B) Quantification of normalized area and solidity in control and allele replacement lines. Normalization was done in relation to the appropriate control (the wild type or NLS4). Letters above the bars indicate statistical groupings. Numbers of plants analyzed: the wild type, 75; wild-type Δaip1 #1, 52; wild-type Δaip1 #2, 74; NLS4 LA, 175; NLS4 LA Δaip1 #1, 100; NLS4 LA Δaip1 #2, 100; NLS4 LA AIP1-mCherry 1, 75; NLS4 LA AIP1-mCherry 2, 75. Error bars indicate se.
(C) RNA expression analysis of control and Δaip1 lines. Representative ethidium bromide–stained gel of RT-PCR analysis using protonemal RNA samples from the indicated lines. Ubiquitin10 is a loading control. Two biological replicates and four technical replicates were performed.
(D) Schematic of the AIP1-mCherry replacement locus. Exons are thick black boxes. Introns are black lines. The stop codon was removed from the end of original coding sequence and moved to the end of the mCherry sequence to generate an in-frame C-terminal fusion protein. Light-gray lines are regions upstream of the start codon (AIP1 5′ region) and downstream of the stop codon (AIP1 3′ region) in the unaltered PpAIP1 locus.
AIP1 knockouts were also generated in the background of a stable moss line, NLS4 LA, which coexpresses a nuclear-localized green fluorescent protein–β-glucuronidase (GFP-GUS) fusion and Lifeact-mEGFP (LA). The GFP-GUS fusion protein serves as an internal reporter for RNA interference (RNAi)–mediated gene silencing, as previously described (Bezanilla et al., 2005), whereas LA is a useful probe for time-lapse imaging of actin in moss (Vidali et al., 2009a). The two independent, stable Δaip1 lines in NLS4 LA were similar to Δaip1 lines generated in the wild-type background (Figures 3A and 3B).
To independently verify whether the Δaip1 lines we analyzed were knockouts, we performed RT-PCR from total RNA extracts of control and Δaip1 lines. While the controls robustly amplified AIP1, we could not detect AIP1 in Δaip1 (Figure 3C), supporting our genotyping results and confirming that these lines are AIP1 knockouts.
AIP1 Is Cytosolic
To investigate the localization of AIP1, we integrated mCherry in frame at the C terminus of the endogenous AIP1 locus (Figure 3D). Stable AIP1-mCherry lines were tested by PCR to ensure proper targeting at both the 5′ and 3′ ends of the locus and for integration of a single copy of the knock-in construct (see Supplemental Figure 2 online). The size and morphology of AIP1-mCherry plants was quantified 7 d after regeneration from protoplasts. We found that AIP1-mCherry lines are indistinguishable in both area and solidity from control lines (Figures 3A and 3B), indicating that AIP1-mCherry is fully functional and does not interfere with tip growth or plant morphology.
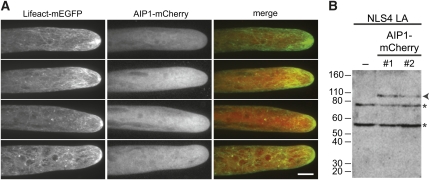
AIP1-mCherry lines were generated in the NLS4 LA background to permit simultaneous imaging of actin and AIP1. Actin is mostly cortical with enrichment near the cell apex (Figure 4A). The apical enrichment is dynamic both in intensity and position (Vidali et al., 2009a). Unlike other actin binding proteins, such as class II formins (Vidali et al., 2009c) and myosin XI (Vidali et al., 2010), AIP1-mCherry does not become enriched at the cell apex (Figure 4A). Instead, AIP1-mCherry remains diffusely cytosolic. This localization is similar to the localization reported for ADF (Augustine et al., 2008). Furthermore, we did not find any obvious association of AIP1 with actin filaments at the cortex (Figure 4A).
Figure 4.
AIP1 Is Diffusely Cytosolic.
(A) Z-projections of four NLS4 LA AIP1-mCherry protonemal cells imaged with spinning disc confocal microscopy. LA (a marker for actin), AIP1-mCherry, and merge of both channels are shown. Bar = 10 μm.
(B) Fifteen micrograms of protonemal cell protein extracts from control and NLS4 LA AIP1-mCherry replacement lines were immunoblotted with a polyclonal dsRed antibody. The arrowhead points to the AIP1-mCherry protein band, while asterisks indicate cross-reactive bands present in all samples.
We performed immunoblotting with a polyclonal dsRed antibody that recognizes mCherry to ensure that the cytosolic localization of AIP1-mCherry is not a result of proteolytic release of the mCherry. We observed the anticipated ~92-kD gene product for full-length AIP1-mCherry in both NLS4 LA AIP1-mCherry lines (Figure 4B).
AIP1 Requires ADF for Function
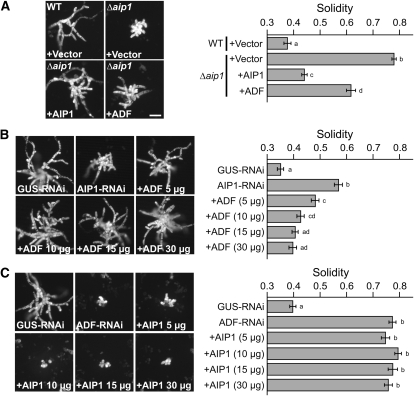
We performed transient complementation to ensure that the Δaip1 phenotype is a result of knockout of the AIP1 locus and not a result of integration into a secondary locus. Δaip1 was transformed with an AIP1 expression plasmid or an empty vector control. In addition, the wild type was transformed with the empty vector as a control for normal growth. Unlike the empty vector control, the AIP1 plasmid restored solidity to nearly wild-type levels when transformed into Δaip1 (Figure 5A), demonstrating the specificity of the AIP1 loss-of-function phenotype.
Figure 5.
AIP1 Functions through ADF.
(A) Representative images of 1-week-old plants imaged with epifluorescence microscopy. On each panel, the top left-hand corner indicates the stable line, and the transformed plasmid is indicated in the bottom left-hand corner. Ten micrograms of plasmid was used in the transformation of each line. The number of plants analyzed is as follows: the wild type (WT) +Vector, 75; Δaip1 +Vector, 100; Δaip1 +AIP1, 75; Δaip1 +ADF, 75.
(B) Representative images of 1-week-old plants imaged with epifluorescence microscopy. AIP1-RNAi was cotransformed with various concentrations of the complementing ADF plasmid, indicated after the plus sign. The number of plants analyzed is 75 for all samples.
(C) Representative images of 1-week-old plants imaged with epifluorescence microscopy. ADF-RNAi was cotransformed with various concentrations of the complementing AIP1 plasmid indicated after the plus sign. The number of plants analyzed is as follows: GUS-RNAi, 75; ADF-RNAi, 75; +AIP1 5 μg, 75; +AIP1 10 μg, 75; +AIP1 15 μg, 50; +AIP1 30 μg, 75.
In (A) to (C), the red channel, representing chlorophyll autofluorescence, was extracted from the color image and converted to gray scale. Solidity values for each rescue are indicated in the graph on the right. Error bars represent se. Letters to the right of the bar indicate statistical groupings. For all images, bar = 100 μm.
AIP1 is known to enhance ADF’s actin filament disassembly activity (Ono, 2003). Thus, we hypothesized that overexpression of ADF might bypass the requirement for AIP1. To test this, we transformed Δaip1 with an ADF expression vector. Interestingly, ADF provided a partial rescue of the Δaip1 phenotype (Figure 5A). Since ADF overexpression partially compensates for loss of AIP1, it is possible that endogenous ADF levels are elevated in the AIP1 knockout lines. To test this, we performed immunoblot analysis of extracts from AIP1 knockout and control lines. We found that, on average, ADF is upregulated two- to fourfold in AIP1 knockouts compared with the wild type (see Supplemental Figure 3 online). These results show that ADF can partially compensate for loss of AIP1, suggesting that AIP1 enhances ADF’s activity by a mechanism partly dependent on ADF.
As a complementary approach to validate that ADF rescues loss of AIP1, we used a previously described transient RNAi complementation assay (Bezanilla et al., 2005). Briefly, we transformed protoplasts expressing nuclear-localized GFP:GUS (NLS4) with an RNAi plasmid that simultaneously silences GUS and a target gene of interest. After 7 d, plants actively silencing the gene of interest can be readily distinguished from nonsilencing plants by identifying plants that lack the nuclear GFP:GUS reporter. We used the coding sequence of the first exon of AIP1 to generate an AIP1-RNAi construct (Figure 1). AIP1-RNAi plants exhibit a similar phenotype to Δaip1 (Figure 5B). The AIP1-RNAi plants have significant morphology defects, as quantified by higher solidity values compared with GUS-RNAi controls (Figure 5B). The AIP1-RNAi morphology defect was rescued by cotransforming AIP1-RNAi with an ADF expression construct in a concentration-dependent manner (Figure 5B).
Since ADF was able to rescue the AIP1 phenotype, we tested the reciprocal experiment to see if AIP1 could compensate for loss of ADF. Using RNAi, ADF was shown to be essential for tip growth and plant viability, and this loss-of-function phenotype was fully rescued by cotransformation with an ADF expression construct (Augustine et al., 2008; Vidali et al., 2009b). We found that cotransformation of ADF-RNAi with an AIP1 expression plasmid was unable to suppress the ADF loss-of-function phenotype (Figure 5C), demonstrating that AIP1 cannot compensate for loss of ADF. Collectively, these results demonstrate that AIP1 works through ADF in vivo, presumably by enhancing ADF activity, and this activity is required to promote normal tip growth.
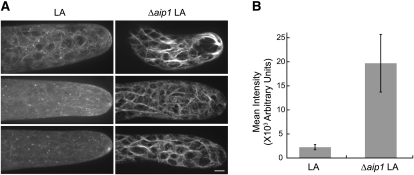
AIP1 Knockout Plants Accumulate Actin Bundles
AIP1 knockdown was reported to induce actin bundle formation in Arabidopsis (Ketelaar et al., 2004). To test whether bundling occurs in Δaip1, we used spinning disc confocal microscopy to image live cells expressing LA. Wild-type cells expressing LA had fine actin filaments throughout the cell cortex, whereas Δaip1 plants had highly bundled cortical actin arrays (Figure 6A). To quantify the amount of bundling, we measured three perpendicular line scans at ~20, 40, and 60 μm from the cell tip. The peaks in the line scan represent actin filaments and/or bundles. Integration of the area under the peak corresponds to the intensity of the actin structure. With this analysis, we observed that Δaip1 cells have actin filament structures that are on average 8.7 times brighter than those of wild-type cells (Figure 6B), demonstrating that Δaip1 cells have highly bundled actin structures.
Figure 6.
Actin Is Highly Bundled in AIP1 Knockout Cells.
(A) Three representative images of LA obtained with spinning disc confocal microscopy from LA wild-type (LA) and LA Δaip1 plant (Δaip1) cells. Note the enhanced bundling of actin filaments in AIP1 knockout cells. Images are maximal projections of 0.5-μm sections spanning the cortex to the medial plane. All images were acquired using the same imaging settings. Bar = 5 μm.
(B) Quantification of the extent of bundling is shown for control (LA) and Δaip1 lines. The amount of bundling was measured by taking three perpendicular line scans at ~20, 40, and 60 μm from the cell tip. The peaks in the line scan represent actin filaments/bundles. Integration of the area under the peak corresponds to the fluorescence intensity of the labeled actin structure. The number of images analyzed is as follows: LA, 8; Δaip1 8. Error bars represent se.
Δaip1 cells containing LA were generated in the NLS4 background. To ensure that the nuclearly localized GFP:GUS fusion does not affect actin filament structure, we also imaged NLS4 cells expressing LA (NLS4 LA). We found that the actin bundling state is indistinguishable from that in wild-type cells (see Supplemental Figure 4 online). Furthermore, we stained actin with fluorescent phalloidin in wild-type and Δaip1 cells and confirmed that Δaip1 cells have highly bundled actin filaments (see Supplemental Figure 5 online). Together, these data suggest that F-actin is stabilized into bundles in the absence of AIP1.
AIP1 and ADF Drive Rapid Actin Dynamics in Vivo
Since AIP1 and ADF work together in vitro to enhance actin filament disassembly, they have been widely regarded to promote the turnover of F-actin in vivo. Interestingly, the contribution of these proteins to F-actin dynamics in live cells has not been directly tested.
To examine F-actin behavior, we imaged cells expressing LA at the cell cortex with spinning disc confocal microscopy or variable angle epifluorescence microscopy (VAEM). We chose the cortical plane for imaging because it contains a population of rapidly rearranging F-actin (Vidali et al., 2009a). For the transient RNAi experiments, there are a limited number of ADF-RNAi plants for each transformation. Furthermore, few RNAi plants are close enough to the cover slip to enable imaging with VAEM. Therefore, spinning disc confocal microscopy was used for ADF-RNAi and GUS-RNAi samples.
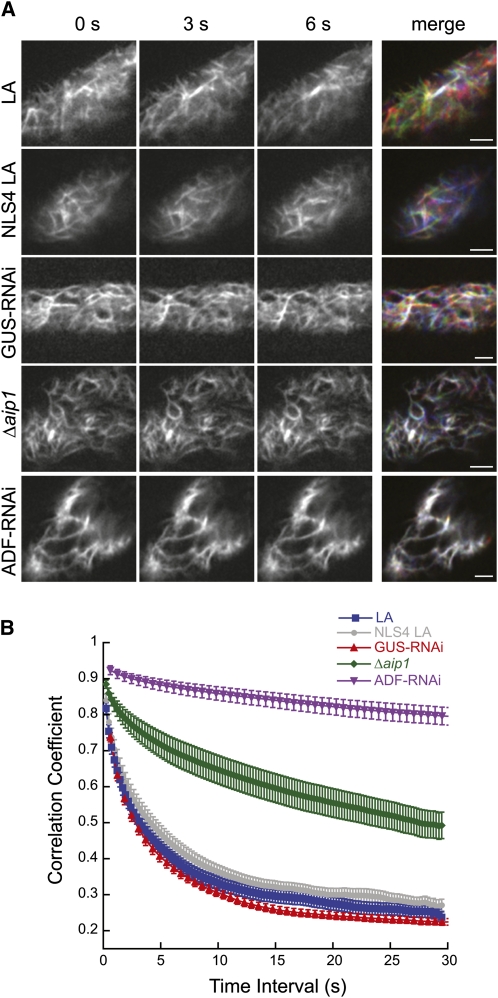
By merging three time points from a time-lapse series, it is possible to visualize changes in cortical F-actin organization. Alterations in F-actin due to polymerization, depolymerization, and/or translocation at any of the time points will appear colored in the merge, whereas F-actin that is static throughout the time course appears white in the merge. In the control lines (NLS4 LA and LA), with or without transformation of GUS-RNAi, we observed robust F-actin dynamics (Figure 7A). By contrast, the F-actin in the NLS4 LA Δaip1 line was more static, and in the NLS4 LA line transformed with ADF-RNAi, the F-actin was almost completely stationary (Figure 7A, stills from Supplemental Movies 1 to 5 online).
Figure 7.
AIP1 and ADF Promote in Vivo Actin Dynamics.
(A) Actin dynamics at the cortex of protonemal cells were visualized by time-lapse spinning disc confocal microscopy (GUS-RNAi and ADF-RNAi) or VAEM (LA, NLS4 LA, and Δaip1) using the actin probe LA. Representative images of actin are shown as grayscale images representing red, green, and blue for the 0-, 3-, and 6-s time points, respectively. The merge combines all time points as separate color channels projected onto one RGB image. White indicates overlap of actin in all time points; color indicates that actin has changed in at least one of the three time points. All images were equivalently adjusted through background subtraction, enhanced contrast, and smoothing in ImageJ. Bars = 2.5 μm.
(B) The correlation coefficient between images was calculated at all temporal spacings (time interval). Low correlation values correspond with higher actin dynamics. The number of plants analyzed is as follows: LA, 18; NLS4 LA, 13; NLS4 LA Δaip1, 24; NLS4 LA GUS-RNAi, 18; NLS4 LA ADF-RNAi, 18. Error bars represent se.
We quantified the changes in F-actin organization in these time series using a previously described algorithm (Vidali et al., 2010) that calculates the correlation of the intensity of the LA signal at all pixel locations between time points. Dynamic F-actin has a low correlation coefficient with subsequent time points. In this analysis, we measured the correlation coefficient over all time intervals (change in time between analyzed frames). The correlation coefficient of F-actin that rapidly rearranges will decay more quickly over the analyzed time intervals compared with a population of static F-actin.
Using this analysis, we found that the control lines (NLS4 LA, LA, and GUS-RNAi) had the lowest correlation coefficient values (Figure 7B), corresponding to rapid F-actin rearrangements. The NLS4 LA line transformed with ADF-RNAi had the highest correlation coefficient, and these values were similar to those observed when treating control plants with the F-actin stabilizing drug jasplakinolide (Vidali et al., 2010). The NLS4 LA Δaip1 line showed intermediate correlation coefficient values with respect to the control and the ADF-RNAi transformed plants. These data demonstrate that, in vivo, AIP1 is important and ADF is absolutely critical for actin dynamics.
The correlation analysis provides a global view of changes in actin organization over time, including growth, shrinkage, and translocation events. To provide insights into the specific changes that affect a reduction in actin turnover, we manually inspected time-lapse image series of the cell cortex acquired with VAEM. F-actin remodeling at the cell cortex in moss is extremely rapid and requires continuous imaging with 30- to 60-ms exposure times to enable tracking of individual filaments. Since we observed little to no changes in the time-lapse acquisitions of ADF-RNAi cells, we focused on analyzing cortical F-actin filament events in the wild type and Δaip1. The correlation (Figure 7B) and bundling (see Supplemental Figure 4 online) analyses showed that LA lines in the wild-type and NLS4 backgrounds were indistinguishable from one another, demonstrating that the nuclearly localized GFP:GUS fusion does not affect F-actin organization or dynamics.
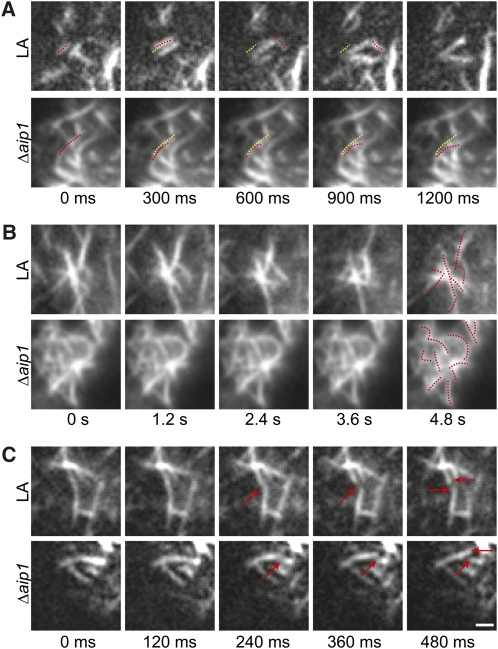
We found that cortical F-actin remodeling is dominated by filament translocations (Figure 8A, stills from Supplemental Movies 6 and 7 online) within the cortical plane as well as into the imaging plane from the cytoplasm. We also observed clusters of filaments, often with a bundled filament in the center, from which many rearrangements were observed (Figure 8B, stills from Supplemental Movies 8 and 9 online). Both translocations and rearrangements around filament clusters occurred faster in control compared with Δaip1 cells. At lower frequencies, we observed elongation, shrinkage, and severing of single filaments. Examples of elongation and shrinkage are shown in Supplemental Movie 8 online. An example of severing is shown in Figure 8C, which show stills from Supplemental Movies 10 and 11 online. Rates of elongation and shrinkage were similar between control and Δaip1 cells (Table 2). Although severing events were uncommon, we found that control cells exhibited 3.52 times more severing events than Δaip1 cells for a given cortical area and time (Table 2). The decrease in severing frequency was also reflected in the number of filament ends undergoing either elongation or shrinkage. Control cells also had 3.58 times more dynamic filament ends compared with Δaip1 cells (Table 2). These data demonstrate that in the absence of AIP1, the cortical F-actin array experiences fewer severing events, which translates to a decrease in the number of available dynamic ends. Ultimately, this may lead to an increase in bundled filaments, since with reduced severing frequency the lifetime of an individual filament will lengthen, enabling time for bundling proteins to access the filament.
Figure 8.
Cortical F-Actin Remodeling Is Slower in the AIP1 Knockout Than in the Wild Type.
(A) Translocation event. Dotted red line shows position of the filament of interest at each time point. Dotted yellow line shows the position of the filament at 0 s.
(B) Rearrangements near a cluster of filaments. Dotted red lines show the position of the filaments at 0 s in the final time point.
(C) Severing event. Red arrows indicate severed filament and filament ends. Bar = 2.5 μm.
Table 2.
F-Actin Dynamic Parameters
| Dynamic Parameters | LA | NLS4 LA Δaip1 |
| Growth rate (μm/s) | 2.32 ± 1.23 | 2.34 ± 1.94 |
| Shrinkage rate (μm/s) | 2.81 ± 2.19 | 3.68 ± 1.91 |
| Severing frequency (events/μm2 s) | 0.00325* ± 0.00155 | 0.000922* ± 0.000609 |
| Number of dynamic ends (ends/μm2 s) | 0.00419** ± 0.00131 | 0.00117** ± 0.000844 |
The cortical region of five cells was imaged with VAEM. Images were analyzed for growth, shrinkage, and severing events. Number of growth events is as follows: LA, 30; Δaip1, 10. Number of shrinkage events is as follows: LA, 16; Δaip1, 5. Standard deviation is reported after the ± sign. The single and double asterisks indicate that the values are significantly different from the wild type with a P value of 0.025 and 0.0036, respectively. Alpha significance level is set at 0.05.
DISCUSSION
Here, we show that AIP1 is required for optimizing expansion of the tip-growing protonemal and rhizoid cells in the moss P. patens. Knockout of the single conserved AIP1 gene results in stunted plants with smaller cells. These plants are viable with no apparent defects in the formation of gametophores. The AIP1 knockout phenotype is most reminiscent of Arp2/3 complex mutants in moss, which are also viable and have reduced tip growth. However, the Arp2/3 complex mutants display more penetrant defects with complete inhibition in the formation of caulonemal cells and rhizoids and mild to severe defects in gametophore development (Harries et al., 2005; Perroud and Quatrano, 2006; Finka et al., 2008). It is interesting that AIP1 (a depolymerizer with ADF) and the Arp2/3 complex (a nucleator) share a similar phenotype. This suggests that Arp2/3 may also be involved in promoting rapid reorganization of F-actin arrays in plants.
With such critical roles in tip growth and plant development, it is intriguing that both ADF (Augustine et al., 2008) and AIP1 are single-copy genes in moss. Other essential proteins critically required for tip growth, such as profilin (Vidali et al., 2007), class II formin (Vidali et al., 2009c), and myosin XI (Vidali et al., 2010), are present in small gene families. Since moss is predominately haploid, having multiple redundant copies of essential genes may be a safeguard in the event of mutation of one of the gene copies or a way to ensure sufficient expression levels. In either case, it is likely that a duplication event occurred in the moss genome (Rensing et al., 2008). Why AIP1 and ADF have remained as single-copy genes remains mysterious. One possible explanation is that genome duplication was incomplete and excluded the ADF and AIP1 genes. Alternatively, gene duplication of ADF and AIP1 resulted in overexpression that was detrimental and was quickly selected against. In support of this, overexpression of ADF and AIP1 has been shown to result in the inhibition of tip growth (Dong et al., 2001; Chen et al., 2002; Ketelaar et al., 2007), further emphasizing that a fine balance of monomeric and F-actin is required to efficiently promote actin-dependent processes such as tip growth.
AIP1 and ADF are critical for tip growth in moss. By comparing the loss-of-function phenotype in moss versus Arabidopsis, it appears that over the course of land plant evolution, AIP1 has become essential for plant growth and development. Strong knockdown of AIP1 results in lethality in Arabidopsis, whereas modest knockdown results in expansion defects in tip-growing and diffusely expanding cells, but not guard cells or trichomes (Ketelaar et al., 2004). In moss, however, AIP1 nulls are viable. The differences in phenotype between Arabidopsis and moss could be accounted for by the fact that ADF is upregulated in moss Δaip1 lines, and higher levels of ADF are sufficient to partially suppress the AIP1 phenotype. Additionally, since AIP1 functions with ADF, the pleiotropic effects observed in Arabidopsis may result from isovariant-specific interactions between the nine vegetatively expressed ADFs (Ruzicka et al., 2007) and the vegetative AIP1-2 (Allwood et al., 2002). By contrast, AIP1 and ADF are single-copy genes in moss, precluding the need to dissect isovariant-specific interactions (Mohri and Ono, 2003). Furthermore, the expansion of both AIP1 and ADF genes over the course of land plant evolution could account for phenotypic differences between AIP1 knockouts in moss compared with Arabidopsis AIP1 RNAi plants. Nevertheless, these studies in moss and Arabidopsis show that AIP1 provides a critical function for growth and development in land plants.
Numerous biochemical and genetic studies demonstrate that AIP1 interacts with ADF (Iida and Yahara, 1999; Okada et al., 1999; Rodal et al., 1999; Allwood et al., 2002), and we provide in vivo evidence that the cooperation of ADF and AIP1 activities is critical for tip growth in moss. We showed that ADF overexpression partially suppresses the AIP1 knockout phenotype. However, AIP1 overexpression cannot overcome loss of ADF. These results demonstrate that ADF and AIP1 coordinate their activities to promote tip growth in moss. Specifically, AIP1 and ADF function in a common pathway where AIP1 requires ADF for its function. This is strongly supported by biochemical data showing that AIP1 alone has minimal effects on actin filaments; however, in combination with ADF it synergistically enhances actin filament disassembly (Aizawa et al., 1999; Okada et al., 1999; Rodal et al., 1999). We also show AIP1-mCherry is diffusely cytosolic, similar to ADF (Augustine et al., 2008). Thus, it is feasible that AIP1 and ADF could interact with one another in vivo. We did not observe AIP1 associated with actin filaments, which is consistent with observations from Narcissus pseudonarcissus where AIP1 decorated actin bundles in dormant pollen grains but became diffusely cytosolic upon germination and pollen tube expansion (Allwood et al., 2002). These results suggest that AIP1 may interact transiently with dynamic F-actin networks.
Although ADF provided a partial rescue of the AIP1 loss-of-function phenotype, overexpression of ADF never fully restored plant morphology in the absence of AIP1. This could be because overexpression of ADF results in suppression of tip growth. Alternatively, it suggests that AIP1 possesses an important and distinct activity from ADF.
Importantly, we show that AIP1 promotes and ADF is absolutely required for F-actin dynamics in plants. Recent characterizations of the cortical actin cytoskeleton in plants revealed that filament severing is the predominant means of disassembling the actin network (Staiger et al., 2009; Smertenko et al., 2010). Therefore, two conserved families of actin filament–severing proteins, ADF/cofilin and villin/gelsolin, were hypothesized to be likely candidates for mediating actin turnover (Staiger et al., 2009). Our loss-of-function study strongly supports a role for both AIP1 and ADF in promoting the rapid reorganization of cortical F-actin in plants. In particular, we found that AIP1 and ADF are critical factors that mediate F-actin translocation, the dominant form of F-actin dynamics in the P. patens cortical array. Since bundled actin filaments are prevalent in both ADF-RNAi (Augustine et al., 2008) and Δaip1 plants, these higher-order actin structures might be a major factor contributing to the inhibition of filament translocation. We also found that AIP1 promotes F-actin severing in vivo. Combined with our genetic data, these findings support a mechanism by which AIP1 enhances actin severing by ADF. By contrast, we found that AIP1 loss of function had no effect on F-actin growth or shrinkage, suggesting that AIP1 is not required for modulating the actin monomer pool. This further supports that AIP1 and ADF regulate actin severing to drive actin dynamics in the cortical F-actin array.
Our results demonstrate that rapid actin cytoskeletal remodeling is essential for tip growth. ADF silencing results in a static cortical F-actin array and a total arrest of tip growth, whereas a reduction in F-actin reorganization in the AIP1 knockout plants leads to a concomitant decrease in the expansion of tip-growing cells. Since the AIP1 knockout lines remain viable despite having impaired F-actin dynamics, these lines will serve as excellent tools for identifying cellular processes in plants that depend on fast F-actin reorganization.
METHODS
Plasmid Construction
Total RNA was isolated from Physcomitrella patens (Gransden isolate) protonemal tissue using the RNeasy plant mini kit (Qiagen), followed by DNase I treatment, according to the manufacturer’s recommendations. Synthesis of cDNA was performed using oligo(dT) primers and Superscript II reverse transcriptase (Invitrogen), according to the manufacturer’s protocol. The predicted AIP1 coding region was amplified with primers (see Supplemental Table 2 online) and cloned into pENT/D-TOPO (Invitrogen) to generate pENT-AIP1. All entry clones in this study were verified by sequencing. To identify the intron-exon borders, the pENT-AIP1 sequence was compared with the genomic sequence (Rensing et al., 2008).
Three-fragment multisite Gateway recombination (Invitrogen) was used to generate the AIP1 knockout construct. To amplify the AIP1 5′ and AIP1 3′ regions, wild-type genomic DNA was used as a template for PCR amplification with primers (see Supplemental Table 2 online). The resistance cassette was generated by amplifying a fragment containing the NOS terminator and hygromycin resistance cassette from pTHUBIGate (Vidali et al., 2007). All primers (see Supplemental Table 2 online) contained the appropriate attB sites, and BP clonase reactions were performed to introduce the PCR products into their respective pDONR vectors to generate the entry clones L1L4-SwaI-AIP-5′, R4R3-NOSter-Lox-Hygro-Lox, and L3L2-AIP-3′-SwaI. The entry clones were recombined into pGEM-gate (Vidali et al., 2009c) using an LR clonase reaction. SwaI sites were engineered at the 5′ and 3′ ends of the knockout construct to linearize the knockout construct before transformation into moss.
The AIP1-mCherry replacement construct was cloned using four-fragment multisite Gateway recombination. The 3′ end of AIP1 spanning the last two exons and intervening intron, but excluding the stop codon was amplified from wild-type genomic DNA using primers (see Supplemental Table 2 online). We amplified mCherry from pRTL2-mCherry (Lee et al., 2008; ABRC, Ohio State University; stock CD3-1062). BP clonase reactions introduced the PCR products into their respective pDONR vectors to generate the entry clones: L1R5-SwaI-AIP-Up-for-3′tagging and L5L4-mCherry-cTer. These clones together with R4R3-NOSter-Lox-Hygro-Lox and L3L2-AIP-3′-SwaI were recombined in an LR clonase reaction with the pGEM-gate destination vector to generate the AIP1-mCherry replacement construct. SwaI restriction sites at the 5′ and 3′ ends of the allele replacement construct were used to linearize the replacement cassette prior to moss transformations.
To generate expression constructs, LR clonase reactions (Invitrogen) recombined the pENT-AIP1 and pENT-ADF (Augustine et al., 2008) clones into the pTH-UbiGate (Vidali et al., 2007) and pMK-UbiGate destination vectors. pMK-UbiGate was generated by modifying pMBL5 (Bezanilla et al., 2003) to include the maize (Zea mays) ubiquitin promoter, Gateway cassette, and NOS terminator.
To generate the AIP1-RNAi construct, the first exon of AIP1 was amplified from genomic DNA and cloned into pENT/D-TOPO. The resulting entry vector was transferred into the destination vector pUGGi (Bezanilla et al., 2005) by an LR clonase reaction to generate AIP1-RNAi. The ADF-RNAi construct is ADF-UTRi (Augustine et al., 2008).
Growth Media
PpNH4 medium [1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.72 mM (PPNH4)2-tartrate, 45 μM FeSO4, 9.93 μM H3BO4, 220 nM CuSO4, 1.966 μM MnCl2, 231 nM CoCl2, 191 ZnSO4, 169 nM KI, and 103 nM Na2MoO4] supplemented with 0.7% agar was used to make growth plates for plant propagation. PpNH4 medium plus 0.7% agar and antibiotics constituted selection plates. Protoplasts were regenerated on plates containing PRMB medium (PpNH4 medium supplemented with 6% mannitol, 10 mM CaCl2, and 0.8% agar). Before plating, protoplasts were resuspended in either 1 mL top agar (PpNH4 medium supplemented with 6% mannitol, 10 mM CaCl2, and 0.3% agar) or 0.5 mL liquid plating medium (PpNH4 medium supplemented with 8.5% mannitol and 10 mM CaCl2). All growth plates were overlaid with cellophane unless otherwise noted.
Isolating Stable Lines and Genotyping
Restriction enzyme–digested AIP1 knockout and AIP1-mCherry replacement constructs were ethanol precipitated, dissolved in TE buffer (10 mM Tris, pH 8, and 1 mM EDTA), and transformed into various stable moss lines. All transformations were performed as previously described (Vidali et al., 2007), with the following modification: protoplasts were transformed at a concentration of 2.0 × 106 protoplasts/mL. Transformations for stable lines were plated in top agar. Plants were regenerated on PRMB plates for 4 d, and transformants were selected by transferring plants to PpNH4 medium supplemented with hygromycin (15 μg/mL). To select for stable integrants, transformants were cycled on and off antibiotic plates for three 1-week intervals. Potential stable integrants were grown on PpNH4 until enough tissue could be acquired for genomic DNA isolation. Plant tissue was transferred to a microcentrifuge tube containing ~15 1.0- to 2.0-mm diameter ZrSiO beads (Next Advance) and 200 μL water. Plants were homogenized in a Bullet Blender (Next Advance) on high speed for 3 min and then mixed with 200 μL of 2× DNA extraction buffer (0.4 M Tris, pH 9.0, 0.8 M LiCl, 50 mM EDTA, and 2% SDS). The homogenate was moved to a new tube, and cell debris was pelleted using a tabletop centrifuge. The supernatant was moved to a new tube, and an equal volume of isopropanol was added to precipitate the DNA. Samples were spun, dried, and resuspended in TE buffer. PCR amplification was performed with Phusion DNA polymerase (Finnzymes) following the manufacturer’s recommendations.
Growth Assays and Cell Wall and Nuclear Staining
Protoplasts were isolated from stable lines as previously described (Vidali et al., 2007). For each line, 20,000 protoplasts were transferred to a microcentrifuge tube. After the protoplasts settled, all but 50 to 100 μL of the supernatant was removed. Plants were resuspended in liquid plating media and plated on PRMB media for 4 d and then transferred to PpNH4 plates for 1, 3, 4, or 17 d. Plants grown for 7 d were imaged with a ×1 lens at ×63 zoom on a fluorescence stereomicroscope (Leica MZ16FA) equipped with a color camera (Leica DF300FX) using the GFP2 filter set (Leica). Bright-field images of 3-week-old plants were acquired with a ×1 lens at ×7.11 zoom on the stereomicroscope (Leica MZ16FA) equipped with a color camera (Leica DF300FX). Alternatively, these plants were transferred to a 12-well plate containing PpNH4 medium without cellophane and grown for an additional 9 weeks. Gametophores from 12-week-old plants were carefully removed from the agar with forceps and laid on their side. Bright-field images were acquired with a ×1 lens at ×7.11 zoom.
Calcofluor (fluorescent brightener 28; Sigma-Aldrich) and DAPI staining was used to determine apical cell length, cell type, and number of cells per plant. To collect plants for staining, 15 mL of water was added to plates containing 5- or 8-d-old plants, and gentle agitation was used to detach plants from the cellophane. Plants were carefully transferred to a microcentrifuge tube by pipetting with a cut pipette tip. After the plants settled in the microcentrifuge tube, all but 50 μL of supernatant was removed. Plants were resuspended in 500 μL of stain solution (100 mM PIPES, pH 6.8, 10 μg/mL calcofluor, 0.2 μg/mL DAPI, 0.1% Nonidet P-40, and 2% paraformaldehyde) and kept in the dark for at least 30 min. Plants plus stain solution were carefully pipetted onto a microscope slide using a cut pipette tip. The slide was covered with a cover slip and sealed with a 1:1:1 mixture of Vaseline, lanolin, and paraffin. Pictures were captured as 24-bit red-green-blue (RGB) images with a ×5 lens at ×30 zoom on the fluorescence stereomicroscope (Leica MZ16FA) equipped with a color camera (Leica DF300FX) and the UV filter set (Leica). The apical cell length of 8-d-old plants was measured in ImageJ using the segmented line tool to manually trace the center of the cell from the cell plate to the apex. The number of cells per plant and number of chloronema/caulonema were counted from images of 5- and 8-d-old plants, respectively.
Caulonemata Dark Growth Assay
To assess caulonemal cell formation, plants from tissue that had been homogenized in water 7 d earlier were placed together in a line ~1 cm long on a cellophane-overlaid plate containing 2% Suc-supplemented PpNH4 medium. The plate was sealed, placed flat in a light-impenetrable black box, and then wrapped in aluminum foil. The box was turned so that the plates were upright with the plants located near the bottom of the plate. After 18 d in the dark, the plates were imaged in bright field with a ×1 lens at ×7.11 zoom on a stereomicroscope (Leica MZ16FA) equipped with a color camera (Leica DF300FX).
RNA Expression Analysis
To ensure that the AIP1 knockout plants do not express AIP1, RNA was isolated from knockout lines 1 week after homogenization. cDNA was prepared from 1 μg of RNA that was quantified using a Nanodrop 1000 (Thermoscientific). Primers for RT-PCR analysis are listed in Supplemental Table 2 online. Amplification of Ubiquitin10 was used as a loading control.
Imaging
Imaging of AIP1-mCherry localization was performed in 35-mm Petri plates with a glass cover slip glued to the bottom. A 1-mL pipette tip (Rainin) placed in the center of the plate over the cover slip was surrounded with 7 to 10 mL of PpNH4 medium. When the medium set, the pipette tip was removed, generating a well that was subsequently filled with 65 to 75 μL of PpNH4 medium, forming a chamber. Small pieces of sample tissue were placed into the middle of the chamber well. Plates were moved into a growth chamber until the plants grew down to the cover slip. Plants were imaged with a Nikon Ti Eclipse inverted microscope equipped with a Yokogawa CSU-X1 spinning disk head and a 512 × 512 Andor iXON electron multiplying charged coupled device (CCD) camera. Plants were imaged with a 488-nm laser, 525/50-nm band-pass filter, and a ×100 oil immersion lens (1.4 numerical aperture; Nikon).
Imaging of cortical F-actin with VAEM and spinning disc confocal microscopy was performed on 1% agar pads as previously described (Vidali et al., 2010). GUS-RNAi and ADF-RNAi plants were transferred to hygromycin-supplemented PpNO3 medium (Vidali et al., 2009a) on day 4 and transferred to agar pads and imaged on days 5 to 6 instead of day 7 to avoid lethality that becomes prevalent with increasing time in ADF-RNAi plants. For all other lines, plants were transferred to agar pads. To image cortical F-actin for quantification of actin dynamics, images of LA were acquired at a single cortical plane. VAEM imaging was performed on a Nikon Ti Eclipse inverted microscope equipped with total internal reflection fluorescence illumination and imaged with a ×60 oil immersion objective (numerical aperture 1.45; Nikon). The ×1.5 optivar was used for all images to increase magnification. Movies were manually inspected and scored for severing, growth, and shrinkage events. Growth and shrinkage events are considered dynamic ends. Change in filament length for calculating growth and shrinkage rates was determined using the straight or segmented line tools in ImageJ. Severing frequency was calculated by the total number of severing events observed in a particular time-lapse series divided by the VAEM imaging area and the total time of the time-lapse series. Similarly, the number of dynamic ends was calculated by dividing the total number of elongating and shortening filaments by the VAEM imaging area and the total time of the time-lapse series. Spinning disc confocal microscopy was performed on a Nikon Ti Eclipse inverted microscope equipped with a Yokogawa CSU-X1 spinning disk head and a 512 × 512 Andor iXON electron multiplying CCD camera. Plants were imaged with a 488-nm laser, 525/50-nm band-pass filter, and a ×100 oil immersion lens (1.4 numerical aperture; Nikon). The ×1.5 optivar was used for all images to increase magnification. Quantification of actin dynamics from LA movies was performed as previously described (Vidali et al., 2010).
To investigate bundling and F-actin organization throughout the cell, we prepared LA, NLS4 LA, and NLS4 LA Δaip1 samples using the 1% agar pad method (Vidali et al., 2010). Optical sections were taken at 0.5-μm intervals from the cortex to the medial plane with a Nikon Ti Eclipse inverted microscope equipped with a Yokogawa CSU-X1 spinning disk head and a 512 × 512 Andor iXON electron multiplying CCD camera. Plants were imaged with a 488-nm laser, 525/50 nm band-pass filter, and a ×100 oil immersion lens (1.4 numerical aperture; Nikon).
Phalloidin staining and imaging were performed as previously described (Vidali et al., 2007).
To quantify the amount of F-actin bundling present in a cell, the total intensity of individual bundles was calculated as follows: Z-stacks acquired with the spinning disk confocal microscope from the cortical section to the medial plane at 0.5-μm intervals were maximally projected. Using ImageJ, the maximal projection was background subtracted (rolling ball radius: 20 pixels). Three line scans perpendicular to the axis of the cell at ~20, 40, and 60 μm from the cell apex were collected. The profile of each scan was imported to the program OriginPro 8.1 for analysis with the peak analyzer function (default parameters). Each individual peak was automatically identified and the area under each peak integrated. The peaks identified in this way correspond to bundles in the cell, and the integrated area under each peak corresponds to the intensity of the corresponding bundle. The integrated values for all the peaks from all three measurements for each cell were averaged. For comparison, independent values obtained from different cells were used.
Protein Extraction and Immunoblotting
For AIP1-mCherry immunoblot, moss protonemal tissue was dried on a paper towel and rapidly frozen in a microcentrifuge tube in liquid nitrogen. Frozen tissue was ground in the microcentrifuge tube using a plastic pestle and then vortexed in grinding buffer (100 mM sodium phosphate, pH 7.0, 10 mM DTT, 20% glycerol, 5 mM EDTA, 1 mM PMSF, and 20 μg/mL leupeptin). For analysis of ADF levels in wild-type and Δaip1 lines, moss protonemal tissue was ground in liquid nitrogen in a mortar with a pestle and then vortexed in extraction buffer (50 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 2 mM PMSF, 20 μg/mL leupeptin, 10 mM EDTA, and 2 mM DTT). All samples were centrifuged at 4°C on a benchtop centrifuge, quantified using a Bradford assay (Bio-Rad), and separated by SDS-PAGE. The protein separation was transferred to a nitrocellulose membrane and immunoblotted for AIP1-mCherry using rabbit anti-dsRed polyclonal antibody (Clontech) or for ADF using rabbit anti-ADF polyclonal antibody (Augustine et al., 2008) and mouse anti-tubulin monoclonal antibody (Sigma-Aldrich). Chemiluminescence emission from horseradish peroxidase fused to polyclonal goat anti-rabbit secondary antibodies was detected on a gel dock system equipped for chemiluminescence detection (Bio-Rad).
Complementation Analyses
In the wild-type and AIP1 knockout lines, transient rescue was performed by cotransformation of 5 μg of pTH-Ubi-GFP (Vidali et al., 2009a) with 10 μg of pMK-Ubi, pMK-Ubi-AIP1, or pMK-Ubi-ADF. The GFP expression plasmid was used as a marker to identify transformed plants. For RNAi complementation analysis in the NLS4 line, 30 μg of AIP1- or ADF-RNAi plasmid were cotransformed with various concentrations of pTH-Ubi-ADF and pTH-Ubi-AIP1, respectively. Transformations were performed as previously described (Augustine et al., 2008), with the following exception. Protoplasts were transformed at 2.0 × 106 protoplasts/mL, grown for 4 d on PRMB medium, and then transferred to PpNH4 plates supplemented with G418 (30 μg/mL) for complementation in the wild type and Δaip1 or hygromycin (15 μg/mL) for complementation in NLS4 lines. On day 7, plants were imaged with a ×1 objective at ×63 zoom on a fluorescence stereomicroscope (Leica MZ16FA) equipped with a color camera (Leica DF300X) using the GFP2 filter set (Leica).
Morphometric Analyses and Statistics
Plant area and solidity were measured as described previously (Vidali et al., 2009b). Briefly, a 24-bit RGB image of 1-week-old plants was manually cropped, and the red channel corresponding to the chlorophyll autofluorescence was separated. Total plant area and solidity were determined from the images. Solidity is the plant area divided by the convex hull area.
Statistical analyses were performed as previously described (Vidali et al., 2007). Analysis of variance for multiple comparisons was done on Kaleidagraph using Tukey's honestly significant difference post hoc tests. The alpha for statistical significance was set to 0.05.
Accession Numbers
The Arabidopsis AIP1-1 and AIP1-2 sequences were acquired from The Arabidopsis Information Resource with the accession numbers At2g01330 and At3g18060, respectively. The accession number for P. patens AIP1 is JF510465 in GenBank.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genotyping AIP1 Knockout Lines.
Supplemental Figure 2. Genotyping AIP1-mCherry Replacement Lines.
Supplemental Figure 3. ADF Protein Levels in Wild-Type and AIP1 Knockout Lines.
Supplemental Figure 4. Bundling Analysis of NLS4 LA and LA Lines.
Supplemental Figure 5. Fluorescent Phalloidin Staining of F-Actin in Wild Type and AIP1 Knockout Lines.
Supplemental Table 1. Plant AIP1 Protein Sequence Comparisons.
Supplemental Table 2. Primers Used in This Study.
Supplemental Movie 1. Dynamic Behavior of Cortical F-Actin in a Representative LA Cell.
Supplemental Movie 2. Dynamic Behavior of Cortical F-Actin in a Representative NLS4 LA Cell.
Supplemental Movie 3. Dynamic Behavior of Cortical F-Actin in a Representative NLS4 LA GUS-RNAi Cell.
Supplemental Movie 4. Dynamic Behavior of Cortical F-Actin in a Representative NLS4 LA Δaip1 Cell.
Supplemental Movie 5. Dynamic Behavior of Cortical F-Actin in a Representative NLS4 LA ADF-RNAi Cell.
Supplemental Movie 6. Example of Filament Translocation Events in the Wild Type (LA).
Supplemental Movie 7. Example of Filament Translocation Events in Δaip1.
Supplemental Movie 8. Example of Rearrangements of Filaments around Bundled Cluster in the Wild Type (LA).
Supplemental Movie 9. Example of Rearrangements of Filaments around Bundled Cluster in Δaip1.
Supplemental Movie 10. Example of Severing Event in the Wild Type (LA).
Supplemental Movie 11. Example of Severing Event in Δaip1.
Supplemental Movie Legends. Text Descriptions of Supplemental Movies.
Acknowledgments
We thank Peter Hepler and members of the Bezanilla lab for careful reading of the manuscript. We thank Graham Burkart and Paula Franco for technical assistance and Ming Li for assistance with spinning disc confocal microscopy. This work was supported by National Science Foundation Grants MCB-0747231 (M.B.), DGE-065412 (R.C.A.), and IOS-1002837 (L.V.) and the David and Lucille Packard Foundation (M.B.). The spinning disc confocal and VAEM microscopes are university facilities funded by a National Science Foundation Major Research Instrumentation Development award (0923318).
AUTHOR CONTRIBUTIONS
R.C.A. and M.B. designed the research and wrote the article. R.C.A. and K.A.P. performed the research. R.C.A., L.V., and M.B. analyzed the data. E.T. contributed new computational tools.
References
- Aizawa H., Katadae M., Maruya M., Sameshima M., Murakami-Murofushi K., Yahara I. (1999). Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells 4: 311–324 [DOI] [PubMed] [Google Scholar]
- Allwood E.G., Anthony R.G., Smertenko A.P., Reichelt S., Drobak B.K., Doonan J.H., Weeds A.G., Hussey P.J. (2002). Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 14: 2915–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Augustine R.C., Vidali L., Kleinman K.P., Bezanilla M. (2008). Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 54: 863–875 [DOI] [PubMed] [Google Scholar]
- Balcer H.I., Goodman A.L., Rodal A.A., Smith E., Kugler J., Heuser J.E., Goode B.L. (2003). Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13: 2159–2169 [DOI] [PubMed] [Google Scholar]
- Bamburg J.R. (1999). Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15: 185–230 [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Pan A., Quatrano R.S. (2003). RNA interference in the moss Physcomitrella patens. Plant Physiol. 133: 470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Perroud P.F., Pan A., Klueh P., Quatrano R.S. (2005). An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol. (Stuttg.) 7: 251–257 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. (2010). Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Chan A.Y., Bailly M., Zebda N., Segall J.E., Condeelis J.S. (2000). Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Wong E.I., Vidali L., Estavillo A., Hepler P.K., Wu H.M., Cheung A.Y. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Niroomand S., Zou Y., Wu H.M. (2010). A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc. Natl. Acad. Sci. USA 107: 16390–16395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D.J., Schild A., Ashton N.W., Hartmann E. (1978). Genetic and physiological studies of the effect of light on the development of moss, Physcomitrella patens. Photochem. Photobiol. 27: 249–254 [Google Scholar]
- Dong C.H., Xia G.X., Hong Y., Ramachandran S., Kost B., Chua N.H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Era A., Tominaga M., Ebine K., Awai C., Saito C., Ishizaki K., Yamato K.T., Kohchi T., Nakano A., Ueda T. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka A., Saidi Y., Goloubinoff P., Neuhaus J.M., Zrÿd J.P., Schaefer D.G. (2008). The knock-out of ARP3a gene affects F-actin cytoskeleton organization altering cellular tip growth, morphology and development in moss Physcomitrella patens. Cell Motil. Cytoskeleton 65: 769–784 [DOI] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304: 743–746 [DOI] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P.A., Pan A., Quatrano R.S. (2005). Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M.K., Lappalainen P. (2005). Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell 16: 649–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K., Yahara I. (1999). Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells 4: 21–32 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Hussey P.J. (2007). Actin organization and root hair development are disrupted by ethanol-induced overexpression of Arabidopsis actin interacting protein 1 (AIP1). New Phytol. 174: 57–62 [DOI] [PubMed] [Google Scholar]
- Konzok A., Weber I., Simmeth E., Hacker U., Maniak M., Müller-Taubenberger A. (1999). DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol. 146: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D.G. (1997). Cofilin promotes rapid actin filament turnover in vivo. Nature 388: 78–82 [DOI] [PubMed] [Google Scholar]
- Lee L.Y., Fang M.J., Kuang L.Y., Gelvin S.B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel T.P., Boujemaa R., Pantaloni D., Carlier M.F. (1999). Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Kunkel J.G., Allwood E.G., Hussey P.J., Hepler P.K. (2006). Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. Plant Cell 18: 2182–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver S.K., Hussey P.J. (2002). The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 3: reviews3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver S.K., Zot H.G., Pollard T.D. (1991). Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 115: 1611–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Calder G., Dolan L. (2007). Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J. Exp. Bot. 58: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C.J., Martiel J.L., Blanchoin L. (2007). Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17: 825–833 [DOI] [PubMed] [Google Scholar]
- Miller D.D., de Ruijter N.C.A., Bisseling T., Emons A.M.C. (1999). The role of actin in root hair morphogenesis: Studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 17: 141–154 [Google Scholar]
- Mohri K., Ono S. (2003). Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Mohri K., Vorobiev S., Fedorov A.A., Almo S.C., Ono S. (2004). Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279: 31697–31707 [DOI] [PubMed] [Google Scholar]
- Okada K., Blanchoin L., Abe H., Chen H., Pollard T.D., Bamburg J.R. (2002). Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 277: 43011–43016 [DOI] [PubMed] [Google Scholar]
- Okada K., Obinata T., Abe H. (1999). XAIP1: A Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 112: 1553–1565 [DOI] [PubMed] [Google Scholar]
- Okada K., Ravi H., Smith E.M., Goode B.L. (2006). Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 17: 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. (2010). Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 188: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152: 1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. (2003). Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: New blades for twisted filaments. Biochemistry 42: 13363–13370 [DOI] [PubMed] [Google Scholar]
- Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. (2007). Actin filament severing by cofilin. J. Mol. Biol. 365: 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud P.F., Quatrano R.S. (2006). The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil. Cytoskeleton 63: 162–171 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Christensen H.E., Ishimaru Y., Dong C.H., Chao-Ming W., Cleary A.L., Chua N.H. (2000). Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124: 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N., Charlton J., Adler P.N. (2007). The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-actin disassembly in pupal epidermal cells. Genetics 176: 2223–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rodal A.A., Tetreault J.W., Lappalainen P., Drubin D.G., Amberg D.C. (1999). Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J., Agnew B.J., Abe H., Bamburg J.R., Mitchison T.J. (1997). Xenopus actin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeria monocytogenes tails. J. Cell Biol. 136: 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka D.R., Kandasamy M.K., McKinney E.C., Burgos-Rivera B., Meagher R.B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52: 460–472 [DOI] [PubMed] [Google Scholar]
- Schaefer D.G., Zrÿd J.P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Deeks M.J., Hussey P.J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123: 3019–3028 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Fay S.N., Franco P., Pattavina K.A., Bezanilla M. (2009b). Rapid screening for temperature-sensitive alleles in plants. Plant Physiol. 151: 506–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Kleinman K.P., Bezanilla M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tüzel E., Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., McKenna S.T., Hepler P.K. (2001). Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12: 2534–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009a). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., van Gisbergen P.A., Guérin C., Franco P., Li M., Burkart G.M., Augustine R.C., Blanchoin L., Bezanilla M. (2009c). Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Yan J., Zhang R., Qu X., Ren S., Chen N., Huang S. (2010). Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22: 3745–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiao Y., Du F., Cao L., Dong H., Ren H. (2011). Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytol. 190: 667–682 [DOI] [PubMed] [Google Scholar]