Actin filaments in the cortical cytoskeleton of epidermal cells undergo rapid turnover that is based on fast polymerization and prolific severing. Here, advanced imaging techniques and reverse-genetic analyses are used to demonstrate that an actin depolymerizing factor (ADF4) contributes to the disassembly of filaments through its stochastic severing activity.
Abstract
Actin filament arrays are constantly remodeled as the needs of cells change as well as during responses to biotic and abiotic stimuli. Previous studies demonstrate that many single actin filaments in the cortical array of living Arabidopsis thaliana epidermal cells undergo stochastic dynamics, a combination of rapid growth balanced by disassembly from prolific severing activity. Filament turnover and dynamics are well understood from in vitro biochemical analyses and simple reconstituted systems. However, the identification in living cells of the molecular players involved in controlling actin dynamics awaits the use of model systems, especially ones where the power of genetics can be combined with imaging of individual actin filaments at high spatial and temporal resolution. Here, we test the hypothesis that actin depolymerizing factor (ADF)/cofilin contributes to stochastic filament severing and facilitates actin turnover. A knockout mutant for Arabidopsis ADF4 has longer hypocotyls and epidermal cells when compared with wild-type seedlings. This correlates with a change in actin filament architecture; cytoskeletal arrays in adf4 cells are significantly more bundled and less dense than in wild-type cells. Several parameters of single actin filament turnover are also altered. Notably, adf4 mutant cells have a 2.5-fold reduced severing frequency as well as significantly increased actin filament lengths and lifetimes. Thus, we provide evidence that ADF4 contributes to the stochastic dynamic turnover of actin filaments in plant cells.
INTRODUCTION
The actin cytoskeleton is a filamentous network that plays a central role in powering a myriad of cellular processes, including the maintenance of cell architecture, cell crawling, and the transport or positioning of organelles (Pollard and Cooper, 2009; Szymanski and Cosgrove, 2009). The actin cytoskeleton undergoes constant rearrangements, as the needs of a cell changes or in response to biotic and abiotic stimuli. The rapid turnover and rearrangements of actin filaments must be regulated in space and time to create a diverse set of actin arrays. Although much has been learned about key regulatory proteins and the assembly of actin filaments in the test tube, a deep understanding of the molecular mechanisms underpinning actin turnover in vivo remains to be fully addressed.
Actin (F-actin) polymerizes at filament ends from a pool of assembly-competent monomers (G-actin). At equilibrium in a test tube and in the absence of regulatory proteins, assembly and disassembly reactions are balanced, leading to a flux of subunits through the polymer in a process known as treadmilling. This turnover process can be enhanced or inhibited by actin binding proteins, including monomer binding proteins, capping proteins, and severing factors. The presence of numerous actin binding proteins in the cytoplasm of cells predicts that actin turnover is precisely choreographed; however, understanding the molecular mechanisms requires imaging cytoskeletal polymers at high temporal and spatial resolution.
Recently, the combination of a minimal set of proteins (a processive formin, profilin, and actin depolymerizing factor [ADF]/cofilin) produced a 155-fold enhancement in the turnover of single actin filaments in vitro and allowed for reconstitution of motility in a simplified system (Michelot et al., 2007; Pavlov et al., 2007; Roland et al., 2008). This turnover by fragmentation was deemed “stochastic dynamics” and demonstrated a clear role for ADF/cofilin in filament disassembly (Michelot et al., 2007; Roland et al., 2008). Moreover, stochastic fragmentation of actin filaments was shown to govern the organization and aging of the dendritic actin filament array in Arp2/3-generated actin comet tails in vitro (Reymann et al., 2011) and was predicted to play a role in yeast actin patch turnover (Berro et al., 2010).
In general, several hypotheses concerning actin filament turnover via ADF/cofilin have been articulated based on observations of filament turnover in vitro as well as from computer-based kinetic simulations. For example, filament disassembly could occur by (1) depolymerization from filament ends (Carlier et al., 1997); (2) turnover by fragmentation of filaments (Andrianantoandro and Pollard, 2006; Chan et al., 2009; Berro et al., 2010; Kueh et al., 2010); or (3) a combination of filament severing and depolymerization, most likely facilitated by the action of other proteins, such as AIP1 (Kueh et al., 2008, 2010; Okreglak and Drubin, 2010). Unfortunately, relatively few direct observations of single actin filament growth and disassembly have been made in vivo (Vavylonis et al., 2008; Staiger et al., 2009; Smertenko et al., 2010); however, it is becoming generally accepted that actin turnover in vivo is dominated by rapid filament elongation and prolific severing, rather than by treadmilling.
The stochastic dynamics of single actin filaments have been observed in the cortical cytoplasm of Arabidopsis thaliana epidermal cells expressing fluorescent actin binding protein reporters (Staiger et al., 2009; Khurana et al., 2010; Smertenko et al., 2010). Two populations of actin filaments, filament bundles and individual filaments, exist in epidermal cells and show remarkably different dynamic properties. Single actin filaments are thin, have lower fluorescence intensity values, and present some challenges in imaging. Nevertheless, using variable-angle epifluorescence microscopy (VAEM), several parameters of their initiation, growth, and turnover have been analyzed quantitatively (Staiger et al., 2009; Smertenko et al., 2010). New actin filaments are synthesized de novo from G-actin subunits in the cytoplasm, from recently severed ends of filaments, and from the side of preexisting filaments or bundles (Staiger et al., 2009; Smertenko et al., 2010). In hypocotyl epidermal cells, these single actin filaments elongate rapidly at rates of 1.7 μm/s and disassemble via prolific severing activity, rather than minus-end shrinkage (Staiger et al., 2009). Although this overall dynamic behavior is conserved in various cell types, the actual rates of filament growth and severing can differ markedly (Smertenko et al., 2010; Wang et al., 2011). Filament turnover by stochastic dynamics is predicted to be regulated by a plethora of actin binding proteins, including ADF/cofilin (Staiger et al., 2009, 2010; Blanchoin et al., 2010; Day et al., 2011). Combining the ability to image single actin filaments in vivo with a large collection of actin binding protein mutants provides an unparalleled opportunity to directly visualize the effect of loss of key regulatory proteins on single actin filament dynamics in living cells. We hypothesize that the severing component of stochastic dynamics is mediated by ADF/cofilin or villin/gelsolin family members in Arabidopsis cells (Blanchoin et al., 2010; Staiger et al., 2010; Day et al., 2011).
In vitro, ADF/cofilins bind to both G- and F-actin with a strong preference for ADP-actin, (Carlier et al., 1997; Blanchoin and Pollard, 1999; Andrianantoandro and Pollard, 2006; Pavlov et al., 2007; Suarez et al., 2011). Historically, ADF/cofilin activity was thought to mediate filament turnover mainly by depolymerization from filament pointed ends (Carlier et al., 1997); however, time-lapse total internal reflection fluorescence microscopy (TIRFM) experiments show conclusively that turnover in vitro occurs predominantly by ADF-mediated severing (Andrianantoandro and Pollard, 2006; Chan et al., 2009; Suarez et al., 2011). The targeted binding of ADF/cofilin to aged regions of an actin filament causes a change in the twist of the filament and lowers the persistence length (McGough et al., 1997; McCullough et al., 2008), which renders the filament more susceptible to breakage events at boundaries between bare and ADF/cofilin-decorated segments (De La Cruz, 2009; Suarez et al., 2011). ADF/cofilin severing activity increases the number of actin filament barbed ends available to capping proteins and stimulates filament disassembly (Andrianantoandro and Pollard, 2006; Reymann et al., 2011).
Plant ADF protein variants show ancient and extreme diversity among their sequences (Ruzicka et al., 2007). Yet, most that have been characterized have the expected biochemical properties in vitro (Gungabissoon et al., 1998, 2001; Ressad et al., 1998; Smertenko et al., 2001; Chen et al., 2004; Schüler et al., 2005; Chaudhry et al., 2007; Khurana et al., 2010). Recombinant Arabidopsis ADF4 is no exception; it binds G-actin, inhibits nucleotide exchange on monomers, and exhibits a 355-fold higher affinity for ADP-loaded actin when compared with ATP-actin (Tian et al., 2009). Similar to other model systems, a direct visualization of the role of ADF-mediated severing in vivo is lacking for plant cells; however, gross rearrangements in actin architecture occur when ADF/cofilin expression levels are altered in Arabidopsis (Dong et al., 2001) and in Physcomitrella patens (Augustine et al., 2008).
Here, we test the hypothesis that plant ADF/cofilin family members contribute to the stochastic dynamic behavior of individual actin filaments in living cells. Using a knockout mutant of Arabidopsis ADF4, we observed that perturbation of the stochastic dynamics of actin filaments leads to marked changes in the cortical actin array. We quantified single actin filament parameters in the adf4 mutant and observed a 2.5-fold to threefold reduction in severing frequency as well as significantly longer filament lengths and lifetimes. Loss of ADF4 also led to quantitatively increased actin filament bundling in cells along the entire length of the hypocotyl. Both phenotypes were rescued by expression of ADF4 behind a constitutive promoter. We present initial evidence for a direct role of severing in modulating the turnover of actin filaments in vivo and provide support for our model of the regulation of stochastic dynamics by specific actin binding proteins.
RESULTS
Multiple ADF Variants Are Expressed in Arabidopsis Hypocotyls
Arabidopsis has a multigene family of ADFs comprising 11 members in four ancient subclasses (Ruzicka et al., 2007). To determine the relative abundance of ADF variant expression in light- and dark-grown hypocotyls, normalized microarray data were obtained from Ma et al. (2005). Subclass I ADFs (ADF1 to ADF4) were the most abundantly expressed variants in hypocotyls, and, with the exception of ADF4, all were expressed more strongly in the dark than in the light (see Supplemental Figure 1A online). Subclass II variants (ADF5 and ADF9) were also modestly expressed in hypocotyls. We chose ADF4 for further analysis due to its reasonable expression level in hypocotyls and because of our previous characterization of a knockout allele for this gene (Tian et al., 2009). To confirm the microarray data, real-time quantitative PCR (qRT-PCR) was performed on 10-d-old light-grown seedlings (see Supplemental Figure 1B online). Plant materials examined included wild-type Columbia-0, the homozygous adf4 mutant, and a rescue line expressing the cDNA of ADF4 driven by the constitutive cauliflower mosaic virus 35S promoter in a mutant background (i.e., 35S:ADF4;adf4). Transcript levels for ADF1 to ADF5 in wild-type seedlings roughly followed the relative expression reported by microarray data, although ADF5 appeared somewhat lower in the wild-type hypocotyls. ADF4 transcript was absent in the homozygous adf4 mutant (see Supplemental Figure 1B online), verifying that this is indeed a knockout (Tian et al., 2009). There was minimal compensation from other highly expressed ADFs in the hypocotyls of adf4 mutant seedlings (see Supplemental Figure 1B online). The 35S:ADF4 construct expressed in the adf4 mutant background resulted in twofold higher ADF4 levels compared with wild-type seedlings (see Supplemental Figure 1B online). The modest level of ADF4 ectopic/overexpression did not lead to any obvious morphological or developmental abnormalities in the rescue plants. This contrasts with a previous report on the overexpression of ADF1 resulting in developmental defects; however, the lines used were in a wild-type genetic background and had a 30- to 50-fold increase in transcript levels (Dong et al., 2001).
Homozygous adf4 Mutant Seedlings Exhibit Several Growth Phenotypes
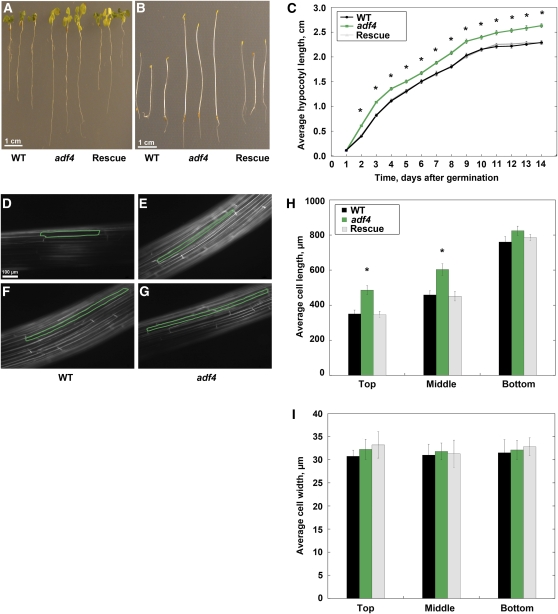
Previously, we observed that adf4 mutant plants were susceptible to the bacterial pathogen Pseudomonas syringae DC3000 expressing the effector protein AvrPphB (Tian et al., 2009). To examine whether the loss of ADF4 resulted in cell and organ growth phenotypes, we grew wild-type, homozygous adf4 mutant, and 35S:ADF4;adf4 rescue seedlings on the same plates under dark- and light-grown conditions. In the light, adf4 seedlings had significantly longer roots compared with the wild type (Figure 1A; see Supplemental Figure 2A online); however, there was no obvious difference in hypocotyl length (Figure 1A).
Figure 1.
Homozygous adf4 Mutant Seedlings Exhibit Several Growth Phenotypes.
(A) Light-grown adf4 seedlings have longer roots compared with controls; however, no hypocotyl phenotype was apparent. Three light-grown seedlings, 10 d after germination, per genotype are shown. Bar = 1 cm. WT, wild type.
(B) Etiolated hypocotyls and roots from homozygous adf4 seedlings are longer than those from wild-type and 35S:ADF4;adf4 (Rescue) lines. Three representative dark-grown seedlings, 10 d after germination, per genotype are shown. Bar = 1 cm.
(C) Etiolated adf4 mutant hypocotyls are significantly longer than wild-type and 35S:ADF4;adf4 rescue seedlings from day 2 onward (denoted by asterisks, P = 0.0001, ANOVA); however, no significant difference in length is present between wild-type and 35S:ADF4;adf4 rescue (P = 0.65 for day 2, t test). Measurements were taken daily from at least 100 seedlings per genotype per day. Values are means ± se.
(D) to (G) Representative images of hypocotyl epidermal cells from wild-type ([D] and [F]) and adf4 mutant ([E] and [G]) seedlings 5 d after germination. Epidermal cells were stained with FM4-64 dye to visualize cell boundaries and imaged with epifluorescence microscopy. Bar = 100 μm.
(D) An axially elongating wild-type epidermal cell located in the top third of the hypocotyl (outlined in green) is shown.
(E) A cell from an adf4 mutant seedling located in the same region as (D) is longer along the axis of expansion than the wild-type cell.
(F) A cell located in the bottom third of the wild-type hypocotyl has nearly completed axial growth.
(G) A cell from an adf4 seedling imaged in the bottom third of the hypocotyl is not different from a wild-type cell imaged in the same region shown in (F).
(H) Epidermal cells from adf4 hypocotyls are significantly longer in the axial dimension than wild-type and 35S:ADF4;adf4 controls (denoted by asterisk, P = 0.0166, ANOVA) during expansion. Knockout adf4 seedlings have significantly longer average epidermal cell lengths in the top (P = 0.0013, ANOVA) and middle (P = 0.0007, ANOVA) thirds of the dark-grown seedling when compared with wild-type and 35S:ADF4;adf4 rescue lines. Cell length values are the mean ± se from n > 100 cells and at least 10 hypocotyls per genotype.
(I) Epidermal cell width is not altered in adf4 mutant hypocotyls. Epidermal cell width values are the mean ± se from n > 100 cells and least 10 hypocotyls per genotype with the wild type shown in black, adf4 shown in green, and 35S:ADF4;adf4 shown in gray.
In the dark, hypocotyls and roots from adf4 seedlings were noticeably longer than for wild-type or rescue line seedlings (Figure 1B). We therefore measured hypocotyl lengths from 2 to 14 d after germination; adf4 mutant hypocotyls were significantly longer than wild-type and 35S:ADF4;adf4 controls (P = 0.0001, analysis of variance [ANOVA]; Figure 1C). This phenotype is consistent with the previous characterization of an Arabidopsis line in which subclass I ADFs were targeted with ADF1 antisense RNA (Dong et al., 2001). Dark-grown roots of adf4 were also significantly longer over a 12-d growth period when compared with wild-type and rescue line roots (see Supplemental Figure 2B online). Because root growth has contributions from both cell division and expansion, the loss of ADF4 could perturb either, or both properties.
Hypocotyl growth occurs predominantly by cell elongation, rather than division, and there is a gradient of axial cell expansion that proceeds in an acropetal direction during development (Gendreau et al., 1997). Therefore, we measured epidermal cell lengths and widths along the long axis of 5-d-old hypocotyls to correlate the organ elongation phenotype with cell expansion. Wild-type epidermal cells located at the top or apical region of the hypocotyl, near the cotyledons, were shorter in the axial dimension (Figure 1D) than cells located at the base of the hypocotyl near the roots (Figure 1F). Homozygous adf4 mutant epidermal cells located near the cotyledons were longer than wild-type cells from the same region (Figure 1E), whereas cells located at the base were not noticeably different from the wild type (Figure 1G). Cell length measurements were binned into three regions (top, middle, and bottom) and plotted as a function of position along the axial gradient of expansion (Figure 1H). Epidermal cells from adf4 mutants were significantly longer than cells from the wild type and 35S:ADF4;adf4 in the top (P value = 0.0013, ANOVA) and middle (P value = 0.0007, ANOVA) regions of the expanding hypocotyl (Figure 1H). However, epidermal cell widths were not altered in adf4 mutant hypocotyls (Figure 1I). Thus, in hypocotyls, the adf4 mutant phenotype comprises longer than normal dark-grown organs. This correlates with premature or faster axial cell expansion in the apical zone of mutant hypocotyls. In contrast with the hypocotyl, epidermal cells from petioles of dark-grown cotyledons were significantly shorter and thinner in the adf4 mutant compared with the wild-type and rescue lines (see Supplemental Figures 3A and B online).
Actin Cytoskeleton Architecture Is Altered in adf4 Epidermal Cells
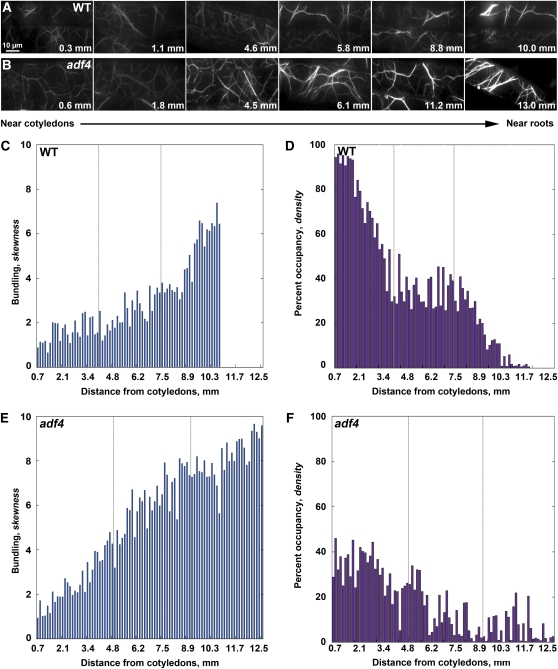
To investigate whether growth phenotypes for the adf4 mutant correlate with an altered actin cytoskeleton, we measured several parameters that relate to cytoskeletal architecture. Snapshots obtained with VAEM (Konopka and Bednarek, 2008) were used to examine the overall arrangement of the actin cytoskeleton in individual epidermal cells from wild-type, adf4 mutant, and rescue line hypocotyls expressing green fluorescent protein (GFP)-fABD2 (Figure 2; see Supplemental Figure 4 online). A montage of micrographs from a representative wild-type seedling showed increased actin filament bundling in the cortical array of dark-grown wild-type cells along the gradient of axial cell expansion (Figure 2A). By contrast, the density or abundance of filaments in the cortical array appeared to decrease as the cells finished expanding near the base of the hypocotyl (Figure 2A). Homozygous adf4 seedlings displayed a similar trend in actin architecture along the gradient; however, the extent of bundling appeared to be more pronounced at the base of the hypocotyl and bundling occurred earlier or closer to the top of the hypocotyl compared with the wild type (Figure 2B).
Figure 2.
Architecture of the Actin Cytoskeleton Is Altered in adf4 Hypocotyl Epidermal Cells.
(A) VAEM micrographs demonstrate increased actin filament bundling and decreased filament density in the cortical array of dark-grown wild-type (WT) epidermal cells. A montage of images from a single representative hypocotyl is displayed, with cells near the cotyledons at the left and cells near the root at the right. Values shown correspond to the distance of each cell from the cotyledons. Bar = 10 μm.
(B) VAEM micrographs of adf4 hypocotyl epidermal cells showing that actin filaments are more bundled and less dense than the wild type (A) in cells located in the lower two-thirds of the hypocotyl. Additionally, bundled actin filaments in cells located near the root in adf4 appear much brighter than bundled filaments in the wild type.
(C) Bundling (skewness) quantitatively increases along the gradient of axial cell expansion in a representative hypocotyl. Skewness values were measured from micrographs for every cell of the hypocotyl shown in (A) and plotted as a function of distance from the cotyledons.
(D) Quantitative analysis of actin filament density along the gradient of axial cell expansion in a representative hypocotyl. Actin filament density decreases in the cortical array of wild-type epidermal cells in the region spanning from cotyledons to root for the hypocotyl shown in (A). Moreover, filament density and filament bundling appear inversely correlated in vivo.
(E) Bundling analysis of the adf4 hypocotyl shown in (B) reveals increased bundling in the cortical actin cytoskeleton. Like the wild type, bundling increases with distance from the cotyledons; however, bundling analysis of adf4 exhibits increased skewness values, indicating more pronounced bundling in the two-thirds of the hypocotyl epidermal cells closest to the roots.
(F) Density analysis of adf4 reveals decreased filament density in the cortical actin cytoskeleton. The decrease in filament density in the adf4 mutant is more pronounced with lower values in the two-thirds of the hypocotyl cells closest to the roots when compared with the wild type.
[See online article for color version of this figure.]
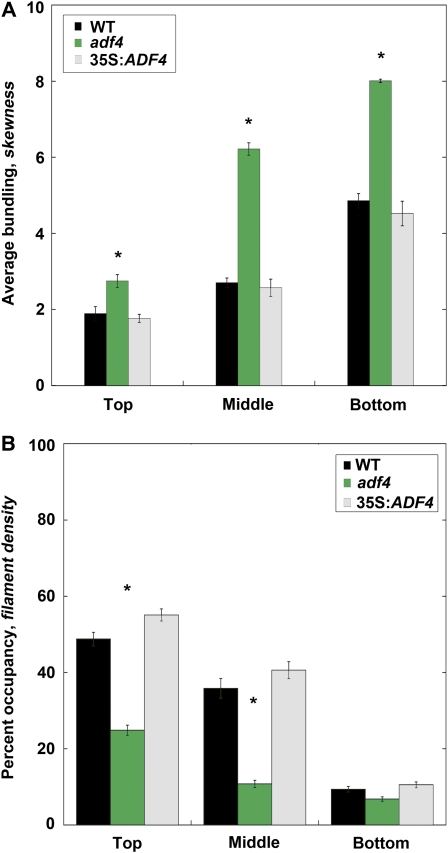
Skewness and density are two statistical parameters that can be used to quantify the extent of actin filament bundling and cytoskeleton density, respectively, in populations of cells (Higaki et al., 2010b). Moreover, the skewness parameter has been validated with a reconstituted system comprising muscle actin filaments and recombinant actin bundling proteins (Khurana et al., 2010; Zhao et al., 2011). To investigate further the relationship between skewness and density, we again used a reconstituted system to measure the density or percentage of occupancy of actin filament arrays generated in the absence and presence of various amounts of recombinant Arabidopsis VILLIN1 (VLN1). We found that skewness values increased with increasing amounts of VLN1, a simple filament bundling protein (Huang et al., 2005; Khurana et al., 2010), whereas percentage of occupancy decreased (see Supplemental Figure 5 online). Thus, our data demonstrate that skewness and density are inversely related when the amount of actin filaments is fixed. We next used these parameters to measure the extent of bundling and percent occupancy along the gradient of axial cell expansion in hypocotyl epidermal cells from representative wild-type (Figures 2C and 2D) and adf4 mutant (Figures 2E and 2F) seedlings. Actin arrays in wild-type epidermal cells became increasingly more bundled (Figure 2C) and less dense (Figure 2D) as one moves down the gradient from actively expanding (near cotyledons) to ceased expanding (near roots). Interestingly, bundling and occupancy measurements appeared inversely correlated in wild-type epidermal cells in vivo. The architecture of the actin cytoskeleton in adf4 was altered, with bundling occurring earlier in the gradient of cell expansion, and the extent of bundling being much greater at the base of the hypocotyl in cells that have finished expanding than in the corresponding cells of the wild type (Figure 2E). Similarly, the percentage of occupancy of filaments was substantially decreased throughout the gradient of axial cell expansion, most notably in the actively expanding cells near the cotyledons (Figure 2F). To quantitatively assess changes in actin architecture in growing cells, we imaged and analyzed actin filament bundling and occupancy in every epidermal cell along a file for more than 10 hypocotyls of each genotype. For ease of comparison, data from cells in the top, middle, and bottom third of hypocotyls were binned (Figure 3). Actin arrays in actively growing (top) adf4 mutant cells were significantly more bundled (Figure 3A) and markedly less dense (Figure 3B) than comparable wild-type cells. Importantly, the 35S:ADF4;adf4 rescue line was not significantly different than the wild type (Figures 3A and 3B; see Supplemental Figure 4 online).
Figure 3.
Actin Arrays in Actively Growing adf4 Cells Are More Bundled and Less Dense Than the Wild Type.
(A) Average bundling was measured and binned into three regions corresponding to the dotted lines in Figures 2C to 2F and Supplemental Figures 4C to 4F online. Knockout adf4 seedlings have significantly elevated average bundling in the top (P = 0.0013, t test), middle (P = 0.0001, t test), and bottom (P = 0.0011, t test) thirds of the dark-grown seedling when compared with wild-type controls. Average bundling values for the 35S:ADF4;adf4 rescue line were not significantly different from the wild type (WT). Values given are means ± se (n = 300 images per region; n = 150 cells per region; n = 10 hypocotyls). Asterisks denote statistical difference by t test.
(B) Average filament density was measured and binned for the same regions as in (A). Knockout adf4 seedlings have significantly decreased filament density in the top (P = 0.0003, t test) and middle (P = 0.0164, t test) thirds of dark-grown seedlings when compared by t test with wild-type controls. Average filament density values for the 35S:ADF4;adf4 rescue line were not significantly different from the wild type. Binned density results in vivo appear inversely related to the binned bundling results in (A). Values given are means ± se (n = 300 images per region; n = 150 cells per region; n = 10 hypocotyls).
[See online article for color version of this figure.]
To examine whether cytoskeletal architecture was altered in adf4 cells from other organs, we measured skewness and density for epidermal cells from 5-d-old dark-grown petioles of the seedling cotyledon. In the smaller cells of adf4 petioles, bundling was significantly increased, whereas percent occupancy was modestly but significantly decreased compared with the wild-type and the rescue line cells (see Supplemental Figures 3C and 3D online). These findings support the conclusion that altered actin filament organization in the adf4 mutant results in an increase in actin bundling and a reduction in the density of the cortical array.
ADF4 Is Partially Responsible for the Severing Component of Stochastic Dynamics in Vivo
The ability to examine single actin filament dynamics in living plant cells offers an unparalleled opportunity for detailed studies of the molecular mechanism of actin turnover in vivo (Staiger et al., 2009; Khurana et al., 2010; Smertenko et al., 2010). Staiger et al. (2009) observed that many individual actin filaments undergo stochastic dynamics, whereby rapid elongation (~1.7 μm/s) at filament plus ends is balanced by prominent severing activity. Based on knowledge of their biochemical properties, we predicted that either villin/gelsolin or ADF family members, or both, contribute to actin filament turnover through their severing activity (Staiger et al., 2009, 2010; Blanchoin et al., 2010).
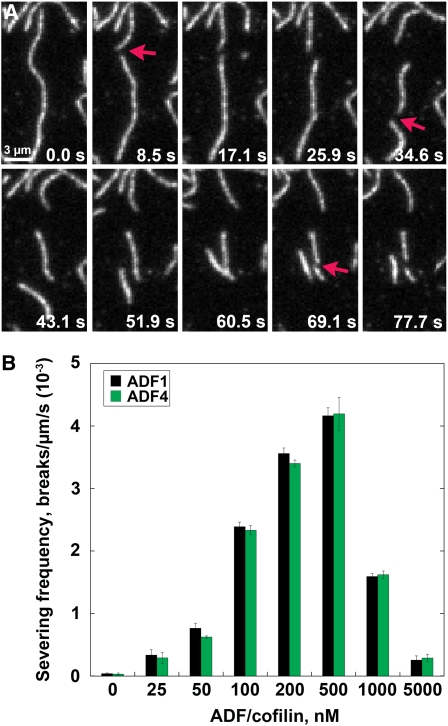
Previously, we demonstrated that recombinant ADF4 binds to G-actin with moderate affinity (Kd value, 0.1 μM) and has a 355-fold preference for ADP-loaded rather than ATP-loaded actin (Tian et al., 2009). To evaluate whether ADF4 is capable of severing actin filaments, we used time-lapse TIRFM and rhodamine-actin filaments (Andrianantoandro and Pollard, 2006; Chan et al., 2009; Khurana et al., 2010). The well-characterized variant, ADF1, was used as a control. Originally, ADF1 was thought to only depolymerize actin filaments from pointed ends (Carlier et al., 1997), but recently its severing activity has been demonstrated directly with TIRFM (Khurana et al., 2010). Analysis of time-lapse series from preformed rhodamine-actin filaments that were perfused with ADF4 showed the generation of numerous breaks in the filament backbone over time (Figure 4A; see Supplemental Movie 1 online). Severing frequency, defined as the number of breaks observed per micron of filament per second, was used as a measure of potency to evaluate multiple experiments at different ADF concentrations. Severing activity increased in a dose-dependent manner in the presence of both ADF4 and ADF1, reaching maximal levels at 500 nM ADF4, with a corresponding rate of 0.0043 ± 0.0003 breaks/μm/s (Figure 4B). In addition, there was a characteristic inhibition of severing at higher concentrations of ADF variants (Andrianantoandro and Pollard, 2006; Chan et al., 2009). However, at each concentration measured, there was no significant difference in severing activity between the two variants (P value = 0.62, ANOVA; Figure 4B). Like ADF1 (Khurana et al., 2010), recombinant ADF4 is therefore capable of disassembling actin filaments via its severing activity.
Figure 4.
Recombinant ADF4 Severs Actin Filaments in Vitro.
(A) Time-lapse TIRFM of 25 nM prepolymerized rhodamine-actin filaments attached to the cover slip of a perfusion chamber. At t = 0 s, 50 nM recombinant ADF4 was perfused into the chamber and imaged at 1.5-s intervals, with every other frame shown in the montage. Time points indicate elapsed time from the start of the experiment. Filaments became fragmented (arrows) over time (see Supplemental Movie 1 online). Bar = 3 μm.
(B) Quantitative analysis of ADF severing frequency. Various concentrations of ADF1 or ADF4 were perfused into chambers containing 25 nM rhodamine-actin filaments. Time-lapse images were recorded at ~1.5- to 3-s intervals with TIRFM. Severing frequency was calculated as the maximum filament length divided by the number of breaks per filament over time (breaks/μm/s). The severing frequency for 50 filaments per concentration was calculated from three independent batches of each protein, with n = 3 replicates per concentration. Means ± sd are shown. No significant differences between ADF1 and ADF4 severing rates were found at any concentration tested (P > 0.05, t test).
To investigate whether the changes in actin architecture observed in adf4 mutant seedlings were the result of altered single actin filament dynamics, we performed time-lapse VAEM on the cortical actin cytoskeleton of hypocotyl epidermal cells undergoing axial elongation (Staiger et al., 2009; Khurana et al., 2010). Arabidopsis epidermal cells have two populations of actin filaments in the cortical cytoplasm: putative single filaments that are extremely dynamic and actin filament cables that are much longer-lived and less dynamic (Figure 5A; Staiger et al., 2009; Khurana et al., 2010). As shown in Figure 5A (see Supplemental Movie 2 online), single actin filaments in wild-type epidermal cells can grow at rates approaching 2 μm/s and are typically disassembled by severing activity. Interestingly, actively expanding epidermal cells from the adf4 mutant showed altered turnover of these single actin filaments, with fewer apparent severing events (Figure 5B; see Supplemental Movie 3 online). We quantified the severing frequency, maximum filament length, maximum filament lifetime, filament origin, and depolymerization and elongation rates in epidermal cells at the apex of 5-d-old hypocotyls of the adf4 knockout mutant,and compared these to values from the wild-type and rescue lines (Table 1). Severing frequency was decreased 2.5-fold in adf4 mutant cells when compared with the wild type. Moreover, the average value for maximum filament length was significantly longer and the maximum filament lifetime was 1.5-fold longer in adf4 mutant cells. Filament elongation rates were slightly lower, as were depolymerization rates; however, neither value for adf4 cells was significantly different from the wild type.
Figure 5.
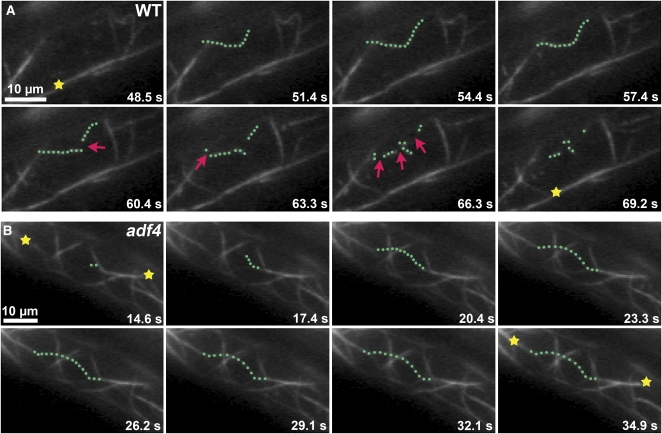
Time-Lapse Imaging of Cortical Actin Filaments in Arabidopsis Epidermal Cells Shows Differences in the Dynamic Behavior between the Wild Type and adf4.
(A) Time-lapse VAEM was used to image the cortical actin cytoskeleton in a dark-grown, 5-d-old wild-type hypocotyl epidermal cell expressing GFP-fABD2, as described previously (Staiger et al., 2009). A representative single actin filament is highlighted (green dots); it grows rapidly and is dismantled by prolific severing (arrows). By contrast, a representative actin filament cable (yellow star) remains relatively stationary throughout the ~20-s elapsed time (see Supplemental Movie 2 online). Bar = 10 μm.
(B) Time-lapse VAEM of a dark-grown, 5-d-old adf4 homozygous seedling expressing GFP-fABD2 shows altered dynamics of individual actin filaments. A representative single actin filament is highlighted (green dots) that grows rapidly but persists throughout the ~20-s elapsed time. More actin filament cables (yellow stars) are present in the adf4 cell than in the wild type (A) (see Supplemental Movie 3 online). Bar = 10 μm.
Table 1.
Comparison of Actin Dynamics Parameters from Wild-Type and Mutant Epidermal Cells of Dark-Grown Hypocotyls
| Stochastic Dynamics Parameters | Wild Type | adf4 | Rescue |
| 5-d-old seedlings | |||
| Elongation rate (μm/s) | 1.6 ± 0.8a | 1.4 ± 0.7* | 1.8 ± 0.8ND |
| Severing frequency (breaks/μm/s) | 0.015 ± 0.010 | 0.006 ± 0.004** | 0.019 ± 0.011* |
| Max. filament length (μm) | 13.6 ± 4.6 | 17.4 ± 5.4** | 13.4 ± 5.5ND |
| Max. filament lifetime (s) | 15.3 ± 8.2 | 23.3 ± 10.8** | 14.3 ± 3.8ND |
| Filament origin (% de novo/end/side) | 28/40/32 | 20/48/32 | 26/38/36 |
| Depolymerization rate (μm/s) | 0.22 ± 0.14 | 0.20 ± 0.12ND | 0.23 ± 0.12ND |
| 11- to 13-d-old seedlings | |||
| Elongation rate (μm/s) | 1.6 ± 0.8a | 2.4 ± 1.0* | 1.7 ± 0.9ND |
| Severing frequency (breaks/μm/s) | 0.018 ± 0.03 | 0.006 ± 0.005** | 0.016 ± 0.011ND |
| Max. filament length (μm) | 11.8 ± 5.5 | 17.6 ± 12.6** | 10.9 ± 5.5ND |
| Max. filament lifetime (s) | 29.6 ± 18.8 | 53.5 ± 32.6** | 39.9 ± 22.1* |
| Filament origin (% de novo/end/side) | 20/25/54 | 28/47/25 | 27/41/32 |
| Depolymerization rate (μm/s) | 0.22 ± 0.11 | 0.21 ± 0.12ND | 0.24 ± 0.08ND |
ND, Not significantly different from wild-type control value by Student’s t test; P value > 0.05. *Significantly different from wild-type control value by Student’s t test; P value ≤ 0.01. **Significantly different from wild-type control value by Student’s t test; P value ≤ 0.001.
Values given are means ± sd, with n > 50 filaments from n > 10 epidermal cells and at least 10 hypocotyls per line. See Methods for details of measurements.
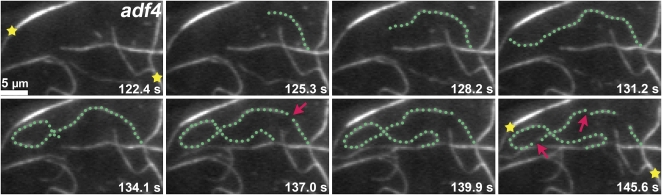
When nonexpanding epidermal cells from the base of 11- to 13-d-old hypocotyls were examined, the trends for most parameters were consistent with the axially expanding cells (Table 1). For example, the adf4 mutant had a threefold reduction in severing frequency, a 1.8-fold increase in maximum filament lifetime, and 1.5-fold increase in filament elongation rate. A representative VAEM time series from an adf4 mutant cell shows the consequences for single filament dynamics; the reduced instances of severing resulted in a maximum filament length of 50 μm for the highlighted filament (Figure 6; see Supplemental Movie 4 online). Values from the 35S:ADF4;adf4 rescue line were not significantly different from wild-type stochastic dynamic parameters, except for a modest but significant enhancement in severing frequency in 5-d-old hypocotyls and maximum filament lifetime in 11- to 13-d-old hypocotyls (Table 1). The latter might be expected from the slight overexpression of ADF4 in the rescue line (see Supplemental Figure 1B online); indeed, the severing frequency was slightly elevated in all cell types examined (Table 1; see Supplemental Table 1 online).
Figure 6.
Time-Lapse Imaging of Cortical Actin Filaments in Arabidopsis adf4 Mutant Epidermal Cells Show Enhanced Growth Rate and Maximum Filament Length.
Time-lapse VAEM of an 11-d-old nonelongating adf4 homozygous mutant epidermal cell shows alterations in single filament dynamics. The representative filament highlighted (green dots) has an average growth rate of 3.4 μm/s and maximum filament length of 50 μm. Few instances of severing (arrows) are apparent and several actin cables remain stationary throughout the montage (stars). Every other consecutive frame is shown (see Supplemental Movie 4 online). Bar = 5 μm.
The literature suggests that in addition to changes in filament turnover with cell age or expansion (Staiger et al., 2009), there may be differences in filament turnover in different cell types (Smertenko et al., 2010; Wang et al., 2011). Therefore, we also measured actin dynamics parameters in epidermal cells from 5-d-old cotyledon petioles. Similar results were obtained for petiole cells, with severing frequency being 2.5-fold reduced and significantly enhanced maximum filament lifetimes and lengths in adf4 mutant compared with wild-type seedlings (see Supplemental Table 1 online). The filament origin parameter showed no substantial differences in trends across all three cell types examined (Table 1; see Supplemental Table 1 online), with roughly equal proportions of growing filaments occurring de novo in the cytoplasm, from the end of preexisting fragments or from the side of filaments or bundles (Staiger et al., 2009). Similarly, depolymerization rates were not altered in the adf4 mutant, irrespective of cell growth status (Table 1). In summary, loss of ADF4 from epidermal cells results in a marked change in actin filament turnover, with a significant reduction in severing activity, longer individual actin filaments, and increased filament lifetimes.
DISCUSSION
In this study, we combined live-cell imaging techniques with reverse-genetic analyses to test a simple model for the regulation of actin filament turnover. Specifically, we dissected the importance of ADF/cofilin activity on the stochastic dynamics of individual actin filament turnover, in general, and on the severing of actin filaments, in particular. Dark-grown Arabidopsis seedlings with reduced ADF4 levels exhibited a hyperelongated hypocotyl phenotype and a corresponding gross disruption of actin cytoskeleton organization, similar to a previous report on ADF1 antisense plants (Dong et al., 2001). Loss of ADF4 resulted in quantifiable changes to the architecture of the cortical actin cytoskeleton in epidermal cells. Most notably, the extent of filament bundling was significantly increased, whereas array density was decreased in the actively growing cells of the adf4 mutant. To understand this phenotype in greater detail, we examined parameters of single actin filament turnover (Staiger et al., 2009; Smertenko et al., 2010). Because recombinant Arabidopsis ADF proteins sever actin filaments in vitro (this study; Khurana et al., 2010), the stochastic severing component of disassembly was predicted to be altered in adf4 mutants. Epidermal cells from the hypocotyls and petioles of adf4 mutants exhibited a 2.5-fold to threefold decrease in severing activity as well as a significant enhancement in maximum filament lengths and filament lifetimes compared with the wild type. All of these phenotypes were reversed upon modest overexpression of ADF4 behind a constitutive promoter in adf4 homozygous mutant plants. These results provide direct evidence for the importance of severing by ADF/cofilin in actin filament turnover by stochastic dynamics in vivo.
Severing Is a Key Mechanism for ADF/Cofilin-Mediated Actin Filament Disassembly
The ADF/cofilin family is considered to be a central regulator of actin filament turnover in eukaryotes (Van Troys et al., 2008; Bernstein and Bamburg, 2010). ADF/cofilins share several in vitro properties, including the ability to bind both G- and F-actin, with a strong preference for ADP-loaded rather than ATP-loaded actin (Carlier et al., 1997; Blanchoin and Pollard, 1999; Andrianantoandro and Pollard, 2006; Pavlov et al., 2007; Suarez et al., 2011). Their activity is concentration dependent; disassembly is favored at low concentrations and nucleation at high concentrations of cofilin (Andrianantoandro and Pollard, 2006; Chan et al., 2009). ADF/cofilin activities are further regulated by pH, phosphoinositide lipids, and other proteins (Okada et al., 2006; Kueh et al., 2008; Gandhi et al., 2009; Berro et al., 2010; Okreglak and Drubin, 2010). Traditionally, based largely on work with Arabidopsis ADF1, ADF/cofilins were thought to disassemble actin filaments by facilitating the depolymerization of ADP monomers from filament pointed ends (Carlier et al., 1997). Briefly, using a nucleotide exchange assay, Carlier et al. (1997) reported a 25-fold increase in the dissociation of subunits from the pointed end of filaments in the presence of ADF1. However, recent TIRFM imaging has shown that single-filament turnover is largely mediated by filament fragmentation, rather than solely by depolymerization (Andrianantoandro and Pollard, 2006; Chan et al., 2009). Moreover, the measured subunit depolymerization rates from individual filaments treated with low concentrations of human or Schizosaccharomyces pombe cofilin were predicted to be too slow to account for the previously observed rates of nucleotide exchange (Andrianantoandro and Pollard, 2006). Several groups have used computer modeling approaches, based on these known biochemical properties, to further dissect the role of this important family of proteins in actin filament turnover, resulting in two main mechanisms for ADF/cofilin’s contribution to filament disassembly, including filament severing and a bursting mechanism that requires the concerted activities of other accessory proteins like coronin and AIP1 (Roland et al., 2008; Berro et al., 2010; Kueh et al., 2010; Sirotkin et al., 2010). Importantly, these modeling results imply that depolymerization alone cannot account for the turnover rates of actin filaments observed originally in vitro.
ADF4 Is Capable of Severing Actin Filaments in Vitro
We used time-lapse TIRF microscopy to show that two plant ADFs, ADF1 and ADF4, are capable of severing actin filaments in vitro. Our severing frequency values indicate that plant ADF1 and ADF4 are somewhat less potent than ADF/cofilins from other eukaryotes (Andrianantoandro and Pollard, 2006; Chan et al., 2009). However, the general properties of the plant ADFs investigated are similar to other eukaryotic ADF/cofilins, including preferential binding to ADP-loaded actin (Chaudhry et al., 2007; Tian et al., 2009), filament severing as the mechanism of turnover (Khurana et al., 2010), and the inability to sever certain types of bundled actin filaments (Huang et al., 2005; Khurana et al., 2010). At least one member of the Arabidopsis ADF family, ADF9, appears to lack or has minimal filament severing activity and is instead a simple filament bundling and stabilizing protein (Tholl et al., 2011). This latter observation emphasizes the importance of performing detailed biochemical analyses prior to interpreting mutant phenotypes or making assumptions about cellular functions. ADF4 lacks filament bundling activity in vitro (J.L. Henty, unpublished data); however, it binds preferentially to ADP-loaded actin monomers (Tian et al., 2009) and severs actin filaments at the same frequency as ADF1 (this study). Thus, Arabidopsis ADFs are predicted to be central players in actin turnover by stochastic dynamics (Blanchoin et al., 2010; Staiger et al., 2010; Day et al., 2011).
Arabidopsis Hypocotyls Are an Ideal Model System for Studying Actin Turnover in Vivo
Epidermal cells from the hypocotyls of Arabidopsis seedlings are an excellent model system for imaging cytoskeletal components and for correlating changes in cytoskeletal architecture or turnover with anisotropic cell expansion (Gendreau et al., 1997; Ehrhardt and Shaw, 2006; Lucas and Shaw, 2008; Staiger et al., 2009). For example, hypocotyl epidermal cells have been used to study the dynamic instability and angle-dependent contact changes in microtubule behavior as well as long-term rotary movements of the entire array (Shaw et al., 2003; Chan et al., 2007; Lucas et al., 2011). The hypocotyl system also features prominently in studies of cortical microtubules as tracks for the guidance of integral membrane protein CesA complexes during the synthesis and oriented deposition of cellulose fibrils in the cell wall (Paredez et al., 2006; Gutierrez et al., 2009). Furthermore, epidermal cells from hypocotyls expressing the actin reporters, GFP-fABD2 or Lifeact-GFP, have been used to quantify mechanisms of actin filament assembly and disassembly. Notably, the ability to visualize single filaments or small bundles by VAEM (Konopka and Bednarek, 2008) has facilitated a detailed analysis of filament growth, elongation and depolymerization rates, severing frequency, and convolutedness (Staiger et al., 2009; Smertenko et al., 2010). These observations, and a deep knowledge of the biochemical and biophysical activities of many plant actin binding proteins, have led to a simple model for the regulation of actin turnover by stochastic dynamics (Staiger et al., 2009; Blanchoin et al., 2010; Day et al., 2011). By combining a large collection of T-DNA insertion mutants and the power of Arabidopsis genetics with advanced imaging modalities, an unparalleled opportunity to dissect the mechanisms that underpin stochastic dynamics exists.
Our results confirm a role for ADF/cofilin in the turnover of filaments by prolific severing activity in vivo. Specifically, we observed marked alterations in stochastic dynamics parameters in adf4 knockout mutant cells, including a significant reduction in severing frequency as well as increased maximum filament lengths and lifetimes. Importantly, depolymerization rates were not different in adf4 mutant cells, consistent with a minor role for this ADF/cofilin in the disassembly of actin by facilitating depolymerization from filament pointed ends. These changes in actin dynamics were observed for actively expanding and nongrowing epidermal cells of the hypocotyl as well as cotyledon petiole cells. Although severing activity was not completely abolished in adf4 knockout cells, this is to be expected since ADFs are present in a multigene family, several of which are expressed in dark-grown hypocotyls (see Supplemental Figure 1A online; Ma et al., 2005). Moreover, other actin binding proteins like the villin/gelsolin/fragmin family are capable of severing filaments in vitro (Khurana et al., 2010; Zhang et al., 2010; Zhang et al., 2011) and are also likely to be expressed in these same cells (Ma et al., 2005). Nevertheless, our findings provide strong evidence that ADF/cofilin contributes to the stochastic dynamic behavior of single actin filaments in plants.
Actin Filament Turnover Is Important for Aspects of Cell Expansion and Elongation
The turnover of individual actin filaments and formation of actin filament bundles are generally considered to be important for anisotropic cell expansion and cellular morphogenesis (Smith and Oppenheimer, 2005; Dhonukshe et al., 2008; Nick et al., 2009; Thomas et al., 2009; Higaki et al., 2010a). Here, we show that actively expanding cells in the apex of adf4 hypocotyls display extensively bundled actin arrays as well as a reduced actin turnover. This implies that increased actin bundling and/or dynamic individual filaments are required for anisotropic cell expansion. The actin cytoskeleton is generally considered to promote cell growth, to contribute to spatially restricted expansion, and to participate in vacuolar morphogenesis as well as transvacuolar strand formation and maintenance. The disruption of actin bundles leads to alteration of plant cell wall thickness, vacuolar shape changes (Higaki et al., 2011), and the loss of transvacuolar strands (Staiger et al., 1994). One possibility is that turgor pressure, the main driving force of plant cell expansion, is altered when vacuolar surface area or morphology are perturbed (Szymanski and Cosgrove, 2009; Higaki et al., 2010a, 2011). The actin cytoskeleton is also thought to be a major contributor to cell expansion and morphology by creating tracks for positioning and/or motility of endomembrane compartments (Smith and Oppenheimer, 2005; Gutierrez et al., 2009; Szymanski and Cosgrove, 2009; Higaki et al., 2010a) and directed delivery of cell wall components (Baskin, 2005; Smith and Oppenheimer, 2005; Szymanski and Cosgrove, 2009). However, the presence of both single and bundled actin filaments throughout the cell cortex where various cargos are delivered during cell expansion (Staiger et al., 2009; Dong et al., 2001) further complicates the involvement of actin in these processes, since little is known about the exact dimensions of F-actin required for specific delivery of various cargos. For example, we do not know at present whether bundle thickness or polarity matter for cargo being trafficked to the actively growing region of a cell. It is perhaps significant to note that myosin motors in animal and S. pombe cells are sensitive to the nature of the tracks on which they transport cargoes (Brawley and Rock, 2009; Clayton et al., 2010), a theme that requires further exploration in plant cells. Loss of ADF4, through altered actin organization and dynamics, could have indirect effects on endomembrane compartment motility or positioning, as well as vacuolar trafficking and morphology. Therefore, altered subcellular trafficking may lead to increased axial cell expansion in the adf4 mutant.
Our data on enhanced actin bundling and reduced filament turnover in hypocotyl epidermal cells are inconsistent with a second model for the role of actin bundles in cell expansion. Primarily based on experiments with monocot coleoptiles and exogenous expression of a bundling factor, it has been proposed that the extent of actin bundling inhibits axial cell expansion (Waller et al., 2002; Nick et al., 2009; Nick, 2010). Furthermore, treatments with the auxin efflux inhibitor triiodobenzoic acid (TIBA) display excessively bundled actin arrays (Dhonukshe et al., 2008), and this correlates with reductions in rice (Oryza sativa) coleoptile growth (Nick et al., 2009; Nick, 2010). Although the exact mechanism of TIBA-induced actin bundling is not known, some component of the cytoskeleton is considered to be the target (Dhonukshe et al., 2008). However, TIBA has no effect on the maximum extent of growth in dark-grown oat (Avena sativa) coleoptiles (Shinkle and Briggs, 1984), suggesting that the bundled state of actin does not necessarily directly correlate with growth. Epidermal cells from petioles of the adf4 mutant also exhibit enhanced bundling; however, these cells are shorter and thinner than the wild type (see Supplemental Figure 3 online), further indicating that the actin bundling status with regard to cell elongation may be more complicated than previously assumed or may be tissue specific. Perhaps the ratio of single actin filaments to actin bundles is important for cell expansion and this has, until recently, resisted detailed analysis due to the lack of quantitative measures for individual actin filaments. Our measurements of actin filament bundling and density in the axially expanding epidermal cells at the apex of the hypocotyl versus the nonelongating cells at the base may address this hypothesis. In wild-type hypocotyls, expanding cells have dense actin arrays and less bundling, whereas filament arrays in nongrowing cells are more bundled and less dense. If F-actin levels remain constant, this suggests that the dynamic interplay between the two filament populations is regulated during cell expansion or organ development.
Based on in vitro data and detailed analysis of the stochastic dynamics parameters of single actin filaments in the adf4 mutant, we hypothesize that plant ADF/cofilins are likely to have direct effects on the turnover of single actin filaments but indirect effects on bundling. Although not quantified, Arabidopsis ADF1 antisense plants appear to have excessively bundled arrays in the hypocotyl, cotyledon petioles, and root hair cells (Dong et al., 2001), indicating that loss of ADF contributes to the actin-based phenotypes reported. Inducible RNA interference lines for ADF2 also result in extremely dense cytoskeletal arrays with prominent bundles in hypocotyl epidermal cells and root vascular cells (Clément et al., 2009). Similarly, inducible suppression of Arabidopsis AIP1, which is thought to function synergistically with ADF/cofilins, leads to excessively bundled arrays but reduced cell expansion (Ketelaar et al., 2004a). Somewhat indirect evidence shows that inducible expression of GFP-mTn leads to enhanced filament bundling and perturbation of root hair growth by inhibiting the activity of ADF/cofilin (Ketelaar et al., 2004b). Biochemical analyses also support a direct role for plant ADF on individual actin filaments rather than bundles because ADF/cofilin cannot disassemble or sever bundles generated by VLN1 (Huang et al., 2005; Khurana et al., 2010) or LIM domain proteins (Thomas et al., 2006). Alternatively, as observed in fission yeast, ADF/cofilin might compete for binding and severing when different populations of actin bundling proteins are bound to filaments (Skau and Kovar, 2010).
Given that adf4 mutant cells exhibit a reduced severing frequency and increased maximum filament lengths and lifetimes, it seems probable that the increase in bundling is a consequence of a disruption in single-filament turnover. Furthermore, since these cells show a quantitative increase in bundling and inverse relationship with filament density, it is likely that the overall amount of actin filaments is not changing in the adf4 mutant. In other words, a lack of increased filament density suggests that there is not more actin present in the adf4 mutant and, therefore, that there is not simply more polymer available to be assembled into bundles. Perhaps the reduction in filament turnover, or the significantly increased filament lifetimes, in the adf4 mutant allows more time for individual actin filaments to make contact with adjacent filaments or bundles and facilitates a “catch and zipper” mechanism, resulting in extensively bundled actin arrays (Michelot et al., 2007; Khurana et al., 2010). Future studies will explore the interrelationship between single actin filaments and bundles in vitro and in vivo and will test the roles for each population in axial cell expansion. It will also be important to test the role of other major actin binding proteins in the stochastic dynamic model for actin turnover.
METHODS
Chemicals
All chemicals were purchased from Sigma-Aldrich unless stated otherwise.
Plant Material/Growth
Homozygous mutant adf4 (GARLIC_823_A11.b.1b.Lb3Fa) was identified and characterized previously (Tian et al., 2009). DNA primers (forward) 5′-gcggtcgacatggctaatgctgcgTcaggaatgg-3′ and (reverse) 5′-GCGGTCGACTTAGTTGACGCGGCTTTTCAAAAC-3′ were used to add SalI restriction enzyme sites (underlined) for cloning the ADF4 open reading frame into pMD1 containing a T7 epitope tag. Homozygous adf4 mutant plants were transformed as previously described (Tian et al., 2009). These lines were crossed to Arabidopsis thaliana Columbia-0 expressing our GFP-fABD2 reporter (Staiger et al., 2009), and homozygotes were recovered in the F2 population. A single-blind experimental design was used to screen Arabidopsis lines for phenotypes objectively. Seeds were surface sterilized and stratified for 3 d at 4°C on 0.5× Murashige and Skoog medium supplemented with 1% Suc. Seedlings were grown under long-day conditions (16 h light/8 h dark) at 21°C. Alternatively, stratified seeds were exposed to 4 h of light, plates triple-wrapped in aluminum foil, and seedlings allowed to germinate in the dark. Arabidopsis seedlings were grown for 2 to 14 d after germination with all three genotypes on the same plates for phenotypic analyses and epidermal cell length and width measurements. Epidermal cell lengths and widths were measured on growing and nongrowing regions of etiolated hypocotyls and on 5-d-old petioles. Five-day-old hypocotyls were incubated in 5 μM FM4-64 dye (Molecular Probes) for 10 min, and then the top third of each hypocotyl below the apical hook, middle, and bottom third closest to the roots were imaged using a ×40/0.95–numerical aperture objective and wide-field fluorescence microscopy. All image measurements were performed in ImageJ (http://rsb.info.nih.gov/ij/).
RNA Extraction and qRT-PCR
Light-grown seedlings (10 d after germination) of wild-type, homozygous mutant adf4, and 35S:ADF4;adf4 rescue lines were flash-frozen and ground to a fine powder in liquid nitrogen. RNA isolation was performed with TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions. Two-step qRT-PCR was performed using 2× SYBR Green master mix (Applied Biosystems), normalized to GAPD transcript levels, and analyzed with Excel software, as described by Khurana et al. (2010). Gene-specific primers for ADF1, ADF2, ADF3, ADF4, and ADF5 (see Supplemental Table 2 online) were used to measure transcript levels. Three biological and technical replicates were performed per gene-specific primer set.
Protein Purification
Recombinant Arabidopsis ADF4 and ADF1 were purified as described previously (Carlier et al., 1997; Tian et al., 2009). Actin from rabbit skeletal muscle was purified by gel filtration chromatography on Sephacryl S-300 (MacLean-Fletcher and Pollard, 1980). ADF concentrations were determined by extinction coefficient 14,340 M−1 cm−1 and 14,690 M−1 cm−1 (Didry et al., 1998; Tian et al., 2009).
Measurement of Severing in Vitro
The ability of purified ADF to sever actin filaments in vitro was determined with a total internal reflection fluorescence microscope equipped with a ×60/1.45–numerical aperture TIRFM PlanApo objective (Olympus). A 1:1 ratio of cold:rhodamine-actin (Cytoskeleton) filaments was polymerized with 1× KMEI at room temperature (Khurana et al., 2010). After 30 min, the 5 μM stock of prepolymerized actin filaments was diluted to 25 nM and adhered to a visualization chamber coated with 5 nM N-ethylmaleimide-myosin. A dose series of 0 to 5 μM ADF1 or ADF4 was used to determine severing activity. Three independent batches of each protein and three technical replications were used per concentration. Filament severing rate was calculated as described by Khurana et al. (2010). Briefly, the maximum length of each filament was measured with ImageJ software (version 1.41) on the first frame following perfusion of ADF and subsequent breaks recorded over time until the filament disappeared.
Time-Lapse Imaging of Actin Filament Dynamics in Vivo
VAEM (Konopka and Bednarek, 2008) was used to analyze the single actin filament dynamics in wild-type, homozygous recessive adf4 mutant, and 35S:ADF4;adf4 rescue line seedlings expressing GFP-fABD2. Dark-grown seedlings were mounted in water and imaged for no more than 15 min at a time. Epidermal cells from the top, middle, and bottom thirds of the hypocotyl were examined. Filament severing frequency, maximum length, filament origin, and elongation rates were calculated according to Staiger et al. (2009). Briefly, severing frequency was determined as described above. Maximum filament length was the longest length of a tracked filament during the course of growth/turnover. Filament origin is given as the percentage of occurrence either de novo, from the side of a bundle or filament, or from the end of a preexisting fragment. The elongation rate was determined by fitting plots of filament length as a function of time with a linear curve in Excel or Kaleidagraph; values obtained were from at least four data points and R2 ≥ 0.95. Maximum filament lifetime was the amount of time a filament was present, from initial filament origin until all pieces of the filament could no longer be tracked. To determine depolymerization rates, kymographs were prepared from growing actin filaments that were computationally straightened and plotted as filament length versus time with a custom plug-in for Image J (Kuhn and Pollard, 2005). Rates were estimated from the slope of a line placed on the kymograph at the presumed pointed end of the growing filament.
Quantitative Analyses of Actin Filament Array Architecture
Skewness analysis, a metric based on the assumption that a population of actin filaments exhibit increased pixel intensities when bundled (Higaki et al., 2010b), was employed to measure the extent of bundling in VAEM micrographs of hypocotyl and petiole epidermal cells. A fixed exposure time and gain setting were selected such that single actin filaments could be seen, but the pixel intensities of higher-order actin structures were not saturated. Micrographs were analyzed in ImageJ using parameters described previously (Higaki et al., 2010b; Khurana et al., 2010). Every epidermal cell along the hypocotyl long axis was imaged with a series of overlapping micrographs. For statistical analysis, raw skewness values were binned into thirds based on position along the hypocotyl. Filament density was calculated as the percent occupancy of GFP-fABD2 signal in each micrograph used for filament bundling analysis (Higaki et al., 2010b), with a few exceptions. Briefly, image threshold settings were set to include all actin filaments and then images were converted to binary black and white images. Because there were no z-series projections and VAEM generates high-contrast images, we did not apply Gaussian blur, high band-pass filter, or skeletonization processing steps. To validate the density measurement, we compared in vitro single actin filaments and reconstituted actin filament bundles generated with various amounts of VLN1 (Khurana et al., 2010). Fifty micrographs per concentration of VLN1 were analyzed as above for the skewness analysis. We also measured density on images of hypocotyl epidermal cells expressing GFP-fABD2. Raw density values were binned into thirds based on length for statistical analysis. At least 500 images of hypocotyl epidermal cells per binned section were collected per genotype, from at least 10 individual seedlings for both measurements.
Statistical Analyses
Mean values, se, sd, and statistical tests were calculated with Microsoft Excel software (version 14.0.2). Statistical significance was assessed by one-tailed Student’s t test with unequal variance and between control and treatment and by ANOVA, as stated in the figure legends.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative under accession number At5g59890 (ADF4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple ADF Variants Are Expressed in Arabidopsis Hypocotyls.
Supplemental Figure 2. Homozygous adf4 Mutant Seedlings Have Longer Roots.
Supplemental Figure 3. Homozygous adf4 Mutant Seedlings Have Altered Petiole Cells.
Supplemental Figure 4. Architecture of the Actin Cytoskeleton Is Not Altered in 35S:ADF4;adf4 Rescue Line.
Supplemental Figure 5. Filament Bundling and Density Are Inversely Correlated in a Reconstituted Bundling Assay.
Supplemental Table 1. Comparison of Actin Dynamics Parameters from Wild-Type and adf4 Mutant Petiole Epidermal Cells.
Supplemental Table 2. Gene-Specific Primers Used for qRT-PCR.
Supplemental Movie 1. Time-Lapse TIRFM of ADF4 Severing in Vitro.
Supplemental Movie 2. Time-Lapse VAEM of a Wild-Type Epidermal Cell.
Supplemental Movie 3. Time-Lapse VAEM of an adf4 Mutant Cell.
Supplemental Movie 4. Time-Lapse VAEM of a Nonelongating adf4 Mutant Cell.
Supplemental Movie Legends. Legends for Supplemental Movies 1 through 4.
Acknowledgments
We thank our colleagues in the Purdue Cytoskeletal Group, especially Dan Szymanski, and at Michigan State University for their continuous support and helpful advice on this project. We are grateful to Miaoying Tian (Boyce Thompson Institute) for generating the initial 35S:ADF4 rescue line. Collaborative activities and joint research between the Staiger and Day laboratories are supported by a U.S. National Science Foundation-Arabidopsis 2010 grant (IOS-1021185). Work in the lab of B.D. is also supported by a National Science Foundation Early CAREER award (IOS-0641319). Work in the Meagher laboratory was supported by a grant from the National Institutes of Health (GM36397). J.L.H. was supported in part by a studentship from the Department of Energy–sponsored Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontiers Research Center (DE-SC0000997). C.J.S. also received partial salary support from C3Bio. The TIRF microscopy facility at Purdue was funded in part by the Bindley Bioscience Center.
AUTHOR CONTRIBUTIONS
J.L.H., L.B., and C.J.S. designed experiments. C.J.S., B.D., and R.B.M. supplied new experimental tools. J.L.H, P.K., and C.J.S. performed experiments. J.L.H., S.W.B., P.K., L.B., and C.J.S. analyzed the data. J.L.H., L.B., and C.J.S. wrote the article.
References
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Augustine R.C., Vidali L., Kleinman K.P., Bezanilla M. (2008). Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 54: 863–875 [DOI] [PubMed] [Google Scholar]
- Baskin T.I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Bernstein B.W., Bamburg J.R. (2010). ADF/cofilin: A functional node in cell biology. Trends Cell Biol. 20: 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berro J., Sirotkin V., Pollard T.D. (2010). Mathematical modeling of endocytic actin patch kinetics in fission yeast: Disassembly requires release of actin filament fragments. Mol. Biol. Cell 21: 2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. (2010). Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D. (1999). Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274: 15538–15546 [DOI] [PubMed] [Google Scholar]
- Brawley C.M., Rock R.S. (2009). Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc. Natl. Acad. Sci. USA 106: 9685–9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.-F., Laurent V., Santolini J., Melki R., Didry D., Xia G.-X., Hong Y., Chua N.-H., Pantaloni D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136: 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Beltzner C.C., Pollard T.D. (2009). Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chaudhry F., Guérin C., von Witsch M., Blanchoin L., Staiger C.J. (2007). Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18: 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Bernstein B.W., Sneider J.M., Boyle J.A., Minamide L.S., Bamburg J.R. (2004). In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry 43: 7127–7142 [DOI] [PubMed] [Google Scholar]
- Clayton J.E., Sammons M.R., Stark B.C., Hodges A.R., Lord M. (2010). Differential regulation of unconventional fission yeast myosins via the actin track. Curr. Biol. 20: 1423–1431 [DOI] [PubMed] [Google Scholar]
- Clément M., Ketelaar T., Rodiuc N., Banora M.Y., Smertenko A., Engler G., Abad P., Hussey P.J., de Almeida Engler J. (2009). Actin-depolymerizing factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 21: 2963–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B., Henty J.L., Porter K.J., Staiger C.J. (2011). The pathogen-actin connection: A platform for defense signaling in plants. Annu. Rev. Phytopathol. 49: 483–506 [DOI] [PubMed] [Google Scholar]
- De La Cruz E.M. (2009). How cofilin severs an actin filament. Biophys. Rev. 1: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 105: 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didry D., Carlier M.-F., Pantaloni D. (1998). Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 273: 25602–25611 [DOI] [PubMed] [Google Scholar]
- Dong C.-H., Xia G.-X., Hong Y., Ramachandran S., Kost B., Chua N.-H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D.W., Shaw S.L. (2006). Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Gandhi M., Achard V., Blanchoin L., Goode B.L. (2009). Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol. Cell 34: 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungabissoon R.A., Jiang C.-J., Drøbak B.K., Maciver S.K., Hussey P.J. (1998). Interaction of maize actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. Plant J. 16: 689–696 [Google Scholar]
- Gungabissoon R.A., Khan S., Hussey P.J., Maciver S.K. (2001). Interaction of elongation factor 1α from Zea mays (ZmEF-1α) with F-actin and interplay with the maize actin severing protein, ZmADF3. Cell Motil. Cytoskeleton 49: 104–111 [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M.C., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Higaki T., Kojo K.H., Hasezawa S. (2010a). Critical role of actin bundling in plant cell morphogenesis. Plant Signal. Behav. 5: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2010b). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Higaki T., Kurusu T., Hasezawa S., Kuchitsu K. (2011). Dynamic intracellular reorganization of cytoskeletons and the vacuole in defense responses and hypersensitive cell death in plants. J. Plant Res. 124: 315–324 [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E.G., Anthony R., Voigt B., Menzel D., Hussey P.J. (2004a). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14: 145–149 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Anthony R.G., Hussey P.J. (2004b). Green fluorescent protein-mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor’s actin depolymerizing activity in vitro. Plant Physiol. 136: 3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196 [DOI] [PubMed] [Google Scholar]
- Kueh H.Y., Brieher W.M., Mitchison T.J. (2010). Quantitative analysis of actin turnover in Listeria comet tails: Evidence for catastrophic filament turnover. Biophys. J. 99: 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh H.Y., Charras G.T., Mitchison T.J., Brieher W.M. (2008). Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 182: 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. (2005). Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88: 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J., Shaw S.L. (2008). Cortical microtubule arrays in the Arabidopsis seedling. Curr. Opin. Plant Biol. 11: 94–98 [DOI] [PubMed] [Google Scholar]
- Lucas J.R., Courtney S., Hassfurder M., Dhingra S., Bryant A., Shaw S.L. (2011). Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell 23: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Sun N., Liu X., Jiao Y., Zhao H., Deng X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean-Fletcher S., Pollard T.D. (1980). Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96: 18–27 [DOI] [PubMed] [Google Scholar]
- McCullough B.R., Blanchoin L., Martiel J.-L., De la Cruz E.M. (2008). Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J. Mol. Biol. 381: 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A., Pope B., Chiu W., Weeds A. (1997). Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 138: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C.J., Martiel J.-L., Blanchoin L. (2007). Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17: 825–833 [DOI] [PubMed] [Google Scholar]
- Nick P. (2010). Probing the actin-auxin oscillator. Plant Signal. Behav. 5: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick P., Han M.-J., An G. (2009). Auxin stimulates its own transport by shaping actin filaments. Plant Physiol. 151: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ravi H., Smith E.M., Goode B.L. (2006). Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 17: 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. (2010). Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 188: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. (2007). Actin filament severing by cofilin. J. Mol. Biol. 365: 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressad F., Didry D., Xia G.-X., Hong Y., Chua N.-H., Pantaloni D., Carlier M.-F. (1998). Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J. Biol. Chem. 273: 20894–20902 [DOI] [PubMed] [Google Scholar]
- Reymann A.-C., Suarez C., Guérin C., Martiel J.-L., Staiger C.J., Blanchoin L., Boujemaa-Paterski R. (2011). Turnover of branched actin filament networks by stochastic fragmentation with ADF/cofilin. Mol. Biol. Cell 22: 2541–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland J., Berro J., Michelot A., Blanchoin L., Martiel J.-L. (2008). Stochastic severing of actin filaments by actin depolymerizing factor/cofilin controls the emergence of a steady dynamical regime. Biophys. J. 94: 2082–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka D.R., Kandasamy M.K., McKinney E.C., Burgos-Rivera B., Meagher R.B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52: 460–472 [DOI] [PubMed] [Google Scholar]
- Schüler H., Mueller A.-K., Matuschewski K. (2005). A plasmodium actin-depolymerizing factor that binds exclusively to actin monomers. Mol. Biol. Cell 16: 4013–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., Ehrhardt D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shinkle J.R., Briggs W.R. (1984). Auxin concentration/growth relationship for Avena coleoptile sections from seedlings grown in complete darkness. Plant Physiol. 74: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin V., Berro J., Macmillan K., Zhao L., Pollard T.D. (2010). Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell 21: 2894–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau C.T., Kovar D.R. (2010). Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr. Biol. 20: 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A.P., Allwood E.G., Khan S., Jiang C.-J., Maciver S.K., Weeds A.G., Hussey P.J. (2001). Interaction of pollen-specific actin-depolymerizing factor with actin. Plant J. 25: 203–212 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Deeks M.J., Hussey P.J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123: 3019–3028 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Oppenheimer D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger C.J., Yuan M., Valenta R., Shaw P.J., Warn R.M., Lloyd C.W. (1994). Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr. Biol. 4: 215–219 [DOI] [PubMed] [Google Scholar]
- Suarez C., Roland J., Boujemaa-Paterski R., Kang H., McCullough B.R., Reymann A.C., Guérin C., Martiel J.-L., De la Cruz E.M., Blanchoin L. (2011). Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 21: 862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D.B., Cosgrove D.J. (2009). Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Tholl S., Moreau F., Hoffmann C., Arumugam K., Dieterle M., Moes D., Neumann K., Steinmetz A., Thomas C. (2011). Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 585: 1821–1827 [DOI] [PubMed] [Google Scholar]
- Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18: 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton 66: 940–957 [DOI] [PubMed] [Google Scholar]
- Tian M., Chaudhry F., Ruzicka D.R., Meagher R.B., Staiger C.J., Day B. (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 150: 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008). Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87: 649–667 [DOI] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O’Shaughnessy B., Pollard T.D. (2008). Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 319: 97–100 [DOI] [PubMed] [Google Scholar]
- Waller F., Riemann M., Nick P. (2002). A role for actin-driven secretion in auxin-induced growth. Protoplasma 219: 72–81 [DOI] [PubMed] [Google Scholar]
- Wang X.-L., Gao X.-Q., Wang X.-C. (2011). Stochastic dynamics of actin filaments in guard cells regulating chloroplast localization during stomatal movement. Plant Cell Environ. 34: 1248–1257 [DOI] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiao Y., Du F., Cao L., Dong H., Ren H. (2011). Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytol. 190: 667–682 [DOI] [PubMed] [Google Scholar]
- Zhao Y., et al. (2011). The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 23: 2314–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]