This work demonstrates the existence of actin within plant mitochondria, and the import of actin into mitochondria is shown in fluorescent-actin transgenic plants and in wild-type cotyledon mitochondria during germination. The findings suggest that the dynamics of mitochondrial actin can regulate cytochrome C release and, consequently, may initiate programmed cell death of the mung bean cotyledons.
Abstract
Here, a large fraction of plant mitochondrial actin was found to be resistant to protease and high-salt treatments, suggesting it was protected by mitochondrial membranes. A portion of this actin became sensitive to protease or high-salt treatment after removal of the mitochondrial outer membrane, indicating that some actin is located inside the mitochondrial outer membrane. The import of an actin–green fluorescent protein (GFP) fusion protein into the mitochondria in a transgenic plant, actin:GFP, was visualized in living cells and demonstrated by flow cytometry and immunoblot analyses. Polymerized actin was found in mitochondria of actin:GFP plants and in mung bean (Vigna radiata). Notably, actin associated with mitochondria purified from early-developing cotyledons during seed germination was sensitive to high-salt and protease treatments. With cotyledon ageing, mitochondrial actin became more resistant to both treatments. The progressive import of actin into cotyledon mitochondria appeared to occur in concert with the conversion of quiescent mitochondria into active forms during seed germination. The binding of actin to mitochondrial DNA (mtDNA) was demonstrated by liquid chromatography–tandem mass spectrometry analysis. Porin and ADP/ATP carrier proteins were also found in mtDNA-protein complexes. Treatment with an actin depolymerization reagent reduced the mitochondrial membrane potential and triggered the release of cytochrome C. The potential function of mitochondrial actin and a possible actin import pathway are discussed.
INTRODUCTION
Bacteria were once thought to be shapeless, disorganized biomolecular sacks but are now known to possess a complex subcellular organization created by cytoskeletal components that are homologous to eukaryotic actin, tubulin, and intermediate filaments (Löwe and Amos, 1998; Jones et al., 2001; van den Ent et al., 2001; Ausmees et al., 2003; Errington, 2003; Carballido-López, 2006; Graumann, 2007). The mitochondrion is a structurally and functionally well-organized organelle. It is reasonable to think that mitochondria also carry cytoskeletons that can regulate mitochondrial morphology and fusion/fission and catalyze mitochondrial DNA (mtDNA) segregation similarly to its ancestors. Indeed, Reyes et al. (2011) demonstrated that β-actin and myosin resided in human mitochondria by applying protease protection and iodixanol gradient analysis, respectively, and also by import studies. They also proved that the β-actin within human mitochondria was associated with mtDNA and could play a role in mtDNA maintenance. In this report, we communicate our findings on the localization and characterization of actin in plant mitochondria as well as the potential function and possible import pathways of plant mitochondrial actin.
The interactions between cytosolic actin and mitochondria have been extensively investigated in both yeast and plants. Extramitochondrial actin cytoskeletons are involved in mitochondrial transport, distribution, morphology, and inheritance (Hermann and Shaw, 1998; Yaffe, 1999; Boldogh et al., 2003; Sheahan et al., 2004; Logan, 2006). Before the report by Reyes et al. (2011), the presence of actin and tubulin in mitochondria and the association of actin with mtDNA have been reported but not confirmed (Etoh et al., 1990; Kuroiwa et al., 1994; Carré et al., 2002; Lo et al., 2002; Dai et al., 2005; Wang and Bogenhagen, 2006). The major problems with these investigations are the lack of mitochondria-targeting presequences in both actin and tubulin and the possible contamination of study samples with sticky actin during mitochondrial purification. The association of cytosolic actin with mtDNA through intermediate proteins has been thoroughly investigated (Hobbs et al., 2001; Meeusen and Nunnari, 2003; Boldogh and Pon, 2006).
Unlike their ancestral bacteria, the problems of mitochondrial morphological dynamics, DNA segregation, organelle division, and macromolecular transportation inside mitochondria of vascular plant are largely unresolved. The actin-like proteins have not been extensively investigated as effectors of the highly dynamic plant mitochondrial architecture in situ or as effectors associated with macromolecular transportation, mtDNA segregation, and mitochondrial fusion/fission. Recently, many reports have suggested that cytosolic proteins may be imported into organelles via multiple routes, in addition to the verified canonical import pathway (Marc et al., 2002; Millar et al., 2006; Neupert and Herrmann, 2007). Cotranslational import, dual targeting import, and the involvement of cytosolic factors during import have been identified in different plant organelles and compartments (Peeters and Small, 2001; Marc et al., 2002; Mackenzie, 2005; Millar et al., 2006; Glaser and Whelan, 2007; Chatre et al., 2009). Therefore, it is conceivable that actin, tubulin, and other cytoskeletal proteins could be imported into mitochondria by an undiscovered mechanism.
In this report, we demonstrate that actin is localized inside mung bean (Vigna radiata) seedling mitochondria. However, prior to germination, actin is hardly detectable within cotyledon mitochondria, and association of cytosolic actin and its gradual import from outside to inside of cotyledon mitochondria occur during seed germination. This transition appears to occur in coordination with the progressive development of the mitochondria from its quiescent state in dormant seeds to an actively respiring organelle in germinating seeds. Mitochondrial actin is localized in the mitochondrial inner membrane, intermembrane space, matrix, and contact sites. The import of fluorescent actin into mitochondria was demonstrated in the transgenic actin:green fluorescent protein (GFP) plant, and the actin moiety appeared to be sufficient to guide actin-GFP into mitochondria. Moreover, filamentous actin was visualized in mung bean mitochondria, while polymeric actin-GFP appeared as filamentous bundle-like structures in mitochondria of transgenic plants. Additionally, Latrunculin B (Lat B), an actin depolymerizing agent, triggered a decrease of the membrane potential and cytochrome C release in isolated mitochondria. Mitochondrial actin is bound to the mtDNA-nucleoprotein complex along with several other proteins, including an actin depolymerizing factor, porin, and adenine nucleotide translocator (ANT). The latter two, which are the constituents of contact site, are bound closely to mitochondrial actin.
RESULTS
Identification of Actin in Mitochondria
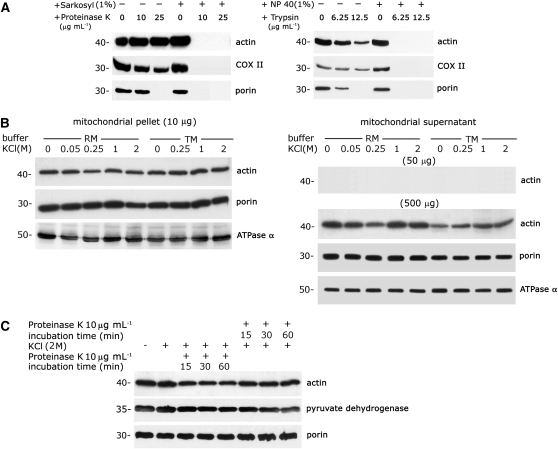
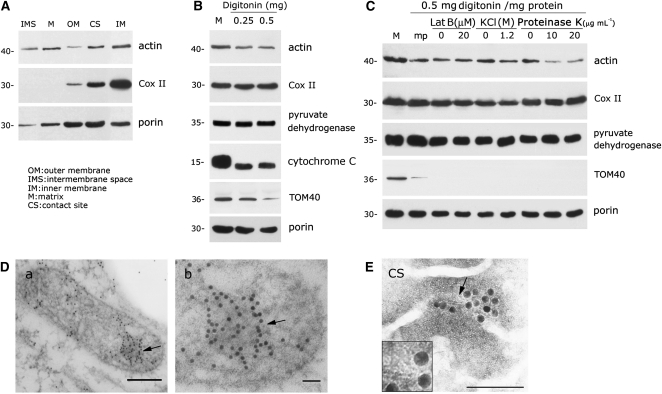
To eliminate actin outside of the mitochondria, purified mitochondria from 3-d-old mung bean seedlings were subjected to progressive proteolysis with serial concentrations of proteinase K (0, 10, and 25 μg mL−1) and trypsin (0, 6.25, and 12.5 μg mL−1). An antiactin antibody cross-reacted with a protein localized in intact mitochondria with and without protease treatment (Figure 1A). The mitochondrial inner membrane protein COX II and outer membrane protein porin were used as controls for mitochondrial integrity. Treatment of intact mitochondria with proteinase K (25 μg mL−1; left section, Figure 1A) or trypsin (12.5 μg mL−1; right section, Figure 1A) removed porin, but the actin and COX II proteins remained intact, as demonstrated by immunoblot analysis. Under these extreme experimental conditions, the proteolytic activity was strong enough to digest the outer membrane protein porin and to slightly digest the inner membrane protein COX II. However, a fraction of mitochondria-localized actin remained protected within the organelle. The resistance of actin, COX II, and porin to protease treatment was abolished if the mitochondria were prelysed with sarkosyl or Nonidet P-40 (Figure 1A).
Figure 1.
A Large Portion of Mitochondria-Localized Actin Is Resistant to Protease and High-Salt Treatments.
(A) A marked proportion of actin is localized to proteinase K– and trypsin-treated mitochondria. Purified mitochondria (5 μg protein) from 3-d-old mung bean seedlings were treated with different concentrations of proteinase K (left section) and trypsin (right section) with or without prelysing with detergent (1% [w/v] sarkosyl or 1% [w/v] Nonidet P-40). Immunoblot analyses with antiactin, anti-COX II (inner membrane protein), and antiporin (outer membrane protein) antibodies were performed after gel transfer.
(B) A large proportion of actin associated with mitochondria is not loosely bound or peripherally bound. Purified mitochondria were treated with various concentrations of KCl (0 to 2 M) in two different buffers (RM and TM). After two consecutive high-speed centrifugations and washing, the stripped mitochondria in the pellet and the dissociated proteins in the supernatant were collected and subjected to immunoblot analysis. Detection of actin in the supernatant was performed with fivefold (50 μg) and 50-fold (500 μg) excesses of protein (referring to the protein in the initial mitochondrial sample) by immunoblot analysis. Immunoblot analysis was performed with antibodies against actin, ATPase α (an inner membrane protein), and porin (an outer membrane protein).
(C) A large fraction of mitochondria-localized actin is resistant to a combined treatment with high salt and proteinase K. Purified mitochondria were subjected to progressive proteolytic digestion either before or after high-salt treatment and were repurified by centrifugation through a sucrose cushion. Antibodies against actin, pyruvate dehydrogenase (a matrix protein), and porin (an outer membrane protein) were used for immunoblot analysis.
Four independent experiments were performed for the above studies. Equivalent results were obtained, and the results of one experiment are presented here.
To remove peripheral proteins associated with the mitochondria, different concentrations of KCl (from 0 to 2 M in two kinds of buffer, RM and TM; see Methods) were applied to freshly purified mitochondria. The results shown in Figure 1B indicate that a large fraction of actin remained associated with the mitochondria after a high-salt treatment with both buffers. Porin and ATPase α (an inner membrane protein) were used as controls. Actin was not released into the supernatant after the salt-stripped mitochondria were removed by centrifugation, though actin was detected in the supernatant when a 50-fold excess of protein (500 μg) was analyzed. This trivial amount of actin was attributed to the mitochondrial debris remaining in the supernatant after centrifugation, as there was no difference in the actin remaining in the supernatant with or without salt treatment.
A large fraction of mitochondria and colocalized actin could not be eliminated by the combination of a high-salt treatment and progressive proteolytic digestion (Figure 1C). Proteolysis, either before or after the high-salt treatment, could not remove mitochondria-associated actin from the purified mitochondria. A matrix protein, pyruvate dehydrogenase, and porin were used as controls for mitochondrial integrity.
To remove possible filamentous actin that continued to be associated with the mitochondrial surface during organelle purification, Lat B treatment of the isolated mitochondria was performed in both F-actin depolymerizing (ADB) and stabilizing (FSB) buffers. No actin was released from the isolated mitochondria under depolymerizing conditions (see Supplemental Figure 1 online). Mitochondrial actin remained in the supernatant when high-salt/proteinase K/Lat B-pretreated mitochondria were lysed before centrifugation (see Supplemental Figure 2 online).
Progressive Import of Actin in Cotyledon Mitochondria during Seed Germination
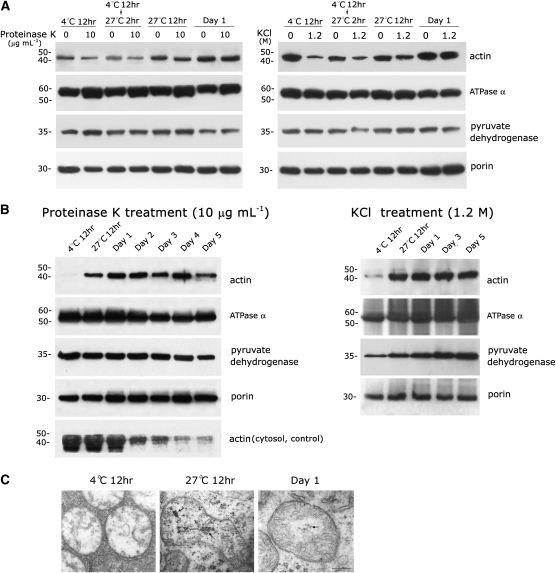
Unlike our results produced using mitochondria from 3-d-old mung bean seedlings described in Figure 1, actin associated with mitochondria harvested from cotyledons (after complete removal of the embryo) of seeds immersed at 4°C for 12 h and at 4°C for 12 h plus 27°C for 2 h were slightly sensitive to proteinase K (Figure 2A, left section) and markedly sensitive to the high-salt treatment (Figure 2A, right section). The sensitivity of cotyledon mitochondrial actin to the proteinase K and KCl treatments declined gradually, and actin from the mitochondria of cotyledons harvested from 1- to 5-d-old seedlings was resistant to both proteinase K and high-salt treatments (Figures 2A and 2B). A considerable amount of loosely bound or peripheral actin outside of the mitochondria could be removed using a high-salt treatment applied to cotyledon mitochondria purified from seeds immersed at 4°C for 12 h and at 4°C for 12 h with an additional 2 h at 27°C. The outer membrane protein porin, the inner membrane protein ATPase α, and the matrix protein pyruvate dehydrogenase were used as controls. Equivalent amounts of mitochondrial outer, inner, and matrix proteins were present in mitochondria obtained from cotyledons of different ages. The progressive accumulation of actin in the mitochondria was observed during cotyledon aging and seed germination (Figures 2A and 2B). Mitochondrial actin was barely detectable in cotyledon mitochondria harvested from seeds immersed at 4°C for 12 h after treatment with proteinase K or high salt, even though the cotyledon cytosolic actin was very abundant at this stage (Figure 2B, left section, compare top row to bottom row). The amount of mitochondrial actin reached a plateau in 1-d-old cotyledons and remained steady until day 4. Actin was always found after proteinase K (Figure 2B, left section) or KCl (Figure 2B, right section) treatment in cotyledon mitochondria that were harvested from seedlings grown for 1 to 5 d at 27°C; by contrast, actin was barely detectable in the cytosol of cotyledons on days 4 and 5 (Figure 2B, left section, bottom row). Apparently, the correlation between mitochondrial actin and cytosolic actin is quantitatively inconsistent during cotyledon development and seed germination. The cotyledon cytosolic actin was appreciably lower in day 4 and 5 seedlings, whereas the level of mitochondrial actin remained about the same at the corresponding stage (Figure 2B). Figure 2C shows the ultrastructures of cotyledon mitochondria in newly germinated seeds. A marked change was found in mtDNA organization during cotyledon mitochondrial development during early seed germination. The mitochondrial chromosome organization changed from nonvisible in cotyledon mitochondria from seedlings immersed at 4°C for 12 h to chromatin-like in cotyledon mitochondria from seedlings immersed at 27°C for 12 h and then to fibril-like in cotyledon mitochondria from 1-d-old seeds (indicated by arrows). This progressively alternating pattern of mtDNA organization undergoes a synchronous process in early cotyledon development during seed germination (see Table 1 in Dai et al., 2005).
Figure 2.
Progressive Import of Actin in Cotyledon Mitochondria during Seed Germination.
(A) Actin associated with mitochondria isolated from mung bean cotyledons (from which the embryo was completely removed) of early germinated seeds was slightly sensitive to proteinase K (left section) and highly sensitive to high-salt (right section) treatment. It became resistant to both treatments when the mitochondria were harvested from cotyledons after 1 d of seed cultivation. Cotyledon mitochondria purified from seeds immersed at 4°C for 12 h, at 4°C for 12 h plus 27°C for 2 h, and at 27°C for 12 h and 1-d-old seedlings were then treated with proteinase K (10 μg mL−1) and KCl (1.2 M) and analyzed by immunoblotting as described in Figure 1.
(B) Progressive import of actin into cotyledon mitochondria during seed germination and stability of actin levels in mitochondria of 1- to 4-d-old cotyledons. Cotyledon mitochondria were purified from seeds immersed at 4°C for 12 h and at 27°C for 12 h and from 1- to 5-d-old seedlings. Mitochondria were treated with proteinase K (10 μg mL−1) and KCl (1.2 M) and analyzed by immunoblotting as described in Figure 1. The presence of cytosolic actin during cotyledon aging and seed germination was also analyzed (bottom row of left section). Two and three independent experiments were performed for the above study in (A) and (B), respectively. Equivalent results were obtained, and only one is presented here.
(C) Ultrastructures of cotyledon mitochondria during early seed germination reveal reorganization of mtDNA from invisible to chromatin-like or fibril-like structures in seeds immersed at 4°C for 12 h and at 27°C for 12 h or 1-d-old seedlings (indicated by arrows), respectively.
Table 1.
Identification of Actin, VDAC, and ADP/ATP Carrier Protein (ANT) as Well-Bound DNA Binding Proteins in Mitochondria from Mung Bean Seedlings
| Accession No. | Protein Name | Mass | Scorea | emPAIb | Coverage (%)c | Peptide Scored | Peptide Sequence |
| Q9SRH5 | VDAC1 | 29,482 | 3,705 | 0.3 | 6.9 | 40.71 | NITTDLK |
| 94.84 | SFFTISGEVDTK | ||||||
| P42054 | VDAC | 29,596 | 1,315 | 2.26 | 12.7 | 40.71 | NITTDLK |
| 72.28 | GPGLYTDIGK | ||||||
| 65.11 | VKGPGLYTDIGK | ||||||
| 56.45 | GELFLGDVNTQLK | ||||||
| 80.46 | KGELFLGDVNTQLK | ||||||
| 58.22 | KGELFLGDVNTQLKNK | ||||||
| O22342 | ADP/ATP carrier protein 1 | 42,264 | 217 | 0.45 | 10.9 | 48.74 | SDGIAGLYR |
| 70.78 | QFNGLVDVYR | ||||||
| 55.7 | LLIQNQDEMIK | ||||||
| 69.61 | YFPTQALNFAFK | ||||||
| P31167 | ADP/ATP carrier protein 1 | 41,589 | 145 | 0.6 | 15 | 40.16 | TAAAPIER |
| 49.6 | GNTANVIR | ||||||
| 49.02 | LQLIVFGK | ||||||
| 62.48 | QFDGLVDVYR | ||||||
| 55.29 | LLIQNQDEMIK | ||||||
| 58.44 | YFPTQALNFAFK | ||||||
| P0C539 | Actin-2 | 41,907 | 234 | 0.32 | 17.8 | 99.11 | SYELPDGQVITIGAER |
| 46.88 | DLYGNIVLSGGSTMFPGIADR | ||||||
| 67.62 | TTGIVLDSGDGVSHTVPIYEGYALPHAILR | ||||||
| P93372 | Actin-66 (fragment) | 37,357 | 138 | 0.23 | 13.7 | 47.96 | SYELPDGQVITIGSER |
| 67.62 | TTGIVLDSGDGVSHTVPIYEGYALPHAILR | ||||||
| P30171 | Actin-97 | 41,870 | 92 | 0.32 | 16.2 | 44.09 | GYSFTTSAER |
| 51.26 | DLYGNIVLSGGTTMFPGIADR | ||||||
| 76.38 | TTGIVLDSGDGVSHTVPIYEGYALPHAILR | ||||||
| Q9LQ81 | Actin-depolymerizing factor 10 | 16,513 | 124 | 0.26 | 8.6 | 74.27 | IFFIAWSPDTSR |
VDAC, ADP/ATP carrier protein, and actin were present in the native well-bound mtDNA fraction (no cross-linking treatment) in four replicate studies. Free proteins, which are not bound to mtDNA, moved out from the well plug (see Supplemental Figure 4B online). A database search for protein identification was performed with MASCOT software against the UniProKB/Swiss-Prot database for Viridiplantae (green plants, 28,773 protein entries). Matched proteins were filtered with a significant threshold (P value) of <0.009 (see Methods).
The protein score from a MASCOT MS/MS search derived from the matched peptide ion scores.
The Exponetially Modified Protein Abundance Index (emPAI), a program integrated in the MASCOT search engine to quantify the relative amount of protein in a mixture based on protein coverage by the peptide matches.
The protein sequence coverage derived from the matched peptides for the protein.
The MASCOT MS/MS ion score for the matched peptide.
Mitochondrial Actin Appears to Possess a Similar Amino Acid Sequence to Cytosolic Actin by LC-MS/MS Analysis
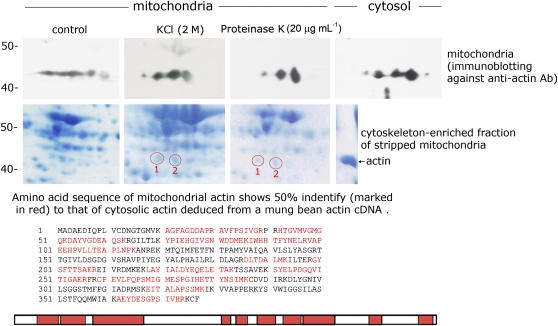
The cytoskeleton-enriched fractions that were isolated from high-salt and proteinase K pretreated mitochondria were analyzed by two-dimensional (2D) gel fractionation. Actin spots that were preidentified by immunoblot analysis (Figure 3, top panel) were then selected for liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) analysis. We compared the deduced amino acid sequence of mung bean cytosolic actin that we reported previously with the obtained mitochondrial amino acid sequence and found 50% identity (Figure 3, bottom panel; identity shown in red). Both spots 1 and 2 (circles) from mitochondrial cytoskeleton-enriched fractions obtained by KCl and proteinase K treatment were analyzed. All four spots showed the same amino acid sequences, but only the sequence of spot 1 (the cytoskeleton-enriched fraction extracted from KCl-treated mitochondria) is presented in the above-mentioned figure. The amino acid sequence of mitochondrial actin appears to be similar to that of cytosolic actin in mung beans.
Figure 3.
Mitochondrial Actin Possesses a Similar Amino Acid Sequence Compared with Cytosolic Actin and Was Identified by LC-MS/MS.
The top panel shows the immunoblot analysis with an antiactin antibody on a 2D gel of separated total mung bean seedling mitochondrial proteins (control [no treatment] or mitochondria treated with 2 M KCl or 20 μg mL−1 proteinase K) and total cytosolic protein. The middle panel shows the Coomassie blue–stained 2D gel fractionation profile of cytoskeleton-enriched fractions isolated from control mitochondria, 2 M KCl-treated mitochondria, and 20 μg mL−1 proteinase K–treated mitochondria (see Methods). Spots 1 and 2 were preidentified as actin based on immunoblot results shown in the top panel. The bottom panel shows the LC-MS/MS analysis of the amino acid sequence of spot 1 in the KCl-treated mitochondria shown in the middle panel. The same amino acid sequence was also found in spot 2 of KCl-treated mitochondria and in spots 1 and 2 of proteinase K–treated mitochondria. The amino acid sequence was compared with an amino acid sequence deduced from the mung bean actin cDNA sequence that we reported previously. The sequences marked in red are the identical sequence fragments between mitochondrial actin and cytosolic actin deduced from a mung bean actin cDNA.
Identification of Fluorescent Actin inside Mitochondria in actin:GFP Transgenic Plants
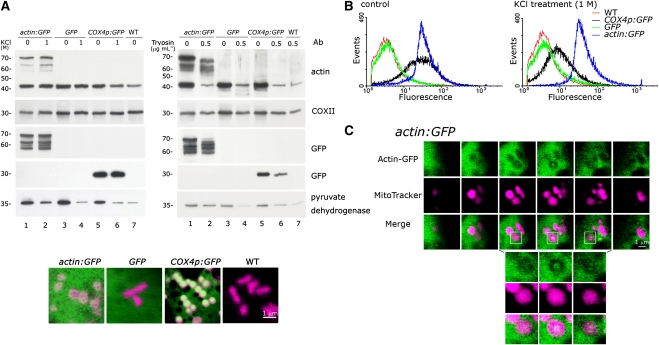
We constructed a transgenic tobacco (Nicotiana tabacum) plant, actin:GFP, which carries a fluorescent actin (using the mung bean actin cDNA) without a mitochondrial presequence and examined the import of the fluorescent actin into plant mitochondria by fluorescent image analysis accompanied by both positive and negative controls. Mitochondria purified from cultured actin:GFP, GFP, and COX4p:GFP (with the yeast COX4 mitochondrial targeting presequence fused to GFP) transgenic tobacco cells were treated with KCl (1 M) and trypsin (0.5 μg mL−1), repurified by cushion washing, and analyzed by immunoblotting (Figure 4A). The presence of the actin-GFP fusion protein (~70 kD) in mitochondria harvested from the actin:GFP cell cultures was detected by immunoblot analysis using either an antiactin or an anti-GFP antibody. Neither KCl nor trypsin treatment removed the actin-GFP fusion protein from actin:GFP mitochondria. However, some degradation occurred during the trypsin treatment. Under the same experimental conditions, GFP was found inside the mitochondria harvested from the COX4p:GFP cultured cells but was not present in mitochondria harvested from GFP transgenic cell cultures, which contained GFP lacking a mitochondrial target sequence. Only mitochondria-targeted GFP and actin-fused GFP resulted in the import of GFP into the mitochondria. Mitochondrial internal actin (42 kD), COX II, and pyruvate dehydrogenase were detected in all three transgenic strains and wild-type cells after KCl and trypsin treatments. Some degradation of actin and pyruvate dehydrogenase due to trypsin treatment is shown in Figure 4A (right section). A single confocal microscopy image of mitochondria from the three transgenic tobacco strains and wild-type cells stained with MitoTracker Red (CM-H2XRos, magenta) is shown at the bottom of Figure 4A.
Figure 4.
An Actin-GFP Fusion Protein without a Mitochondrial Targeting Presequence Is Imported into Mitochondria in Transgenic actin:GFP Tobacco Cells.
(A) Immunoblot analysis was used to demonstrate that actin-GFP is present in the mitochondria of actin:GFP transgenic cells. Mitochondria isolated from transgenic actin:GFP, COX4p:GFP, GFP, and wild-type cultured tobacco cells were treated with 1 M KCl or 0.5 μg mL−1 trypsin. Immunoblot analysis, as described in Figure 1, was performed after the treated mitochondria were washed on a Suc cushion. Three independent replicates were performed, and equivalent results were obtained. A single confocal microscopy image of mitochondria in the three transgenic tobacco cell lines and wild-type cells stained with MitoTracker Red (CM-H2XRos, magenta) is shown at the bottom. The cells used for the live image analysis were from 2-d-old cell cultures after transfer to fresh medium. Bar = 1 μm.
(B) Flow cytometric analysis of mitochondria isolated from tobacco cell cultures of wild-type (WT) and the GFP, COX4p:GFP, and actin:GFP transgenic plants. Purified mitochondria were also treated with 1 M KCl prior to flow cytometry. For each experiment, 50,000 mitochondria were analyzed. Three independent replicates were performed, and similar results were obtained.
(C) The actin-GFP fusion protein without a mitochondrial targeting presequence was imported into mitochondria in the actin:GFP transgenic cells and was visible by confocal microscopy. A clear actin-GFP fluorescent patch localized in the center of the mitochondrion (stained with MitoTracker Red, CM-H2XRos, indicated in magenta) is shown in the box. The top section contains six continuous sections of an actin:GFP cell. The bottom three images present enlargements of the region corresponding to the boxed area in the top section. Optical sections were made at 0.5-μm intervals, and the pinhole aperture was set to an optical thickness of 1 μm in this study. The cells used for the live image analysis were from a 2-d-old cell culture after transfer to a fresh medium. Bar = 1 μm.
More than 10 independent experiments were performed, and equivalent results were observed in each experiment.
The flow cytometry data shown in Figure 4B indicate that the isolated mitochondria from both COX4p:GFP and actin:GFP cultured tobacco cells exhibit GFP fluorescence; mitochondria isolated from cultures of GFP and wild-type cells did not show GFP fluorescence. After KCl treatment followed by cushion washing, both actin:GFP and COX4p:GFP mitochondria retained their GFP fluorescence. A significant decrease in GFP fluorescence was observed in the COX4p:GFP mitochondria. The actin:GFP mitochondria consistently remained at approximately the same fluorescence intensity after KCl treatment (Figure 4B, right section). Mitochondria isolated from actin:GFP tobacco cell cultures were extremely sensitive to protease treatment (shown in Figure 4A; as described above). The mitochondrial membrane was damaged at even moderate concentrations of protease, and it was therefore difficult to perform an accurate flow cytometric analysis of protease-treated mitochondria.
After staining with MitoTracker Red (CM-H2XRos, magenta), mitochondria from actin:GFP cultured cells were examined by confocal microscopy. A region of actin-GFP fusion protein localized in the center of a mitochondrion in cultured transgenic actin:GFP tobacco cells is indicated by the boxes in Figure 4C. Six continuous serial cell sections (0.5 μm per section under pinhole set at 1-μm optical thickness) are shown in the top section of Figure 4C. The bottom section of Figure 4C shows three enlarged continuous sections that correspond to the boxed area in the top section. A merged image of the actin-GFP and MitoTracker Red fluorescence (in white) clearly shows that the actin-GFP is localized at the center of the mitochondrion in the middle section of the three enlarged continuous sections. We found that denser green actin-GFP fluorescence appeared in the mitochondria of older tobacco cell cultures much more frequently than in the mitochondria of younger cultured cells. All of the mitochondria with dense green actin-GFP fluorescence displayed perfect merging between actin-GFP and MitoTracker (see Supplemental Figure 3 online).
Localization of Mitochondrial Actin in Different Mitochondrial Subcompartments
Following hypotonic treatment, differential speed centrifugation, and Suc gradient fractionation, mitochondrial subfractions corresponding to the outer membrane, intermembrane space, inner membrane, matrix, and contact sites were obtained and examined by immunoblot analysis. Figure 5A shows that actin was present in the fractions corresponding to the intermembrane space, matrix, inner membrane, and contact sites and was detected in small quantities in the outer membrane of the mitochondria fraction obtained from high-salt pretreatment followed by cushion washing.
Figure 5.
Biochemical Analysis of the Localization of Mitochondrial Actin in Different Mitochondrial Subcompartments.
(A) Immunoblot analysis indicates that mitochondrial actin resides in the inner membrane, matrix, contact sites, and, to a lesser extent, in the intermembrane space and in the outer membrane.
(B) The presence of actin in mitoplasts after most of the mitochondrial outer membrane was removed by different concentrations of digitonin. The mitoplast fraction after treatment with 0.25 to 0.5 mg digitonin per mg mitochondrial protein was purified as described in Methods, and immunoblot analysis was then performed. M indicates mitochondria.
(C) Mitoplast actin is slightly sensitive to high-salt treatment and highly sensitive to proteinase K treatment. Purified mitoplasts (marked as mp) isolated from digitonin-treated mitochondria were subjected to high salt (1.2 M), proteinase K (10 and 20 μg mL−1), or Lat B (20 μM) treatments. Immunoblot analysis was then performed after the mitoplasts were repurified on a Suc cushion. M indicates mitochondria.
Studies in (A) to (C) were repeated three to four times, and equivalent results were obtained each time.
(D) Immunogold labeling of an antiactin antibody indicates that actin resides in the mitochondrial matrix. Arrows in (a) and (b) indicate the gold particles representing actin inside mitochondrion ([b] is the enlargement area of [a] indicated by arrow); 5-nm gold labeling was applied in this experiment. Bars = 200 nm in (a) and 50 nm in (b).
(E) Immunogold-labeled negative staining ultrastructure analysis on contact sites purified by fractionation through two sequential Suc gradients (see Methods) revealed that gold labeling (18-nm gold) of an antiactin antibody appears coincident with ~5-nm filaments that pass across the mitochondrial membrane of the contact site. The inset box is the enlarged area indicated by the arrow. Bar = 200 nm.
Studies in (D) and (E) were repeated more than six and two times, respectively, and similar results were obtained each time.
An immunoblot analysis of mitoplasts purified from mitochondria treated with different concentrations of digitonin is presented in Figure 5B. We found that digitonin treatments higher than 0.5 mg per mg of mitochondria caused severe damage to the mitochondrial membranes. Therefore, we selected 0.5 mg digitonin per mg mitochondria for mitoplast isolation. A significant reduction in the intermembrane space protein cytochrome C was found in purified mitoplasts. Outer membrane proteins, including a small amount of Tom-40 and considerable amounts of porin, remained within mitoplasts harvested after digitonin treatment. Under our experimental conditions, most mitochondria that were converted into mitoplasts demonstrated a drastic loss of cytochrome C and Tom-40 but maintained steady levels of inner membrane and matrix proteins (COX II and pyruvate dehydrogenase, respectively). Sizable amounts of mitochondrial actin loss accompanied the removal of the mitochondrial outer membrane (cf. lane M, mitochondrion, to lane mp, mitoplast, in Figure 5C). However, considerable amounts of actin remained within mitoplasts purified from digitonin-treated mitochondria (Figures 5B and 5C). The results in Figure 5C also demonstrate that mitoplast (mp) actin is not sensitive to Lat B, slightly sensitive to high-salt treatment, and markedly sensitive to proteinase K treatment. Mitoplast actin, but not mitochondrial actin, was sensitive to proteinase K and KCl treatments. The levels of the inner membrane protein COX II and the matrix protein pyruvate dehydrogenase remained constant in mitoplasts after Lat B, KCl, and proteinase K treatments. The outer membrane protein porin was consistently associated with mitoplasts after treatment with different concentrations of digitonin. In situ immunogold labeling of thin sections of mung bean mitochondria supported the localization of actin in the matrix (Figure 5D). Gold particles representing actin often colocalized with denser areas in the mitochondrial matrix under electron microscopy examination (arrow). A contact site repurified from a secondary linear Suc gradient was used for an immunogold label-negative staining analysis. Gold particles representing actin appear to be colocalized with ~5-nm globular filament-like structures (Figure 5E). Moreover, these filament-like structures appear to pass across the mitochondrial membrane of the contact site.
Existence of Actin Filaments and Filamentous Bundle-Like Actin Patches in Mitochondria
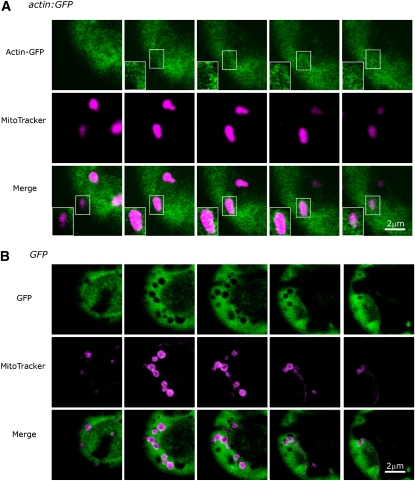
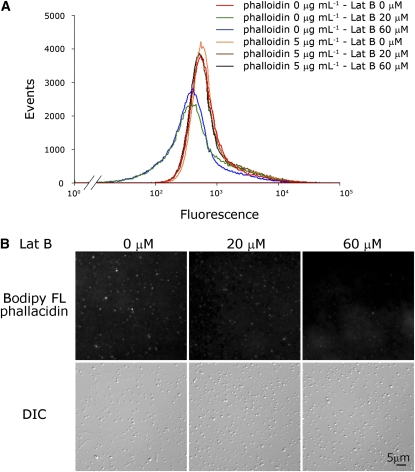
Under confocal microscopy, the optical continuous sections of actin:GFP protoplast cells shown in Figure 6A indicate that some actin-GFP fusion proteins are apparently arranged in filamentous bundle-like patches in actin:GFP mitochondria. Due to the size limitation of mitochondria, there is a degree of uncertainty regarding whether each and every actin-GFP fluorescent patch is completely situated inside the mitochondria. Nevertheless, the fact that more than 10 independent critical experiments yielded essentially the same result suggests that filamentous actin-GFP proteins exist in the mitochondria of actin:GFP transgenic tobacco plants. In addition, there is no GFP fluorescent patch was found in control transgenic plant GFP (Figure 6B). Furthermore, the actin appears to have polymerized into polymers in purified mung bean mitochondria, as demonstrated by staining of high salt–stripped mitochondria with F-actin–specific fluorescent phallacidin (Figure 7). The flow cytometric analysis shown in Figure 7A indicates that mung bean mitochondria can be stained by the F-actin–specific dye and that this fluorescence intensity decreases with serial concentrations of Lat B treatment in ADB buffer (cf. green and blue profiles to the red profile, respectively, in Figure 7A). Stabilizing mitochondrial F-actin via the addition of 5 μM phalloidin prior to Lat B treatment rescued the effect of Lat B on mitochondrial F-actin and hence retained the mitochondrial phallacidin fluorescence intensity in the flow cytometric analysis (the brown and black profiles shown in Figure 7A). The fluorescence images shown in Figure 7B provide the same conclusion (i.e., that some actin in purified mung bean mitochondria exist in a filamentous form and can be detected by the F-actin–specific dye). Depolymerization of F-actin by Lat B may reduce the fluorescence intensity of mitochondria stained by fluorescent phallacidin during image analysis. It is interesting to note that the fluorescence intensity of filamentous actin in mitochondria is highly diverse among individual organelles.
Figure 6.
The Existence of Filamentous Actin-GFP or Filamentous Bundle-Like Actin Patch in actin:GFP Mitochondria as Visualized by Confocal Microscopy.
(A) Five continuous sections of a protoplast cell from 12-d-old leaves of sterile transgenic actin:GFP tobacco plants are shown. Some actin-GFP fusion proteins (with no exogenously inserted mitochondrial targeting sequence) are apparently arranged in a filamentous bundle within mitochondria (stained with MitoTracker Red, CM-H2XRos, indicated in magenta). Inset boxes exhibit spots or filamentous bundle-like patches of merged actin-GFP and MitoTracker fluorescence (in white) in serial sections of a single mitochondrion.
(B) Five continuous optical sections of a protoplast cell from a leaf of a 12-d-old GFP transgenic tobacco plant, shown as a control for (A).
Figure 7.
F-Actin in Mitochondria, as Indicated by Staining with the F-Actin–Specific Fluorescent Dye Phallacidin.
(A) Isolated mitochondria washed briefly with 1 M KCl followed by various concentrations of Lat B treatment with or without 5 μM phalloidin pretreatment. Flow cytometric analysis indicates that KCl stripped mitochondria can be stained by the F-actin–specific dye FL phallacidin (red curve) and that this fluorescence is reduced by treatment with the F-actin depolymerization reagent Lat B (20 and 60 μM, respectively, shown in the green and blue curves). The stabilization of mitochondrial F-actin by phalloidin (5 μg mL−1) rescues the effect of Lat B on mitochondrial F-actin and retains the mitochondrial phallacidin fluorescence intensity (brown and black curves) in flow cytometric analyses. For each experiment, 500,000 mitochondria were analyzed.
(B) Fluorescence images indicate that the fluorescence of mitochondrial filamentous actin stained by fluorescent phallacidin was decreased caused by Lat B (20 and 60 μM) treatment. This result provides the same conclusion as that in (A) and indicates that actin in mitochondria exists in a filamentous form and that Lat B treatment decreases the fluorescence of FL phallacidin-stained mitochondria. Differential interference contrast (DIC) images in the bottom row represent individual mitochondrion existing in each image of the top row correspondingly.
Depolymerization of Mitochondrial Actin by Lat B Induces Mitochondrial Membrane Potential Loss and Cytochrome C Release
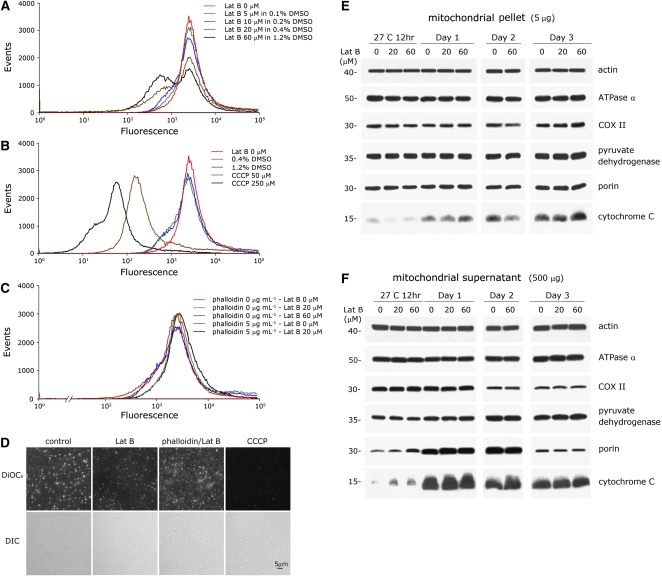
Mitochondrial membrane potential loss caused by treatment with F-actin depolymerizing reagent Lat B in ADB buffer was found in a large portion of the mitochondrial population (Figure 8A; compare the brown and black profiles to the red profile). Two distinct subpopulations were found in Lat B–treated mitochondria: the subpopulation that was sensitive to Lat B showed a reduced membrane potential, and the other subpopulation was resistant to Lat B treatment and retained its membrane potential. The degree of mitochondrial membrane potential loss, both in quality and quantity, was correlated with the concentration of Lat B applied. The control analyses for the mitochondrial membrane potential alteration experiments, application of DMSO (which was the Lat B solvent) or CCCP in ADB buffer, are shown in Figure 8B. Treatment of Lat B on mitochondria in F-actin stabilizing buffer FSB, with or without addition of phalloidin, shown no mitochondrial membrane potential alteration caused by Lat B treatment (Figure 8C). Five to 10 μM Lat B treatment does not cause a marked alteration of membrane potential in mitochondria. The fluorescence images in Figure 8D display the effect of Lat B in reducing the mitochondrial membrane potential in isolated mitochondria in ADB buffer. As shown in Figure 8A, the Lat B treatment reduced the mitochondrial membrane potential, and this effect can be rescued by treatment with phalloidin prior to Lat B treatment in ADB buffer.
Figure 8.
Depolymerization of Mitochondrial Actin by Lat B Induces Mitochondrial Membrane Potential Loss and Triggers the Release of Cytochrome C.
Purified mitochondria harvested from 1-d-old cotyledons treated with 1 M KCl. The mitochondrial membrane potential was assessed using the fluorescent probe DiOC6, followed by flow cytometric analysis.
(A) and (B) Mitochondrial membrane potential loss caused by various concentrations of Lat B treatment in ADB buffer. The control treatment of DMSO (in ADB buffer) on mitochondria is shown in (B). CCCP (50 and 250 μM) treatment of mitochondria was used as a positive control and is also shown in (B).
(C) Mitochondria in F-actin stabilizing buffer FSB, with or without phalloidin pretreatment, showed no effect of Lat B on the mitochondrial membrane potential.
(D) Fluorescence images of isolated mitochondria treated with Lat B (60 μM) in ADB buffer. The Lat B treatment reducing the mitochondrial membrane potential is detected by staining of mitochondria by DiOC6, and this effect can be rescued by pretreatment with phalloidin (5 μM) prior to Lat B (60 μM) treatment. CCCP, as a positive control, reduced the mitochondrial membrane potential completely. Three independent replicates were performed for (A) to (C), and eight independent repeats were conducted for (D). Similar results were obtained among the repeats. Differential interference contrast (DIC) images in the bottom row in (D) represent individual mitochondrion existing in each image of the top row correspondingly.
(E) and (F) Depolymerization of mitochondrial actin dynamics by Lat B triggers the release of cytochrome C in cotyledon mitochondria. Cotyledon mitochondrial samples were isolated and purified from mung bean seeds/seedlings that had been germinating for 12 h, 1 d, 2 d, and 3 d at 27°C. Purified mitochondrial samples were treated with 0, 20, or 60 μM Lat B in TM buffer for 30 min at 25°C. Immunoblot analysis of the mitochondrial pellet and supernatant was performed following the procedures described in Figure 1B. Detectable increases of cytochrome C released into the supernatant were observed in 12 h to day 2 cotyledon mitochondria caused by Lat B treatment in comparison to the control ([F], bottom row). This effect was not observed in mitochondria from 3-d-old seedlings. This study was repeated three times, and equivalent results were obtained each time. Samples of 12 h, day 1, and day 3 mitochondria are analyzed on the same PAGE gel, and the day 2 samples were analyzed on a separate gel.
Immunoblot analyses of the mitochondrial pellet and supernatant, respectively, following the Lat B treatments on cotyledon mitochondria of different ages indicated that depolymerization of mitochondrial actin dynamics by Lat B triggers the release of cytochrome C from mung bean mitochondria. Detectable increases in the release of cytochrome C caused by Lat B treatment were observed in mitochondria of 12-h to 2-d-old cotyledons compared with the control (with no Lat B treatment; Figure 8F, bottom row). The same effect of Lat B on the release of cytochrome C was not observed in mitochondria from 3-d-old seedlings.
Identification of Actin Binding to mtDNA by LC-MS/MS Analysis of a Native mtDNA-Protein Complex Obtained from an Agarose Plug after PFGE Fractionation
Purified mitochondria pretreated with high salt (1.2 M KCl) were lysed completely and fractionated by pulsed-field gel electrophoresis (PFGE). An mtDNA/mtDNA-nucleoprotein complex remained in a well-bound fraction in the original agarose plug (see Supplemental Figure 4A online) and was processed for LC-MS/MS analysis without cross-linking. Well-bound (wb) mtDNA did not migrate out of the sample well. It has been suggested that an intermediate of replicating mtDNA in yeast and plants is a rolling circle and/or a multifiber network similar to bacteriophage T4-replicating molecules. Either of these replicating molecules are likely causes of the immobility of plant mtDNA in PFGE analysis (Maleszka et al., 1991; Oldenburg and Bendich, 1998; Dai et al., 2005). Of the ~70 to 90 proteins identified, actin, porin, and ADP/ATP carrier proteins were repeatedly associated with well-bound mtDNA (Table 1). Because the amount of completely dissolved mitochondrial sample applied to each plug for PFGE analysis was limited, the quantity of the mtDNA/mtDNA-nucleoprotein complex obtained for each LC-MS/MS analysis was very low (see Methods). Regardless, actin, porin, and ADP/ATP binding proteins were repeatedly identified in each analysis from four independent studies.
DISCUSSION
The issue whether actin resides inside plant mitochondria is particularly difficult to address. There are good reasons for this is being so. First, no formal genetics is available in those plant species befitting molecular/cellular research on mitochondria. Equally difficult is the extrinsic technical challenge in obtaining large enough samples of adequate purity for conducting such research, while the size of a typical punctate mitochondrion is close to the limit of optical resolution.
Since actin is the most abundant protein in most cells, to demonstrate unambiguously that actin exists in mitochondria, it is critical to remove loosely bound and peripheral actin associated with the mitochondrial surface systematically. To this end, throughout this study, intact mitochondria samples were subjected to vigorously controlled high-salt and/or protease treatment prior to analysis. The protease treatment applied was adjusted to be strong enough to digest completely all mitochondria-associated actin and the inner/outer membrane proteins of the lysed mitochondrial samples. Even the outer membrane protein porin was degraded under these vigorous experimental conditions (for both proteinase K and trypsin), whereas a considerable amount of mitochondrial actin remained intact (Figure 1A). We conclude that this fraction of actin resides inside mitochondria protected from degradation by the mitochondrial membrane. Combining a high salt pre- and post-treatment with protease treatment did not alter the situation (Figure 1C). Boldogh et al. (1998) clearly showed that peripheral actin associated with isolated yeast mitochondria can be dislodged by high-salt treatment; this peripheral actin is ATP sensitive, and reversible actin binding activity toward mitochondria could be recovered after the ATP was removed.
Unlike in isolated yeast mitochondria, no appreciable amount of actin was released from mung bean seedling mitochondria into the supernatant after treatment with high salt (Figure 1B). The same was true for treatment with Lat B, an F-actin depolymerizing agent (see Supplemental Figure 1 online). These results indicate that either peripheral actin was lost during the mung bean mitochondrial purification procedure or that mitochondria of 3-d-old mung bean seedlings possess no peripheral actin or loosely bound actin. Stabilization of F-actin by phalloidin during mitochondrial purification did not alter these results. However, if mitochondria were pretreated with high salt, proteinase K, and Lat B and then lysed, actin was found in the supernatant, indicating that actin came from inside the mitochondria (see Supplemental Figure 2 online). On the other hand, a considerable amount of actin associated with young cotyledon mitochondria could be stripped away by high-salt or proteinase K treatment (Figure 2A). Interestingly, young cotyledon mitochondria-associated actin gradually became resistant to both protease and KCl treatments as the cotyledons aged during seed germination. In addition, the amount of KCl/protease-resistant actin markedly increased in 1-d-old and older cotyledon mitochondria (Figures 2A and 2B). Our previous report demonstrates that young cotyledon mitochondria from 4°C for 12 h or 4°C for 12 h followed by 27°C for 12 h immersed seeds and from seeds germinated for 1 d undergo a synchronous development in terms of their DNA organization. mtDNA converted from invisible to chromatin-like and then fibril-like as the cotyledon aged (Figure 2C; Table 1 in Dai et al., 2005), probably reflecting the progressive mitochondrial gene expression that takes place during this period.
The respiration activity of cotyledon mitochondria gradually increased during seed germination; it became readily detectable at day 1 and peaked at day 3 (see Supplemental Figure 5 online). Taken together, these results prompted us to postulate that the progressive decline of mitochondrial actin’s sensitivity to high-salt and protease treatments during seed germination may have resulted from the import of loosely bound/peripheral actin into the mitochondria. This change may occur in concert with the conversion of cotyledon mitochondria from a quiescent organelle in dormant seeds to an active organelle in germinating seeds. The inconsistency of mitochondrial versus cytosolic actin levels in developing cotyledons during seed germination (Figure 2B, left panel) suggests that mitochondrial actin and cytosolic actin may play independent roles in their cellular environments. Our previous report also showed that there was little protein left in the cytoplasm of 4- to 5-d-old mung bean cotyledons (Egorova et al., 2010). However, cotyledon mitochondria in these cells maintained a considerable amount of actin (Figure 2B). This phenomenon suggests that, during the process of programmed cell death (PCD) that occurs in mung bean seedling cotyledons, the persistent function of mitochondrial actin but not cytosolic actin may be necessary until the very end of the orderly cell death process.
Using immunoblotting, flow cytometry, and confocal microscopic analysis, we found that, in actin:GFP cultured tobacco cells, the actin-GFP fusion protein without a mitochondrial targeting presequence localized in mitochondria (Figure 4). As shown in Figure 4A, similar to mitochondria-targeted GFP, actin-fused GFP, but not GFP alone, was detected in mitochondria after protease and KCl treatment. Moreover, by flow cytometry, the intensity of GFP fluorescence detected in the mitochondrial sample isolated from transgenic actin:GFP tobacco cells was approximately the same as that from mitochondria-targeted GFP (COX4p:GFP) tobacco cells (Figure 4B). These results suggest that the cytosolic actin moiety may have guided the actin-GFP fusion protein into the mitochondria by an unknown mechanism.
Due to the limitation of resolution, microscopy detection of the presence of a protein/protein complex inside 1- to 2-μm punctate mitochondria is a technically challenging task. Thus, a set of extremely stringent experimental conditions was applied throughout our live image analysis (see Methods). Critical replicates were analyzed for both actin:GFP and control plants (COX4p:GFP and GFP) to confirm our results shown in Figures 4C and 6.
The analysis of mitochondrial actin in different mitochondrial subcompartments revealed that actin is present in the intermembrane space, inner membrane, matrix, and contact sites (Figure 5A). However, we cannot rule out the possibility of cross-contamination of different subcompartments during fractionation. Actin associated with mitoplasts (isolated by treating intact mitochondria with digitonin to remove their outer membrane) was sensitive to protease treatment and slightly sensitive to high-salt treatment (Figure 5C). This result suggests that a significant fraction of mitochondrial actin is located on the inner mitochondrial membrane that encapsulates the mitoplasts, while the remaining protease-insensitive actin resides in the matrix. This interpretation is consistent with the result of in situ immunogold labeling of thin mitochondrial sections. This experiment revealed that heavy gold particles representing actin often colocalized with denser areas in the mitochondrial matrix, as observed by electron microscopy examination (Figure 5D, arrow). This population of actin may correspond to the fluorescent actin patches seen by confocal microscopy (Figure 4C).
Immunogold actin labeling was also detected across the membrane of the contact site by phosphotungstic acid negative staining (Figure 5E). Some filaments, ~5 nm in width, that were composed of a globular protein-like structure colocalized with gold particles representing actin (see inset area indicated by the arrow in Figure 5E). This result suggests that actin filaments may be imported into the mitochondria via the contact site by a yet unidentified mechanism.
Actin-GFP fusion proteins organized into filamentous bundle-like patches were visualized inside mitochondria by confocal microscopy (Figure 6). The presence of filamentous actin in mitochondria was further demonstrated by biochemical analysis in mung bean mitochondria (Figure 7). To confirm that actin-GFP fusion protein molecules were capable of forming filamentous polymers, the polymerized filamentous network of actin-GFP in leaf epidermal cells and in chloroplasts are presented in Supplemental Figures 6 and 7 online, respectively. The presence of F-actin in mitochondria strongly suggests that this actin polymer may possess a functional role in mitochondria, similar to the F-actin found in other organelles.
Without using any protein cross-linkers, we demonstrated that mitochondrial actin was bound to well-bound mtDNA, which represents the mtDNA-nucleoprotein complex still organized similarly to its organelle form following PFGE fractionation of lysed intact mitochondria embedded in an agarose plug. Among the 70 to 90 proteins identified by LC-MS/MS analysis, porin and ADP/ATP carrier protein (ANT) repeatedly appeared to bind to well-bound mtDNA (Table 1). Notably, an actin-depolymerizing factor was also found in this native mtDNA-nucleoprotein complex. This interaction of mitochondrial actin with mtDNA is consistent with our previous report (Dai et al., 2005). Direct association of actin with mtDNA has also been shown in human mitochondria, suggesting that actin inside human mitochondria plays a role in mtDNA maintenance (Reyes et al., 2011).
Several other investigators have also reported the direct and indirect association or binding of actin to mtDNA (Boldogh et al., 2003; Dai et al., 2005; Wang and Bogenhagen, 2006). Instead of the possibility that internal mitochondrial actin is associated with mtDNA, Boldogh et al. (2003) concluded that mtDNA was linked to cytosolic actin through the mitochondrial protein complex, the mitochore, and that this interaction between mtDNA and actin is critical for mtDNA inheritance (Boldogh et al., 2003). Furthermore, the well-bound DNA fraction after PFGE is believed to be an intermediate of replicating mtDNA in yeast and plants (Maleszka et al., 1991; Oldenburg and Bendich, 1998; Dai et al., 2005). The actin bound to the well-bound mtDNA shown in Table 1 strongly suggests that actin is directly or indirectly involved in the segregation or inheritance of newly replicated mtDNA near the contact site, which connects the inner and outer membranes (Figure 5E; Boldogh et al., 2003; Meeusen and Nunnari, 2003; Dai et al., 2005). The appearance of actin-depolymerizing factors among the mtDNA binding proteins suggests that ADF may play a role in the regulation of gene expression in mitochondria. It was reported previously that ADFs may affect nuclear gene expression in Arabidopsis thaliana (Ruzicka et al., 2007; Burgos-Rivera et al., 2008).
Moreover, the coimmunoprecipitation data presented in Supplemental Figure 8 online demonstrate that porin and ANT are closely associated with mitochondrial actin. Given that porin and ANT are the constituents of the contact site, this reinforces a similar finding derived from the electron microscopy immunogold labeling experiment (Figure 5E). Taken together, these results suggest that the contact site may be involved in the import of actin from the cytosol, and actin dynamics at the contact site may affect the opening and closure status of the mPTP. Actin has been shown to be capable of modulating the gating process in Neurospora crassa voltage-dependent anion channel (VDAC) (Xu et al., 2001).
It is generally accepted that the alteration of mitochondrial membrane permeability may facilitate the release of cytochrome C and other apoptogenic proteins and could consequently trigger apoptosis or PCD (Kroemer et al., 2007). Our data support and reinforce this view. Beginning 12 h after seed imbibition at 27°C, a considerable amount of actin was associated with cotyledon mitochondria (Figures 2A and 2B). Following an in vitro Lat B treatment of cotyledon mitochondria isolated at this time (12 h after seed imbibition), we detected an increase of cytochrome C release (Figure 8F). Significantly, a caspase-like protease activity also emerged at this time in cotyledon tissue, while nuclear DNA fragmentation began ~12 h later (Egorova et al., 2010). We also observed a decrease in membrane potential induced by treatment with Lat B in day 1 cotyledons (Figure 8A). Our findings suggest that the dynamics of mitochondrial actin could trigger cytochrome C release, consequently initiate PCD/apoptosis, and play a functional role in PCD/apoptosis modulation. The dynamics of cytosolic actin involved in yeast apoptosis has been studied by Gourlay et al. (2004). Instead of cytosolic actin, we suggest that interior mitochondrial actin signaling may play a role in regulating mitochondrial cytochrome C release and initiation the PCD/apoptosis in mung bean cotyledons.
It is interesting to note that only a subpopulation of cotyledon mitochondria exhibited a decreased membrane potential in response to Lat B–induced mitochondrial actin depolymerization (Figure 8A), and, presumably, cytochrome C is released only from this subpopulation of mitochondria. If this presumption is correct, then our results subtly differ from a well-reasoned idea. Commenting in a review article, Reed (1997) distinguished apoptosis from necrosis by suggesting that only a small fraction of cytochrome C escaped into the cytosol to trigger an orderly apoptotic cell death process, whose progression may require continued oxidative phosphorylation sustained by cytochrome C remaining in the mitochondria. Here, Reed most likely implied that cytochrome C was released from all mitochondria rather than a subpopulation of mitochondria in the cell.
Our previous study (Egorova et al., 2010) showed that during seed germination, the cotyledon underwent PCD from day 1 to day 5 and the cotyledon mitochondria kept approximately the same amount of cytochrome C throughout, while respiration activity was elevated from its lowest level observed at 12 h at 4°C imbibition to the highest level at day 3 and subsequently declined (see Supplemental Figure 5 online). Additionally, the amount of cytosolic cytochrome C did not markedly change during this period. No ruptured mitochondria were found in day 3 and younger cotyledon cells by electron microscopy analysis in situ (Egorova et al., 2010). This latter result is in contrast with an earlier report of rupture of the outer mitochondrial membrane in apoptosis (Vander Heiden et al., 1997).
It is generally accepted that caspase-dependent apoptosis is triggered by a mitochondrial outer membrane permeabilization or compromised mitochondrial outer membrane integrity resulting in the release of soluble protein, including cytochrome C from intermembrane space. However, there is still no clear consensus as to how such compromised mitochondrial outer membrane integrity arises mechanistically. Pore formation involving VDAC homo-olimers, VDAC-Bax hetero-oligomers, and Bax homopolymers have been suggested (Rostovtseva et al., 2005; Martinou and Youle, 2011). Moreover, apoptotic mechanisms may differ in different organisms.
Taken together, these results suggest that mitochondrial actin may be involved in mitochondrial DNA segregation and mitochondrial division. Our results also imply that the dynamics of mitochondrial actin regulate the release of cytochrome C and consequently initiate PCD/apoptosis. We recognize that our work has just laid the groundwork for further exploration. It is our hope that in-depth investigations will be forthcoming to confirm and extend the exact role of actin in mitochondria and to elucidate the mechanistic details of actin import.
METHODS
Plant Materials and Growth Conditions
Mung bean used in this study was Vigna radiata cv Tainan No. 5, and the tobacco used for transgenic plants was Nicotiana tabacum W 38. The growth conditions for mung bean were 27°C in the dark unless otherwise indicated. The growth conditions for transgenic tobacco plant were 16 h light/8 h dark at 26°C.
Transgenic Plants
Transgenic GFP and COX4p:GFP (with an N-terminal mitochondrial targeting presequence from yeast COX4) plants were described previously (Lo et al., 2004). Actin:GFP was constructed using similar procedures. A complete actin cDNA gene fragment isolated from mung bean (V. radiata, TN-5) was ligated to the 5′-end of GFP and then into the pKYLX71:35S2 plasmid (a kind gift from A. Hunt, University of Kentucky). Primers used for actin were U1, 5′-CCGCTCGAGATGGCCGATGCTGAGGAT-3′ (underlining indicates the for XhoI site), and L1, 5′-CCCAAGCTTAAAGCATTTCCTGTGTAC-3′ (underlining indicates the HindIII site); primers used for GFP were U1, 5′-CCGCTCGAGAAGCTTATGGTGAGCAAGGGCGAG-3′ (underlining indicates the XhoI and HindIII sites), and L1, 5′-GCTCTAGAGTCGCGGCCGCTTTACTT-3′ (underlining indicates the XbaI site). The amplified actin fragment was digested by XhoI and HindIII, and the amplified GFP fragment was digested with HindIII and XbaI and then ligated to the vector pKYLX71, which was digested with XhoI and XbaI. Tobacco cells were used for transformation.
Mitochondria and Mitoplast Preparations
Mung bean mitochondria were prepared from 3-d-old etiolated mung bean seedlings and purified on a Suc gradient as described previously (Dai et al., 1998). Immunoblot analysis with the ribulose-1,5-bis-phosphate carboxylase/oxygenase L antibody was routinely performed to verify the purity of the isolated mitochondria. Mitochondria from tobacco cell cultures were purified using this procedure, with minor modification by doubling the cell blending speed. Mitoplasts were purified as described by Donahue et al. (2001).
Cotyledon mitochondria were isolated from cotyledons of different ages after the embryos were carefully and completely removed at 4°C in mitochondrial isolation buffer (Dai et al., 1998). The mitochondria used in this study were harvested from cotyledons of seeds immersed in water at 4°C for 12 h, at 4°C for 12 h plus 27°C for 2 h, at 27°C for 12 h, and at 27°C for 12 h followed by 12 h of growth in vermiculite (1-d-old and 2- to 5-d-old seedlings).
Protoplast Isolation
Protoplasts were isolated from 12-d-old sterile transgenic plants. Fully expanded leaves were cut into 0.1- to 0.5-cm2 pieces in a liquid medium containing 0.5% cellulase (Onozuka-R), 0.06% macerozyme (Yakult), 0.1% BSA, and 0.55 M mannitol and were incubated overnight at 26°C in the dark with gentle shaking. Subsequently, this mixture was passed through a sterile stainless steel mesh sieve (100-μm mesh size) followed by a 5-min centrifugation at 107g, 25°C. The protoplast pellet was then gently dissolved in 2 mL 0.55 M mannitol. After 2 mL of 0.55 M Suc was added to the bottom of the above protoplast mixture in a centrifuge tube, centrifugation was conducted at the same speed. Intact protoplasts were collected from the interphase and were transferred into a new tube. The protoplasts were washed once with 5 mL 0.55 M Suc. Purified protoplasts were resuspended in 0.55 M mannitol for further examination.
Preparation of Suspension Cells
Callus cultures were initiated from 1-month-old transgenic plant leaves. Leaf segments (0.5 to 1.0 cm) were placed on Murashige and Skoog medium supplemented with 2 mg L−1 of 2,4-D, 2.5 g L−1 phyta agar, 0.2 g L−1 kanamycin, and 30 g L−1 Suc in the dark for callus induction. The pH of this medium was adjusted to 5.7. For the initiation of suspension cultures, ~1 g of callus clumps were transferred to 25 mL liquid medium with 1 mg L−1 naphthaleneacetic acid, 0.5 mg L−1 kinitin, 0.2 g L−1 kanamycin, and 30 g L−1 Suc in 125-mL flasks shaking at 125 rpm in complete darkness at 26°C. After 7 to 14 d of incubation, the cells were passed through a sterile stainless steel mesh sieve (mesh size 1 mm) and transferred to solid Murashige and Skoog medium.
High-Salt, Protease, and Lat B Treatments of Purified Mitochondria or Mitoplasts
Various concentrations (0 to 2 M) of KCl in two buffers (RM and TM buffers) were used to treat the mitochondria. Mitochondria (1 mg mL−1) were treated in RM buffer (0.6 M sorbitol, 20 mM HEPES, pH 7.4, 2 mM MgCl2, protease inhibitor cocktail [Roche], and 1 mM PMSF) on ice for 15 min (Boldogh and Pon, 2001) or TM buffer (0.25 M Suc, 20 mM Tris-HCl, pH 7.2, protease inhibitor cocktail [Roche], and 1 mM PMSF) on ice for 45 min. Because our experimental results differed from those reported by Boldogh and Pon (2001), we applied the TM buffer in parallel for a double verification. After centrifugation at 20,000g for 20 min at 4°C, both the supernatant and pellet were collected. The supernatant protein was desalted and concentrated using Centricon 30 columns (Millipore). If the supernatant was not used for analysis, the reaction mixture was centrifuged through a Suc cushion (25% [w/v] in 20 mM HEPES-KOH, pH 7.4, protease inhibitor cocktail [Roche], and 1 mM PMSF) to recover mitochondria from the pellet. Protease treatments of the mitochondria were performed in the buffer described by Chaumont et al. (1990) (20 mM HEPES, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM KH2PO4, 10 mM malate, 1 mM DTT, and 0.25 M mannitol) with minor modifications on treatment time. Mitochondria (1 mg mL−1) with or without KCl pretreatment were treated with proteinase K at 4°C for 15 min or with trypsin at 37°C for 30 min. For the protease treatment of lysed mitochondria, Nonidet P-40 (1% [w/v]) or sarkosyl (1% [w/v]) was added prior to the reaction.
Mitochondria suspended in FSB buffer (2 mM Tris-HCl, pH 8.0, 4 mM MgCl2, 50 mM KCl, 0.5 mM ATP, 1 mM EGTA, and 0.25 M Suc; modified from Boldogh et al. [1998]) with or without 5 μg mL−1 phalloidin were incubated for 30 min on ice. Mitochondria were pelleted and then treated with various concentrations of Lat B in F-actin nonprotection buffer, ADB (2 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, and 0.25 M Suc), and FSB buffer, respectively. The pellet and supernatant were then collected by centrifugation at 20,000g twice for 20 min each (see Supplemental Figure 1 online).
Mitochondrial Subfractionation
Purified mitochondria stripped by high salt (2 M KCl) were suspended in the hypotonic buffer HBH (10 mM HEPES-KOH, pH 8.0, 68 mM Suc, 0.25 mM DTT, 0.2 mM CaCl2, 0.2 mM ATP, 0.1 mM PMSF, and 0.1 mg mL−1 leupeptin) at a mitochondria concentration of 10 mg mL−1 and stored on ice for subfractionation. Mitochondrial subfractionation followed the procedures described by Hoppel et al. (2002) with minor modifications. The highly purified contact sites obtained by two sequential Suc gradient fractionations also followed the methods of Hoppel et al. (2002). The HBH buffer system described above was applied throughout the subfractionation process.
Immunoblot Analysis
Proteins were separated by SDS-PAGE and were then transferred to polyvinylidene fluoride membranes (Millipore). Actin, COX II, ATPase subunit α, pyruvate dehydrogenase, cytochrome C, TOM 40, and porin (VDAC) were identified by incubating the membranes with antiactin (obtained by immunizing rabbits with recombinant mung bean actin in this laboratory: cDNA of mung bean actin was amplified with primers U1, 5′-GGATCCGAATTCATGGCCGATGCTGAG-3′, and L1, 5′-GTCGACCTCGAGTTAAAA GCATTTCCT-3′, for EcoRI and XhoI sites, respectively, and then cloned into pGEX-6P-1 [GE] for protein expression followed by actin protein purification and rabbit immunization) or clone C4 (the antiactin antibody of clone C4 [Boehringer Mannheim], used only in Figures 1 and 5D; see Supplemental Figure 1 online), anti-COX II (a gift from T. Mason, University of Massachusetts), anti-porin, anti-ATPase α (GTMA), antipyruvate dehydrogenase (GTMA), anticytochrome C (BD Pharmingen), anti-GFP (Millipore), or anti-TOM 40 (Santa Cruz) antibodies, respectively. Immunoprotein complexes were detected with the Amersham ECL PlusWestern Blotting Detection System (GE Healthcare). Both self-immunized antiactin antibodies used in our laboratory (after repurification) and the purchased clone C4 antiactin antibody were highly specific for actin in different plant samples (mung bean and tobacco) as well as in different tissue samples (cotyledon, leaf, and stem). Cytosolic proteins were obtained by grinding the cotyledons into powder in liquid nitrogen, dissolving the powder in TM buffer, maintaining the samples at −80°C, and using the samples within 7 d.
Fine Structure and Immunolocalization Analysis
The fine structure analysis and the immunolocalization in tissue sections were performed as described previously (Dai et al., 1998, 2005).
PFGE Fractionation of the mtDNA/mtDNA-Nucleoprotein Complex
Purified mitochondria (2 mg) pretreated with 1 M KCl were completely lysed with 1% (w/v) sarkosyl and 1% (w/v) Nonidet P-40. An aliquot of lysate (0.5 mL) was then mixed with an equal volume of 1.5% (w/v) low melting point agarose (stored at 42°C), and the mixture was pipetted into a mold and allowed to set on ice for 10 min. The plugs were then loaded into the well and subjected to PFGE analysis (Dai et al., 2005). PFGE was performed with a 30- to 60-s pulse time (at a ratio of A:B = 1) and 150 V (5 V/cm) for 10 h in 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA) at 13°C. Gels were then stained with ethidium bromide. The well-bound fraction (remaining in the plug) was collected for further LC-MS/MS analysis.
LC-MS/MS Analysis
Untreated mung bean seedling mitochondria or mitochondria treated with 2 M KCl or 20 μg mL−1 proteinase K were used for 2D gel electrophoresis. Mitochondrial actin was detected by immunoblot analysis with an antiactin antibody. Cytosolic protein was harvested from a total extract of mung bean seedlings. The mitochondrial cytoskeleton-enriched fraction was the pellet obtained after centrifugation at 26,000g from purified mitochondria lysed with 1% (w/v) each of Triton X-100 and DOC and pretreated with 2 M KCl or 20 μg mL−1 proteinase K. Protein spots were used for LC-MS/MS analysis following the procedures described by Shevchenko et al. (1996) and Heazlewood and Millar (2007).
The protocol used for trypsin digestion of proteins in agarose gels was primarily adapted from the method described by Wilm and Mann (1996), with some modifications. Briefly, the well-bound mtDNA/mtDNA nucleoprotein complex in the agarose plug was manually excised and cut into small pieces. The gel pieces were placed in Eppendorf tubes, washed with 25 mM NH4HCO3 solution for 20 min, and then dried in a vacuum centrifuge. The reduction of proteins with DTT was performed at room temperature for 2 h and then alkylation with iodoacetamide was performed. The gel pieces were then washed and dried in a vacuum centrifuge before trypsin digestion. An aliquot of 12.5 ng μL−1 trypsin (sequencing grade; Promega) in 25 mM NH4HCO3 was added to cover all gel pieces, and the mixture was incubated for 12 to 16 h at 37°C. To recover the tryptic peptides, an ~50-μL aliquot of solution containing 5% (w/v) formic acid and 50% (w/v) acetonitrile was added to the gel pieces, agitated for 1 h, and moved into a new tube. The recovery was then repeated with solution containing 2.5% (w/v) formic acid and 50% (w/v) acetonitrile for 1 h. The resulting two elutions were combined, and the tryptic peptides were purified using a PepClean C-18 spin column (Pierce). The dried pellet was redissolved in 10 to 20 μL 0.1% (w/v) formic acid for LC-MS/MS analysis.
The database search for protein identification is described as follows. The acquired MS/MS data were analyzed using an in-house Mascot search program (v. 2.2.06; Matrix Science) against the UniProtKB/Swiss-Prot Viridiplantae (green plants, 28,773 protein entries) database (release-2010_02) downloaded from the website ftp://ftp.uniprot.org/. The peak list data (DTA) files used for database searches were generated from Xcalibur raw files using Mascot Daemon with the incorporated extract_msn.exe program (BioWorks Rev. 3.3.1 SP1; Thermo Scientific). The parameters used for DTA conversion were as follows: a minimum mass of 600, a maximum mass of 4500, a group scan of 1, a minimum group count of 1, and a grouping tolerance of 1.4 atomic mass units (u). The protein sequences in the database were searched with trypsin/P (cleavage allowed in sequence Lys/Arg-Pro) digestion at both ends. Two missed cleavages, fixed modifications of carbamidomethylation at Cys, and variable modifications of oxidation at Met were allowed. The DTA files were searched with 1.5 D of precursor peptide mass tolerance (average mass) and 0.8 D of MS/MS mass tolerance. A decoy database search was performed, and a significant threshold (P) of ~0.009% (w/v), an expectation cutoff value of 0.05, and bold red requirements were then used to filter the resulting peptides to obtain more confident protein hits.
Flow Cytometric Analysis of Isolated Mitochondria
The GFP associated with mitochondria harvested from cell cultures of transgenic actin:GFP, GFP, and COX4p:GFP lines with and without high-salt pretreatment was analyzed with an Elite ESP flow cytometer (Beckman and Coulter; Figure 4B) using a 35-mW 488-nm argon laser for excitation. Fluorescence was measured at 488 to 550 nm.
For mitochondrial F-actin and mitochondrial membrane potential analyses, a Beckman Coulter MoFlo Height Performance cell sorter was used after KCl-stripped mitochondria were stained with Bodipy FL phallacidin and DiOC6 (3′,3′ dihexyloxacarbocyanine iodide), respectively. The Bodipy Fl phallacidin and DiOC6 treatment consisted of 5 units mL−1 and 500 nM in TM buffer for 3 min, respectively. The treatment conditions for each sample were strictly controlled under exactly the same conditions.
Live Imaging of Cultured Transgenic Plant Cells
Transgenic actin:GFP, COX4p:GFP, GFP, and wild-type control tobacco cells were stained with 250 nM MitoTracker Red CM-H2XRos (Molecular Probes) for 30 min at room temperature. Images were collected with a Zeiss LSM510 meta confocal microscope (Carl Zeiss MicroImaging) and a C-Aprochromat ×63/1.2-W objective (Carl Zeiss MicroImaging). GFP was excited using a 488-nm argon laser, and the fluorescence signal was collected with a 505- to 530-nm band-pass filter. The fluorescence of MitoTracker was excited using the 543-nm line of a He-Ne laser, and the fluorescence signal was collected with a 565- to 615-nm band-pass filter. Optical sections were made at 0.5-μm intervals, and the pinhole aperture was set at an optical thickness of 1 μm. The analysis conditions for each experiment were carefully controlled. The necessary replicates were performed on both actin:GFP and control plants (COX4p:GFP and GFP, positive and negative controls, respectively).
Fluorescent images analyses on isolated mitochondrial membrane potential and on mitochondrial filamentous actin were performed after staining with 500 nM DiOC6 for exactly 10 to 15 s and 10 units mL−1 FL phallacidine for exactly 10 to 15 s, respectively. Fluorescent images for analysis of mitochondria were taken using a Zeiss Axio-Z1 microscope. The experiments were independently repeated eight times; therefore, comparable conclusions can be drawn for each.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number 143208 for actin cDNA from mung bean.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Resistance of Mitochondrial Actin to the F-Actin Depolymerizing Reagent Latrunculin B.
Supplemental Figure 2. Mitochondrial Actin Remained in Supernatant after Mitochondria Were Lysed with 1% (w/v) Sarkosyl.
Supplemental Figure 3. Actin-GFP Fusion Protein without a Mitochondrial Targeting Presequence Is Imported into Mitochondria in Older actin:GFP Transgenic Cultured Cells.
Supplemental Figure 4. Well-Bound mtDNA/mtDNA-Nucleoprotein Complexes after PFGE Fractionation Used for LC-MS/MS Analysis.
Supplemental Figure 5. Comparison of Respiratory Activity among Mitochondria Isolated from Different Aged Cotyledons during Mung Bean Seed Germination.
Supplemental Figure 6. Networks of Actin-GFP Fusion Protein Filaments Seen in the Cytoplasm of an actin:GFP Transgenic Tobacco Plant.
Supplemental Figure 7. Filament Network of an Imported Actin-GFP Fusion Protein (with No Exogenously Inserted Chloroplast Targeting Sequence) in Chloroplasts of Transgenic Tobacco Cells.
Supplemental Figure 8. Interaction between Mitochondrial Actin and Porin/ANT as Demonstrated by Immunoprecipitation Analysis.
Acknowledgments
This work was supported by research grants from the National Science Council and Academia Sinica, Republic of China. A.A. was supported by a National Science Council postdoctoral fellowship. We thank Wann-Neng Jane for his help with the electron microscopy study. We acknowledge the technical services provided by the Flow Cytometry Core Facility of the Scientific Instrument Center, Academia Sinica in Institute of Plant and Microbial Biology–Academia Sinica.
AUTHOR CONTRIBUTIONS
Y.-S.L., N.C., L.-J.H., and A.A. performed research. G.-Y.J. contributed new analytic tools. T.-N.W. contributed new computational tools. H.D. designed the research, analyzed data, and wrote the article. K.-S.C. designed the research and analyzed data.
References
- Ausmees N., Kuhn J.R., Jacobs-Wagner C. (2003). The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 115: 705–713 [DOI] [PubMed] [Google Scholar]
- Boldogh I., Vojtov N., Karmon S., Pon L.A. (1998). Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J. Cell Biol. 141: 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I.R., Nowakowski D.W., Yang H.C., Chung H., Karmon S., Royes P., Pon L.A. (2003). A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell 14: 4618–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I.R., Pon L.A. (2001). Assaying actin-binding activity of mitochondria in yeast. Methods Cell Biol. 65: 159–173 [DOI] [PubMed] [Google Scholar]
- Boldogh I.R., Pon L.A. (2006). Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta 1763: 450–462 [DOI] [PubMed] [Google Scholar]
- Burgos-Rivera B., Ruzicka D.R., Deal R.B., McKinney E.C., King-Reid L., Meagher R.B. (2008). ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 68: 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R. (2006). The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70: 888–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré M., André N., Carles G., Borghi H., Brichese L., Briand C., Braguer D. (2002). Tubulin is an inherent component of mitochondrial membranes that interacts with the voltage-dependent anion channel. J. Biol. Chem. 277: 33664–33669 [DOI] [PubMed] [Google Scholar]
- Chatre L., Matheson L.A., Jack A.S., Hanton S.L., Brandizzi F. (2009). Efficient mitochondrial targeting relies on co-operation of multiple protein signals in plants. J. Exp. Bot. 60: 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F., O’Riordan V., Boutry M. (1990). Protein transport into mitochondria is conserved between plant and yeast species. J. Biol. Chem. 265: 16856–16862 [PubMed] [Google Scholar]
- Dai H., Lo Y.S., Jane W.N., Lee L.W., Chiang K.S. (1998). Population heterogeneity of higher-plant mitochondria in structure and function. Eur. J. Cell Biol. 75: 198–209 [DOI] [PubMed] [Google Scholar]
- Dai H., Lo Y.S., Litvinchuk A., Wang Y.T., Jane W.N., Hsiao L.J., Chiang K.S. (2005). Structural and functional characterizations of mung bean mitochondrial nucleoids. Nucleic Acids Res. 33: 4725–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue R.J., Razmara M., Hoek J.B., Knudsen T.B. (2001). Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. FASEB J. 15: 635–644 [DOI] [PubMed] [Google Scholar]
- Egorova V.P., Zhao Q., Lo Y.S., Jane W.N., Cheng N., Hou S.Y., Dai H. (2010). Program cell death of the mung bean cotyledon during seed germination. Bot. Stud. 51: 439–449 [Google Scholar]
- Errington J. (2003). Dynamic proteins and a cytoskeleton in bacteria. Nat. Cell Biol. 5: 175–178 [DOI] [PubMed] [Google Scholar]
- Etoh S., Matsui H., Tokuda M., Itano T., Nakamura M., Hatase O. (1990). Purification and immunohistochemical study of actin in mitochondrial matrix. Biochem. Int. 20: 599–606 [PubMed] [Google Scholar]
- Gourlay C.W., Carpp L.N., Timpson P., Winder S.J., Ayscough K.R. (2004). A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 164: 803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser E., Whelan J. (2007). Import of nuclear-encoded mitochondrial proteins. In Plant Mitochondria, Logan D.C., (Oxford, UK: Blackwell; ), pp. 97–128 [Google Scholar]
- Graumann P.L. (2007). Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 61: 589–618 [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Millar A.H. (2007). Arabidopsis mitochondrial proteomics. Methods Mol. Biol. 372: 559–571 [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Shaw J.M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14: 265–303 [DOI] [PubMed] [Google Scholar]
- Hobbs A.E., Srinivasan M., McCaffery J.M., Jensen R.E. (2001). Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppel C., Kerner J., Turkaly P., Minkler P., Tandler B. (2002). Isolation of hepatic mitochondrial contact sites: Previously unrecognized inner membrane components. Anal. Biochem. 302: 60–69 [DOI] [PubMed] [Google Scholar]
- Jones L.J., Carballido-López R., Errington J. (2001). Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T., Ohta T., Kuroiwa H., Shigeyuki K. (1994). Molecular and cellular mechanisms of mitochondrial nuclear division and mitochondriokinesis. Microsc. Res. Tech. 27: 220–232 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Brenner C. (2007). Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87: 99–163 [DOI] [PubMed] [Google Scholar]
- Lo Y.S., Hsiao L.J., Jane W.N., Charng Y.C., Dai H., Chiang K.S. (2004). GFP-targeted mitochondria show heterogeneity of size, morphology, and dynamics in transgenic Nicotiana tabacum L. plants in vivo. Int. J. Plant Sci. 165: 949–955 [Google Scholar]
- Lo Y.S., Wang Y.T., Jane W.N., Hsiao L.J., Chen O.L.F., Dai H. (2002). The presence of actin-like filaments in higher plant mitochondria. Bot. Bull. Acad. Sin. 44: 19–24 [Google Scholar]
- Logan D.C. (2006). Plant mitochondrial dynamics. Biochim. Biophys. Acta 1763: 430–441 [DOI] [PubMed] [Google Scholar]
- Löwe J., Amos L.A. (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature 391: 203–206 [DOI] [PubMed] [Google Scholar]
- Mackenzie S.A. (2005). Plant organellar protein targeting: a traffic plan still under construction. Trends Cell Biol. 15: 548–554 [DOI] [PubMed] [Google Scholar]
- Maleszka R., Skelly P.J., Clark-Walker G.D. (1991). Rolling circle replication of DNA in yeast mitochondria. EMBO J. 10: 3923–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. (2002). Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou J.C., Youle R.J. (2011). Mitochondria in apoptosis: bcl-2 family members and mitochondrial dynamics. Dev. Cell 21: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., Nunnari J. (2003). Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Whelan J., Small I. (2006). Recent surprises in protein targeting to mitochondria and plastids. Curr. Opin. Plant Biol. 9: 610–615 [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. (2007). Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Oldenburg D.J., Bendich A.J. (1998). The structure of mitochondrial DNA from the liverwort, Marchantia polymorpha. J. Mol. Biol. 276: 745–758 [DOI] [PubMed] [Google Scholar]
- Peeters N., Small I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541: 54–63 [DOI] [PubMed] [Google Scholar]
- Reed J.C. (1997). Cytochrome c: Can’t live with it—can’t live without it. Cell 91: 559–562 [DOI] [PubMed] [Google Scholar]
- Reyes A., et al. (2011). Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 39: 5098–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtseva T.K., Tan W., Colombini M. (2005). On the role of VDAC in apoptosis: Fact and fiction. J. Bioenerg. Biomembr. 37: 129–142 [DOI] [PubMed] [Google Scholar]
- Ruzicka D.R., Kandasamy M.K., McKinney E.C., Burgos-Rivera B., Meagher R.B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52: 460–472 [DOI] [PubMed] [Google Scholar]
- Sheahan M.B., Rose R.J., McCurdy D.W. (2004). Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 37: 379–390 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- van den Ent F., Amos L.A., Löwe J. (2001). Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Chandel N.S., Williamson E.K., Schumacker P.T., Thompson C.B. (1997). Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91: 627–637 [DOI] [PubMed] [Google Scholar]
- Wang Y., Bogenhagen D.F. (2006). Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 281: 25791–25802 [DOI] [PubMed] [Google Scholar]
- Wilm M., Mann M. (1996). Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68: 1–8 [DOI] [PubMed] [Google Scholar]
- Xu X., Forbes J.G., Colombini M. (2001). Actin modulates the gating of Neurospora crassa VDAC. J. Membr. Biol. 180: 73–81 [DOI] [PubMed] [Google Scholar]
- Yaffe M.P. (1999). The machinery of mitochondrial inheritance and behavior. Science 283: 1493–1497 [DOI] [PubMed] [Google Scholar]