This study reports on the tight functional interplay between cotranslational N-terminal methionine excision (NME) and the thylakoid FtsH protease complex, highlighting that correctly NME-processed N-termini of D1 and D2 polypeptides of photosystem II are indispensable for both FtsH-mediated protein quality control and repair-related degradation of D1 and D2.
Abstract
N-terminal methionine excision (NME) is the earliest modification affecting most proteins. All compartments in which protein synthesis occurs contain dedicated NME machinery. Developmental defects induced in Arabidopsis thaliana by NME inhibition are accompanied by increased proteolysis. Although increasing evidence supports a connection between NME and protein degradation, the identity of the proteases involved remains unknown. Here we report that chloroplastic NME (cNME) acts upstream of the FtsH protease complex. Developmental defects and higher sensitivity to photoinhibition associated with the ftsh2 mutation were abolished when cNME was inhibited. Moreover, the accumulation of D1 and D2 proteins of the photosystem II reaction center was always dependent on the prior action of cNME. Under standard light conditions, inhibition of chloroplast translation induced accumulation of correctly NME-processed D1 and D2 in a ftsh2 background, implying that the latter is involved in protein quality control, and that correctly NME-processed D1 and D2 are turned over primarily by the thylakoid FtsH protease complex. By contrast, inhibition of cNME compromises the specific N-terminal recognition of D1 and D2 by the FtsH complex, whereas the unprocessed forms are recognized by other proteases. Our results highlight the tight functional interplay between NME and the FtsH protease complex in the chloroplast.
INTRODUCTION
Most proteins are modified at their N termini by enzymes that attach or remove small molecules; these events are known to regulate many crucial cellular processes. Such modifications may reflect or affect the status, fate, and function of the protein (Giglione et al., 2004; Walling, 2006; Meinnel and Giglione, 2008). The earliest N-terminal modification corresponds to cotranslational excision of the initiating Met (Schmitt et al., 1996), with approximately two thirds of the protein in a given proteome processed this way (Martinez et al., 2008). Dedicated N-terminal Met excision (NME) machinery has been found in all organisms and in all cell compartments in which protein synthesis occurs: cytoplasm, mitochondria, and plastids. In the cytosol, the initiating Met of nascent proteins is unformylated and is processed directly by Met aminopeptidase (METAP). By contrast, in the chloroplasts and mitochondria, the initiating Met is originally N-formylated. Before a dedicated organellar METAP can act, deformylation of the ~80 chloroplast-encoded open reading frames by a prokaryotic-like peptide deformylase (PDF) is required (Giglione et al., 2004; Meinnel et al., 2006). Two PDFs have been identified in plants: mitochondrion-targeted PDF1A and PDF1B, which are targeted to both mitochondria and plastids (Giglione et al., 2000; Dirk et al., 2001). Only PDF1A has been found in humans and other animals (Giglione et al., 2000; Nguyen et al., 2003; Serero et al., 2003; Lee et al., 2004), and a PDF3 was identified in trypanosomatids (Bouzaidi-Tiali et al., 2007). Note that the many proteins encoded by the nuclear genome, which are further imported in the plastids, are not processed by the chloroplastic NME (cNME) but rather by a dedicated transit stromal peptidase.
The NME process has long been considered constitutive. Interestingly, accumulating evidence supports both transcriptional and posttranscriptional control of NME activity (Giglione et al., 2004). In plants, PDF1B expression is strongly induced in developing flowers and leaves, whereas both PDF1B and PDF1A are stress-responsive genes in Arabidopsis thaliana (Giglione et al., 2004). An epigenetic increase in At - PDF1A levels, which leads to partial targeting to the chloroplast, was observed in a pdf1b genetic background (Giglione et al., 2003). By contrast, At-PDF1B fully compensates for the lack of At-PDF1A in a pdf1a knockout mutant. The reason for such intricate physiological regulation of NME enzymes and its influence on global NME processes remain poorly understood. However, evidence suggests that NME, regardless of the cellular compartment involved, is essential in all organisms; thus, its regulation must be important (Giglione et al., 2004).
The NME pathway has been linked to targeting proteins for degradation (Meinnel et al., 2006). Genetic and pharmacological studies revealed that inhibition of NME activity in vivo leads to the destabilization of several central proteins of either plastid or mitochondrial proteomes (Giglione et al., 2003; Bouzaidi-Tiali et al., 2007; Moon et al., 2008; Escobar-Alvarez et al., 2010). Moreover, organellar NME inhibition induces a cascade of events that impair the biogenesis of the entire compartment as well as the development of the whole organism. This effect was attributed to the extreme sensitivity of certain essential plastid-encoded proteins to the lack of NME (Giglione and Meinnel, 2001; Serero et al., 2003). For example, the core photosystem II (PSII) reaction center proteins D1 and D2 are more rapidly degraded on retention of the N-terminal Met (N-Met) (Giglione et al., 2003). In addition, recent bioinformatics and proteomics analyses have revealed that most cytosolic proteins undergoing N-terminal ubiquitination retain their N-Met and that many proteins with N-Met residues are poorly accumulated (Meinnel et al., 2005; Martinez et al., 2008). Furthermore, the cytoplasmic NME is essential for normal growth and development of Arabidopsis. NME orchestrates a crosstalk between two fundamental signaling pathways that are frequently deregulated under pathological conditions: thiol status and proteolysis (Ross et al., 2005; Frottin et al., 2009). In all studied compartments, the developmental defects induced by NME inhibition seem to be caused by increased cellular proteolytic activity, primarily induced by an increase in the number of proteins targeted for rapid degradation (Frottin et al., 2009). Although increasing evidence supports the close connection between NME and protein degradation, nothing is known about the associated proteolytic machinery that works downstream of the cNME process. Studies in Chlamydomonas reinhardtii suggest that N-terminal–unprocessed D1 and D2 components of PSII are degraded via a caseinolytic protease P (ClpP)-independent proteolytic pathway (Giglione et al., 2003). In addition to Clp, the plastid contains many other proteases, including FtsH, Lon, and Deg (Adam and Clarke, 2002; Adam et al., 2006; Sakamoto, 2006; Huesgen et al., 2009; Kato et al., 2009), that might be involved in this process. For instance, in vitro and in vivo studies have implicated the thylakoid FtsH protease in the degradation of PSII subunits during its repair after photoinhibition (Lindahl et al., 2000; Bailey et al., 2002; Sakamoto et al., 2002; Komenda et al., 2006; Kato et al., 2007; Kato et al., 2009; Kato and Sakamoto, 2009; Komenda et al., 2010), as well as in the biogenesis of chloroplasts (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002; Sakamoto et al., 2003; Zhang et al., 2010).
Because NME and the FtsH protease complex act on common substrates, and because both affect chloroplast biogenesis, we challenged the hypothesis that cNME enzymes and the thylakoid FtsH complex operate sequentially in the same enzymatic cascade. To this end, we crossed the pdf1b mutant with the ftsh2 mutant, which contains reduced levels of the thylakoid FtsH protease complex (Sakamoto, 2003; Zaltsman et al., 2005b). Our results support the molecular basis of a tight functional interplay between NME enzymes and the thylakoid protease FtsH complex.
RESULTS
Chloroplast Development Promoted by FtsH Depends on Prior Action of cNME
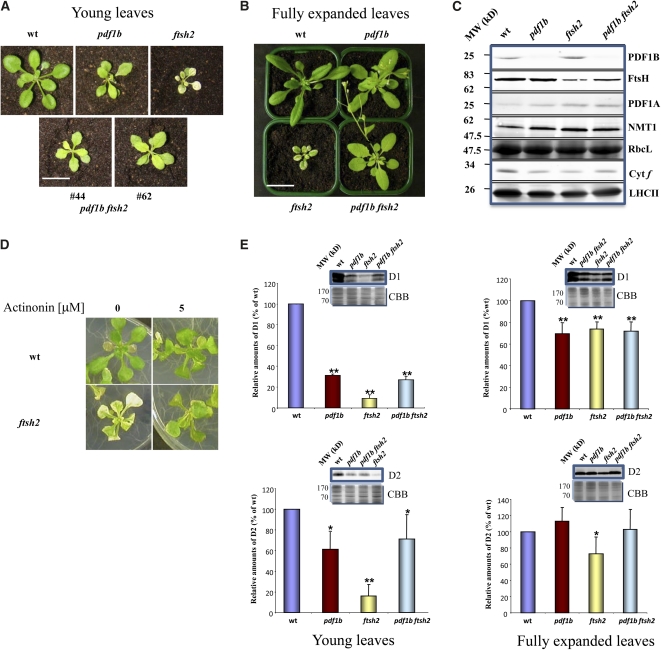
We have previously shown that when N-formyl-Met is not properly processed, either because of a mutation in the PDF1B gene or inhibition of the enzyme by a specific PDF inhibitor (such as actinonin), the steady state level of PSII core proteins (e.g., D1 and D2), is lower than in controls. This is due to an increased rate of degradation of the D1/D2 proteins (Giglione et al., 2003); however, this “protein quality control” is not mediated by the stromal Clp protease. In search of a protease responsible for this degradation, we crossed the ftsh2 (also known as var2) knockout mutant, which features reduced levels of the thylakoid FtsH protease complex (Sakamoto, 2003), with the well-characterized mutants of the organellar NME pathway pdf1b and pdf1a (Giglione et al., 2003). T2 BASTA-resistant seedlings were genotyped by PCR (see Supplemental Figure 1 online), and homozygous double mutants were selected. Surprisingly, in all pdf1b ftsh2 double mutants (isolated from more than 10 independent genetic crosses) grown at either 50 or 100 μE m−2 s−1, the variegated phenotype of ftsh2 was entirely suppressed, and these double mutants closely resembled the single mutant pdf1b both in size and pigmentation (Figures 1A and 1B; see Supplemental Figure 2A online). Immunoblot analysis using specific antibodies confirmed the expected absence of PDF1B in the single pdf1b and double pdf1b ftsh2 mutants (Figure 1C). Three proteins, with different compartmental localizations, were analyzed as controls (PDF1A, RbcL, and NMT1 for mitochondria/chloroplast, chloroplast, and cytoplasm localization, respectively) (Figure 1C). No variation in either PDF1A or NMT1 levels was observed in pdf1b, ftsh2, and double pdf1b ftsh2 mutants. The somewhat lower levels of these proteins in the wild type are likely due to the fact that the wild type accumulates a higher level of the large subunit of RuBisco (RbcL), the most abundant protein in plant cells. Other chloroplast controls, such as cytochrome f and the light-harvesting complex of PSII (LHCII), displayed the same behavior as RbcL. Quantification of these bands showed changes of comparable magnitude. Interestingly, in the ftsh2 background, the level of PDF1B was upregulated compared with the wild type, suggesting that the FtsH complex operates downstream of PDF1B in a common pathway. Immunoblots were obtained with an antibody recognizing all four isomers of the thylakoid FtsH complex (i.e., FtsH1, FtsH2, FtsH5, and FtsH8) (Zaltsman et al., 2005b). Thus, the faint band in the ftsh2 sample represents the remaining FtsH complex, featuring FtsH1, FtsH5, and FtsH8, and not a residual accumulation of FtsH2 in the mutant line. Why the double mutant contains more FtsH than the single ftsh2 mutant remains unclear, but it might be another manifestation of the strong interrelation between these pathways.
Figure 1.
Genetic Analysis of the Double Mutant pdf1b ftsh2 Places PDF1B Upstream of the FtsH Pathway.
(A) Photographs of 3-week-old plants corresponding to the wild type (wt), pdf1b, and ftsh2 single mutants and the double mutant pdf1b ftsh2, grown for 1 week in soil in a greenhouse under standard light conditions. The mutant progenies of pdf1b, ftsh2, pdf1b ftsh2, and the wild type were obtained from the self-pollination of heterozygous pdf1b PDF1B ftsh2 FtsH2 generated by crossing the single pdf1b and ftsh2 mutants (two different pdf1b ftsh2 double mutants are shown). Homozygosis was confirmed by PCR analysis of genomic DNA (see Supplemental Figure 1 online).
(B) Photographs of 6-week-old plants corresponding to the wild type, pdf1b, and ftsh2 single mutants and the double mutant pdf1b ftsh2, grown for 4 weeks in soil in a greenhouse under standard light conditions. The mutant progenies of pdf1b, ftsh2, pdf1b ftsh2, and the wild type were obtained as described in (A).
(C) Immunoblot analysis of proteins isolated from 30 mg of 2-week-old seedlings corresponding to the wild type, pdf1b, and ftsh2 single mutants and the double mutant pdf1b ftsh2 grown as described in (B). Total proteins (25 μg) were probed using anti-PDF1B, anti-FtsH, anti-PDF1A, anti-NMT1, anticytochrome f, anti-LHCII, and anti-RbcL antibodies. MW, molecular weight.
(D) Photographs of 3-week-old plants corresponding to the wild type and ftsh2 single mutant were grown on 1% Suc medium (see Methods) in the absence or presence of 5 μM actinonin in a growth chamber at 22°C, 16 h of daylight, and a light intensity of 100 μE m−2 s−1.
(E) Immunoblot analysis was performed using anti-D1 and anti-D2 antibodies to quantify the protein levels in the wild type, pdf1b, and ftsh2 single mutants and the double mutant pdf1b ftsh2 under standard light conditions. Membrane proteins were extracted from 3-week-old (A) and fully expanded leaves (B). Signals from each immunoblot were quantified and normalized using Quantity One 1-D Analysis Software (Bio-Rad, see Methods). The protein level in the wild type was taken as 100%. Values are means of three biological replicates and 9 to 11 technical replicates, and error bars indicate sd. Asterisks indicate the error probability as performed with a two-tailed Student’s t test (*, <0.05; **, <0.01). Representative immunoblots for each condition are shown. The CBB-stained gels are shown just below their respective immunoblot.
Bar in (A) = 0.5 cm; bar in (B) = 1 cm.
[See online article for color version of this figure.]
Suppression of the variegated phenotype of ftsh2 by genetic knockout of PDF1B implied that chemical inhibition of this enzyme should have a similar effect on the ftsh2 mutant. Indeed, seedling growth on plates containing 5 μM actinonin, a well-characterized chloroplast PDF-specific inhibitor (Serero et al., 2001), led to the disappearance of variegation of the ftsh2 mutant (Figure 1D). By contrast, pdf1b and pdf1b ftsh2 mutants could not survive in the presence of 0.5 μM actinonin (see Supplemental Figure 3 online), further highlighting the requirement of PDF activity in the chloroplast. The dramatic effect of either PDF1B mutation or inhibition of its activity on the variegated phenotype of ftsh2 raised the question of whether a knockout of pdf1a would have the same effect. Homozygous mutants among the progeny of a cross between pdf1a and ftsh2 were identified by PCR (see Supplemental Figure 1B online), but they were visually indistinguishable from ftsh2 (see Supplemental Figure 2B online). This suggests that pdf1a and ftsh2 do not operate in the same pathway, which agrees with their different localizations: chloroplast for FtsH2 (Sakamoto et al., 2003) and mitochondria for PDF1A (Giglione et al., 2000). Taken together, these results strongly suggest that, in the context of chloroplast biogenesis, the thylakoid FtsH protease complex operates downstream of PDF1B. Because PDF inhibition does not affect the rate of protein synthesis (Giglione et al., 2003), the suppression effect associated with PDF inhibition shown here is therefore induced by a mechanism clearly uncoupled from protein synthesis inhibition (Park and Rodermel, 2004; Koussevitzky et al., 2007; Miura et al., 2007; Yu et al., 2008; Zhang et al., 2009; Liu et al., 2010).
Characterization of D1 and D2 Accumulation in Young and Fully Expanded Leaves
Because of the suggested interplay between PDF1B and the FtsH complex, and because the D1 and D2 subunits of PSII are known substrates of both the NME process and FtsH, we tested how D1 and D2 levels are affected by altered levels of both PDF1B and the FtsH complex. The overall degree of variegation has been shown to depend on environmental conditions and leaf developmental stage (Zaltsman et al., 2005a; Sakamoto et al., 2009). Moreover, in the pdf1b background, accumulation of D1 and D2 increases during development. This is a consequence of PDF1A protein relocalization to the chloroplast, which is induced by an epigenetic increase of its expression in the pdf1b background (Giglione et al., 2003). In this context, we investigated the accumulation of D1 and D2 polypeptide chains in young and fully expanded leaves (3- and 6-week-old, respectively). Immunoblot analyses on all four genetic backgrounds revealed that, consistent with previous reports (Bailey et al., 2002; Giglione et al., 2003; Kato et al., 2009), D1 and D2 levels in the young leaves of all three mutants were lower than in the wild type, with ftsh2 most strongly affected (Figure 1E). Here again, similar to the visual phenotypes, the pdf1b mutation in the background of ftsh2 restored D1 and D2 levels to those of the single pdf1b mutant.
In fully expanded leaves, D1 levels in the three mutant backgrounds were higher than those observed in young leaves, but lower than in the wild type (Figure 1E, right panels). D2 levels in the pdf1b and pdf1b ftsh2 variants were similar to those in the wild type. The D2 levels in fully expanded leaves of ftsh2 were higher than those in young leaves but still lower than in the wild type (Figure 1E).
Chemical Inhibition of Chloroplastic Deformylation Triggers Identical Effects on D1 and D2 Accumulation Promoted by pdf1b Knockout in a ftsh2 Background
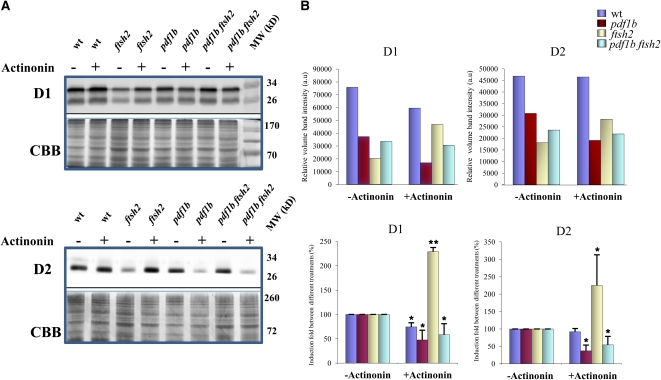
The higher levels of D1 and D2 observed in the double mutant pdf1b ftsh2 compared with the single mutant ftsh2 implied that chemical inhibition of PDF1B activity by actinonin should have a similar effect on the ftsh2 mutant. We thus performed an immunoblot analysis to monitor the steady state levels of D1 and D2 in the four genotypes, grown in the presence of actinonin concentrations able to affect any of the genotypes either positively or negatively (Figure 2; see Supplemental Figures 3A and 3B online). In the wild type, D1 and D2 levels were unaffected at low concentrations of actinonin (<5 μΜ). At actinonin concentrations of 5 to 10 μΜ, the level of remaining D1 (but not D2) was reduced slightly (~75% of the control) (Figure 2B). The protein level was progressively reduced concomitantly with an increased concentration of actinonin for both D1 and D2 (see Supplemental Figure 3B online). Progressive reduction of both D1 and D2 levels as a function of actinonin concentration has been already reported in Giglione et al. (2003). These data are in full agreement with the expectation that the wild-type plants in the presence of actinonin mimic the situation in the pdf1b mutant. A 0.25-μΜ concentration of actinonin (instead of 50 μM for the wild type), which almost completely blocks deformylation in both the pdf1b and pdf1b ftsh2 backgrounds, led to reduced accumulation of both D1 and D2 (Figure 2B; see Supplemental Figure 3B online). In the absence of FtsH2 and the presence of actinonin, the levels of NME-unprocessed D1 and D2 polypeptide chains were similar in the double mutant pdf1b ftsh2 and in the single mutant pdf1b. However, the levels of NME-unprocessed D1 and D2 polypeptide chains were clearly higher in the double mutant pdf1b ftsh2 than in the single mutant pdf1b in the presence of 0.5 μM actinonin (see Supplemental Figure 3B online). These data suggest that accumulation of the NME-unprocessed forms of D1 and D2 partially require FtsH2 (see Supplemental Figure 4A online). In agreement, both D1 and D2 levels increased more than twofold when the ftsh2 mutant was treated with actinonin at a concentration that suppresses variegation (Figure 2B). As expected, D1 and D2 levels in the ftsh2 mutant at the actinonin concentration that induces greening (up to 5 μM) approached those of the untreated double mutant pdf1b ftsh2.
Figure 2.
Partial Chemical Inhibition of Chloroplastic Deformylation Suppresses the Variegated Phenotype in the ftsh2 Background.
(A) Membrane proteins of each genotype were isolated from 3-week-old seedlings grown in the absence or presence of 5 μΜ actinonin for the wild type (wt) and ftsh2 mutant and 0.25 μΜ actinonin for the pdf1b and pdf1b ftsh2 mutants (see Supplemental Figure 3A online); 25 μg membrane proteins were separated by 12% SDS-PAGE, and accumulation of D1 and D2 was estimated by immunoblot analysis. The CBB-stained gels are shown just below their respective immunoblots. The stained portions of the gels were used to calculate the normalization coefficient for the equal loading of the samples.
(B) Relative volume intensities of D1 and D2 for one representative immunoblot after normalization are shown in arbitrary units (a.u) (Top). Data show the induction fold of D1 and D2 between different treatments, generated from four biological replicates. The values after normalization are reported as a percentage of the value of D1 or D2 corresponding to each genotype in the absence of actinonin, taken as 100% (error bars indicate sd) (Bottom). Asterisks indicate the error probability as performed with a two-tailed Student’s t test (*, <0.05; **, <0.01).
[See online article for color version of this figure.]
FtsH-Dependent Accumulation of D1 and D2 Polypeptides, with Partial Inhibition of Protein Synthesis, Is Mainly Associated with Prior Action of the NME Process
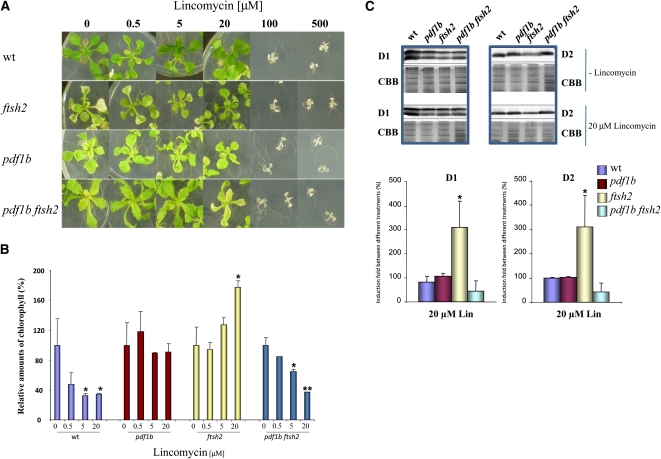
To characterize the relationship between NME and FtsH further, we investigated the effect of inhibiting chloroplast translation in the variants using lincomycin, a known inhibitor of translation elongation in prokaryotes. Increasing amounts of lincomycin can be used to adjust levels of protein synthesis and, as a result, the coupling between protein synthesis and degradation in the various genetic backgrounds. Lincomycin at concentrations of 100 μM or higher, which completely block protein synthesis in organelles, has a lethal effect on all seedlings, whereas no effect was observed on the wild-type, pdf1b, and pdf1b ftsh2 mutants at concentrations of 20 μM or lower (Figure 3A). By contrast, sublethal concentrations of lincomycin suppressed the variegation phenotype of ftsh2, as previously reported for other chloroplast protein-synthesis inhibitors (Figure 3A; see Supplemental Figure 5A online) (Yu et al., 2008). These data are consistent with the differences in chlorophyll content detected in the four genotypes as a function of lincomycin concentration (Figure 3B; see Supplemental Figure 5B online), in which opposite tendencies were observed in the ftsh2 and the pdf1b ftsh2 mutants.
Figure 3.
Under Standard Light Conditions, Reduced Chloroplast Translation Induces Accumulation of the Correctly NME-Processed D1 and D2 Solely in the Absence of FtsH2.
(A) Wild type (wt), pdf1b and ftsh2 single mutants, and the double mutant pdf1b ftsh2 were grown on 1% Suc medium (see Methods) in the absence or presence of increasing concentrations of lincomycin in a growth chamber at 22°C, 16 h of daylight, and a light intensity of 100 μE m−2 s−1 for 3 weeks.
(B) Chlorophyll content obtained from seedlings in (A). Total chlorophyll content was determined spectroscopically and was normalized to fresh weight. Total chlorophyll content for each variant in the absence of lincomycin was taken as 100%. Values are means of three biological replicates, and error bars indicate sd. Asterisks indicate the error probability as performed with a two-tailed Student’s t test (*, <0.05; **, <0.01).
(C) Immunoblot analysis of the accumulation of membrane proteins D1 and D2 from each genotype in (A) in the absence or presence of 20 μM lincomycin. Total membrane proteins (25 μg) were separated by 12% SDS-PAGE. Each value in the absence of lincomycin corresponding to each genotype was taken as 100%. Values correspond to two independent experiments each consisting of 6 to 9 biological replicates originating from the measurement performed from independent leaves. Error bars indicate sd. Asterisk indicates the error probability as performed with a two-tailed Student’s t test (*, <0.05). Representative immunoblots for each condition are shown. The CBB-stained gels used for normalization are shown just below their respective immunoblot.
[See online article for color version of this figure.]
We next used immunoblot analysis to investigate D1 and D2 accumulation with increasing concentrations of lincomycin in all genotypes. High concentrations of the protein-synthesis inhibitor (up to 50 μM) induced a proportionally reduced accumulation of D1 and D2 in the wild type, pdf1b, and pdf1b ftsh2 mutants (see Supplemental Figure 5C online). Reduced accumulation of D1 and D2 was also observed in the ftsh2 mutants treated with high concentrations of lincomycin, whereas a different behavior was observed at low concentrations of the drug. For instance, in all mutant plants grown under standard light conditions (100 μE m−2 s−1), reduction in protein synthesis by long exposure of the plants to 20 μM lincomycin, which is the concentration that maximally suppresses the variegated phenotype in the ftsh2 mutant, did not induce significant changes in D1 or D2 polypeptide accumulation in the wild type, pdf1b, or pdf1b ftsh2 mutants compared with untreated plants (Figure 3C). By contrast, partial inhibition of protein synthesis by lincomycin resulted in an approximately threefold increase in levels of both D1 and D2 in the ftsh2 mutant. When NME and the FtsH complex are fully active, as in the wild type, partial reduction of protein synthesis does not affect D1 or D2 accumulation. This is also the case when either the NME pathway is affected or both proteolytic processes (i.e., NME and protein degradation by the FtsH complex) are inhibited, as in the pdf1b and pdf1b ftsh2 mutants, respectively. Conversely, when the N-Met of D1 is correctly processed by NME in the absence of FtsH2, D1 accumulates to high levels; however, the protein synthesis is constantly partially inhibited. These data suggest that when protein synthesis is slowed, the N-terminally processed D1 and D2 polypeptide chains are primarily degraded by the FtsH complex, which ensures their quality control (see Supplemental Figure 4B online). Under the same conditions but without deformylation, accumulation of N-formyl D1 or D2 is independent of the level of the FtsH complex (Figure 3C; see Supplemental Figure 4B online). Therefore, accumulation of the unprocessed form of D1 and D2 might be controlled by proteases other than the FtsH complex.
Taken together, these results indicate that accumulation of the unprocessed forms of D1 and D2 is controlled by several chloroplast proteases, including FtsH2, whereas accumulation of the correctly processed forms of D1 and D2 is mainly controlled by the FtsH complex (see Discussion).
Immediate Arrest of Protein Synthesis Rapidly Decreases D2 Levels in the ftsh2 Mutant under Dark Conditions
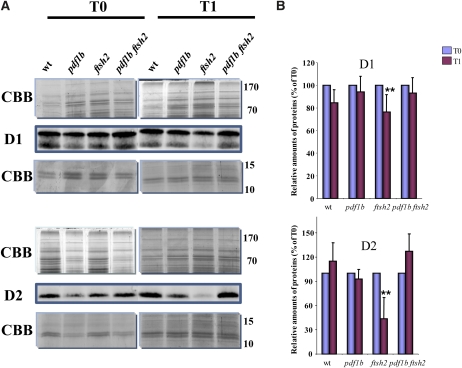
In vivo degradation of D1 and D2 can be efficiently and reliably monitored by incubating detached leaves with their petioles in a high concentration of lincomycin for short times (e.g., between 1 and 6 h) (Bailey et al., 2002; Kato et al., 2009). This procedure—unlike pulse labeling with radioactive amino acid, which is the commonly used approach to study protein turnover—has been used successfully to test the fate of D1 and D2 proteins under photoinhibitory conditions (Bailey et al., 2002). Thus, we used this procedure to analyze D1 and D2 degradation in vivo in our four genotypes. Incubation of detached leaves from 6-week-old plants grown in the greenhouse with 5 mM lincomycin either in the dark or under low irradiation (10 to 20 μE m−2 s−1) for 1 h induced a strong reduction of D2 and a lesser reduction of D1 only in the ftsh2 mutant (Figures 4A and 4B). Indeed, after blocking protein synthesis, we observed ~50% degradation of D2 that was correctly processed at its N terminus, degradation that seemed to be driven by the lack of FtsH2 expression. The sensitivity of D2 to the ftsh2 mutation was not observed when the N-terminal region of the protein was unprocessed, suggesting that FtsH is probably not involved in the stability of unprocessed D2 in the dark, in line with results from complementary experiments (Figures 2 and 3).
Figure 4.
Protein Synthesis Inhibition Causes a Rapid Decrease of D2 in the ftsh2 Mutant under Dark Conditions.
(A) Representative protein immunoblots of the PSII core polypeptides from the different genotypes with anti-D1 and anti-D2 antibodies. Membrane proteins were extracted from untreated leaves after removal of leaf tissue from plants grown in the greenhouse (T0). Similar leaves were incubated via their petioles in 5 mM lincomycin for 1 h in the dark. After this incubation time (T1), some of these leaves were used to extract membrane proteins that were subjected to immunoblot analysis. wt, wild type.
(B) Signals from immunoblot analyses as in (A) were quantified and normalized as reported in Figure 3. Data were obtained from five biological replicates and between 10 to 15 technical replicates. To compare the relative levels of D1 and D2 proteins, the value at T0 in each genotype was set to 100%. Error bars indicate sd. Asterisks indicate the error probability as performed with a two-tailed Student’s t test (**, <0.01).
[See online article for color version of this figure.]
Sensitivity of pdf1b ftsh2 to High Light
Apart from the obvious but unexplained role of the FtsH protease complex in plastid differentiation, it is now well established that it participates in the PSII repair cycle by degrading subunits damaged by oxidative stress (Lindahl et al., 2000; Bailey et al., 2002; Sakamoto et al., 2002; Kato et al., 2009). Inhibition of chloroplastic deformylation also affects the repair cycle of PSII in C. reinhardtii (Giglione et al., 2003). The possible crosstalk between PDF1B and FtsH prompted us to characterize the susceptibility of the pdf1b single and double mutants to photoinhibition. We grew the four genotypes at either 50 or 500 μE m−2 s−1 for 6 weeks. Under low light, the double mutant was slightly smaller than the wild type but indistinguishable from pdf1b (Figure 5A). By contrast, under high light, the double mutant was pale green but not variegated; bigger than ftsh2 yet smaller than pdf1b (Figure 5A; see Supplemental Figure 6 online). To complement these observations, we monitored PSII activity in the plants grown under low light. After dark adaptation, PSII activity in pdf1b and pdf1b ftsh2 was similar to that in the wild type (Figure 5B, black columns). ftsh2 grown under these conditions demonstrated lower PSII activity. After exposing these plants to high light (500 μE m−2 s−1) for 30 min, PSII activity in the wild type had decreased, as expected from photoinhibited plants (Figure 5B, white columns). The ftsh2 mutant demonstrated higher sensitivity to high light, as reported previously, but pdf1b behaved just like the wild type. Notably, the double mutant was less sensitive than ftsh2 plants to photoinhibition but more sensitive than pdf1b and the wild type (Figure 5B). These results explain why, after prolonged exposure to high light, the double mutant is bigger than ftsh2 but smaller and paler than the wild-type or the single pdf1b mutant (Figure 5A). Moreover, the lack of PDF1B not only rescues the leaf variegation in ftsh2, but also partially allows it to cope better with high light intensity. This ability is also correlated with the higher level of the FtsH complex found in the double mutant compared with ftsh2 (Figure 1C).
Figure 5.
NME Inhibition Suppresses ftsh2 Sensitivity to Photoinhibition under Short-Term Exposure to High Light.
(A) Four-week-old plants corresponding to the wild type (wt), the pdf1b and ftsh2 single mutants, and the double mutant pdf1b ftsh2, grown for 2 weeks on soil in the greenhouse, were transferred to a growth chamber at 22°C, 16 h of daylight, and a light intensity of 20 μE m−2 s−1 (Top), then the pots were transferred to an equivalent chamber with a light intensity of 500 μE m−2 s−1 for another 2 weeks (Bottom). Photographs are taken at the same time on 8-week-old plants.
(B) For the photoinhibition experiments, 6-week-old plants corresponding to different variants grown in the greenhouse under standard light conditions were used. PSII activity, monitored as Fv/Fm, was measured after dark adaptation (dark columns), then again after exposing the different variants to high light (500 μE m−2 s−1) for 30 min (white columns). The values are means ± se (error bars) of eight independent experiments of each genotype.
[See online article for color version of this figure.]
Taken together, our results suggest that the mutation in pdf1b suppresses not only the variegation in the ftsh2 mutant but also its sensitivity to photoinhibition under conditions of short exposure to high light. Under prolonged exposure, the double mutant showed higher sensitivity than the wild type and the pdf1b mutant and only partially suppressed developmental defects of the ftsh2 mutation.
Correctly Processed N-Terminal D1 and D2 Are Exclusively Degraded by FtsH under Photoinhibitory Conditions
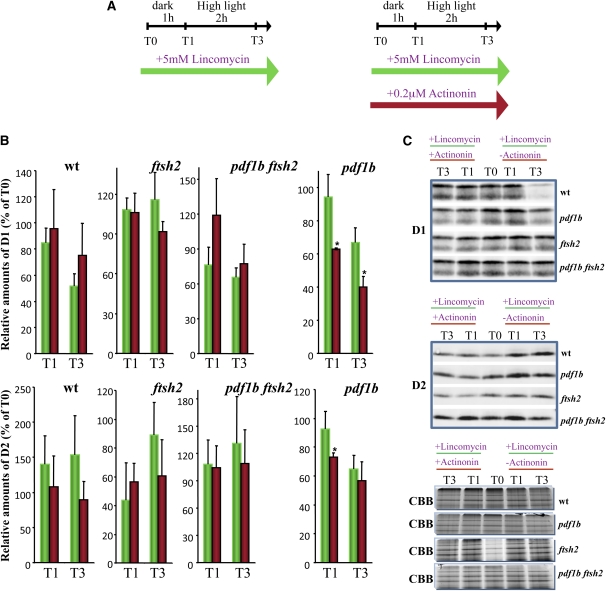
Next, we examined D1 and D2 degradation under short exposure to high light. The scheme of the experiment is shown at the top of Figure 6A. Detached leaves, which had been incubated for 1 h with 5 mM lincomycin in the dark, were exposed to high light (500 μE m−2 s−1) for an additional 4 h in the presence of the drug. In agreement with previous reports (Bailey et al., 2002; Kato et al., 2009), the level of D1 in the wild type decreased by 40% after 2 h under high light but remained stable in the ftsh2 mutant (Figures 6B and 6C); the levels of the protein in the two genotypes remained unchanged for the next 2 h. D1 degradation induced by high-light treatment in the pdf1b single mutant was comparable with the wild type, which is consistent with their similar sensitivity to photoinhibition (Figure 5B). By contrast, in the double mutant pdf1b ftsh2, we observed mixed behavior in the wild type, pdf1b mutant, and ftsh2 mutant. Indeed, D1 was not degraded in the pdf1b ftsh2 mutant under short-term photoinhibition, suggesting involvement of the FtsH complex in the degradation of nonprocessed D1. In the pdf1b ftsh2 mutant, prolonged high-light exposure induced a decrease in D1, most likely via the induction of other proteases, including other components of the FtsH complex (see Supplemental Figure 4C online). This suggestion was further supported by repeating the photoinhibition experiments in the presence and absence of 0.2 μM actinonin, the lowest concentration known to affect the phenotype of pdf1b and pdf1b ftsh2 mutants and to block chloroplast deformylation completely in the two genotypes (Figures 7A to 7C). Under these conditions, D1 degradation was increased only in the pdf1b mutant, strongly affecting D1 accumulation when its synthesis was hindered at the ribosomal level in the dark. An increase in D1 turnover was observed after exposure to high-light conditions (Figures 7B and 7C). This effect was not seen in the double mutant, further confirming participation of the FtsH protease complex in degradation of the unprocessed form of D1. Similar but less pronounced effects were observed with D2.
Figure 6.
In Vivo Analysis of D1 and D2 Degradation Reveals a Rapid Decrease of D2 in the ftsh2 Mutant under Dark Conditions.
The ability to degrade D1 and D2 polypeptides in vivo following light-induced damage was assayed by incubating 6-week-old detached leaves of each variant in the absence or presence of 5 mM lincomycin. Three different biological replicates and 9-11 technical replicates were used for each genotype. Leaves from T0 were exposed to high light (μE m2 s1) for 4 additional hours prior to protein preparation.
(A) Experimental setting.
(B) Protein immunoblot analyses were performed for each point and genotype using anti-D1 and anti-D2 antibodies (see also Figures 4 and 7 for the immunoblots corresponding to the dark samples [T0]). Representative immunoblots for each condition and genotype are shown. The upper CBB-stained gels used for normalization are shown to the right of the respective immunoblots. wt, wild type.
(C) Signals of the immunoblot analysis were quantified and normalized as described in Methods. To compare the relative levels of D1 and D2 protein, the value at time T0 in each genotype was set to 100%. Data were obtained from five biological replicates and between 10 to 15 technical replicates. Asterisks indicate the error probability as performed with a two-tailed t test (*, <0.05; **, <0.01).
[See online article for color version of this figure.]
Figure 7.
Under Photoinhibitory Conditions, the Contribution of FtsH2 to the Degradation of Fully Unprocessed N-Terminal D1 and D2 Is Irrelevant for D2 and Minor for D1.
The ability to degrade the D1 and D2 polypeptides in vivo following light-induced damage was assayed by incubating 8-week-old detached leaves of each variant in the presence of 5 mM lincomycin and in the absence and presence of 0.2 mM actinonin (red bars). Three different biological replicates and 9-11 technical replicates were used for each genotype.
(A) Experimental setting.
(B) Immunoblot analysis was performed using anti-D1 and anti-D2 antibodies to quantify the protein levels in the wild type (wt), pdf1b, and ftsh2 single mutants and the double mutant pdf1b ftsh2. Signals were quantified and normalized as described in Methods. To compare the relative levels of D1 and D2, the value at time T0 in each genotype was set to 100%. Data were obtained from five biological replicates and between 10 to 15 technical replicates. Error bars indicate SD. * indicates an error probability (two-tailed t test) less than 0.05.
(C) Representative immunoblots for each condition and genotype are shown. The equivalent upper gels used for normalization (see Methods) are shown.
[See online article for color version of this figure.]
DISCUSSION
Until very recently, the physiological function of the cotranslational NME process was unknown. It was first suggested that NME might be involved in regulating protein turnover (Arfin and Bradshaw, 1988), and recent data provide strong support for this hypothesis. In particular, most of these data point to maintenance of the N-Met as a protein destabilization signal. Consequently, removal of the first Met should yield a set of proteins with high stability (Meinnel et al., 2005). Moreover, a combination of bioinformatics and proteomics analyses indicate that proteins containing an unprocessed and unmodified N-Met are short-lived (Ciechanover and Ben-Saadon, 2004; Meinnel et al., 2006). Thus, unprocessed proteins in each proteome may be naturally unstable per se and tagged for rapid degradation. Retention of the N-Met in proteins that usually undergo NME can also result in acceleration of their degradation, which implies that NME is a regulated process and that the fraction of proteins undergoing NME may vary spatially and temporally. Indeed, NME is tightly regulated, and both transcriptional and posttranscriptional mechanisms have been observed (Giglione et al., 2004). This study attempts to demonstrate a direct link between physiologically downregulated NME and reduced half-life of proteins that retain their N-Met. Therefore, identification of the physiological conditions modulating NME levels is important to identify the actual pool of proteins whose lifespan is affected by N-Met retention and their role under these physiological conditions.
Several pieces of indirect evidence corroborate this assumed relationship between NME and protein half-life. In this context, retention of the initiator Met in mutants of glutathione-S-transferase expressed in the yeast cytosol or of β-glucuronidase in plants drastically reduces their half-life in vivo (Sawant et al., 2001; Chen et al., 2002). Inhibition of cNME induced by disruption of the PDF gene and/or drug-specific PDF inactivation decreases the steady state levels of PSII D1 and D2 polypeptides and further destabilizes the complex and the whole compartment both in A. thaliana and C. reinhardtii (Giglione et al., 2003). This effect can be fully recapitulated by site-directed substitutions altering N-Met cleavage of the corresponding plastid target D2 protein. Very recently, it was shown that cytoplasmic NME downregulation (i.e., partial retention of the N-Met) in Arabidopsis triggers an increase in cellular proteolytic activity, initiated by an increase in the size of the pool of proteins suitable for processing (Frottin et al., 2009). This deregulation of proteolysis, driven by the increase in the free amino acid pool and NADPH depletion, perturbs the glutathione redox state and ultimately leads to the plant developmental defects observed when cytoplasmic NME is impaired. Although these studies clearly show that an N-Met may act as a signal for degradation, they also show that the N-Met, even if necessary, is not sufficient per se to confer short half-life to a given protein. Indeed, in both studied cases (chloroplast and cytoplasm), downregulation of the NME process did not affect the stability of most proteins; only a few became highly sensitive to the retention of their N-Met, such as the PSII D1 and D2 proteins. Thus, other features/modifications of the proteins on or close to their N-Met may participate in the acceleration of protein degradation. Indeed, Varshavsky and coworkers recently demonstrated that N-terminally acetylated Met residues can act as a degradation signal for several cytosolic yeast proteins via the Doa10 ubiquitin ligase (Hwang et al., 2010). This suggests that the major role of this acetylated N-degron probably involves quality-control mechanisms and regulation of protein stoichiometries. Those authors proposed a model in which this acetylated N-degron mediates assembly-regulated protein degradation by being accessible when the protein is free but inaccessible when the protein is assembled with its counterparts. They also proposed that in the chloroplast, given the similarity of the N-formyl and N-acetyl groups, the formyl-Met functions as a formyl-dependent degradation signal. In contrast with this proposal, our previous data exclude the possibility of the formyl group acting as an additional signal for degradation (Giglione et al., 2003), and other as yet unidentified elements might be important for accelerating the turnover of proteins that are normally subjected to the NME process.
In the cytoplasm of eukaryotes, N-terminal acetylated or nonacetylated Met residues may be specific signals for protein degradation, ultimately triggered by the 26S proteasome (Meinnel et al., 2005; Martinez et al., 2008; Hwang et al., 2010). By contrast, nothing is known about the degradation machinery pertinent to retention of the N-Met in the chloroplast. Plastids do not possess 26S proteasomes but instead carry prokaryote-related ATP-dependent proteolytic machineries, with Clp, FtsH, and Lon being the major enzymes for processive degradation (Adam and Clarke, 2002; Sakamoto, 2006). The overall architecture of the Clp protease complex resembles that of the multimeric 26S proteasome (Zheng et al., 2002; Sjögren et al., 2006). In elucidating which chloroplastic proteases are involved in the degradation of the PSII subunits sensitive to NME inhibition, we showed that this process does not involve the Clp protease (Giglione et al., 2003). Here, we also demonstrate that the FtsH protease complex is not directly responsible for accelerated degradation of PSII subunits when they retain their N-Met. Surprisingly, our genetic analysis reveals that PDF1B acts upstream of the FtsH protease in a common pathway. The developmental defects observed in plants containing lower levels of the thylakoid FtsH complex, because of the loss of the FtsH2 protein, are abrogated by inhibition of NME. Here, we show that degradation of the D1 and D2 components of PSII by the metalloprotease FtsH requires the earlier action of the NME process. Our results are in line with recent studies in Synechocystis sp PCC6803 that also identified an important physiological role for the exposed N-terminal tail of D1 in degradation (Komenda et al., 2007). Using mutagenesis in combination with functional assays, Komenda et al. highlighted the involvement of the exposed N-terminal tail of D1 in the selective degradation of damaged D1 during PSII repair in the cyanobacterium, suggesting an attractive model in which FtsH complexes first recognize the N terminus of damaged D1 and then degrade the damaged protein in a highly processive reaction. The question of how FtsH complexes recognize the N-terminal tail of D1 has not been answered yet. Previous studies showed that translation of D1 and D2 is arrested in the mRNA elongation phase (Klein et al., 1988; van Wijk and Eichacker, 1996; Colón-López and Sherman, 1998; Herranen et al., 2001; Tyystjärvi et al., 2001). Interestingly, we found that dark conditions associated with protein synthesis inhibition caused a rapid decrease of D2 only in the ftsh2 background, because no change was detected in the wild-type, pdf1b, or pdf1b ftsh2 backgrounds. This effect has not been investigated in the ftsh2 genetic background. Indeed, many reports have addressed the relationship between D1 degradation and photoinhibition, but few have addressed the relationship between D1 degradation and low light intensity. Furthermore, there is no hypothesis on the mechanism(s) that determine D2 instability, particularly in the dark. In this context, our data suggest that, in the ftsh2 background, the turnover of N-correctly processed D2 is highly accelerated, probably because of the absence of FtsH2. We propose two nonexclusive hypotheses: i) the absence of FtsH2, which leads to lower levels of the FtsH complex, increases the proteolytic activity of other chloroplastic proteases; and ii) the FtsH complex is a chaperone for newly synthesized proteins. This second hypothesis is further supported by the chaperone activity demonstrated for the bacterial and mitochondrial FtsH homologs (Shirai et al., 1996; Suzuki et al., 1997; Schumann, 1999; Sakamoto, 2006). We provide experimental evidence that correctly NME-processed N termini of D1 and D2 are indispensable for both FtsH-mediated quality control of the proteins and repair-related degradation of D1 and D2. Our results do not exclude the possibility that partially redundant pathways of D1 and particularly D2 degradation might exist, as previously proposed (Haussühl et al., 2001; Huesgen et al., 2009). However, our findings indicate that FtsH-mediated D1 quality control is always dependent on the cotranslational removal of the formyl group followed by excision of the first Met by the NME process. When the NME process is inhibited, the nonprocessed PSII subunits are recognized primarily by proteases other than the FtsH complex (Figure 8). Indeed, previous studies have pointed to the involvement of other protease(s) in degradation of unassembled D1 protein (Komenda et al., 2010).
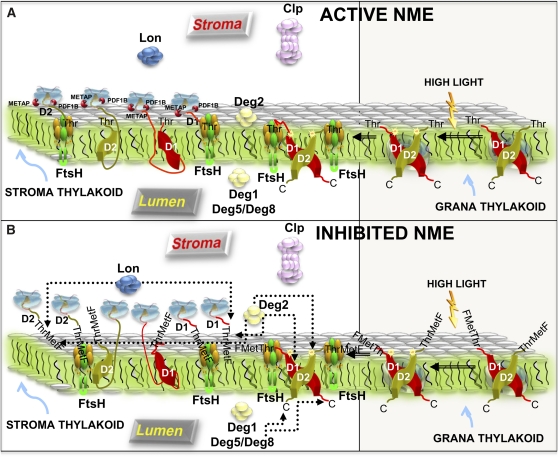
Figure 8.
Model Recapitulating D1 and D2 Degradation by Different Chloroplastic Proteases as a Function of the NME Process.
Both cartoons show the architecture around the thylakoid membranes and the possible interplay between all chloroplastic proteases (Lon, ClpP, Deg, or FtsH) and D1 and D2 at the level of the PSII complex or the ribosome as a function of the NME process (PDF and METAP) and under normal or photoinhibition conditions (referred to as “High Light” in the cartoon). Note that FtsH also can be located in the thylakoid grana (Yoshioka et al., 2010).
(A) Independent of the conditions, correctly NME-processed D1 and D2 components of PSII are turned over primarily by the FtsH metalloprotease complex.
(B) When the NME process is inhibited, the NME non-processed PSII subunits starting now with a N-terminal Formyl-Met-Threonine (indicated as FMetThr) appear to be recognized and degraded primarily by proteases other than the FtsH complex (dotted arrows).
[See online article for color version of this figure.]
In evaluating the relationship between the NME process and protein degradation in chloroplasts, the thylakoid FtsH hexameric complex cannot be ignored. It is composed of two types of subunits, each encoded by a set of duplicated genes (FtsH1 and FtsH5 in type A, FtsH2 and FtsH8 in type B). Within a pair, the genes are redundant (Yu et al., 2004; 2005), but two types of subunits must be present for the complex to accumulate, because the absence of any pair of the redundant genes is fatal (Zaltsman et al., 2005b). Strong and weak leaf variegation are associated with mutations in FtsH2 and FtsH5, respectively, consistent with their differential abundance. In the absence of either of these proteins, the level of the complex is reduced (Sakamoto, 2003). Thus, all attributes of the ftsh2 (var2) mutant derive from lower levels of the thylakoid FtsH complex rather than from the loss of FtsH2 itself. In this respect, successful complementation of the var2 mutant with a proteolytically inactive variant of FtsH2 suggests that, at least for the purpose of proper chloroplast biogenesis, the FtsH complex can accommodate some proteolytically inactive subunits (Zhang et al., 2010). The remaining active subunits of FtsH8, FtsH5, and FtsH1 are apparently sufficient for normal function. In this regard, these data are reminiscent of the stromal and cyanobacterial Clp proteases, which contain ClpR subunits that are inactive homologs of the active ClpP subunits (Andersson et al., 2009; Olinares et al., 2011). Note that these subunits are upregulated in the double mutant, as previously shown in the single mutant (Zaltsman et al., 2005b).
At present, all suppressor genes of the ftsh-mediated variegated phenotype in Arabidopsis but one (see above) (Zhang et al., 2010) have been obtained by second-site suppressor screens (Park and Rodermel, 2004; Koussevitzky et al., 2007; Miura et al., 2007; Yu et al., 2008; Zhang et al., 2009; Liu et al., 2010). Unfortunately, the possible connection between the proteins encoded by the identified suppressor genes and the FtsH protease complex in the thylakoid remains unclear. A connection between several suppressors and PDF1B apparently lies in the importance of ribosome-associated biogenesis factors in assisting protein translation (Giglione et al., 2009). Full integration between chloroplast protein synthesis, proteolysis, and the various mutant suppressors awaits clarification.
This report highlights a direct link between the NME requirement and control of protein turnover in the chloroplast, where a specific protease recognizes the N terminus of NME substrates. This link seems to be so crucial that it governs normal chloroplast development. Future work will focus on the mechanistic details of the recognition of different N termini, modified or not, by the respective proteases in the chloroplast.
METHODS
Materials
All chemicals were purchased from Sigma-Aldrich Chimie. Lincomycin was dissolved in water, and actinonin was dissolved in methanol.
Plant Material
The original single T-DNA insertion lines were described and characterized previously (Giglione et al., 2003; Zaltsman et al., 2005b). Both pdf1a and pdf1b lines were derived from the Wassilewskija-2 ecotype (Giglione et al., 2003), whereas the ftsh2 mutant was in the Columbia ecotype (Sakamoto et al., 2003). To eliminate any putative effect relative to the original background, all wild-type, single mutant, and double mutant Arabidopsis thaliana plants were obtained by screening the F2 generation originating from crossing the single mutants pfd1a or pf1b with ftsh2. Cosegregation between the insertions and their respective phenotypes was verified. Several independently obtained plants of the F2 generation were systematically used, the phenotypes were checked, and protein levels were measured under different conditions.
Plant Growth
Seeds were sterilized and sown on 0.5× Murashige and Skoog (Sigma-Aldrich Chimie) medium supplemented with 0.8% (w/v) agarose (Difco) and 1% (w/v) Suc (Sigma-Aldrich Chimie) unless otherwise stated. Petri dishes were incubated in the dark for 48 h at 4°C and then transferred to a growth chamber. Next, 2-week-old plantlets corresponding to plants with two cotyledons and two small emerging true leaves were planted on soil and transferred to the greenhouse and grown at 22°C with 16 h of daylight (100 μE m−2 s−1) for up to 5 weeks. The different Arabidopsis lines were propagated under greenhouse conditions, as previously described (Giglione et al., 2000).
Chlorophyll and PSII Activity Measurements
Two-week-old seedlings were harvested and weighed. Leaves were ground in liquid nitrogen, and chlorophyll was extracted in 1 mL of 80% acetone. After 5 min of centrifugation at 4°C, absorbance was measured at 647 and 664 nm. For PSII activity, 6-week-old plants of the four genotypes, grown under 50 μE m−2 s−1, were dark-adapted for 10 min before or after 30 min exposure to high light intensity (1300 μE m−2 s−1). After dark adaptation, chlorophyll fluorescence parameters (F0 and Fm) were measured at room temperature, and PSII activity values (Fv/Fm) were determined.
Protein Analysis
A. thaliana leaves were frozen in liquid nitrogen and disrupted in an MM 300 mixer mill (Qiagen). The procedure described by Schägger and von Jagow (1991) was used to extract membrane proteins. Protein concentration was determined using the Bio-Rad protein assay kit, and the extracts were analyzed by SDS-PAGE or immunoblot. D1 and D2 subunits are hydrophobic proteins strongly attached to the thylakoid membrane, for which extraction procedures have been already reported to be critical and influenced by many factors (Adir and Ohad, 1988; Zhang et al., 1999). In this context, we carefully repeated each experiment using both technical and biological replicates. Moreover, given the best performance with respect to sensitivity, dynamic range, linearity, and signal-to-noise ratio, the ECL Plex system together with Pharos FX Molecular Imager System (Bio-Rad) were chosen for immunoblot analysis. For this, 25 μg of membrane proteins were separated by 12% SDS-PAGE. After electrophoresis, the proteins were transferred to a low-fluorescent polyvinylidene difluoride membrane (GE Healthcare). Blots were blocked immediately after transfer in 5% (w/v) ECL Advance blocking reagent (GE Healthcare) in 1× PBS with 0.3% (v/v) Tween-20 for 2 h at room temperature with agitation or overnight at 4°C. The blots were then incubated with the specific primary antibody used at dilutions of 1:1000 to 1:10000, depending on the antibody, overnight at 4°C. The antisera against PDF1A, PDF1B, NMT1, cytochrome f, LHCII, and RbcL were described previously (Giglione et al., 2003), and the antibodies against both D1 and D2 were provided by Agrisera. The blots were washed several times before incubation with the secondary antibody (ECL Plex goat anti-rabbit IgG-Cys5; GE Healthcare) at a dilution of 1:1000 for 2 h. The membranes were washed before drying and scanned for Cy5 on the Pharos FX Molecular Imager System (Bio-Rad). Imaging was performed using a 635-nm laser with a 695-nm band-pass filter. The images were then analyzed, and the intensity of bands was quantified using Quantity One software (Bio-Rad) according to the manufacturer’s instructions. In this case, to quantify each band, we used the relative volume intensity of the band that corresponded to the total signal intensity within a defined area. Background subtraction was performed automatically by the software. Two strategies were used to normalize the immunoblot signals, and both gave the same results. In the first, each volume band intensity was normalized using the normalization coefficient obtained from the relative intensities of both RbcL and NMT1 (two known markers for chloroplast and cytoplasm compartments, respectively, whose levels are constant). In the second strategy, before proteins were transferred from gels to polyvinylidene difluoride membranes, the upper region (between 170 and 70 kD) and the lower region (between 15 and 10 kD) of the gels were cut, stained with Coomassie Brilliant Blue (CBB) solution (40% [v/v] methanol, 10% [v/v] acetic acid and 0.025% [w/v] CBB R-250), and scanned with the GS800 Imager System (Bio-Rad). The densitometry of the lanes was used as a standard for the densitometric data from the bands on the immunoblot. Relative volume intensities of immunoblot bands are shown as arbitrary units, and the same values are reported as percentages of the value of D1 or D2 corresponding to each genotype in the absence of actinonin, taken as 100%. Dealing with D1, where two bands could be identified as reported by the supplier, we measured the intensity each of the two bands separately and obtained similar results with either one or both band intensities added together. Experiments using statistical analyses were reported using sd. Statistical analyses were run with R software (http://cran.cict.fr/).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At PDF1A, At1g15390.1; At PDF1B, At5g14660.1; At FTSH2, At2g30950.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Isolation of pdf1b ftsh2 and pdf1a ftsh2 Null Mutants from F2 Progeny of the Cross between pdf1b or pdf1a and ftsh2 Homozygous Mutants.
Supplemental Figure 2. Thylakoid FtsH Protease Complex Operates Exclusively Downstream of PDF1B.
Supplemental Figure 3. Chemical Inhibition of Chloroplastic Deformylation Triggers Identical Effects on ftsh2 Mutant Promoted by pdf1b Knockout in ftsh2 Background.
Supplemental Figure 4. Summary Representation on the Accumulation of D1 and D2 in the Wild Type, pdf1b, ftsh2, and pdf1b ftsh2.
Supplemental Figure 5. Partial Inhibition of Chloroplast Translation by Lincomycin Rescues the Variegated Phenotype in the ftsh2 Background.
Supplemental Figure 6. Effect of Long High Light Exposure on ftsh2 and pdf1b ftsh2 Mutant Leaves.
Acknowledgments
Z.A. was supported by a Centre National de la Recherche Scientifique grant during his sabbatical stay in France. F.F. was supported by a PhD grant from the French Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche as part of Ecole Doctorale ED145 (University Paris-Sud) and by the Association pour la Recherche sur le Cancer Thesis studentship.
AUTHOR CONTRIBUTIONS
Z.A. and C.G. designed the experiments. Z.A., C.E., F.F., and C.G. performed the experiments. Z.A, C.G., and T.M. analyzed data. Z.A., C.G., and T.M. wrote the article.
References
- Adam Z., Clarke A.K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7: 451–456 [DOI] [PubMed] [Google Scholar]
- Adam Z., Rudella A., van Wijk K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Adir N., Ohad I. (1988). Structural properties of the D1 and surrounding photosystem II polypeptides as revealed by their interaction with cross-linking reagents. J. Biol. Chem. 263: 283–289 [PubMed] [Google Scholar]
- Andersson F.I., et al. (2009). Structure and function of a novel type of ATP-dependent Clp protease. J. Biol. Chem. 284: 13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfin S.M., Bradshaw R.A. (1988). Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry 27: 7979–7984 [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P.J., Horton P., Mullineaux C.W., Robinson C., Mann N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Bouzaidi-Tiali N., Giglione C., Bulliard Y., Pusnik M., Meinnel T., Schneider A. (2007). Type 3 peptide deformylases are required for oxidative phosphorylation in Trypanosoma brucei. Mol. Microbiol. 65: 1218–1228 [DOI] [PubMed] [Google Scholar]
- Chen M., Choi Y., Voytas D.F., Rodermel S. (2000). Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Chen S., Vetro J.A., Chang Y.H. (2002). The specificity in vivo of two distinct methionine aminopeptidases in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 398: 87–93 [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Ben-Saadon R. (2004). N-terminal ubiquitination: More protein substrates join in. Trends Cell Biol. 14: 103–106 [DOI] [PubMed] [Google Scholar]
- Colón-López M.S., Sherman L.A. (1998). Transcriptional and translational regulation of photosystem I and II genes in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 180: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirk L.M., Williams M.A., Houtz R.L. (2001). Eukaryotic peptide deformylases. Nuclear-encoded and chloroplast-targeted enzymes in Arabidopsis. Plant Physiol. 127: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Alvarez S., Gardner J., Sheth A., Manfredi G., Yang G., Ouerfelli O., Heaney M.L., Scheinberg D.A. (2010). Inhibition of human peptide deformylase disrupts mitochondrial function. Mol. Cell. Biol. 30: 5099–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F., Espagne C., Traverso J.A., Mauve C., Valot B., Lelarge-Trouverie C., Zivy M., Noctor G., Meinnel T., Giglione C. (2009). Cotranslational proteolysis dominates glutathione homeostasis to support proper growth and development. Plant Cell 21: 3296–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C., Meinnel T. (2001). Organellar peptide deformylases: Universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci. 6: 566–572 [DOI] [PubMed] [Google Scholar]
- Giglione C., Vallon O., Meinnel T. (2003). Control of protein life-span by N-terminal methionine excision. EMBO J. 22: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C., Boularot A., Meinnel T. (2004). Protein N-terminal methionine excision. Cell. Mol. Life Sci. 61: 1455–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C., Fieulaine S., Meinnel T. (2009). Cotranslational processing mechanisms: Towards a dynamic 3D model. Trends Biochem. Sci. 34: 417–426 [DOI] [PubMed] [Google Scholar]
- Giglione C., Serero A., Pierre M., Boisson B., Meinnel T. (2000). Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 19: 5916–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussühl K., Andersson B., Adamska I. (2001). A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 20: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranen M., Aro E.M., Tyystjärvi T. (2001). Two distinct mechanisms regulate the transcription of photosystem II genes in Synechocystis sp. PCC 6803. Physiol. Plant. 112: 531–539 [DOI] [PubMed] [Google Scholar]
- Huesgen P.F., Schuhmann H., Adamska I. (2009). Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Res. Microbiol. 160: 726–732 [DOI] [PubMed] [Google Scholar]
- Hwang C.S., Shemorry A., Varshavsky A. (2010). N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2009). Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kato Y., Miura E., Matsushima R., Sakamoto W. (2007). White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 144: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. (2009). The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.R., Mason H.S., Mullet J.E. (1988). Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J. Cell Biol. 106: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J., Knoppová J., Krynická V., Nixon P.J., Tichý M. (2010). Role of FtsH2 in the repair of photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn(4) cluster. Biochim. Biophys. Acta 1797: 566–575 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C.W., Tichy M., Nixon P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J., Tichy M., Prásil O., Knoppová J., Kuviková S., de Vries R., Nixon P.J. (2007). The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystis sp PCC 6803. Plant Cell 19: 2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Stanne T.M., Peto C.A., Giap T., Sjögren L.L., Zhao Y., Clarke A.K., Chory J. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63: 85–96 [DOI] [PubMed] [Google Scholar]
- Lee M.D., She Y., Soskis M.J., Borella C.P., Gardner J.R., Hayes P.A., Dy B.M., Heaney M.L., Philips M.R., Bornmann W.G., Sirotnak F.M., Scheinberg D.A. (2004). Human mitochondrial peptide deformylase, a new anticancer target of actinonin-based antibiotics. J. Clin. Invest. 114: 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Spetea C., Hundal T., Oppenheim A.B., Adam Z., Andersson B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu F., Rodermel S. (2010). An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A., Traverso J.A., Valot B., Ferro M., Espagne C., Ephritikhine G., Zivy M., Giglione C., Meinnel T. (2008). Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8: 2809–2831 [DOI] [PubMed] [Google Scholar]
- Meinnel T., Giglione C. (2008). Tools for analyzing and predicting N-terminal protein modifications. Proteomics 8: 626–649 [DOI] [PubMed] [Google Scholar]
- Meinnel T., Peynot P., Giglione C. (2005). Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie 87: 701–712 [DOI] [PubMed] [Google Scholar]
- Meinnel T., Serero A., Giglione C. (2006). Impact of the N-terminal amino acid on targeted protein degradation. Biol. Chem. 387: 839–851 [DOI] [PubMed] [Google Scholar]
- Miura E., Kato Y., Matsushima R., Albrecht V., Laalami S., Sakamoto W. (2007). The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S., Giglione C., Lee D.Y., An S., Jeong D.H., Meinnel T., An G. (2008). Rice peptide deformylase PDF1B is crucial for development of chloroplasts. Plant Cell Physiol. 49: 1536–1546 [DOI] [PubMed] [Google Scholar]
- Nguyen K.T., Hu X., Colton C., Chakrabarti R., Zhu M.X., Pei D. (2003). Characterization of a human peptide deformylase: Implications for antibacterial drug design. Biochemistry 42: 9952–9958 [DOI] [PubMed] [Google Scholar]
- Olinares P.D., Kim J., van Wijk K.J. (2011). The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 1807: 999–1011 [DOI] [PubMed] [Google Scholar]
- Park S., Rodermel S.R. (2004). Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. USA 101: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S., Giglione C., Pierre M., Espagne C., Meinnel T. (2005). Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 137: 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. (2003). Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 78: 1–9 [DOI] [PubMed] [Google Scholar]
- Sakamoto W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Tamura T., Hanba-Tomita Y., Murata M., Sodmergen (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7: 769–780 [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Uno Y., Zhang Q., Miura E., Kato Y., Sodmergen (2009). Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol. 50: 2069–2083 [DOI] [PubMed] [Google Scholar]
- Sawant S.V., Kiran K., Singh P.K., Tuli R. (2001). Sequence architecture downstream of the initiator codon enhances gene expression and protein stability in plants. Plant Physiol. 126: 1630–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Schmitt E., Guillon J.M., Meinnel T., Mechulam Y., Dardel F., Blanquet S. (1996). Molecular recognition governing the initiation of translation in Escherichia coli. A review. Biochimie 78: 543–554 [DOI] [PubMed] [Google Scholar]
- Schumann W. (1999). FtsH—a single-chain charonin? FEMS Microbiol. Rev. 23: 1–11 [DOI] [PubMed] [Google Scholar]
- Serero A., Giglione C., Meinnel T. (2001). Distinctive features of the two classes of eukaryotic peptide deformylases. J. Mol. Biol. 314: 695–708 [DOI] [PubMed] [Google Scholar]
- Serero A., Giglione C., Sardini A., Martinez-Sanz J., Meinnel T. (2003). An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway. J. Biol. Chem. 278: 52953–52963 [DOI] [PubMed] [Google Scholar]
- Shirai Y., Akiyama Y., Ito K. (1996). Suppression of ftsH mutant phenotypes by overproduction of molecular chaperones. J. Bacteriol. 178: 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L., Stanne T.M., Zheng B., Sutinen S., Clarke A.K. (2006). Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C.K., Rep M., van Dijl J.M., Suda K., Grivell L.A., Schatz G. (1997). ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 22: 118–123 [DOI] [PubMed] [Google Scholar]
- Takechi K., Sodmergen, Murata M., Motoyoshi F., Sakamoto W. (2000). The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 41: 1334–1346 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi T., Herranen M., Aro E.M. (2001). Regulation of translation elongation in cyanobacteria: Membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol. Microbiol. 40: 476–484 [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Eichacker L. (1996). Light is required for efficient translation elongation and subsequent integration of the D1-protein into photosystem II. FEBS Lett. 388: 89–93 [DOI] [PubMed] [Google Scholar]
- Walling L.L. (2006). Recycling or regulation? The role of amino-terminal modifying enzymes. Curr. Opin. Plant Biol. 9: 227–233 [DOI] [PubMed] [Google Scholar]
- Yoshioka M., Nakayama Y., Yoshida M., Ohashi K., Morita N., Kobayashi H., Yamamoto Y. (2010). Quality control of photosystem II: FtsH hexamers are localized near photosystem II at grana for the swift repair of damage. J. Biol. Chem. 285: 41972–41981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Park S., Rodermel S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Yu F., Park S., Rodermel S.R. (2005). Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Liu X., Alsheikh M., Park S., Rodermel S. (2008). Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Feder A., Adam Z. (2005a). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—implications for thylakoid formation and photosystem II maintenance. Plant J. 42: 609–617 [DOI] [PubMed] [Google Scholar]
- Zaltsman A., Ori N., Adam Z. (2005b). Two types of FtsH protease subunits are required for chloroplast biogenesis and Photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Kato Y., Zhang L., Fujimoto M., Tsutsumi N., Sodmergen, Sakamoto W. (2010). The FtsH protease heterocomplex in Arabidopsis: Dispensability of type-B protease activity for proper chloroplast development. Plant Cell 22: 3710–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., van Wijk K.J., Aro E.M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274: 16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wei Q., Wu W., Cheng Y., Hu G., Hu F., Sun Y., Zhu Y., Sakamoto W., Huang J. (2009). Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidopsis. Plant J. 58: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A.K. (2002). Characterization of Chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol. Plant. 114: 92–101 [DOI] [PubMed] [Google Scholar]