This work demonstrates that an ATG1/13 kinase complex analogous to such complexes regulating autophagy in yeast and metazoans exists in Arabidopsis. The ATG1/13 complex is central to autophagic induction under nutrient-limiting conditions when it also becomes a target of autophagic turnover.
Abstract
Autophagy is an intracellular recycling route in eukaryotes whereby organelles and cytoplasm are sequestered in vesicles, which are subsequently delivered to the vacuole for breakdown. The process is induced by various nutrient-responsive signaling cascades converging on the Autophagy-Related1 (ATG1)/ATG13 kinase complex. Here, we describe the ATG1/13 complex in Arabidopsis thaliana and show that it is both a regulator and a target of autophagy. Plants missing ATG13 are hypersensitive to nutrient limitations and senesce prematurely similar to mutants lacking other components of the ATG system. Synthesis of the ATG12-ATG5 and ATG8-phosphatidylethanolamine adducts, which are essential for autophagy, still occurs in ATG13-deficient plants, but the biogenesis of ATG8-decorated autophagic bodies does not, indicating that the complex regulates downstream events required for autophagosome enclosure and/or vacuolar delivery. Surprisingly, levels of the ATG1a and ATG13a phosphoproteins drop dramatically during nutrient starvation and rise again upon nutrient addition. This turnover is abrogated by inhibition of the ATG system, indicating that the ATG1/13 complex becomes a target of autophagy. Consistent with this mechanism, ATG1a is delivered to the vacuole with ATG8-decorated autophagic bodies. Given its responsiveness to nutrient demands, the turnover of the ATG1/13 kinase likely provides a dynamic mechanism to tightly connect autophagy to a plant’s nutritional status.
INTRODUCTION
Like other eukaryotes, plants employ sophisticated mechanisms to recycle intracellular constituents needed for growth, development, and survival under nutrient-limited conditions. Autophagy is emerging as an important recycling route by which cytoplasmic material is sequestered in vesicles and subsequently delivered to the vacuole for breakdown (Thompson and Vierstra, 2005; Bassham, 2009; Rabinowitz and White, 2010). These vesicles can be assembled de novo from cup-shaped precursors called phagophores (or isolation membranes) with the resulting double membrane–bound autophagosomes then fusing with the tonoplast to release the internal vesicle into the vacuole as an autophagic body (macroautophagy). They can also be formed by invagination of the tonoplast to pinch off autophagic bodies directly into the vacuolar lumen (microautophagy). The autophagic bodies and their cargo are then degraded by an array of vacuolar hydrolases followed by transport of the products back into the cytosol for reuse. A derivative of the AUTOPHAGY-RELATED (ATG) system called the cytoplasm-to-vacuole (CVT) pathway is also employed by yeast (and possible other eukaryotes) to selectively sequester functional oligomeric cargo in the vacuole (Xie and Klionsky, 2007).
In plants, autophagy is upregulated under nutrient-limiting conditions to help replenish internal supplies of fixed nitrogen (N) and carbon (C) to support continued biosynthesis and energy production and during developmentally programmed cell death and senescence to encourage nutrient remobilization (Doelling et al., 2002; Thompson and Vierstra, 2005; Bassham, 2009; Reyes et al., 2011). It also promotes survival during pathogen invasion by helping orchestrate the hypersensitive response, whereby host plants undergo localized cell death to discourage pathogen spread (Liu et al., 2005; Hofius et al., 2009; Yoshimoto et al., 2009; Lenz et al., 2011). While it was first thought to be nonspecific, recent studies have identified routes for selective autophagy (Xie and Klionsky, 2007; Noda et al., 2008; Behrends et al., 2010). Such selectivity likely provides a critical housekeeping function by removing damaged chloroplasts, mitochondria (mitophagy), and ribosomes (ribophagy), scavenging free porphyrins, clearing unwanted peroxisomes (and possibly glyoxysomes) as their available substrate pools change (pexophagy), degrading ubiquitylated aggregates too large for the 26S proteasome, and possibly even sequestering pathogens that invade the cytosol (Ishida et al., 2008; Wada et al., 2009; Hillwig et al., 2011; Johansen and Lamark, 2011; Vanhee et al., 2011; Youle and Narendra, 2011).
Thus far, over 36 ATG proteins have been described that drive the autophagic process, identified primarily from the analysis of yeast (Saccharomyces cerevisiae) autophagy defective mutants (Ohsumi, 2001; Suzuki et al., 2007). They have since been cataloged based on their hierarchical interactions and/or activities into four core functional groups (Xie and Klionsky, 2007; Behrends et al., 2010; Suzuki and Ohsumi, 2010). (1) The ATG1/13 kinase complex initiates autophagosome formation in response to nutrient demands sensed by an assemblage of upstream regulators. (2) The ATG9/2/18 transmembrane complex is proposed to promote phagophore expansion by shuttling membrane from multiple sources such as the endoplasmic reticulum, trans-Golgi network, and mitochondria. (3) A class III phosphatidylinositol (PI) 3-kinase complex containing ATG6 (Beclin 1 in mammals) mediates vesicle nucleation. And finally, (4) a conjugation pathway decorates the phagophore membrane with the ubiquitin fold proteins ATG8 and ATG12 attached to the lipid phosphatidylethanolamine (PE) and the ATG5 protein, respectively. Similar to ubiquitylation, both ATG8 and ATG12 are activated by a common E1 ATG7 and transferred to their respective E2s ATG3 and ATG10 before final linkage to their substrates. The ATG12-ATG5 conjugate then combines with ATG16 to generate the properly localized E3 activity that assembles and integrates lipidated ATG8 into the expanding phagophore membrane. Emerging data indicate that the ATG8-PE adduct not only aids in vesicle expansion and fusion but also helps direct selective autophagy by providing a docking site for shuttle proteins that recruit specific proteins, protein complexes, and organelles as cargo (Xie and Klionsky, 2007; Mizushima, 2010). Many of the yeast ATG factors nucleate together in a large perivacuolar complex called the preautophagosomal structure (PAS), which then drives phagophore development after becoming tethered to the phagophore membrane by the ATG17 scaffold protein (Suzuki et al., 2001, 2007).
Studies from yeast identified the ATG1/13 kinase complex as a key positive regulator of autophagy (Suzuki et al., 2007; Mizushima, 2010). The core components are the Ser/Thr kinase ATG1 and its accessory proteins ATG11, ATG13, ATG17, ATG29, and ATG31. ATG13 binds to and stimulates the kinase activity of ATG1, whereas ATG17 and ATG11 help scaffold the entire complex to the PAS and phagophore during nonselective autophagy (ATG17) and CVT and selective mitophagy (ATG11) (Suzuki et al., 2007; Suzuki and Ohsumi, 2010). The activity of the ATG1/13 complex is regulated by various upstream kinase cascades that affect the interaction strength among the subunits and the kinase activity of ATG1. One of the key negative effectors is the Target of Rapamycin (TOR) kinase (Rabinowitz and White, 2010). TOR dampens autophagy under nutrient-rich conditions by hyperphosphorylating ATG13, which decreases its affinity for ATG1. Starvation-induced inactivation of TOR leads to rapid dephosphorylation of ATG13, tighter binding of ATG13 with ATG1, and then ATG1-dependent phosphorylation of itself and a number of other factors that drive phagophore expansion and enclosure (Mizushima, 2010).
Recent studies showed that a related but mechanistically distinct ATG1/13 complex exists in metazoans, which includes thus far an ortholog of ATG1 (Unc-51-like kinase1 [ULK1]), mATG13, a likely ortholog of ATG17 (Focal Adhesion Kinase Family-Interacting Protein of 200 kD [FIP200]), and ATG101, a component not found in the yeast particle (Chang and Neufeld, 2009; Mizushima, 2010). In contrast with the yeast ATG1/13 complex, the assembly of ULK1 into the mammalian complex is nutrient and TOR independent (Ganley et al., 2009; Hosokawa et al., 2009b). As in yeast, mammalian ATG13 promotes the interaction between ULK1/2 and FIP200 (Jung et al., 2009), but by contrast, ULK1 can also directly interact with FIP200 (Hara et al., 2008; Ganley et al., 2009). Instead of functioning as an ATG1 on/off switch by affecting reversible ATG13 binding, TOR-directed phosphorylation of ATG13 may have a more complex role in animals, possibly by influencing the stability of the ATG1/13 complex, its interaction network, and/or its target specificity (Chang and Neufeld, 2009).
Prior studies by us and others using Arabidopsis thaliana as the model have identified a mechanistically similar ATG autophagic system in plants (Thompson and Vierstra, 2005; Bassham, 2009). Components characterized to date include the entire ATG8/12 conjugation system (Doelling et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Fujioka et al., 2008; Phillips et al., 2008; Chung et al., 2010) and components of the PI3 kinase complex (Liu et al., 2005) and the ATG9/2/18 membrane shuttling complex (Hanaoka et al., 2002; Xiong et al., 2005; Inoue et al., 2006). Notably, whereas the current collection of Arabidopsis atg mutants senesce prematurely and are hypersensitive to nutrient-limiting conditions, they are for the most part phenotypically normal and fertile under nutrient-rich conditions, indicating that the ATG system is not essential to plant growth and development. Using green fluorescent protein (GFP)-ATG8 fusions or vacuolar dyes as reporters, the possible identification of autophagosomes and the dynamics of the resulting autophagic bodies have also been described (Yoshimoto et al., 2004; Contento et al., 2005; Thompson et al., 2005; Chung et al., 2010). As expected, the accumulation of GFP-ATG8–decorated autophagic bodies is enhanced by nutrient stress and blocked by mutations that disrupt ATG8/12 modification.
To help understand how autophagy is regulated by various nutritional signals in plants, we have begun to characterize the ATG1/13 kinase complex from Arabidopsis. Here, we describe the ATG1 and ATG13 subunits and show their regulation by starvation. Whereas ATG13 is encoded by a pair of genes (ATG13a and ATG13b), ATG1 is encoded by a four-gene family with one isoform representing a previously unidentified higher plant-specific truncation encompassing just the kinase domain. Double null mutants eliminating ATG13 are phenotypically similar to previously described atg mutants; they are viable and fully fertile but display accelerated senescence and a hypersensitivity to nutrient limitations. Whereas formation of the ATG12-ATG5 and ATG8-PE conjugates proceeds normally in homozygous atg13a atg13b plants, the mutants are impaired in forming autophagic bodies, thus placing the ATG1/13 kinase complex near the step(s) that encloses autophagosomes. Surprisingly, levels of the ATG1a and ATG13a phosphoproteins are dynamically regulated in planta, with fixed C and N limitations dramatically enhancing their turnover by an autophagy-dependent mechanism. We propose that this breakdown likely provides another layer of control to regulate autophagy in response to a plant’s nutritional status.
RESULTS
The Arabidopsis Genes Encoding ATG1 and ATG13
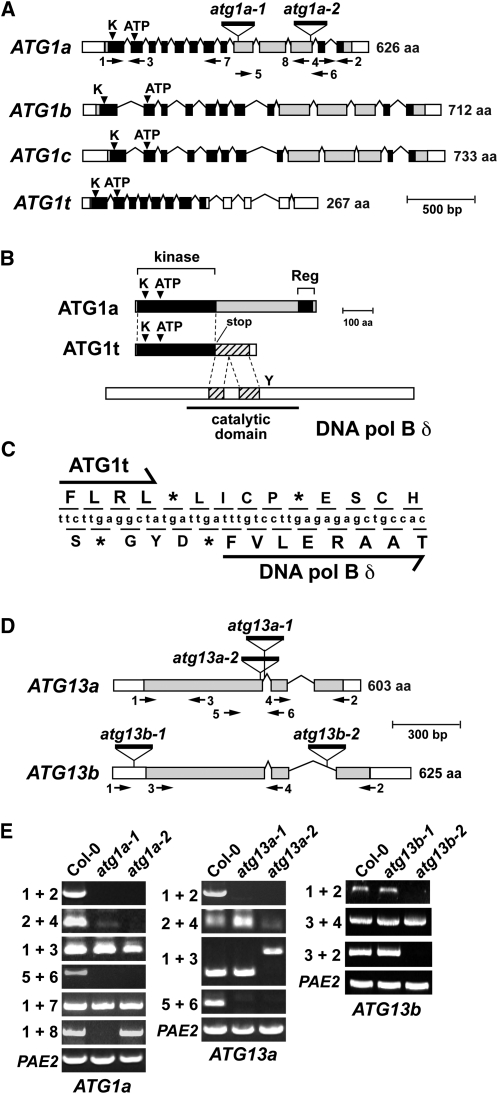
To begin to define the organization of the ATG1/13 kinase complex and its role(s) during plant autophagy, we exhaustively searched the Arabidopsis Columbia-0 (Col-0) ecotype genome by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/blast.cgi) for genes related to those encoding the known subunits in the yeast and metazoan complexes (Xie and Klionsky, 2007; Mizushima, 2010). Similar to metazoans, we failed to find close relatives of yeast ATG11, ATG29, and ATG31 but found possible orthologs of ATG1, ATG13, ATG17/FIP200, and ATG101. In agreement with Hanaoka et al. (2002), we identified three full-length ATG1 genes (designated ATG1a [At3g61960], ATG1b [At3g53930], and ATG1c [At2g37840]) and two full-length ATG13 genes (designated ATG13a [At3g49590] and ATG13b [At3g18770]). Although the overall amino acid sequence identities of ATG1a-c are low when compared with yeast ATG1, both reciprocal BLAST searches of the yeast genome using the Arabidopsis sequences as queries and the clustering of amino acid sequence conservation within regions of potential catalytic importance imply strongly that they are functional orthologs (Figure 1A; see Supplemental Figure 1 online). In addition to the canonical ATG1 forms, we found a previously unidentified truncation designated ATG1t. Upon reannotation of the current gene model for ATG1t present in The Arabidopsis Information Resource database (http://www.Arabidopsis.org) using its cDNA sequence for comparison, we discovered that this locus includes the coding region for the kinase domain but is missing much of the 3′ exonic region present in ATG1a-c (see below). cDNAs for all ATG1 and ATG13 loci were either found within the Arabidopsis EST database or generated by RT-PCR, indicating that each gene is transcriptionally active.
Figure 1.
Structure and Genetic Analysis of Arabidopsis ATG1 and ATG13 Gene Families.
(A) Gene diagrams of the ATG1 gene family. White and shaded boxes indicate UTRs and coding regions, respectively. Black boxes identify the N-terminal kinase and C-terminal regulatory domains. The catalytic site Lys (K) and ATP binding site in the kinase domain are indicated by the arrowheads. Lines denote introns. Triangles mark T-DNA insertion sites and are labeled with allele names. The positions of primers used for RT-PCR are located by the arrows. aa, amino acids.
(B) Comparison of the ATG1a and ATG1t transcript structures. The coding region for ATG1t terminates immediately distal to the kinase domain (black box) and is followed out of frame by a portion of the transcript encoding the DNA polymerase B δ subunit (cross-hatched box). Y locates the catalytic Tyr required for δ subunit activity. aa, amino acids.
(C) Nucleotide and derived amino acid sequence of the ATG1t gene surrounding the junction of the ATG1t transcript with a portion of DNA polymerase B δ coding sequence. The asterisks denote stop codons.
(D) Gene diagrams of the ATG13 gene family. White and shaded boxes indicate UTRs and coding regions, respectively. Lines denote introns. Triangles mark T-DNA insertion sites and are labeled with allele names. The positions of primers used for RT-PCR are located by the arrows. aa, amino acids.
(E) RT-PCR analysis of the atg1a, atg13a, and atg13b T-DNA insertion mutants. Total RNA from wild-type or homozygous mutant plants was subjected to RT-PCR using the indicated primer pairs in (A) and (D). RT-PCR with PAE2-specific primers was used to confirm the analysis of equal amounts of RNA.
All four Arabidopsis ATG1 genes have identical intron/exon boundaries within their shared regions, implying that they evolved from a common ancestor (Figure 1A). ATG1b and ATG1c in particular likely arose from a whole-genome duplication within the Arabidopsis lineage based on their locations within syntenic duplication blocks on chromosomes III and II, respectively. Similar to the yeast ortholog, the ATG1a-c and ATG1t genes were predicted to encode an ~260-residue Ser/Thr protein kinase domain at their N termini (see Supplemental Figure 1 online). The kinase regions share 47 to 65% amino acid sequence identity within the Arabidopsis ATG1a-c/ATG1t family and are 26 to 39% identical to the comparable region within yeast ATG1. Despite these low identities, the regions include all 11 subdomains conserved within the Ser/Thr/Tyr kinase superfamily (Hanks and Hunter, 1995) as well as the conserved Cys (subdomain II) and the ATP binding motif (subdomain V) required for the phosphotransferase activity of the yeast ortholog (Kamada et al., 2000) (see Supplemental Figure 1 online). Downstream of the kinase domain in ATG1a, ATG1b, and ATG1c is a long variable stretch with poor sequence conservation followed by a reasonably conserved region (32 to 45% identical to yeast ATG1) near the C terminus (Figure 1A). Yeast ATG13 and ATG17 bind ATG1 via this C-terminal region; the interaction with ATG13 in particular enhances the kinase activity of ATG1 (Kamada et al., 2000; Cheong et al., 2008).
Scans of available plant genome sequences identified canonical ATG1 orthologs in numerous other species, including various angiosperms (both eudicot and monocot) and the bryophytes Physcometrilla patens and Selaginella moellendorffii, strongly suggesting that ATG1 proteins are present throughout the plant kingdom (see Supplemental Figures 1 and 2 and Supplemental Data Set 1 online). Invariably, multiple canonical ATG1 loci were identified in each species, including three in maize (Zea mays), four in popular (Populus trichocarpa), and two each in rice (Oryza sativa), S. moellendorffii, and P. patens. The angiosperm proteins separated phylogenetically from their bryophyte relatives into “a” and “b/c” subclades that contained Arabidopsis ATG1a and ATG1b/c, respectively (see Supplemental Figure 2 online). Consequently, we propose that the first terrestrial plants contained a single ATG1 locus that duplicated and diverged early during seed plant evolution.
In contrast with ATG1a-c, the reannotated Arabidopsis ATG1t gene encodes only the kinase domain with an in-frame stop codon immediately distal to its coding region (Figures 1B and 1C; see Supplemental Figure 3 online). By comparing the 3′ region of the ATG1t genomic sequence with cDNAs generated by RT-PCR from whole-seedling mRNA, we also discovered an unusually organized and surprisingly long 3′ untranslated region (UTR) following the stop codon. Even after removing the four identified introns, a 494-bp 3′ UTR remained before the polyadenylation site. Interestingly, we detected a 169-codon opening reading frame just 1 bp downstream of the ATG1t stop codon that partially matched the coding region for two internal segments from the catalytic region in the DNA polymerase B δ subunit (residues 401 to 562) (Figure 1B). The first 133 residues of the predicted polypeptide are 51% identical to the δ subunit sequence, while the last 29 residues are unrelated. This δ subunit sequence is out-of-frame relative to that for ATG1t and does not begin with an ATG initiation codon, strongly suggesting that it is neither cotranslated with ATG1t nor translated by itself from the mature ATG1t mRNA (Figure 1C). Given that the predicted δ subunit sequence is missing the conserved Tyr critical for DNA polymerase B activity, the truncated δ subunit polypeptide would be catalytically inactive even if expressed. Sequence analysis of 21 independent cDNAs generated with RNA isolated from various Arabidopsis tissues identified the same transcript arrangement, suggesting that this region does not undergo alternative splicing.
From scans of other eukaryotic genomes, we found genes encoding proteins related in organization to Arabidopsis ATG1t in numerous angiosperms and the gymnosperm Pinus taeda, but not in S. moellendorffii, P. patens, yeast, or any animal genomes (see Supplemental Figure 2 online). In fact, amino acid sequence alignments revealed that these seed plant-specific truncations terminate either at or close to the same position, ranging from three to 16 residues downstream of an invariant Phe at the end of the kinase domain (see Supplemental Figure 3 online). The ATG1t isoforms also clustered phylogenetically apart from ATG1a and ATG1b/c clades based on alignment of just the kinase domains (see Supplemental Figure 2 online), suggesting that these variants arose early during seed plant evolution. However, sequences 3′ from the translation termination site are not related within the ATG1t subfamily and showed no evidence for the DNA polymerase B δ coding sequence found at the 3′ UTR of Arabidopsis ATG1t. Consequently, we propose that ATG1t likely represents a novel adaptation to the ATG1 kinase family in seed plants and that the unique organization of the Arabidopsis ATG1t locus arose recently by an inadvertent transposition of DNA polymerase δ subunit gene fragment(s) into this locus.
The Arabidopsis ATG13a and ATG13b genes have identical intron/exon boundaries (Figure 1D), and the corresponding proteins are mostly colinear along their entire length of 603 and 625 residues, respectively (see Supplemental Figure 4 online). The predicted coding regions share 32/42% amino acid sequence identity/similarity with each other but only 13/21% and 14/20% identity/similarity with yeast ATG13. ATG13 proteins are collectively unrelated in sequence to any other proteins analyzed thus far, thus precluding in silico hypotheses about their potential activities. Patches of reasonable conservation were found between the Arabidopsis and yeast orthologs; unfortunately, these patches did not correspond to any known sequence motifs or protein folds, leaving their importance unclear. One clear distinction is that the poly-Gln track found in the middle of yeast ATG13 (Funakoshi et al., 1997) is absent in its Arabidopsis counterparts.
Like ATG1, orthologs of ATG13 are present in a number of angiosperms (both eudicot and monocot), P. taeda, P. patens, and S. moellendorffii, implying strongly that ATG13 proteins are present throughout the plant kingdom (see Supplemental Figure 5 and Supplemental Data Set 2 online). Phylogenetic analysis suggests that the evolution of the ATG13 family paralleled that for ATG1 (i.e., the first terrestrial plants likely contained a single ATG1 locus that duplicated and diverged early during the emergence of seed plants). Notably, seed plants typically contain two or more ATG13 loci, which separate phylogenetically into “a” and “b” subclades apart from the single ATG13 genes present in bryophytes (see Supplemental Figure 5 online).
ATG1 and ATG13 Interact with Each Other in Possible Autophagic Structures
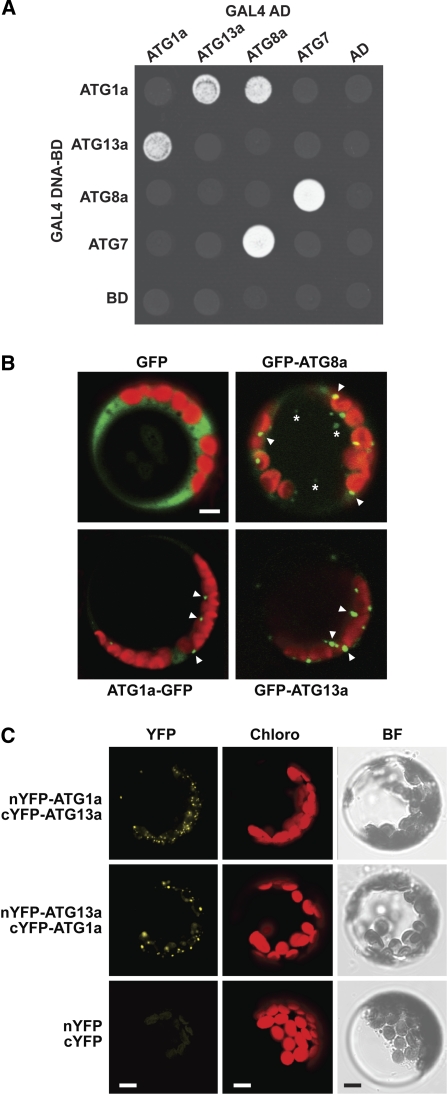
Prior studies with yeast ATG1 and ATG13 showed that the pair reversibly associates depending on nutritional status to form an active kinase complex (Kamada et al., 2000; Mizushima, 2010). To confirm that the Arabidopsis pair also interacts, we tested for binding by yeast two-hybrid analysis (Y2H). As shown in Figure 2, directed Y2H assays using ATG1a and ATG13a fused to the C terminus of either the GAL4 DNA-activation domain or GAL4 DNA binding domain showed strong binding as measured by positive growth on selection medium. Growth was nearly as robust as that for the ATG8a/ATG7 pair, which forms a stable, thioester-linked intermediate during the ATP-dependent activation of ATG8 by ATG7 (Doelling et al., 2002; Fujioka et al., 2008). Surprisingly, we also detected an interaction between ATG1a and ATG8a in one of the paired orientations (Figure 2A). As ATG8 appears to provide a docking platform to selectively recruit cargo into autophagosomes (Noda et al., 2008; Behrends et al., 2010), this interaction may be relevant to the intracellular dynamics of ATG1 (see below).
Figure 2.
ATG1a and ATG13a Interact and Associate with Autophagic Compartments.
(A) Y2H analysis of ATG1-ATG13 interactions. Full-length ATG1a and ATG13a expressed as N-terminal fusions to either the GAL4 DNA activator (AD) or binding domains (BD) were coexpressed in yeast on selection medium containing 3-amino-1,2,4-triazole and lacking Trp, Leu, and His. The known interaction between ATG7 and ATG8a was included as a positive control. AD and BD indicated empty AD and BD plasmids.
(B) Localization of ATG1a and ATG13a to autophagy-type compartments in Arabidopsis protoplasts. Free GFP and GFP fused to ATG8a, ATG1a, or ATG13a were transiently expressed in leaf protoplasts and their localization observed 18 h after transformation by fluorescence confocal microscopy. Green, GFP fluorescence; red, chlorophyll fluorescence. Arrowheads locate possible autophagosomes in the cytoplasm. Asterisks mark potential autophagic bodies inside the vacuole.
(C) BiFC analysis of ATG1a and ATG13a in Arabidopsis. The split nYFP and cYFP reporters simultaneously expressed alone or fused to the N terminus of ATG1a and ATG13a were detected by fluorescence confocal microscopy in leaf protoplasts 18 h after transformation. Images obtained from YFP and chlorophyll (Chloro) fluorescence and bright-field (BF) microscopy are shown. Bars = 5 μm.
To test further whether Arabidopsis ATG1 and ATG13 associate with autophagic structures, we transiently expressed ATG1a and ATG13a as GFP fusions in Arabidopsis leaf protoplasts and examined their location by fluorescence confocal microscopy. Whereas the protoplasts accumulated free GFP in a diffuse pattern throughout the cytoplasm and nucleus, the GFP-ATG8a fusion accumulated in ~1- to 2-μm punctate regions in the cytoplasm and more weakly in the vacuole (Figure 2B). Based on mutant analyses of the Arabidopsis ATG8 pathway (Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010) and studies with yeast ATG8 (Suzuki et al., 2001), these structures likely represent autophagosomes and autophagic bodies, respectively. Similarly sized puncta could be seen for GFP fused to ATG1a and ATG13a. However, most of the fluorescent foci appeared to be confined to the cytoplasm, suggesting that the bulk of the ATG1/13 kinase complex at steady state is bound to phagophores/autophagosomes (Figure 2B).
Proof that ATG1a and ATG13a directly interact with each other in planta was provided by colocalization of the two proteins using bimolecular fluorescence complementation (BiFC) of the split yellow fluorescent protein (YFP) (Citovsky et al., 2006). Whereas simultaneous expression of the N- and C-terminal halves of YFP failed to reconstitute intact YFP and generate a fluorescent signal in Arabidopsis leaf protoplasts following transient coexpression, such BiFC fluorescence could be detected by confocal microscopy when ATG1a and ATG13a were fused in either arrangement to the C termini of nYFP or cYFP (Figure 2C). In agreement with the transient expression of full-length GFP fusions, split YFP expression of the ATG1a/ATG13a pair displayed a punctate pattern that was restricted to the cytoplasm, further implying that ATG1a and ATG13a colocalize to phagophore/autophagosome-related structures.
Expression Patterns of ATG1 and ATG13
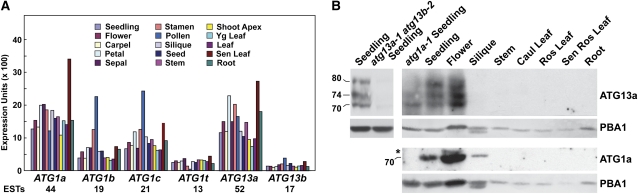
Measurement of transcript levels in the Genevestigator DNA microarray (https://www.genevestigator.ethz.ch/), EST (www.Arabidopsis.org/), and Massively Parallel Signature Sequencing databases (www.mpss.udel.edu/at/) and the DNA microarray data set of Ma et al. (2005) showed that each member of the ATG1 and ATG13 families is widely expressed in Arabidopsis (Figure 3A; data not shown). Normalized mRNA levels for ATG1a and ATG13a were consistently higher than their paralogs under most situations, a pattern also supported by EST counts. Notably, ATG1a and ATG13a were most highly expressed in senescing leaves (Figure 3A), in agreement with a role for autophagy during leaf senescence (Doelling et al., 2002; Hanaoka et al., 2002; Chung et al., 2009, 2010). Conversely, ATG1b and ATG1c were most highly expressed in pollen.
Figure 3.
Expression Patterns of ATG1 and ATG13.
(A) Transcript abundance patterns for the ATG1 and ATG13 families obtained from the Genevestigator database (https://www.genevestigator.ethz.ch/). The EST number for each locus is listed at the bottom.
(B) Immunoblot detection of the ATG1a and ATG13a proteins in crude extracts from various Arabidopsis tissues dissected from soil-grown plants. Immunoblot analysis with anti-PBA1 antibodies was performed to assess protein loading. The migration positions of ATG1a and the various ATG13a species, as confirmed by their absence in extracts from atg1a-1 and atg13a-1 atg13b-2 plants, are indicated. The asterisk identifies a protein nonspecifically cross-reacting with the anti-ATG1a antibodies. Ros, rosette; Sen, senescing; Yg, young.
The accumulation of the ATG1a and ATG13a proteins was investigated by immunoblot analyses using antibodies generated against full-length recombinant polypeptides. In accordance with the limited sequence similarity within the Arabidopsis clades (see above), these antibodies appear to be isoform specific as the anti-ATG1a antibodies failed to recognize ATG1t and the anti-ATG13a antibodies failed to recognize ATG13b (see Supplemental Figure 6 online). Cross-reaction of the anti-ATG1a antibodies with ATG1b and ATG1c was not tested due to their poor expression in Escherichia coli.
Using anti-ATG1a antibodies, we could readily detect in wild-type plants, but not in atg1a mutant plants, a 70-kD species close in size to the 69-kD molecular mass predicted for ATG1a (Figure 3B). When anti-ATG13a antibodies were used, a diffuse ladder of three and sometimes four species was observed in wild-type plants, which was absent in plants mutated for both ATG13a and ATG13b. Their apparent molecular masses ranged from 70 to 80 kD, indicating that a heterogeneous collection of ATG13a proteins accumulate (Figure 3B). This heterogeneity does not arise from alternative splicing as judged by sequence analysis of numerous ATG13a mRNAs amplified by RT-PCR but, as shown below, appears to be generated by phosphorylation of the 66-kD translation product. When the antibodies were used to detect ATG1a and ATG13a in various Arabidopsis tissues, we found high levels in seedlings and flowers but low or undetectable levels in other tissues, including senescing leaves that should have contained the highest transcript amounts (Figure 3B). Taken together, there appears to be a strong disconnection between mRNA and protein abundance for both ATG1a and ATG13a, implying that posttranscriptional mechanism(s) strongly affect their accumulation.
Identification of Mutants Inactivating ATG1a, ATG13a, and ATG13b
To help dissect genetically the functions of the ATG1/13 complex, we assembled a collection of T-DNA insertional mutants that compromise expression of ATG1a and the ATG13a/b pair (Figures 1A and 1D). The exact positions of the T-DNA insertions can be found in see Supplemental Figures 1 and 4 online. RT-PCR and RNA gel blot analyses of homozygous mutant lines confirmed that five of the six mutations (atg1a-1, atg1a-2, atg13a-1, atg13a-2, and atg13b-2) block accumulation of the full-length mRNAs (Figure 1E; see Supplemental Figures 7A to 7C online). In several cases, RT-PCR products could be generated at low levels downstream of the T-DNA insertion site (atg1a-1 and atg13a-2), which likely represent transcription driven from a cryptic promoter within the T-DNA sequence (Zheng et al., 2011). For the atg13a-2 allele, strong accumulation of a partial transcript was evident by RNA gel blot analysis, suggesting that this exonic insertion would direct the misregulated production of a substantially truncated polypeptide. A novel transcript that was ~500 bp larger than expected was detected for atg13a-2 when the upstream region was amplified by RT-PCR (Figure 1E). It contained ATG13a sequence and sequence corresponding to the T-DNA, suggesting that the T-DNA rearranged the front end of the ATG13a locus upon insertion. Comparison of the ATG1a, ATG13a, and ATG13b mRNA levels in the various mutant backgrounds failed to detect any changes in transcript abundance from the unaffected loci, suggesting that expression of the complex is not feedback regulated (see Supplemental Figures 7A to 7C online). Regardless of whether a transcript was detected or not, homozygous atg1a-1, atg13a-1, and atg13a-2 plants failed to accumulate the corresponding proteins, indicating that these mutants likely represent null alleles (Figure 3B; see below). Although not yet confirmed due to the lack of anti-ATG13b antibodies, we expect based on transcript analyses that the atg13b-2 allele also fails to accumulate the complete ATG13b protein.
The exception within our mutant collection was atg13b-1, which harbors a T-DNA in the 5′ UTR of ATG13b. ATG13b transcripts encompassing the entire coding region were found in homozygous mutant seedlings by both RT-PCR and RNA gel blot analysis, indicating that expression of the full-length protein was possible (see Supplemental Figures 7A and 7C online). In fact, it is likely that the T-DNA insertion abnormally upregulates ATG13b transcription as its mRNA levels were consistently higher in homozygous atg13b-1 seedlings.
Plants Missing ATG13 Senesce Early and Are Hypersensitive to Nutrient Deprivation
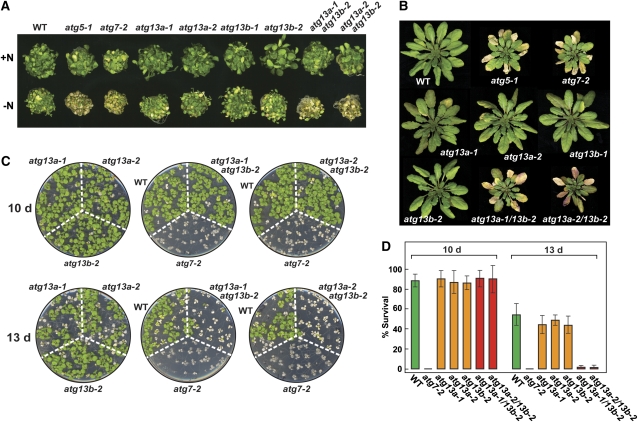
Homozygous single null mutants affecting ATG1a, ATG13a, and ATG13b resembled the wild type and were fertile when grown in nutrient-rich conditions, indicating that none of the corresponding proteins alone are essential to the Arabidopsis life cycle. However, like mutants fully eliminating other ATG components (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Liu et al., 2005; Thompson et al., 2005; Xiong et al., 2005; Inoue et al., 2006; Phillips et al., 2008; Chung et al., 2010), the atg13a atg13b double mutants displayed accelerated leaf senescence and a hypersensitivity to nutrient-limiting conditions. Similar to atg5-1 and atg7-2 plants, atg13a-1 atg13b-2 and atg13a-2 atg13b-2 plants grew poorly and became more chlorotic when grown in N-deficient medium compared with the wild type or any of the atg13a and atg13b single mutants (Figure 4A). The atg13a atg13b double mutants, like atg5-1 and atg7-2, also senesced faster when grown on soil in a short-day (SD) photoperiod (Figure 4B).
Figure 4.
atg13a atg13b Plants Display Phenotypes Characteristic of Autophagy Mutants.
Lines tested include wild-type (WT) Col-0, homozygous atg5-1, atg7-2, atg13a-1, atg13a-2, atg13b-1, atg13b-2 single mutants, and atg13a-1 atg13b-2 and atg13a-2 atg13b-2 double mutants
(A) Sensitivity to N starvation. Seedlings were grown for 1 week on MS+N liquid medium and then transferred to N-rich (+N) or N-deficient (–N) media for an additional 2 weeks.
(B) Accelerated senescence. Plants were soil grown at 22°C in an SD photoperiod for 10 weeks.
(C) Sensitivity to fixed-C starvation. Plants were grown under LD on MS-containing agar without added Suc for 2 weeks, transferred to darkness for 10 or 13 d, and then allowed to recover for 12 d in LD.
(D) Percentage of plant survival after the fixed-C starvation shown in (C). Each bar represents the average survival (±sd) of three replicates examining at least 15 seedlings each.
Using a recently developed assay to sensitively measure the effects of fixed-C deprivation (Chung et al., 2010), we found that atg13a atg13b seedlings are also hypersensitive to inadequate fixed C. Here, 14-d-old plants were grown in a long-day photoperiod (LD) on agar medium without added Suc, transferred to darkness for 10 or 13 d, and then allowed to recover in LD. As demonstrated in Figures 4C and 4D, atg7-2 plants are highly sensitive to this regime. Most, if not all, of the mutant plants died after either 10 or 13 d in darkness compared with ~15 and 46% mortality for wild-type plants, respectively. A similar hypersensitivity was seen for the atg13a-1 atg13b-2 and atg13a-2 atg13b-2 double mutant combinations but not for the atg13a-1, atg13a-2, or atg13b-2 single mutants. After 13 d in darkness, nearly all the double mutants died, while the survival rates of the single mutants were comparable to the wild type. However, after 10 d in darkness, the atg13a atg13b double mutants behaved like the wild type, indicating that the loss of ATG13 does not compromise autophagy as strongly as mutants disrupting ATG8/12 conjugation (Figures 4C and 4D).
It should be emphasized that the atg13a and atg13b single mutants behaved like the wild type under all conditions tested, implying that the corresponding genes have redundant functions. Thus, while the expression of ATG13a may be ~3 times higher than ATG13b (Figure 3A), either gene is sufficient to supply ATG13 protein under most conditions. A similar redundancy may exist for the ATG1a-c/ATG1t family. Even though ATG1a is the best expressed locus in the family, its two apparent null alleles (atg1a-1 and atg1a-2) were without apparent phenotypic consequences. Clearly, the analysis of combinatorial mutants affecting the entire ATG1 family will be needed to address the phenotypic roles of this subunit fully.
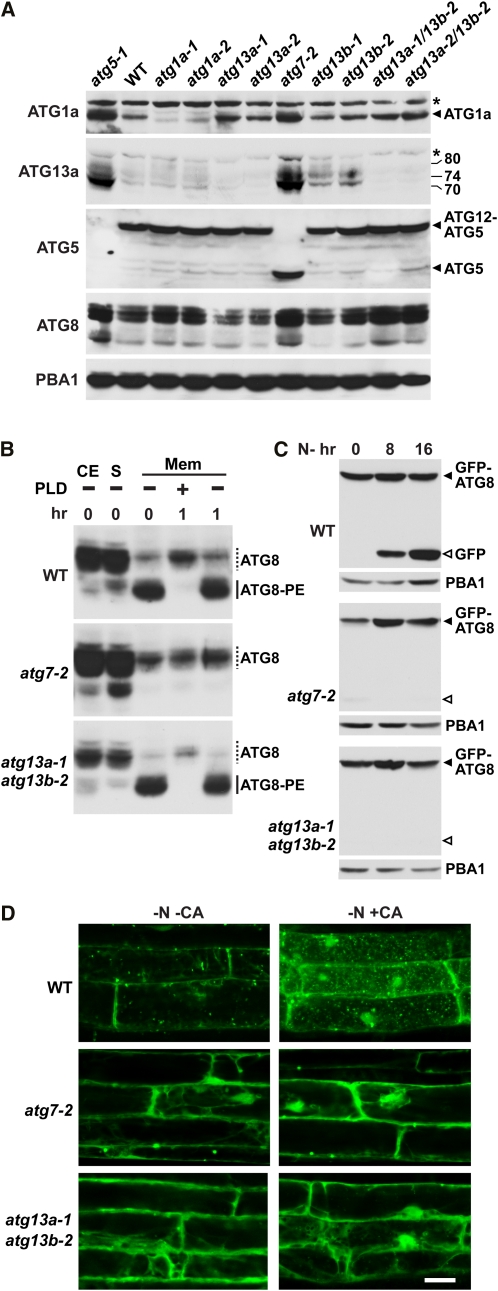
Loss of ATG13 Blocks Autophagic Body Deposition but Not ATG8 and ATG12 Conjugation
ATG-dependent autophagy is driven by assembly of the ATG12-ATG5 and ATG8-PE adducts, the latter of which helps capture specific targets and then encloses the autophagosome. Similar to the situation in yeast (Suzuki et al., 2007), ATG13 (and the ATG1a isoform) is not necessary for synthesis of the ATG12-ATG5 conjugate in Arabidopsis. As shown in Figure 5A, both atg1a null alleles, the single atg13a-1, atg13a-2, and atg13b-1 mutants, and the atg13a atg13b double mutant combinations did not dampen accumulation of the 50-kD ATG12-ATG5 adduct. This is in contrast with other atg mutants, such as atg7-2 where ATG5 conjugation is blocked concomitant with the appearance of the free ATG5 protein at 40 kD (Thompson et al., 2005; Phillips et al., 2008; Figure 5A).
Figure 5.
atg13a atg13b Mutants can Generate ATG12-ATG5 and ATG8-PE Adducts but not Autophagic Bodies
(A) Immunoblot detection of ATG1a, ATG13a, ATG8, ATG5, and the ATG12-ATG5 conjugate in single and double homozygous seedlings bearing mutations in ATG1a, ATG5, ATG7, ATG13a, and ATG13b. Arrowheads locate ATG1a, ATG5, and the ATG12-ATG5 conjugate. The apparent molecular masses of the various forms of ATG13a are indicated. Asterisks identify proteins nonspecifically cross-reacting with the anti-ATG1a or ATG13a antibodies. Immunoblot analysis with anti-PBA1 antibodies was used to confirm equal protein loading. WT, wild type.
(B) Immunoblot detection of the ATG8-PE adducts in membrane fractions. CE, crude seedling extract prior to fractionation; S, soluble fraction obtained after centrifugation; Mem, membrane fraction obtained after centrifugation and solubilized in Triton X-100. Fractions were treated for 1 h at 37°C with PLD prior to SDS-PAGE in 6 M urea. Dashed lines, free ATG8; solid lines, ATG8-PE adducts.
(C) Detection of free GFP generated by vacuolar breakdown of GFP-ATG8a. Seven-day-old mutant and wild-type seedlings expressing GFP-ATG8a were exposed to N-deficient medium for the indicated time before extraction. Crude extracts were subjected to SDS-PAGE and immunoblot analysis with anti-GFP antibodies. GFP-ATG8a and free GFP are located by the closed and open arrowheads, respectively. Anti-PBA1 antibodies were used to confirm equal protein loading.
(D) atg13a atg13b mutant plants cannot generate autophagic bodies. Seven-day-old mutant and wild-type seedlings expressing GFP-ATG8a were exposed to N-deficient medium with or without CA for 16 h and then visualized by fluorescence confocal microscopy. Bar = 10 μm
[See online article for color version of this figure.]
The lack of ATG13 also did not impair ATG8 lipidation. Unlike atg5-1 and atg7-2 mutants that have elevated levels of total ATG8 protein, presumably due to impaired vacuolar turnover of ATG8 when autophagy is blocked (Thompson et al., 2005; Chung et al., 2010), the atg13a atg13b double mutants have near normal levels (Figure 5A). To assess the lipidation status of this ATG8 pool, we collected the total membrane fraction from seedlings and then assayed for the adducts by immunoblot analysis of the solubilized material before or after phospholipase D (PLD) pretreatment. Lipidated ATG8 can be readily detected by this assay in wild-type membranes based on their faster migration during SDS-PAGE in the presence of urea and their sensitivity to PLD (Figure 5B; Chung et al., 2010). Whereas the ATG8-PE adducts were absent in atg7-2 membranes, they were abundant in wild-type and atg13a-1 atg13b-2 membranes (Figure 5B).
Following lipidation, ATG8 is transported along with autophagosomes into the vacuole where it can now be seen bound to autophagic bodies (Figure 2B). This transport can be visualized in intact plants expressing GFP-ATG8a by the appearance of small fluorescent vacuolar puncta, whose frequencies increase upon nutrient deprivation and/or treatment with the autophagy inhibitor concanamycin A (CA) that stabilizes autophagic bodies or substantially decrease in ATG8 lipidation mutants, such as atg7-2 (Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010). Consequently, vacuoles from wild-type root cells but not those from atg7-2 roots accumulate large numbers of autophagic bodies when subjected to N starvation followed by CA exposure (Figure 5D). Surprisingly, the atg13a-1 atg13b-2 roots, like atg7-2 roots, failed to accumulate autophagic bodies decorated with GFP-ATG8a even though they could lipidate ATG8 (Figure 5D).
Upon entry into the vacuole, we and others showed previously that GFP-ATG8a-PE is digested during autophagic body turnover to release free GFP in a soluble, relatively stable form (Meiling-Wesse et al., 2002; Chung et al., 2010). This free vacuolar GFP accumulates to high levels when autophagy is accelerated by nutrient starvation and thus represents a second reporter for ATG8-PE transport into vacuoles. As shown in Figure 5C, free GFP was generated from the GFP-ATG8a fusion in wild-type plants under N deprivation but was not in atg7-2 and atg13a-1 atg13b-2 plants treated similarly. Taken together, our data likely place ATG13 specifically, and the ATG1/13 complex more generally, after the events that assemble the phagophore and generate the ATG12 and ATG8 adducts, but before the autophagosome enters the vacuole in Arabidopsis.
Levels of the ATG1/13 Kinase Complex Are Strongly Regulated by Autophagy
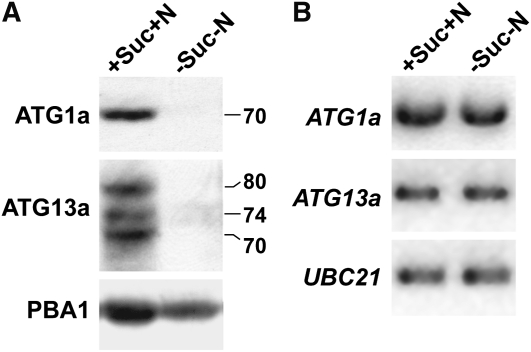
During our analysis of the ATG1a and ATG13a proteins, we noticed that the abundance of both often failed to correlate with their transcript abundance (Figure 3). Their levels were also consistently higher in various atg mutant backgrounds (e.g., atg5-1 and atg7-2; Figure 5A) but consistently lower when plants were subjected to N or fixed-C deprivation. As an example of the latter, both ATG1a and the multiple forms of ATG13a were readily detected in green seedlings grown with added N and 2% Suc but were nearly undetectable 24 h after removing these nutrients (Figure 6A). These drops in protein abundance were not observed for ATG7 nor the ATG12-ATG5 conjugate (see Supplemental Figure 8 online) and appeared unrelated to expression of the ATG1a and ATG13a genes given that the levels of the corresponding mRNAs remained constant during this short starvation regime (Figure 6B). Together, it appears that the level of the ATG1/13 complex is specifically and strongly regulated posttranscriptionally by nutrition.
Figure 6.
ATG1a and ATG13a Protein but Not Transcript Levels Disappear during Fixed-C/N Limitation.
Five-day-old wild-type seedlings were either kept in fixed-C/N-rich (+Suc+N) medium or switched to fixed-C/N-limiting medium (−Suc−N) under continuous light and then incubated for 1 d before extraction of total protein and RNA.
(A) ATG1a and ATG13a protein levels detected by immunoblot analysis. Anti-PBA1 antibodies were used to confirm equal protein loading. The apparent molecular masses of ATG1a and the three ATG13a isoforms are indicated.
(B) ATG1a and ATG13a mRNA levels as detected by RT-PCR. RT-PCR with UBC21-specific primers was used to confirm the analysis of equal amounts of total RNA.
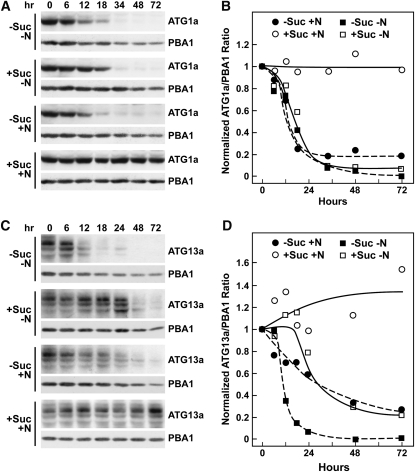
To examine the kinetics of ATG1a/ATG13a protein disappearance, we incubated light-grown wild-type seedlings in liquid +Suc+N medium and then transferred the plants for varying lengths of time to fresh +Suc+N medium or to media missing either N (+Suc-N), Suc (−Suc+N), or both (−Suc−N). The −Suc media was supplemented with an equivalent amount of mannitol to maintain the osmotic potential. Whereas the levels of ATG1a and ATG13a remained steady over a 72-h time course in +Suc+N medium, their levels dropped precipitously when either N or Suc was omitted, such that after 72 h the two proteins were barely detectable (Figures 7A and 7C). For ATG1a, the kinetics of this decrease were indistinguishable for all three starvation combinations following an apparent lag of ~6 h, which presumably reflected the time needed to deplete the seedlings of internal fixed-C and/or N reserves (Figure 7B). The apparent half-life of the ATG1a protein was calculated to be ~9 h once the decline commenced. Whereas similar kinetics were observed for ATG13a, the lag time and response to the three starvation conditions differed. The most dramatic loss and shortest half-life (~6 h) was observed using the −Suc−N medium, with the +Suc−N and −Suc+N media eliciting intermediate responses (Figures 7C and 7D). During the same deprivation time course, the abundance of the ATG1a and ATG13a mRNAs did not vary (see Supplemental Figure 9 online), implying that the drop in protein abundance was caused by decreased translation and/or increased proteolysis rather than decreased transcription.
Figure 7.
ATG1a and ATG13a Proteins Rapidly Disappear during Fixed-C/N Limitation.
Five-day-old wild-type seedlings grown in LD were either kept in +Suc+N medium or switched to +Suc−N, −Suc+N, or −Suc−N media for the indicated times before extraction of total protein.
(A) and (C) Immunoblot analysis of the starvation time course with anti-ATG1a (A) and ATG13a antibodies (C). Each lane contains an equivalent amount of tissue fresh weight. Immunoblot analyses with anti-PBA1 antibodies were used to confirm near equal protein loading.
(B) and (D) Quantitation of ATG1a (B) and ATG13a (D) disappearance normalized using the signals from PBA1.
Interestingly, the relative abundance of the various ATG13a isoforms varied during the decline, suggesting that there is a dynamic interplay among these species during fixed-C/N limitation. For example, in +Suc+N medium, the highest molecular mass 80-kD species was most abundant throughout the time course (Figure 7C). However, in −Suc−N and −Suc+N media, a 74-kD intermediate ATG13a species became more prevalent toward the end of the starvation time course, while in +Suc−N medium, the lowest molecular mass, 70-kD isoform became more prevalent (Figure 7C).
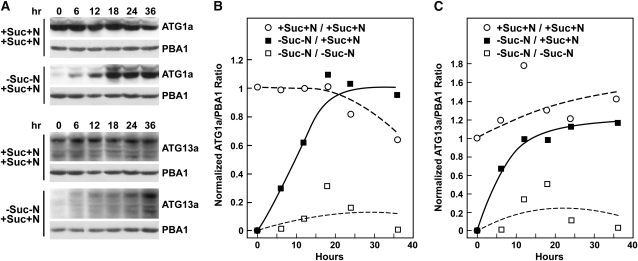
Refeeding experiments showed that the loss of ATG1a and ATG13a induced by fixed-C/N limitation is rapidly and fully reversible. When seedlings grown without Suc and N for 36 h were exchanged into +Suc+N medium, both proteins reappeared rapidly and reached levels equivalent to those in nonstarved plants within ~18 h for ATG1a and ~24 to 36 h for ATG13a (Figure 8). For ATG13a, the predominant form that accumulated after refeeding was the 80-kD species and not the 70-kD species (Figure 8A). As with the starvation-induced declines, the refeeding-induced increases in the ATG1a and ATG13a proteins were not paralleled by simultaneous increases in the corresponding mRNAs (see Supplemental Figure 9 online), suggesting that increased translation and/or decreased protein breakdown rather than increased transcription drove the recovery.
Figure 8.
ATG1a and ATG13a Proteins Rapidly Reappear after Suc/N-Limited Plants Are Refed.
Five-day-old wild-type seedlings were either kept in +Suc+N medium or switched to −Suc−N for 36 h and then switched back to +Suc+N medium for the indicated times before extraction of total protein.
(A) Immunoblot analysis of the refeeding time course with anti-ATG1a or ATG13a antibodies. Each lane contains an equivalent amount of tissue fresh weight. Immunoblot analyses with anti-PBA1 antibodies were used to confirm near equal protein loading.
(B) and (C) Quantitation of ATG1a (B) and ATG13a (C) reappearance normalized using the signals from PBA1.
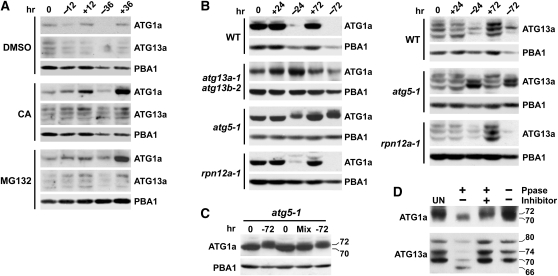
To help identify the mechanism(s) responsible for the drop in ATG1a and ATG13a levels during fixed-C/N limitation, we examined their accumulation in the presence of inhibitors or mutant backgrounds that perturb either the 26S proteasome or autophagy, the two dominant proteolytic routes in plants (Bassham, 2009; Vierstra, 2009). Here, plants were grown for 5 d in +Suc+N medium, transferred to fresh +Suc+N medium or –Suc−N medium for an additional 12 or 36 h, and then measured for the abundance of ATG1a and ATG13a. As shown in Figure 9A, both ATG1a and ATG13a were barely detectable after 36 h without added Suc and N with a marked drop seen after 12 h. Simultaneous addition of CA or the 26S proteasome inhibitor MG132 attenuated the –Suc−N–induced decline, suggesting initially that both proteolytic routes are involved. The inhibitors also increased ATG1a and ATG13a levels in well-fed plants (+Suc+N) (Figure 9A). This increase could be blocked by simultaneous addition of the translation inhibitor cycloheximide (5 μM), suggesting that ATG1a/ATG13a turnover also happens under nutrient-rich conditions but at a slower rate.
Figure 9.
Turnover and Phosphorylation States of ATG1a and ATG13a Proteins Are Regulated by Autophagy.
(A) to (C) Effect of inhibitors and mutants on the turnover of ATG1a and ATG13a during starvation. Five-day-old seedlings of the various genetic backgrounds were either kept in +Suc+N medium (+) or switched to –Suc−N medium (−) at t = 0 and then incubated for the indicated times before extraction of total protein. ATG1a and ATG13a proteins levels were detected by immunoblot analysis with anti-ATG1a and ATG13a antibodies. Immunoblot analyses with anti-PBA1 antibodies were used to confirm near equal protein loading. Each lane contains an equivalent amount of tissue fresh weight.
(A) Effect of various inhibitors. CA (0.5 μM), MG132 (50 μM), or an equivalent volume of DMSO were added at t = 0.
(B) Effect of various mutations. WT, wild type.
(C) Fixed-C/N limitation alters the SDS-PAGE mobility of ATG1a. Extracts from plants before and after 72 h of −Suc−N limitation were electrophoresed in adjacent lanes or mixed (Mix). The apparent molecular masses of the two ATG1a species are indicated.
(D) Effect of λ protein phosphatase on the SDS-PAGE mobilities of ATG1a and ATG13a. For analysis of ATG1a, atg5-1 plants were incubated for 72 h in –Suc−N medium. For analysis of ATG13a, wild-type plants were grown in +Suc+N medium. Extracts were treated for 1 h with λ phosphatase (Ppase) with or without the phosphatase inhibitor PhosSTOP and then subjected to immunoblot analysis with anti-ATG1a and anti-ATG13a antibodies. UN, untreated extracts.
Further analyses of mutants that impair the 26S proteasome or block autophagy demonstrated that autophagy is more directly involved. Whereas the decline in ATG1a and ATG13a during 24 or 72 h of Suc/N limitation proceeded normally in the 26S proteasome mutant rpn12a-1, which compromises proteasome integrity (Smalle et al., 2002), their levels were dramatically stabilized in the strong autophagy mutant atg5-1 (Thompson et al., 2005; Phillips et al., 2008) (Figure 9B). The transcript levels of both ATG1a and ATG13a were unaffected by the atg5-1 mutation, indicating that the stabilization reflected a posttranscriptional event and not atg5-1-mediated induction of the two genes (see Supplemental Figure 7D online). A moderate stabilization of ATG1a was also seen in the weaker autophagy mutant combination atg13a-1 atg13b-2 described here (Figure 9B).
Similar to the profile changes during fixed-C/N limitations, we observed a dramatic change in the ladder of ATG13a species when their turnover was blocked by eliminating ATG5. Here, two intermediate species (~76 and 72 kD) accumulated in atg5-1 seedling within 24 h in –Suc−N medium that were different from those observed before Suc and N removal (Figure 9B). Intriguingly, a subtle but reproducible upshift in ATG1a mobility during SDS-PAGE was observed as well in atg5-1 plants exposed to −Suc−N medium (Figures 9B and 9C). Interestingly, no such upshift in ATG1a was apparent in atg13a-1 atg13b-2 plants (Figure 9B), implying that ATG13 is required for the modification.
ATG1a and ATG13a Are Reversibly Modified Phosphoproteins
The variable SDS-PAGE mobilities of Arabidopsis ATG1a and ATG13a were consistent with a reversible modification of the two proteins by phosphorylation as in other eukaryotes (Chang and Neufeld, 2009; Mizushima, 2010). We confirmed this possibility by examining the effects of λ protein phosphatase on their electrophoretic patterns with or without inclusion of the phosphatase inhibitor PhosSTOP. The λ phosphatase treatment of ATG13a extracted from nonstarved wild-type seedlings substantially reduced the levels of the 80-, 74-, and 70-kD species via a reaction that was blocked by PhosSTOP, concomitant with the appearance of a new species at 66 kD that matched the expected mass of the unmodified ATG13a polypeptide (Figure 9D). A similar but more subtle effect of λ phosphatase was also observed for ATG1a extracted from Suc and N-starved atg5-1 seedlings. Here, the electrophoretic pattern of ATG1a appeared to shift from the higher mobility 72-D form to the lower mobility 70-kD form upon incubation with phosphatase (Figure 9D). It is not yet known where the phosphates are added to ATG1a or ATG13a. Using the predictions provided by KinasePhos (http://kinasephos.mbc.nctu.edu.tw/), a number of possible attached sites were identified in ATG1a-c (see Supplemental Figure 1 online).
ATG1a Associates with Autophagic Bodies during Turnover
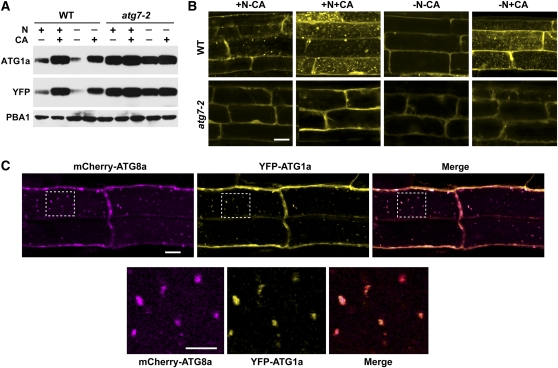
The facts that ATG1a and ATG13a are degraded under nutrient-limiting conditions by an autophagic route and bind to autophagic-like cytosolic structures made it likely that the complex is eventually broken down in the vacuole in association with autophagic bodies. To demonstrate that ATG1a enters the vacuole, we generated transgenic plants expressing YFP-ATG1a either in the wild-type or atg7-2 backgrounds and examined their localization by fluorescence confocal microscopy under either N-rich or N-deficient conditions. Immunoblot analysis of the seedlings with either anti-ATG1a or YFP antibodies readily detected the intact YFP-ATG1a fusion (Figure 10A). Importantly, the turnover of YFP-ATG1a mirrored that of endogenous ATG1a, being stabilized by CA or the atg7-2 mutation and destabilized by N starvation, indicating that the YFP moiety did not interfere with the natural dynamics of ATG1a (Figure 10A). From microscopy analyses of root tip cells, we demonstrated that YFP-ATG1a enters the vacuole and associates with small vesicles. These vesicles were readily evident in wild-type cells after CA treatment with or without N but were undetectable in atg7-2 cells under all conditions and undetectable in N-limited wild-type cells without CA exposure, consistent with the rapid turnover of ATG1a in wild-type plants deprived of N (Figure 10B).
Figure 10.
ATG1a Associates in the Vacuole with ATG8a-Decorated Autophagic Bodies.
(A) Levels of YFP-ATG1a protein are reduced by N starvation–induced autophagy. Wild-type (WT) and atg7-2 seedlings were grown for 6 d on N-rich agar medium and then transferred to N-rich (+N) or N-deficient (–N) liquid media without or with 0.5 μM CA (+CA) for 16 h. YFP-ATG1a protein levels were measured by immunoblots with anti-ATG1a and anti-YFP antibodies. Immunoblot analysis with anti-PBA1 antibodies was used to confirm equal protein loading.
(B) YFP-ATG1a associates with autophagic bodies in a process that requires ATG7. Seven-day-old wild-type and atg7-2 seedlings were exposed to N-containing or N-deficient liquid media with or without 0.5 μM CA. After 16 h, the root tip cells were imaged by fluorescence confocal microscopy. Bar = 10 μm.
(C) YFP-ATG1a and mCherry-ATG8a colocalize in autophagic bodies. Seven-day-old wild-type plants expressing both reporters were grown on +Suc−N medium and then treated for 8 h with 0.5 μM CA. Root tip cells were imaged by fluorescence confocal microscopy for YFP and mCherry. Merged represents the superimposition of the two images. Bar = 10 μm. Higher magnifications of the vacuolar lumen (outline by dashed lines) demonstrating colocalization of the two proteins on autophagic bodies are shown below. Bar = 5 μm.
To confirm that the YFP-ATG1a–decorated vesicles are autophagic bodies, we attempted to colocalize YFP-ATG1a with mCherry fused to the ATG8a (Yoshimoto et al., 2004; Thompson et al., 2005). Simultaneous imaging of YFP and mCherry found nearly complete coincidence of these reporters in N-treated root tip cells exposed to CA (Figure 10C). Analysis of roots expressing YFP-ATG1a and mCherry-ATG8a alone demonstrated that the overlapping signals were not caused by fluorescence of one reporter contaminating the other’s signal (see Supplemental Figure 10 online). Taken together, we conclude that ATG1a (and likely ATG13a) becomes associated with autophagic vesicles during their turnover under fixed-C/N-limiting conditions and are eventually recycled in the vacuole in association with autophagic bodies.
DISCUSSION
Genetic analyses of the autophagic system in yeast and metazoans have identified the ATG1/13 kinase complex as a key regulator of autophagy, which is controlled, at least in part, by the upstream TOR kinase that responds to nutrient availability (Xie and Klionsky, 2007; Mizushima, 2010). Here, we demonstrate that a similar ATG1/13 kinase complex exists in Arabidopsis, which is also central to autophagic induction under nutrient-limiting conditions. Beyond sequence similarities, we confirmed that the Arabidopsis ATG1 and ATG13 proteins assemble into an analogous complex by demonstrating that (1) ATG1a and ATG13a interact with each other in vitro and in vivo, (2) ATG1a and ATG13a localize to autophagic bodies in the vacuole, (3) eliminating ATG13 blocks the delivery of ATG8a-decorated autophagic bodies to the vacuole, and (4) loss of ATG13 generates phenotypes similar to those generated by other mutants (atg5-1 and atg7-2) shown to substantially compromise the Arabidopsis ATG system (Doelling et al., 2002; Thompson et al., 2005; Chung et al., 2010). Our identification of ATG1 and ATG13 genes in a variety of eudicots, monocots, gymnosperms, and bryophytes implies that a structurally similar complex is widely distributed throughout the plant kingdom and thus represents a centrally conserved regulator of stress-induced autophagy.
The yeast ATG1/13 kinase complex also includes ATG11 (CVT-specific), ATG17 (autophagy-specific), ATG29, and ATG31 (Mizushima, 2010), while the partially characterized metazoan counterparts include to date the ATG17 ortholog FIP200 and the ATG101 protein not found in yeast (Chang and Neufeld, 2009; Hosokawa et al., 2009a; Mercer et al., 2009). Such distinctions imply that the composition of the complex can vary among eukaryotes, which may reflect different roles and/or methods of regulation. Thus far, we identified by genome searches likely orthologs of FIP200/ATG17 and ATG101 in Arabidopsis and other plants with no evidence yet for the existence of ATG11, ATG29, and ATG31, suggesting that the plant ATG1/13 complex more closely resembles the metazoan forms. The absence of an obvious ATG11 sequence may imply further that the CVT pathway, which is exploited to deliver oligomeric vacuolar proteins across the yeast tonoplast (Johansen and Lamark, 2011), does not operate in plants.
The main difference thus far between the plant and yeast ATG systems is the extensive redundancy of the pathway. For example, whereas yeast encodes single ATG8 and ATG12 proteins, Arabidopsis synthesizes nine and two distinct isoforms, respectively (ATG8a-i and ATG12a/b; Doelling et al., 2002; Hanaoka et al., 2002), with evidence for nonredundant expression patterns and possible functions (Contento et al., 2004; Sláviková et al., 2005; Phillips et al., 2008; Chung et al., 2010). Such increased complexity has also been observed for the maize and rice ATG systems where multiple isoforms of ATG8 are evident (Chung et al., 2009).
This increased genetic complexity also extends to the ATG1/13 complex. Whereas yeast encodes a single ATG1 kinase subunit, Arabidopsis and most, if not all, other seed plants and the several metazoans analyzed express three or more ATG1 isoforms (this report; Mizushima, 2010). For example, mammals express two canonical ULK proteins (ULK1 and ULK2) in addition to three variants (ULK3, ULK4, and STK36) that have strikingly different C-terminal sequences coupled to the conserved kinase domain (Mizushima, 2010). Thus far, only ULK1 and ULK2 have been demonstrated to activate autophagy. Phylogenetic analysis of the plant ATG1 family suggests that three distinct types (a, b/c, and t) emerged early during seed plant evolution. Whereas the ATG1a and ATG1b/c paralogs in Arabidopsis have slightly different expression patterns, the normal phenotype of atg1a plants implies that that these more prototypic forms have redundant activities.
The most unique isoform is ATG1t, which appears to be seed plant specific with obvious relatives present in eudicots, monocots, and gymnosperms but absent in the bryophytes. ATG1t members contain just the kinase domain and end abruptly just a few amino acids downstream of the last conserved kinase motif (IX). Given that the C-terminal region of ATG1 provides the docking site for ATG13 and ATG11/ATG17/FIP200 (Cheong et al., 2008; Hara et al., 2008; Chan et al., 2009), the sequence differences evident in mammalian ULK3, ULK4, and STK36 and plant ATG1t could impart novel interaction networks, activities, and/or substrate preferences to the kinase particles assembled with these variants. For plants in particular, we can also imagine that the ATG1a and b/c isoforms specifically partner with the ATG13a and b isoforms to create two separate sets of ATG1/13 particles. Based on expression data alone, ATG1a/ATG13a and ATG1b-c/ATG13b pairs are predicted. Clearly, a detailed genetic analysis of the Arabidopsis ATG1 family is necessary to address potential nonredundancy, along with characterization of the kinase activity presumed to be associated with the ATG1t isoform. An intriguing possibility is that ATG1t works independently of ATG13 to direct a more constitutive autophagic route immune to TOR regulation.
Studies with the yeast and metazoan ATG1/13 kinase complexes have implicated them in the PAS assembly and subsequent biogenesis of the phagophore (Suzuki et al., 2001, 2007; Mizushima, 2010). Similarly, we found that Arabidopsis ATG13 (and likely the entire ATG1/13 complex) is essential for the deposition of ATG8-decorated autophagic bodies into the vacuole but surprisingly not for the preceding steps needed to synthesize the ATG12-ATG5 and ATG8-PE adducts. This hierarchy might place the plant complex after assembly of the PAS and initiation of the phagophore but before delivery of the autophagosome to the vacuole. Such a position implies that the assembly of the phagophore and its decoration with ATG8-PE is independent of ATG1 and ATG13 but that final enclosure and/or delivery of the autophagosome to the vacuole are under their control. Our observations that atg13a atg13b plants are less compromised phenotypically compared with those abrogating ATG8/12 conjugation (e.g., atg7-2 and atg5-1) implies that the ATG1/13 kinase complex is not essential to PAS assembly but instead might act as a modulator influencing autophagic flux.
In yeast and metazoans, both ATG1/ULK and ATG13 have been shown to be phosphoproteins, with the extent of phosphorylation dramatically regulated by nutritional state through the action of TOR and other kinases and ATG1/ULK autophosphorylation (Chang and Neufeld, 2009; Hosokawa et al., 2009b; Jung et al., 2009). Arabidopsis ATG1 and ATG13 are likely regulated by a similar mechanism. Similar to the observations of Chang and Neufeld (2009) for Drosophila melanogaster ATG13, we detected by SDS-PAGE a ladder of multiple ATG13a species whose abundance changes dynamically during nutrient deprivation, with the higher apparent molecular mass species representing phosphorylated forms based on phosphatase sensitivity assays. A similar but more subtle shift in electrophoretic mobility and comparable phosphatase sensitivity were observed for ATG1a as well. If we assume that the higher and lower mobility species represent hyper- and hypophosphorylated forms, the phosphoregulation of Arabidopsis ATG1a and ATG13a has striking parallels to that in yeast (Kamada et al., 2000; Cheong et al., 2008; Mizushima, 2010). The hyperphosphorylated forms of ATG13a appeared most abundant in fixed-C/N-rich conditions, in agreement with the situation in yeast where TOR-dependent hyperphosphorylation of ATG13 blocks autophagy when nutrients are readily available. Conversely, the slightly higher mobility species seen here for Arabidopsis ATG1a, which presumably represents its (hyper)phosphorylated state, was most obvious under fixed-C/N deprivation, in agreement with the situation in yeast where TOR inactivation triggers ATG13 dephosphorylation which in turn stimulates ATG1 autophosphorylation. Clearly, in-depth phosphoproteomic analyses of ATG1a and ATG13a are now needed to appreciate the interplay between their phosphorylation states and a plant’s nutritional status.
The most surprisingly finding of our study was the discovery that the levels of ATG1a and ATG13a but not other ATG proteins (ATG7 and ATG12-ATG5) are dynamically regulated by nutrient availability, indicating that the kinase complex is both a regulator and a specific target of autophagy. Following a short lag period, which we presume represents the time required to diminish available pools of fixed-C and N, both proteins rapidly disappear and then rapidly reappear when the plants are refed. The transfer of ATG1a to the vacuole in association with autophagic bodies was even seen in well-fed plants, implying that turnover of the complex occurs in nutrient-rich conditions and is then accelerated during nutrient stress in concert with the greater accumulation of autophagic bodies under these conditions. A similar phenomenon has not yet been reported for the yeast and metazoan ATG1/ULK and ATG13 proteins, nor is it known whether the other ATG1 and ATG13 isoforms in Arabidopsis are similarly regulated, leaving it unclear whether this proteolytic regulation represents a conserved and central mechanism. Whereas all our data point to a dominant role of proteolysis in reducing ATG1/13 protein levels during nutrient deprivation, we cannot yet rule out a simultaneous inhibition of translation for the corresponding mRNAs. Such regulation is consistent with the role of yeast and mammalian TOR in dampening translation during starvation via increased phosphorylation of both ribosomal subunits and translation regulators (Zoncu et al., 2011). Although the precise roles of TOR in plants remain unclear, the phenotypes of null and weak alleles affecting the single TOR gene in Arabidopsis suggest a deep involvement in nutrient stress, translation, and autophagy (Menand et al., 2002; Deprost et al., 2007; Liu and Bassham, 2010).
At first glance, rapid turnover of the ATG1a/13a complex during nutrient stress appears counterintuitive given that this positive autophagic regulator should be most in demand under starvation conditions. Its breakdown could represent the inevitable consequence of the assembled PAS becoming fixed to the developing phagophore and then captured during autophagosome enclosure. Alternatively, starvation-induced turnover could represent a deliberate negative feedback mechanism to remove activated ATG1/13 kinase complexes after they stimulate autophagosome assembly. Such turnover could reset autophagic induction by requiring the incorporation of freshly activated ATG1/13 kinase during each round of PAS and phagophore assembly. Consequently, the changes in phosphorylation status of ATG1 and ATG13 may have a dual purpose (i.e., enhancing their activities and subsequently directing their autophagic turnover). It will be interesting to determine whether the abundance of other PAS-associated ATG components, such as the PI3 kinase complex (Behrends et al., 2010; Suzuki and Ohsumi, 2010), are also subjected to this dynamic regulation.
We are uncertain how ATG1 and ATG13 become tethered to the autophagosome before vacuolar delivery. One possibility is that the entire PAS is removed due to its association with the phagophore membrane through the ATG17/FIP200 scaffold. Another possibility, based on our demonstration that ATG1a and ATG8 can directly interact (Figure 2A), is that the ATG1/13 complex is recruited to the phagophore before enclosure by binding to ATG8-PE decorating the membrane surface. Thus, ATG1/13 could be transported to the vacuole using the same mechanism employed by the expanding class of shuttle proteins that dock cargo to the autophagosome via the ATG8 interface (Noda et al., 2008; Behrends et al., 2010).
METHODS
Identification and Phylogenetic Analysis of ATG1 and ATG13 Genes
The Arabidopsis thaliana Col-0 ecotype genomic and EST databases were searched by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/blast.cgi) for potential ATG1 and ATG13 orthologs using the yeast protein sequences as queries. Full-length cDNAs were obtained either from ABRC (ATG1a, ATG1b, and ATG1c) or amplified from cDNA synthesized from Arabidopsis Col-0 leaf total RNA (ATG1t, ATG13a, and ATG13b) using gene-specific primers. (All primers used in this study are listed in Supplemental Table 2 online.) The PCR products were cloned into the TOPO vector by recombination according to the manufacturer’s instructions (Invitrogen) and verified by sequencing. These constructs were used later as entry vectors to make the plasmids for antibody production, Y2H assays, and transgenic plant construction. Intron/exon boundaries were identified by comparing the cDNA sequences with their respective genomic DNA sequences. Synteny was predicted using the http://chibba.agtec.uga.edu/duplication/ website.
For phylogenetic analyses, ATG1, ATG13, and related sequences were downloaded from the National Center for Biotechnology Information or Department of Energy Joint Genome Initiative Phytozome databases (http://www.phytozome.net/). Their GenBank accession numbers or Phytozome locus names are listed in Supplemental Table 1 online. The kinase domains in the ATG1 family were predicted by PFAM (http://pfam.sanger.ac.uk) and were used for phylogenetic analysis. ATG1 and ATG13 protein sequences were aligned by ClustalX (Chenna et al., 2003) under the default settings and compared by MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003) using the proportion of invariable sites γ rate model and the mixed amino acid model as described (Ulijasz et al., 2009). The program was run for 1,000,000 generations, and the first 25% (337,500 generations) was discarded as “burn in” after checking that the log likelihood values had reached a plateau. The strict consensus tree was displayed with TreeView (Page, 1996).
Plant Materials and Growth Conditions
The atg1a-1 (SAIL_27_A05), atg1a-2 (SALK_054351), atg13a-1 (SALK_065240), atg13a-2 (SALK_044831), and atg13b-1 (SALK_129206) T-DNA insertion mutants were obtained from the SIGnAL collection in the Col-0 background (Alonso et al., 2003). The atg13b-2 (GK-510F06) T-DNA insertion allele in the Col-0 background was obtained from the GABI-Kat T-DNA insertion collection (http://www.gabi-kat.de) (Rosso et al., 2003). The insertion position of each mutation was verified by genomic PCR using a 5′ gene-specific primer in combination with a T-DNA left border–specific primer. Primers used throughout this study are listed in Supplemental Table 2 online. Each mutant was backcrossed three times to wild-type Col-0 before phenotypic analysis. The atg5-1, atg7-2, and rpn12a-1 mutants in the Col-0 background were as previously reported (Smalle et al., 2002; Thompson et al., 2005; Chung et al., 2010).
Arabidopsis seeds were vapor-phase sterilized, soaked in water at 4°C for 2 d, and then sown on Gamborg's B5 agar (Sigma-Aldrich) containing 2% Suc except where noted. The plates were incubated at 21°C under LD (16 h light/8 h dark) or SD (8 h light/16 h dark) photoperiods. Plants exposed to LD were transferred to soil after 2 weeks. All liquid-grown seedlings were exposed to continuous light. For assessing the effects of fixed-C/N limitation, liquid media were prepared with either full-strength Murashige and Skoog salts (MS) (Sigma-Aldrich) or MS micronutrient salts (Sigma-Aldrich) amended with 3 mM CaCl2, 1.5 mM MgSO4, 1.25 mM KH2PO4, 5 mM KCl, and 2 mM MES-KOH, pH 5.7 (MS-N). These media were supplemented with 2% Suc (+Suc), 1% mannitol (−Suc), and/or the inhibitors CA (0.5 μM) (Wako Chemicals) and MG132 (N-(benzyloxycarbonyl)-Leu-Leu-Leu-al) (50 μM) (Sigma-Aldrich) at the indicated times. Both the control and treated seedlings were washed three times in fresh medium at t = 0 for all time courses.
Effects of N limitation on plant growth and survival were determined according to Phillips et al. (2008). Briefly, 1-week-old seedlings grown under continuous light in MS liquid medium plus 2% Suc were transferred to MS-N liquid medium plus 2% Suc for two additional weeks. Fixed-C limitation of seedlings on agar medium was performed according to Chung et al. (2010). Recovery was scored by obvious regreening and additional growth of the seedlings after an additional 12 d in LD.
Y2H Assays
Assays for protein–protein interactions by Y2H were conducted as previously described (Gingerich et al., 2007) with minor modifications. Full-length cDNAs encoding ATG1a, ATG13a, ATG7, and ATG8a in the TOPO entry vectors were recombined into either the pDEST22 GAL4 activation domain plasmid or the pDEST32 GAL4 binding domain plasmid using the LR Clonase reaction (Invitrogen). Both plasmids were transformed into yeast strain MaV203. Protein–protein interactions were identified by growth after 2 d on medium lacking His, Leu, and Trp and containing 25 mM 3-amino-1,2,4-triazole, according to the manufacturer’s recommendations (Invitrogen).
RT-PCR and RNA Gel Blot Analyses
RNA was isolated from liquid-grown or soil-grown plants using Trizol reagent (Invitrogen) and then treated with DNase RQ1 (Promega). The first-strand for each cDNA was synthesized by Superscript II reverse transcriptase (Invitrogen) in combination with the reverse primers described in Supplemental Table 2 online. RT-PCR was then accomplished for 35 cycles using this template, Ex-Taq polymerase (TaKaRa), and the reverse primers in combination with forward primers (see Figures 1A and 1D for primer positions). The 26S proteasome subunit PAE2 gene was used as a RT-PCR internal control. Quantitative RT-PCR was performed with MyiQ5 two-color real-time PCR detection system using iQ SYBR green supermix (Bio-Rad). Relative transcript abundance of target genes was determined by the comparative threshold cycle method using reactions with the Arabidopsis UBC9 transcript as the internal control (Chung et al., 2009).
For RNA gel blot analysis, total RNA was isolated with the TRIzol regent (Invitrogen) according to Smalle et al. (2002). 32P-labeled riboprobes were synthesized with T7, SP6, or T3 RNA polymerase using the RiboprobeR system and the linearized pGEMT (Promega) or pBluescript (Stratagene) cDNA constructs for ATG1a, ATG1b, ATG13a, ATG13b, and TUB1. Membranes were hybridized overnight at 68°C and washed according to Smalle et al. (2002) prior to autoradiography.
Construction of Transgenic Plants
A transgene expressing full-length ATG1a as a C-terminal fusion to YFP and downstream of the cauliflower mosaic virus 35S promoter was constructed by recombining the ATG1a full-length cDNA in the TOPO entry vector into the pEarleyGate 104 Gateway vector (Earley et al., 2006). Lines expressing mCherry as an N-terminal fusion to ATG8a were generated using the mCherry cDNA (Nelson et al., 2007) PCR amplified and inserted into the KpnI/AscI site of the pMDC99 plasmid (Curtis and Grossniklaus, 2003) upstream of the Arabidopsis UBQ10 promoter. The resulting pMDC99-AtUBQ10p-mCherry plasmid was used as a destination vector in an LR recombinase (Invitrogen) reaction with the entry vector pENTR/D-TOPO-AtATG8a containing the Arabidopsis ATG8a cDNA (Phillips et al., 2008). The YFP and mCherry constructs were sequence verified, introduced into Agrobacterium tumefaciens strain GV3101, and then transformed into wild-type Col-0 plants by the floral dip method. Homozygous plants for each transgene were identified in the T3 generation by either genomic PCR, antibiotic resistance, immunoblot analysis with anti-GFP antibodies, and/or by fluorescence confocal microscopy. Plants expressing YFP-ATG1a and/or mCherry-ATG8a in the various atg mutant backgrounds were created by crossing.
Protein Isolation and Immunoblot Analyses
Total protein was isolated from liquid-grown or soil-grown plants by homogenization in 2:1 (volume to fresh weight) SDS-PAGE sample buffer (125 mM Tris-HCl, pH 6.8, 5% SDS, 20% glycerol, and 10% 2-mercaptoethanol). Clarified extracts were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore) for immunoblot analysis using alkaline phosphatase–labeled or peroxidase-labeled goat anti-mouse or goat anti-rabbit immunoglobulins (Kirkegaard and Perry Laboratories) for detection. Extract volumes were adjusted to reflect either equal protein or equal fresh weight as indicated. Quantification of immunoblot signal strength was determined with NIH ImageJ (http://rsb.info.nih.gov/ij/) using the signal from PBA1 to normalize protein loads. All bands related to ATG13a were incorporated into the quantitation. The ATG8-PE adducts were detected in membrane-enriched fractions by the method of Chung et al. (2010) following separation by SDS-PAGE in the presence of 6 M urea. The Triton X–solubilized fractions were incubated with 250 units/mL of Streptomyces chromofuscus PLD (Enzo Lifesciences) for 1 h at 37°C prior to electrophoresis.
Antibodies against ATG1a and ATG13a were generated against full-length recombinant proteins bearing an N-terminal 6His tag. The full coding regions for the proteins were transferred from the TOPO entry vector into the Gateway pDEST17 vector (Invitrogen) through the LR reaction and expressed in E. coli BL21 cell upon induction of the cells with 1 mM isopropyl β-d-1-thiogalactopyranoside for 3 h. Insoluble fractions containing the recombinant proteins were resuspended in SDS-PAGE sample buffer containing 8 M urea and subjected to SDS-PAGE. Gel pieces containing the recombinant proteins (~2 mg) as identified by Coomassie Brilliant Blue staining were homogenized and injected directly into rabbits (Polyclonal Antibody Service). Antibodies against of ATG8a, ATG5, ATG7, and PBA1 were as described (Smalle et al., 2002; Doelling et al., 2002; Thompson et al., 2005; Phillips et al., 2008). GFP was detected with a cocktail of two mouse anti-GFP monoclonal antibodies purchased from Roche Applied Science. YFP was detected with a mouse anti-GFP monoclonal antibody purchased from Miltenyi Biotec.
λ Protein Phosphatase Treatments
Liquid-grown wild-type or atg5-1 plants were homogenized in 1.25:1 (volume to fresh weight) extraction buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM DTT, 0.01% Brij35, 1 mM MnCl2, 10 mM Na2EDTA, and 0.1% Triton X-100) supplemented with 1 mM phenylmethylsulfonyl fluoride and Complete Plant Protease Inhibitor Cocktail (Sigma-Aldrich). Clarified extracts were incubated with λ protein phosphatase (40 units μL−1) (New England Biolabs) with our without addition of a 1× concentration of the phosphatase inhibitor PhosSTOP (Roche) for 30 min at 30°C. The reactions were quenched by adding an equal volume of 2× SDS-PAGE sample buffer and heating to 95°C for 5 min.
Protoplast Isolation and Fluorescence Confocal Microscopy
Protoplasts were generated from 2-week-old seedling leaf tissue by the method of Lee et al. (2003) and suspended in 1.5 mM MES-KOH, pH 5.6, 150 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 5 mM mannitol. Full-length coding regions for GFP, GFP-ATG8a, ATG1a-GFP, and GFP-ATG13a were placed under the control of the 35S promoter in the plasmids pMDC43 or pMDC83 (Curtis and Grossniklaus, 2003). For BiFC assays, full-length cDNAs for ATG1a and ATG13a were placed after the N-terminal half of EYFP in the pSAT4-DEST-n(1-174)EYFP-C1 vector or after the C-terminal half of EYFP in the pSAT5-DEST-c(175-end)EYFP-C1(B) vector through LR reactions, respectively (Citovsky et al., 2006). The purified plasmids were introduced into the protoplasts by polyethylene glycol–mediated transformation (Lee et al., 2003) and observed by fluorescence microscopy 18 h later. Transgenic lines stably expressing GFP-ATG8a, YFP-ATG1a, and/or mCherry-ATG8a were grown on MS agar medium containing 1% Suc under LD condition. Five to six days after germination, seedlings were transferred to fresh medium or to MS-N liquid medium containing 1% Suc, with or without the addition of CA or an equivalent volume of DMSO. After a 16-h incubation, root cells were observed using a Zeiss 510-Meta laser scanning confocal microscope. GFP and chlorophyll fluorescence were detected with the band-pass 500- to 530-nm and ChS 650- to 670-nm filter sets, respectively. YFP fluorescence was visualized in the single-track mode with the band-pass 500- to 530-nm IR filter while irradiated with 488-nm light. For simultaneous detection of YFP and mCherry in the multitrack mode, plants were irradiated with 488- or 543-nm light, and the emission signals were collected with the band-pass 500- to 550-nm IR or 565- to 615-nm IR filters, respectively. All images represented single optical sections with the focal points varied to discriminate between vesicles and strands. Images were processed with the LSM510 image browser (Zeiss) and either NIH ImageJ (http://rsb.info.nih.gov/ij/) or Adobe Photoshop CS4.
Accession Numbers
The accession numbers for the ATG1 and ATG13 genes described in this article can be found in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of the Plant ATG1 Family with ATG1 from S. cerevisiae.
Supplemental Figure 2. Phylogenetic Analyses of the ATG1 Family in Plants.
Supplemental Figure 3. Alignment of the Sequence Surrounding the C-Terminal End of Plant ATG1t Proteins Compared with Plant and Yeast Representatives of ATG1a.
Supplemental Figure 4. Amino Acid Sequence Alignment of the Plant ATG13a Protein Family with ATG13 from S. cerevisiae.
Supplemental Figure 5. Phylogenetic Analyses of the ATG13 Protein Family in Plants.
Supplemental Figure 6. Anti-ATG1a and ATG13a Antibodies Show High Specificity for Their Respective Antigens.
Supplemental Figure 7. ATG1a, ATG13a, and ATG13b Transcript Abundance in Various atg Mutant Backgrounds.
Supplemental Figure 8. Levels of ATG7, ATG5, and the ATG12-ATG5 Conjugate Are Not Affected by Fixed-C/N Limitation.
Supplemental Figure 9. Suc and N Limitations Do Not Affect ATG1a and ATG13a Transcript Levels.
Supplemental Figure 10. Selective Detection of mCherry-ATG8a and YFP-ATG1a in Transgenic Arabidopsis Plants by Fluorescence Confocal Microscopy.
Supplemental Table 1. Abbreviations, Species Names, and Data Sources for Various Members of the ATG1/13 Families Used for the Phylogenetic Analysis.
Supplemental Table 2. Oligonucleotide Primers Used in This Study.
Supplemental Data Set 1. Text File of the ATG1 Sequence Alignment Used in Supplemental Figure 2.
Supplemental Data Set 2. Text File of the ATG13 Sequence Alignment Used in Supplemental Figure 5.
Acknowledgments
We thank Robb Stankey, Allison Phillips, Yangrong Cao, and Andrew F. Bent for technical help and reagents. This work was supported by a grant from the USDA–Agriculture and Food Research Institute program (2008-02545) to R.D.V. and a Thailand National Science Foundation predoctoral fellowship to A.S.
AUTHOR CONTRIBUTIONS
A.S., F.L., and R.D.V. designed the research. A.S. and F.L. performed the research. T.C. contributed materials and technical information. A.S., F.L., and R.D.V. analyzed the data and wrote the article.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bassham D.C. (2009). Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta 1793: 1397–1403 [DOI] [PubMed] [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. (2010). Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y., Longatti A., McKnight N.C., Tooze S.A. (2009). Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.Y., Neufeld T.P. (2009). An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D.J. (2008). The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- Contento A.L., Kim S.J., Bassham D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolaï M., Bedu M., Robaglia C., Meyer C. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Noda N.N., Fujii K., Yoshimoto K., Ohsumi Y., Inagaki F. (2008). In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283: 1921–1928 [DOI] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura A., Noda T., Ohsumi Y. (1997). Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192: 207–213 [DOI] [PubMed] [Google Scholar]
- Ganley I.G., Lam H., Wang J., Ding X., Chen S., Jiang X. (2009). ULK1-ATG13-FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284: 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich D.J., Hanada K., Shiu S.H., Vierstra R.D. (2007). Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19: 2329–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S.K., Hunter T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596 [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig M.S., Contento A.L., Meyer A., Ebany D., Bassham D.C., Macintosh G.C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. USA 108: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H.T., Mattsson O., Jørgensen L.B., Jones J.D.G., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., et al. (2009b). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. (2009a). Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of RUBISCO and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.S., Cho H.S., Yoon G.M., Ahn J.W., Kim H.H., Pai H.S. (2003). Interaction of NtCDPK1 calcium-dependent protein kinase with NtRpn3 regulatory subunit of the 26S proteasome in Nicotiana tabacum. Plant J. 33: 825–840 [DOI] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830 [DOI] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Ma L., Sun N., Liu X., Jiao Y., Zhao H., Deng X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K., Barth H., Thumm M. (2002). Ccz1p/Aut11p/Cvt16p is essential for autophagy and the CVT pathway. FEBS Lett. 526: 71–76 [DOI] [PubMed] [Google Scholar]
- Menand B., Desnos T., Nussaume L., Berger F., Bouchez D., Meyer C., Robaglia C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer C.A., Kaliappan A., Dennis P.B. (2009). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5: 649–662 [DOI] [PubMed] [Google Scholar]
- Mizushima N. (2010). The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22: 132–139 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008). Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2001). Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Page R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J.D., White E. (2010). Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Chung T., Holding D., Jung R., Vierstra R.D., Otegui M.S. (2011). Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell 23: 769–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y.L., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Yang P., Babiychuk E., Kushnir S., Durski A., Vierstra R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. (2010). Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 584: 1280–1286 [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Vierstra R.D. (2005). Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Ulijasz A.T., Cornilescu G., von Stetten D., Cornilescu C., Velazquez Escobar F., Zhang J., Stankey R.J., Rivera M., Hildebrandt P., Vierstra R.D. (2009). Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J. Biol. Chem. 284: 29757–29772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23: 785–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. (2007). Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., Narendra D.P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Miller N.D., Lewis D.R., Christians M.J., Lee K.H., Muday G.K., Spalding E.P., Vierstra R.D. (2011). AUXIN UP-REGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol. 156: 1878–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D.M. (2011). mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]