Conserved, plant-specific regions of Arabidopsis RIN4 are shown to contribute to its regulation of plant-innate immunity. Also, derivatives of RIN4 that disrupt its normal tethering to the plasma membrane are hyperactive suppressors of immunity, leading to a model in which a pathogen virulence protein proteolytically cleaves RIN4, thereby releasing an active fragment from the membrane.
Abstract
RPM1-interacting protein 4 (RIN4) is a multifunctional Arabidopsis thaliana protein that regulates plant immune responses to pathogen-associated molecular patterns (PAMPs) and bacterial type III effector proteins (T3Es). RIN4, which is targeted by multiple defense-suppressing T3Es, provides a mechanistic link between PAMP-triggered immunity (PTI) and effector-triggered immunity and effector suppression of plant defense. Here we report on a structure–function analysis of RIN4-mediated suppression of PTI. Separable fragments of RIN4, including those produced when the T3E AvrRpt2 cleaves RIN4 and each containing a plant-specific nitrate-induced (NOI) domain, suppress PTI. The N-terminal and C-terminal NOIs each contribute to PTI suppression and are evolutionarily conserved. Native RIN4 is anchored to the plasma membrane by C-terminal acylation. Nonmembrane-tethered derivatives of RIN4 activate a cell death response in wild-type Arabidopsis and are hyperactive PTI suppressors in a mutant background that lacks the cell death response. Our results indicate that RIN4 is a multifunctional suppressor of PTI and that a virulence function of AvrRpt2 may include cleaving RIN4 into active defense-suppressing fragments.
INTRODUCTION
Plants rely on a layered defense system to fend off pathogen attack. Among the active responses are those emanating from two branches of the plants innate immune system: pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Chisholm et al., 2006; Jones and Dangl, 2006). PTI is a first line of defense activated upon recognition of MAMPs (microbe-associated molecular patterns) or PAMPs, which are ubiquitous structural elements of molecules essential to the microbial lifestyle. Perception of PAMPs by pattern recognition receptors (PRRs) leads to the elicitation of PTI. The prototype PRR in Arabidopsis thaliana is FLS2, which is the receptor for a polypeptide PAMP (flg22) present in the flagellin protein of some bacteria (Felix et al., 1999). PTI from activated FLS2 and other PRRs includes generation of reactive oxygen species (ROS), activation of MAP kinases, production of the plant hormones SA and ethylene, transcriptional reprogramming, and cell wall fortification marked by callose deposition (Chinchilla et al., 2007; Tsuda et al., 2008). Various PAMPS, including flg22, Ef-Tu, and chitin, induce activation of a common set of genes, indicating that signals from PRRs may converge on a limited set of pathways (Zipfel et al., 2006; Wan et al., 2008). PTI typically does not cause a hypersensitive response (HR) or localized cell death, but nonetheless effectively combats potentially pathogenic microbes.
Pathogens counter this first line of defense by delivering PTI-suppressing virulence effectors. Suppression of PTI is hypothesized to be a key step in the evolution of pathogenicity and can lead to a diseased state also known as effector-triggered susceptibility (ETS) (Jones and Dangl, 2006). Gram-negative bacteria use type III secretion systems (T3Ss) to deliver defense-suppressing type III effectors (T3Es) into the cytosol of plant cells (Alfano and Collmer, 2004). Individual bacteria deliver a repertoire of T3Es that move to a variety of subcellular locations and perturb various host targets (da Cunha et al., 2007). Many T3Es have been shown to suppress PTI, including limiting callose deposition and enhancing the growth of T3S-deficient bacteria (Guo et al., 2009).
To counter ETS, R-genes mediate recognition of pathogen-encoded effectors and activate ETI. The prototypical R-proteins are composed of a central nucleotide binding site and C-terminal leucine-rich repeats (McHale et al., 2006). These intracellular R-proteins function like receptors that either interact with effectors directly or perceive effectors indirectly via their perturbations of host targets (Mackey and McFall, 2006). ETI typically produces a robust defense response that potently restricts the growth of microbes and frequently elicits a HR. Current data support models in which differences between the outputs of ETI and PTI are quantitative rather than qualitative (Maleck et al., 2000; Tao et al., 2003; Jones and Dangl, 2006; Shen et al., 2007).
RIN4 (RPM1-interacting protein 4) is a multifunctional protein that links PTI, ETS, and ETI. RIN4 is a negative regulator of PTI (Kim et al., 2005b). Arabidopsis plants lacking or inducibly expressing RIN4 display enhanced or suppressed flg22-induced callose deposition, respectively. Pseudomonas syringae pv tomato strain DC3000 (PstDC3000) is pathogenic on tomato and Arabidopsis, but the T3S-deficient hrcC mutant fails to grow, because it is unable to deliver PTI-suppressing T3Es (Hauck et al., 2003). Thus, the ability of hrcC to grow is a useful proxy for suppression of PTI. The hrcC mutant grows to reduced or elevated levels in Arabidopsis plants lacking or inducibly expressing RIN4, respectively.
Numerous T3Es target RIN4 as part of their attempt to cause ETS. The T3Es AvrRpm1, AvrB, and AvrRpt2 each suppress flg22-induced callose deposition and promote the growth of hrcC (Kim et al., 2005b; Shang et al., 2006). AvrRpm1 and AvrB each induce phosphorylation of RIN4, whereas AvrRpt2 proteolytically clips RIN4 into three pieces (Mackey et al., 2002; Axtell et al., 2003; Mackey et al., 2003; Chisholm et al., 2005; Kim et al., 2005a; Takemoto and Jones, 2005). RPM1-induced protein kinase (RIPK) is a receptor-like cytoplasmic kinase that contributes toT3E-induced phosphorylation of RIN4 (Liu et al., 2011). It has proven difficult to determine how perturbation of RIN4 by AvrRpm1, AvrB, and AvrRpt2 regulates the PTI-suppressing function of RIN4, because each of these T3Es has additional virulence targets inside plant cells (Belkhadir et al., 2004; Lim and Kunkel, 2004). Targeting of RIN4 by HopF2, a T3E with ADP-ribosyltransferase activity, is required for its virulence activity in Arabidopsis, further demonstrating the bona fide role of RIN4 in defense regulation (Wang et al., 2010; Wilton et al., 2010).
Perturbation of RIN4 by T3Es mediates the induction of ETI by two Arabidopsis R-proteins, RPM1 and RPS2. Despite lacking predicted transmembrane domains, RIN4, RPM1, and RPS2 each localize to the plasma membrane (Boyes et al., 1998; Axtell and Staskawicz, 2003; Takemoto and Jones, 2005). Palmitoylation of C-terminal cysteines mediates membrane anchoring of RIN4 (Kim et al., 2005a). RIN4 interacts with both RPM1 and RPS2, perhaps targeting them to the membrane, and prevents ectopic activation of both R-proteins (Mackey et al., 2002; Axtell and Staskawicz, 2003; Mackey et al., 2003; Belkhadir et al., 2004; Day et al., 2005). RPM1 responds strongly to AvrRpm1 and AvrB and weakly to AvrRpt2, whereas RPS2 responds strongly to AvrRpt2 and weakly to AvrRpm1 and AvrB (Kim et al., 2009). The activation of RPM1 by AvrB, and partially by AvrRpm1, is mediated through the phosphorylation of amino acid T166, a residue targeted by RIPK (Chung et al., 2011; Liu et al., 2011). Thus, each of these R-proteins seems to induce defense responses of different strengths in response to different perturbations of RIN4. In addition to these PTI-, ETS-, and ETI-related functions of RIN4 that are manifested when bacteria invade the apoplast of the plant leaf, RIN4 also negatively regulates preinvasion defense via interaction with (H+)–ATPases that promote reopening of stomata after perception of bacterial PAMPs (Liu et al., 2009).
We performed a structure–function analysis of postinvasion defense regulation by RIN4. We show that separable fragments of RIN4, each containing a plant-specific nitrate-induced (NOI) domain (Pfam: PF05627), suppress PTI. Notably, these results include two derivatives of RIN4 that correspond to the two primary AvrRpt2 cleavage products (ACP2 and ACP3) of RIN4. We show that separable fragments, each containing an NOI domain, contribute to PTI suppression by RIN4. Phylogenetic analysis indicates that distinct N-terminal NOI (N-NOI) and C-terminal NOI (C-NOI) domains have been maintained in RIN4 homologs across the plant lineages. We show that RIN4 derivatives that are not properly targeted to the plasma membrane, because they lack the C-terminal region or the cysteines in the C-terminal region, elicit a cell death response. In a triple mutant background that prevents the cell death response, we demonstrate that nonmembrane-tethered derivatives are more potent suppressors of PTI, indicating that membrane targeting of RIN4 limits its ability to suppress PTI and that AvrRpt2 may enhance PTI suppression by liberating a hyperactive fragment of RIN4 from the membrane. Collectively, our results indicate that both NOI-containing fragments of RIN4 contribute to negative regulation of PTI and that cleavage of RIN4 by AvrRpt2 produces two active PTI-suppressing fragments, including one that is no longer tethered to the plasma membrane.
RESULTS
RIN4 Contains Separable Domains Capable of Suppressing PTI
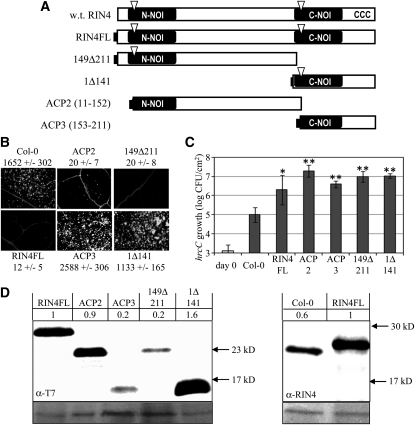
Our initial goal was to identify the protein region(s) of RIN4 that mediate suppression of PTI. To this end, we generated transgenic Arabidopsis lines that inducibly express RIN4 derivatives, including the N-terminal two thirds (149Δ211), C-terminal one third (1Δ141), and the two major AvrRpt2 proteolytic products consisting of AA 11 to 152 (ACP2) and AA 153 to 211 (ACP3) in wild-type Columbia-0 ecotype (Col-0) plants (Figure 1A). All the RIN4 derivatives in Figure 1 and throughout this study, including full-length RIN4 (RIN4FL), have an N-terminal T7 tag to permit uniform assessment of their expression. Also, for all derivatives, two or more independent transgenic lines with single homozygous insertions were produced and tested, and data from single representative lines are presented.
Figure 1.
RIN4 Contains Separable Domains with Distinct Defense Suppressing Activities.
(A) Schematic diagram of the 211 amino acid RIN4 protein and its derivatives. RIN4 contains two conserved NOI domains (black bars), two AvrRpt2 cleavage sites (triangles), and C-terminal cysteines required for acylation and membrane targeting of RIN4 (CCC, amino acids 203, 204, and 205). Full-length RIN4 (RIN4FL) and other derivatives all contain an N-terminal T7-tag (black rectangle). AvrRpt2 cleaves RIN4 between amino acids 10/11 and 152/153 and the derivatives ACP2 and ACP3 (AvrRpt2-cleavage products) consist of RIN4 amino acids 11 to 152 and 153 to 211, respectively. w.t., wild type.
(B) and (C) Callose induced by flg22 infiltration (B) and growth of hrcC (C) in Col-0 and lines of Col-0 inducibly expressing the indicated RIN4 derivatives. Expression was induced with 20 μM dexamethasone 48 h before infiltration with (B) 30 μM flg22 or (C) 1 × 105 CFU/mL of hrcC. Numbers in (B) are average and sd of the number of callose deposits per 1.1 mm2. Bacterial numbers and sd (C) are plotted at the time of infiltration (day 0) in Col-0 and after 4 d. *P = 0.008; **P < 0.0002 [two-tailed Student’s t test for comparison with Col-0 plants].
(D) Immunoblots with anti-T7 antisera to compare levels of RIN4 derivatives and with anti-RIN4 antisera to compare levels of native RIN4 and RIN4FL. Numbers are band intensities with the signal in plants expressing RIN4FL normalized to 1. Panels below are ponceau stains for total protein loading.
The effect of inducibly expressed RIN4 derivatives on PTI signaling was tested in two assays: callose deposition in response to flg22 infiltration and growth of the hrcC mutant of PstDC3000. Expression of the RIN4 derivatives was induced by spraying 4- to 5-week-old plants with dexamethasone (Dex). At 48 h after induction, leaves were syringe-infiltrated with either 30 μM flg22 or 105 colony-forming units (CFU) per mL of hrcC bacterial suspension. Callose deposition was measured by aniline blue staining at 16 h after flg22 infiltration (Figure 1B). Growth of hrcC bacteria was measured at 4 d postinfiltration (Figure 1C). Similar to RIN4FL, the N-terminal fragments of RIN4 (ACP2 or 149Δ211) potently suppressed callose deposition and enhanced the growth of hrcC. The C-terminal fragments of RIN4 (ACP3 or 1Δ141) failed to suppress flg22-induced callose deposition but still permitted the hrcC mutant to grow to high levels. Thus, these derivatives are all capable of suppressing effective defense against hrcC, but only the N-terminal fragments suppressed flg22-induced callose deposition.
Immunoblotting was used to assess the expression of the RIN4 derivatives (Figure 1D). Immunoblots detecting the T7 epitope were used to compare expression levels of the derivatives relative to one another, and blots detecting RIN4 were used to compare expression levels of RIN4FL with that of native RIN4 in nontransgenic Col-0. The anti-RIN4 blot shows that the overall level of RIN4 protein (native RIN4 plus RIN4FL) in the RIN4FL line was only modestly elevated (approximately twofold) relative to the level of RIN4 in Col-0. Although the ACP3 and 149Δ211 derivatives accumulated to only ~20% the levels of RIN4FL, each of these derivatives was able to suppress PTI. We also examined the accumulation of ACP2 and ACP3 from native RIN4 after delivery of AvrRpt2 during P. syringae infection (see Supplemental Figure 1 online). ACP3 accumulated detectably and remained membrane-associated, consistent with a previous report (Kim et al., 2005a). ACP2 is only weakly detected by our RIN4 polyclonal sera, because the major epitope(s) are in the C-NOI. Despite this limitation, we detected accumulation of ACP2 released from native RIN4 by AvrRpt2 at 3.5 and 6 h after bacterial infiltration (see Supplemental Figure 1 online). Because PAMP-induced responses occur rapidly (e.g., even the “late” induction of callose by flg22 is apparent by 4 h after infiltration [Kim et al., 2005b]), the ACP2 and ACP3 fragments released by AvrRpt2 from native RIN4 persist long enough to suppress PTI.
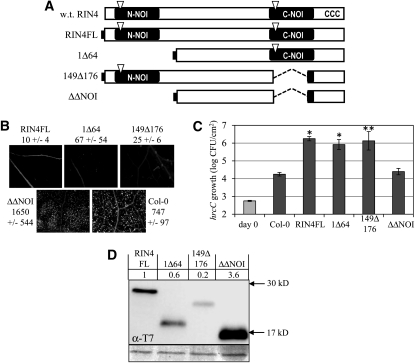
Fragments Containing N- and C-NOI Regions Redundantly Contribute to PTI Suppression by RIN4
Figure 1 showed that separable fragments of RIN4 are individually capable of suppressing PTI. Because each of the derivatives tested in Figure 1 contains a plant-specific NOI domain (Figure 1A), we speculated that the NOI domains contribute to PTI suppression by RIN4. To determine the role of the N-NOI and C-NOI, we constructed lines expressing RIN4 derivatives lacking N-NOI (1Δ64), C-NOI (149Δ176), or both (1Δ64 and 149Δ176 = ΔΔNOI) (Figure 2A). The 1Δ64 and 149Δ176 derivatives, each with one NOI, retained the ability to suppress flg22-induced callose deposition (Figure 2B) and to promote growth of the hrcC mutant (Figure 2C). On the contrary, the ΔΔNOI derivative lacking both NOIs was unable to suppress PTI in either assay (Figures 2B and 2C). The inability of ΔΔNOI to suppress PTI was not the result of poor expression of this derivative (Figure 2D), but might result from disrupted protein structure. The ability of 149Δ176 to promote growth of hrcC was variable between assays, possibly because of the low expression level of this derivative. Figure 2C shows an example of those experiments in which this derivative was as active as RIN4FL, whereas in other experiments, it supported growth of hrcC to levels intermediate between RIN4FL and the nontransgenic Col-0 plants.
Figure 2.
NOI Domains Are Required for Defense-Suppressing Activity of RIN4.
(A) Schematic diagram of RIN4 derivatives as in Figure 1. ΔΔNOI lacks amino acids 1 to 64 and 149 to 176. w.t., wild type.
(B) and (C) Callose induced by flg22 infiltration (B) and growth of hrcC (C) in Col-0 and lines of Col-0 inducibly expressing the indicated RIN4 derivatives as in Figure 1. *P < 0.005; **P < 0.05 [two-tailed Student’s t test for comparison with Col-0 plants].
(D) Immunoblots with anti-T7 antisera to compare levels of RIN4 derivatives as in Figure 1.
The N- and C-terminal fragments of RIN4 differ in their ability to suppress flg22-induced callose deposition (Figure 1). Derivatives lacking either NOI individually (1Δ64 and 149Δ176) suppress flg22-induced callose deposition, so differences between the NOI domains are not sufficient to account for differences in suppression of callose (Figure 2). One possible explanation is that sequences located between the two NOIs, which are part of the N-terminal fragments (149Δ211 and ACP2), are required for suppressing flg22-induced callose deposition. To examine this possibility, we tested the PTI suppression of RIN4 derivatives lacking residues between the NOIs (65Δ91, 94Δ111, and 112Δ141) (see Supplemental Figure 2A online). In these derivatives, all of the residues from AA 65 to AA 141 except two Gly (AA 92 and 93) were deleted; however, each derivative was able to suppress callose deposition and enhance growth of hrcC relative to nontransgenic Col-0 plants (see Supplemental Figures 2B and 2C online). Thus, none of these intervening sequences are essential for suppression of PTI. All three internal deletion derivatives were inducibly expressed and membrane-localized as predicted (see Supplemental Figure 3 online). Growth of hrcC in plants expressing 65Δ91 was intermediate between Col-0 plants and plants expressing RIN4FL, possibly because of the low expression level of 65Δ91 (see Supplemental Figure 3 online). Because sequences between the NOIs were not essential for suppression of callose deposition, an alternative explanation for the difference between the N- and C-terminal derivatives is that the N-terminal derivatives are not membrane-associated (see Supplemental Figure 3 online), which enhances the defense-suppressing activity of RIN4 derivatives (see below).
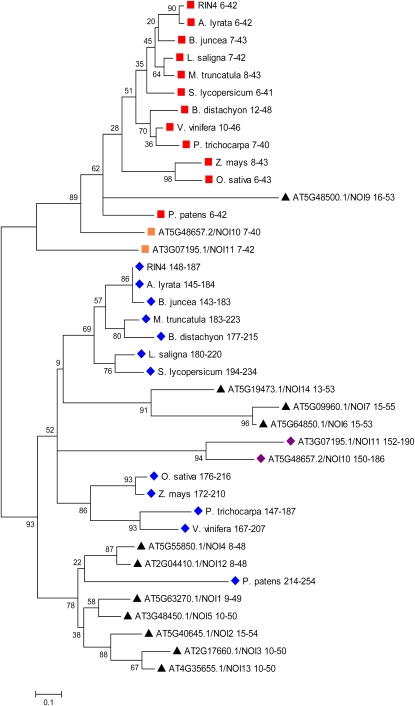
The N- and C-NOIs Are Evolutionarily Distinct
Based on the hypothesized role of the N-NOI and C-NOI domains in the suppression of PTI (Figure 2), we took a phylogenetic approach to examine the relationship of the NOIs from RIN4, other NOI-containing proteins of Arabidopsis, and the closest RIN4 homologs (by protein Blast) from various plant lineages, including several monocots and moss. The NOI sequences, starting with the AvrRpt2 cleavage sites (Chisholm et al., 2005) were aligned by ClustalW (see Supplemental Figure 4 and Supplemental Data Set 1 online). Only two of the examined NOIs, the C-NOIs of the Arabidopsis proteins NOI10 (At5G48657.2) and NOI11 (At3G07195.1), lack predicted cleavage sites for AvrRpt2. Based on this alignment, the evolutionary history of the NOI domains was inferred by the neighbor-joining method (Figure 3). Remarkably, the N-NOI domains (red squares) and C-NOI domains (blue diamonds) of RIN4 homologs clustered into separate clades, indicating that the two domains have remained distinct since before the divergence of moss and vascular plants (~400 million years ago). In addition to amino acid sequence differences, a prominent distinguishing feature between NOIs from the two clades is the difference in spacing between the AvrRpt2 cleavage site and the more C-terminal–conserved residues (see Supplemental Figure 4 online). The distinctness of the two clades is even more pronounced than what is represented in Figure 3, because the tree was constructed using the complete deletion option, in which all positions containing gaps and missing data, including the spacing difference, were eliminated from the data set. Thus, although both contribute to PTI suppression by RIN4, the N-NOI and C-NOI are evolutionarily and perhaps functionally distinct.
Figure 3.
The N-NOI and C-NOI of RIN4 Are from Evolutionarily Distinct Clades.
The NOI domains from RIN4, its closest BLAST homolog from a variety of plants, and other NOI-containing proteins from Arabidopsis (NOI1 to NOI14) were analyzed. The NOI domains were aligned by ClustalW, and their evolutionary relationship was inferred by the Neighbor-Joining method using MEGA4. The confidence probability (×100) that the interior branch length is greater than 0, as estimated using the bootstrap test (1000 replicates), is shown next to the branches. Branch lengths represent evolutionary distances. Number ranges are the amino acid positions of each NOI within its respective protein. The N-NOIs and C-NOIs from RIN4 homologs from different plant species are indicated by red squares and blue diamonds, respectively. Orange squares and purple diamonds indicate the N-NOIs and C-NOIs from Arabidopsis NOI proteins with a structure similar to RIN4 (NOI10 and NOI11). Black triangles indicate the NOIs from Arabidopsis proteins with only a single NOI domain. All NOIs start with an AvrRpt2 cleavage site, with the exception of the C-NOIs of NOI10 and NOI11 from Arabidopsis, which have a Pro insertion and Trp substitution predicted to disrupt cleavage by AvrRpt2. NOI8 (At5G18310) is excluded from the analysis, because ambiguous gene models exist in TAIR9. RIN4 is At3G25070. For other Arabidopsis proteins containing NOIs, the AGI numbers are shown. RIN4 homologs are from the following plant species: A. lyrata, B. juncea, M. truncatula, L. saligna, S. lycopersicum, P. trichocarpa, V. vinifera, O. sativa, Z. mays, B. distachyon, and P. patens.
The NOI domains of other NOI-containing proteins from Arabidopsis fall into both clades (Figure 3). Two of these NOI-proteins (NOI10 and NOI11) have domain architectures similar to RIN4 with N- and C-NOI domains. The C-NOIs of NOI10 and NOI11 lack predicted AvrRpt2 cleavage sites. The N-NOIs (orange squares) and C-NOIs (purple diamonds) of NOI10 and NOI11 are in the same clades as the N-NOI and C-NOI of RIN4, respectively. The other Arabidopsis proteins contain single NOIs (black squares in Figure 3) that, with the exception of NOI9, are in the C-NOI clade.
Structure of the N Terminus Regulates PTI Suppression by RIN4
An N-terminal deletion (1Δ31) disrupted the PTI-suppressing function of RIN4; the 1Δ31 derivative failed to suppress flg22-induced callose deposition or promote the growth of hrcC (see Supplemental Figure 5 online). Specific residues in the first 31 amino acids of RIN4 are not essential for PTI suppression, because the 1Δ64 derivative retained the ability to suppress PTI (Figure 2; see Supplemental Figure 5 online). Expression levels do not account for the difference, because 1Δ31 and 1Δ64 accumulated to similar levels (see Supplemental Figure 5D online). Interestingly, the 1Δ31 derivative retains some activity. When expressed as a transgene under control of the native RIN4 promoter in rpm1 rps2 rin4 RPM1-myc plants (Boyes et al., 1998; Belkhadir et al., 2004), 1Δ31 restored RPM1-myc–mediated HR in response to AvrRpm1 or AvrB (see Supplemental Figure 6 online). Using the same expression method, we have shown that 1Δ141 supports RPM1 function and that the C-NOI (AA 149 to 176) is required for RPM1 function (Chung et al., 2011).
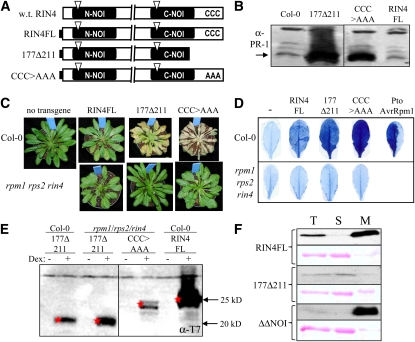
Nonmembrane-Tethered Derivatives of RIN4 Elicit Cell Death in Col-0
Arabidopsis RIN4 has three cysteines (AA 203, 204, and 205) near its C terminus that mediate palmitoylation and membrane targeting of the protein and that are key for its ability to suppress RPS2 activity (Day et al., 2005; Kim et al., 2005a). We looked for the presence of similar sequences in the RIN4 homologs and Arabidopsis NOI-containing proteins from Figure 3. Remarkably, all of these proteins have one to three cysteines within the final 12 AA of their C termini, and the cysteines are closely preceded and/or followed by aromatic AAs, especially phenylalanines (see Supplemental Figure 7 online). The phenylalanines following the three cysteines at the C terminus of RIN4 were shown to be essential for membrane localization (Takemoto and Jones, 2005). These data indicate that most or all of these NOI-containing proteins are likely acylated and membrane-tethered through a process similar to that which targets RIN4 (Kim et al., 2005a). Thus, membrane localization may be generally relevant to the functions of NOI-containing proteins.
To examine how membrane tethering affects the function of RIN4, we examined RIN4 derivatives with mutations in the C-terminal region that disrupt the acylation site (177Δ211 and CCC>AAA) (Figure 4A). Expression of 177Δ211 and CCC>AAA elicited PR-1 expression and a macroscopic cell death response in Col-0 (Figures 4B and 4C). Trypan blue staining revealed that RIN4FL also induces cell death in Col-0, albeit to a lesser extent than 177Δ211 and CCC>AAA (Figure 4D). The cell death phenotype observed was not a result of these derivatives being expressed to a higher level than RIN4FL. In fact, consistent with earlier results (Kim et al., 2005a), 177Δ211 and CCC>AAA accumulated to significantly lower levels than RIN4FL in Col-0 (Figure 4E; see Supplemental Figure 3 online). Subcellular fractionation confirmed that all of the derivatives predicted to be nonmembrane-tethered were in the soluble fraction as predicted (Figure 4F; see Supplemental Figure 3 online).
Figure 4.
Nonmembrane-Tethered Derivatives of RIN4 Cause a Cell Death Response in Col-0.
(A) Schematic representation of RIN4FL and derivatives as in Figure 1. CCC>AAA has RIN4 Cys203/204/205 substituted by Ala203/204/205. w.t., wild type.
(B) Immunoblot showing PR-1 expression in Col-0 and transgenic plants expressing RIN4FL, CCC>AAA, and 177Δ211 at 48 h after Dex treatment.
(C) CCC>AAA and 177Δ211 cause a cell death–like response in Col-0 but not in rpm1 rps2 rin4. Pictures show plant phenotypes 72 h after RIN4FL or the indicated derivatives were induced by Dex in either Col-0 or rpm1 rps2 rin4. Macroscopic cell death was evident in plants expressing either CCC>AAA or 177Δ211 in Col-0, whereas mild symptoms were observed in Col-0 plants expressing RIN4FL. Cell death symptoms were not observed for any of the RIN4 derivatives in rpm1 rps2 rin4 plants.
(D) Trypan blue staining of leaves from plants shown in (C). Leaves of plants were collected 72 h after Dex spray and were stained with trypan blue. Consistent with results obtained in (C), CCC>AAA and 177Δ211 show strong cell death in Col-0 but not in rpm1 rps2 rin4.
(E) Anti-T7 immunoblot showing that CCC>AAA and 177Δ211 accumulate to similar levels in Col-0 and rpm1 rps2 rin4 plants 48 h after mock (−) or Dex (+) spray. Col-0 plants expressing the CCC>AAA derivative are not included, because the tissue is completely collapsed by 48 h.
(F) Total protein extracts (T) of Col-0 leaves expressing indicated derivatives of RIN4 were separated into soluble (S) and microsomal (M) fractions and were subjected to anti-T7 immunoblotting. The microsomal fractions are overloaded by approximately fivefold relative to the total and soluble fractions. Ponceau red staining (shown below T7 blots) demonstrates that RuBisCo partitions into the soluble fraction.
We sought to test the PTI-suppressing activity of nonmembrane-tethered derivatives of RIN4 in the absence of cell death. We hypothesized that the defense and cell death response elicited by expression of nonmembrane-tethered RIN4 derivatives was the result of activation of RPM1 and/or RPS2 or of interaction with native RIN4. Thus, we constructed new transgenic lines that inducibly express RIN4FL, 177Δ211, or CCC>AAA in the rpm1 rps2 rin4 triple mutant background. Unlike in Col-0, expression of RIN4FL, 177Δ211, and CCC>AAA caused no cell death response in the triple mutant (Figures 4C and 4D). This result does not distinguish between whether the cell death induced by nonmembrane-tethered derivatives of RIN4 in Col-0 is dependent on RIN4, RPM1, and/or RPS2.
Nonmembrane-Tethered Derivatives Are Hyperactive Suppressors of PTI
Cleavage of RIN4 by AvrRpt2 has been equated to disappearance of RIN4 (Mackey et al., 2003; Day et al., 2005; Kim et al., 2005a; Kim et al., 2005b). However, it is difficult to explain why a T3E would eliminate a negative regulator of PTI. We hypothesized that AvrRpt2 does not simply eliminate RIN4, but instead produces fragments of RIN4 that actively suppress PTI. Indeed, ACP2 and ACP3, which accumulate after cleavage of native RIN4 by AvrRpt2 (see Supplemental Figure 1 online), were each capable of potently suppressing PTI when inducibly expressed (Figure 1).
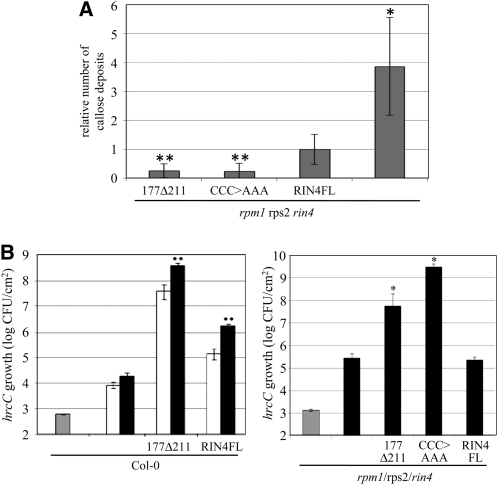
Using the lines expressing RIN4FL, 177Δ211, and CCC>AAA in the rpm1 rps2 rin4 background, we compared the ability of these nonmembrane-tethered RIN4 derivatives to suppress PTI in the absence of cell death. First, we examined their ability to suppress flg22-induced callose deposition (Figure 5A). SALK T-DNA insertions, including the rin4 mutation, cause frequent posttranscriptional gene silencing of the Dex responsive transcription factor in the Dex-inducible system (Geng and Mackey, 2011). We took advantage of this fact to examine the capacity of RIN4 derivatives to suppress callose deposition when expressed at low levels, which revealed differences in the activity of the individual derivatives. Multiple plants for each line were grown, sprayed with Dex, and subsequently infiltrated with flg22. In addition to processing the flg22-infiltrated leaves for callose staining, noninfiltrated leaves from the same plants were harvested for anti-T7 immunoblotting. The results of the immunoblots were used to select individual plants with comparable, low levels of expression of RIN4FL, 177Δ211, and CCC>AAA (see Supplemental Figure 8 online). Among the selected plants, those expressing the nonmembrane-tethered derivatives of RIN4 produced fewer flg22-induced callose deposits than those expressing RIN4FL (Figure 5A). Figure 1 shows that nonmembrane-tethered derivatives of RIN4 suppress flg22-induced callose deposition as well as RIN4FL when expressed at much lower levels. Figure 5A shows that the derivatives suppress callose deposition more efficiently than RIN4FL when the derivatives and RIN4FL are expressed at comparable low levels.
Figure 5.
Derivatives of RIN4 Suppress PTI More Potently When Not Membrane-Tethered.
(A) Expression of RIN4 derivatives in rpm1 rps2 rin4 plants was induced by spraying with Dex 48 h before infiltration with 30 μM flg22. Callose numbers were counted from plants expressing comparable levels of RIN4FL, CCC>AAA, and 177Δ211 proteins (see Supplemental Figure 8 online). The results shown are averaged from three independent biological replicates, with the average number of callose deposits induced in plants expressing RIN4FL in each experiment normalized to 1. Error bars represent sd. *, P = 0.001; **P < 0.0001 [two-tailed Student’s t test for comparison with rpm1 rps2 rin4 plants expressing RIN4FL].
(B) Expression of RIN4 derivatives in Col-0 or rpm1 rps2 rin4 plants was induced by spraying with Dex 48 h before infiltration with 105 CFU/mL of hrcC. Growth data in rpm1 rps2 rin4 was obtained from plants expressing similar, low levels of RIN4FL and derivatives (see Supplemental Figure 9 online). Numbers of hrcC were determined 0 (gray bars), 2 (white bars), and 4 (black bars) d after infiltration. Error bars represent sd. *P < 0.05 [two-tailed t test for comparison with rpm1 rps2 rin4 plants]; **P < 0.005 [two-tailed Student’s t test for pairwise comparison with Col-0 plants].
Nonmembrane-tethered derivatives of RIN4 also potently promote the growth of hrcC. In Col-0 plants, expression of 177Δ211 caused hrcC to grow to a level two orders of magnitude higher than did RIN4FL (Figure 5B), despite the fact that it does not accumulate as well (Figure 4E). Col-0 plants expressing CCC>AAA were not included, because they undergo rapid cell death within 36 h of Dex spray. To assess the effect of inducible expression of RIN4 derivatives in the absence of cell death, we used the lines in rpm1 rps2 rin4 plants (Figure 5B). RIN4FL did not enhance the growth of hrcC in the rpm1 rps2 rin4 triple mutant background, possibly because of low levels of expression resulting from posttranscriptional gene silencing. Despite similar low levels of expression (see Supplemental Figure 9 online), 177Δ211 and CCC>AAA still enabled hrcC to grow to high levels. Collectively, the growth and callose data in Figure 5 indicate that inducibly expressed RIN4 more potently suppresses PTI when it is not tethered to the plasma membrane.
To learn more about the PTI-suppressing activity of RIN4FL and nonmembrane-tethered derivatives of RIN4, we examined the effect of RIN4FL, 177Δ211, and CCC>AAA on early PTI responses (see Supplemental Figures 10 and 11 online). The accumulation of reactive oxygen is a very early response to flg22 (Felix et al., 1999) that depends on ethylene signaling (Mersmann et al., 2010). We used a luminol-based assay to examine the induction of ROS induced by flg22. To avoid ROS production associated with cell death in Col-0, we used the lines expressing these derivatives in the rpm1 rps2 rin4 triple mutant. In addition to Arabidopsis plants expressing RIN4 derivatives, rpm1 rps2 rin4 plants were included as a positive control, and fls2 plants were included as a negative control. After measuring flg22-induced ROS, immunoblots were done to examine plants for expression levels of the RIN4 derivatives (see Supplemental Figure 9 online). Traces from selected plants were then used to determine the peak value of ROS accumulation (sample traces from individual plants are shown in Supplemental Figure 10A online), and data from five biological replicates were normalized with the average peak values in rpm1 rps2 rin4 set to one and combined (see Supplemental Figure 10B online). Although slight decreases in the peak accumulation of ROS relative to rpm1 rps2 rin4 were observed with the RIN4 derivatives, these decreases were of a much lower magnitude than the decrease in flg22-induced callose deposition in the same plants (Figure 5). Furthermore, the decreases in ROS in plants expressing RIN4 derivatives were not significant relative to the rpm1 rps2 rin4 control plants (two-tailed t test P values for all were >0.07). Thus, inducible expression of RIN4 derivatives, including the potent nonmembrane-tethered derivatives, did not efficiently suppress PTI-induced ROS accumulation.
We also tested the effect of RIN4 derivatives on early, PTI-induced gene expression. Leaves of plants induced to express RIN4FL, 177Δ211, and the inactive ΔΔNOI, as well as Col-0 and fls2 control plants, were infiltrated with flg22 or H20. After 2 h, quantitative real-time PCR was used to measure FRK1 and MYB51 transcript levels (see Supplemental Figure 11 online). In Col-0 plants and ΔΔNOI-expressing plants, each transcript accumulated to higher levels after infiltration with flg22 than with water. Each transcript remained expressed at low levels after flg22 infiltration into fls2 plants. These findings confirm that both genes are flg22-induced. In plants expressing RIN4FL, MYB51 expression was elevated in water-infiltrated plants, and expression of both FRK1 and MYB51 were induced by flg22 relative to water. In plants expressing 177Δ211, expression of both genes was elevated in water-infiltrated plants, and no further increase was observed in flg22-infiltrated plants. Collectively, these data indicate that RIN4FL fails to suppress rapid PTI-induction of ROS and gene expression. Similar to RIN4FL, the nonmembrane-tethered 177Δ211 derivative fails to suppress ROS. However, in plants expressing 177Δ211, flg22 did not induce expression of FRK1 and MYB51 above their already elevated levels.
DISCUSSION
To better understand how RIN4 links ETI, ETS, and PTI, we performed a structure–function analysis of postinvasion defense regulation by RIN4 (data summarized in Supplemental Figure 12 online). We show that separable fragments of RIN4—the N-terminal two thirds and the C-terminal one third of the protein, including two derivatives of RIN4 that correspond to the two primary AvrRpt2 cleavage products (ACP2 and ACP3)—are capable of suppressing PTI. Notably, ACP2 and ACP3 can be detected from native RIN4 after cleavage by bacterially delivered AvrRpt2. Thus, RIN4 contains nonoverlapping fragments with defense-suppressing activity, each containing a plant-specific NOI domain. The N-NOI and C-NOI domains of RIN4 belong to evolutionary clades that have remained distinct from each other since ~400 million years ago. However, at the level of our assays, these domains contribute redundantly to defense suppression; derivatives lacking either the N-NOI or C-NOI (1Δ64 or 149Δ176) effectively suppress callose deposition and promote the growth of T3S-deficient bacteria, whereas a derivative lacking both is inactive. Nonetheless, it seems likely that the N-NOI and C-NOI will differ in their function. Indeed, the C-NOI includes a fragment of RIN4 that interacts with AvrB (Desveaux et al., 2007) and is required for AvrRpm1- and AvrB-induced activation of RPM1(Chung et al., 2011; Liu et al., 2011). The Arabidopsis genome encodes two proteins that, similar to RIN4, have two NOI domains and 12 proteins with a single NOI domain. The importance of the NOIs for defense suppression by RIN4 highlights the potential role of these other NOI-containing proteins in regulating plant defense.
N- and C-terminal fragments of RIN4 differ in their ability to suppress callose deposition. Both 1Δ141 and ACP3 promote the growth of hrcC but are unable to suppress callose deposition. This is in contrast with 149Δ211 and ACP2, which promote the growth of hrcC and suppress callose deposition. Several possible explanations might account for this difference between the N- and C-terminal fragments of RIN4. One is that the threshold for suppressing callose deposition is more stringent than the threshold for promoting growth of the hrcC mutant bacteria. A second possible explanation is a difference in the activity of the N-NOI and C-NOI domains of RIN4. However, derivatives of RIN4 lacking either NOI (1Δ64 or 149Δ176) retain the ability to suppress callose as well as to promote growth of T3S-deficient bacteria. A third possible explanation is that there is a function for the sequences between the NOIs that are included in 149Δ211 and ACP2 and are absent in 1Δ141 and ACP3. Although these intervening sequences do not contain any residues absolutely required to suppress callose deposition, their absence might affect RIN4 function through structural perturbation. A fourth possible explanation to account for the difference in PTI suppression between N- and C-terminal fragments of RIN4 is that the N-terminal fragments are not tethered to the plasma membrane, which enhances their PTI-suppressing activity.
The subcellular localization of RIN4 dramatically affects its activity. Derivatives of RIN4 lacking the C-terminal cysteines are no longer membrane-tethered. In Col-0, inducible expression of these nonmembrane-tethered derivatives causes a cell death response. Derivatives lacking the C terminus are also potent suppressors of PTI in Col-0. However, it is unclear how the cell death response affects our PTI readouts. To circumvent this problem, we expressed nonmembrane-tethered derivatives in an rpm1 rps2 rin4 triple mutant and observed that they no longer caused cell death. In this background, the nonmembrane-tethered derivatives suppress PTI. In fact, assays for growth of hrcC mutant bacteria and callose deposition revealed that these derivatives suppress PTI more potently than does RIN4FL. Thus, anchoring of RIN4 to the plasma membrane limits its defense-suppressing capacity. Release of RIN4 via cleavage by AvrRpt2, or by an endogenous protease (Luo et al., 2009), could thus release a hyperactive defense-suppressing fragment of RIN4. Notably, the nonmembrane-tethered derivative 177Δ211 prevents flg22 from further elevating expression of FRK1 and MYB51. However, interpretation of this result is difficult, because expression of both genes is already elevated in plants expressing 177Δ211. Release from the plasma membrane could enhance the ability of fragments of RIN4 to suppress defense transcription downstream of cytoplasmic and nuclear localized MPK4 (Cui et al., 2010). Further work is necessary to determine the subcellular location and mode of action by which the nonmembrane-tethered derivatives act to promote cell death and suppress PTI.
A structural model generated by the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) on the basis of multiple-threading alignments by LOMETS and iterative TASSER simulations (Zhang, 2008) predicted a globular structure for RIN4 (see Supplemental Figure 13 online). In addition to the I-TASSER model, modeling RIN4FL using four other top-ranked prediction servers in the CASP8 and CASP9 experiments also yielded globular structures (see Supplemental Figure 14 online). The robustness of the I-TASSER model was supported by Ramachandran plot (see Supplemental Figure 15 online), in which 96.7% of the residues were in favored and allowed regions, and only 3.3% of residues were in disallowed regions (Morris et al., 1992). We favor the I-TASSER model, because it fits with available data regarding the structure and function of RIN4 and because I-TASSER was the top-ranked server in the CASP7, CASP8, and CASP9 experiments. First, the N-terminal two thirds and the C-terminal one third of the protein fold into separate domains (see Supplemental Figure 13A online), consistent with the functionality of each of these domains in isolation (Figure 1). Second, residues of the C-NOI that are critical for interaction with AvrB (Desveaux et al., 2007) and function of RPM1 (Chung et al., 2011; Liu et al., 2011), as well as comparable residues in the N-NOI, are surface-exposed (see Supplemental Figures 13B and 13C online). Third, the C-terminal cysteines required for acylation and membrane targeting of RIN4 (Kim et al., 2005a; Takemoto and Jones, 2005) and the AvrRpt2 cleavage sites in N-NOI and C-NOI (Chisholm et al., 2005) are all exposed (see Supplemental Figures 13B, 13D, and 13E online). An interesting feature of the model is that the two NOIs interact extensively with one another (see Supplemental Figure 13B online). Physical contact between NOIs could contribute to the observed intermolecular interactions between RIN4 homologs of soybeans (Selote and Kachroo, 2010). We hypothesized that the inability of 1Δ31 to suppress PTI resulted from misfolding. To test this idea, we obtained structural predictions of 1Δ31 and 1Δ64 from I-TASSER. Indeed, 1Δ31 is predicted to be mostly unstructured, whereas significant structure is restored in 1Δ64 (see Supplemental Figure 16 online). 1Δ64 retains some secondary structure elements that are present in RIN4FL and are absent in 1Δ31, including an α helix containing Gly127-Lys128. Although 1Δ31 is predicted to be predominantly unstructured, the C terminus including the C-NOI domain is predicted to resemble that of RIN4FL at the secondary structure level (see Supplemental Figure 16C online). Hence, the in vivo and modeling results are consistent with the partially structured C terminus of 1Δ31 being sufficient to support RPM1 function, whereas the lack of broader structure precludes PTI suppression (see Supplemental Figures 5 and 6 online).
Our work provides new ideas about the targeting of RIN4 by T3Es. We have recently shown that AvrRpm1 and AvrB induce phosphorylation of RIN4 T166, and phosphorylation of this residue elicits RPM1 activation (Chung et al., 2011). T3E-induced phosphorylation of T166 is performed, at least in part, by RIPK (Liu et al., 2011). NOI domains have a conserved Thr or Ser at the position comparable with T166 in RIN4. The hypothesis that the activity of NOI proteins is controlled, at least in part, via phosphorylation of this residue is supported by the role of T166 phosphorylation in control of RPM1 activation (Chung et al., 2011; Liu et al., 2011). We hypothesize that phosphorylation of RIN4 on T166 will enhance its defense-suppressing activity. Also of interest is how AvrRpt2 might enhance the defense-suppressing activity of RIN4. AvrRpt2 is a protease that cleaves RIN4 at two locations. It seemed counterintuitive that AvrRpt2 cleaves a negative regulator of PTI. However, rather than simply inactivating or eliminating RIN4, our data support a model in which cleavage by AvrRpt2 produces active fragments of RIN4. Indeed, the two main cleavage products produced by AvrRpt2, ACP2 and ACP3, can be detected after cleavage of native RIN4 by AvrRpt2 and are potent defense suppressors when inducibly expressed. These findings, along with the prior observations that native RIN4 negatively regulates PTI in the absence of any T3E-mediated perturbation (Kim et al., 2005b) and that RIN4 is a required target of the virulence activity of HopF2 (Wilton et al., 2010), support the hypothesis that RIN4 is a bona fide virulence target of T3Es rather than a decoy (van der Hoorn and Kamoun, 2008). Similar to RPM1 being activated by AvrRpm1- or AvrB-induced phosphorylation of RIN4, RPS2 may respond to AvrRpt2-induced cleavage product(s) of RIN4. In particular, cleavage-induced release of ACP2 from the membrane may be a key step in activation of RPS2. Thus, as predicted by the “guard hypothesis,” T3E-induced perturbations of RIN4 that suppress PTI may also serve as elicitors of ETI in plants expressing RPM1 or RPS2.
METHODS
Plants and Growth Conditions
Arabidopsis thaliana Col-0 wild-type plants were used in this study. The mutants generated from Col-0 plants were as follows: rps2-101C has a stop codon at amino acid 235 of RPS2 (Bent et al., 1994); rpm1-3 has a stop codon at amino acid 87 of RPM1 (Grant et al., 1995); rin4 has a T-DNA insertion after amino acid 146 of RIN4 (Mackey et al., 2003); the rpm1 rps2 rin4 triple mutant was identified by marker-assisted breeding (Belkhadir et al., 2004). Plants were grown on Metro Mix 360 (Sun Gro Horticulture) at 25°C/16°C under an 8-h-light/16-h-dark cycle.
DNA Manipulation and Generation of Transgenic Plants
All T7-tagged RIN4 derivatives were produced by overlap extension PCR with full length RIN4 cDNA as the template (Aiyar et al., 1996). Overlap extension PCR products were digested with NcoI (in the extreme N terminus overlapping ATG in the T7-tag) and XbaI (at the extreme C terminus after the stop codon) and cloned downstream from the RIN4 native promoter in pMAC100c (a PUC19-based vector containing 1340 bp of RIN4 promoter from Col-0 up to the naturally occurring NcoI site at the RIN4 ATG).
To generate plants inducibly expressing RIN4 derivatives, the encoding fragments (without the RIN4 native promoter) were subcloned from pMAC100c vectors into the Dex-inducible binary vector, PTA7002 (Aoyama and Chua, 1997). The resulting plasmids were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Transgenic plants were generated by vacuum-assisted floral dipping (Clough and Bent, 1998) of flowering Col-0 or rpm1 rps2 rin4 plants into GV3101 carrying individual plasmids. Transgenic progeny were selected by growth on Gamborg’s B5 (Invitrogen) agar plates containing 20 μM hygromycin B (Sigma-Aldrich). Independent lines with single insertion loci, as determined by 3:1 segregation of hygromycin B resistance in the T2 generation, were selected and propagated to homozygosity. Expression of FL:RIN4 and RIN4 derivatives was induced by spraying plants with 20 μM Dex (Sigma-Aldrich) plus 0.005% Silwet L-77 solution (Lehle Seeds).
Bacterial Growth Assays
The hrcC T3S-deficient mutant of PstDC3000 (formerly hrpH) was used in this study (Yuan and He, 1996). A suspension of 105 CFU/mL of hrcC in 10 mM MgCl2 was pressure-infiltrated with a needleless syringe into the leaves of 4- to 5-week-old plants. After the infiltration, the leaves were allowed to dry (~4 h), and the plants were kept in 100% humidity (by covering with a clear dome) for the remainder of the experiment. For each measurement, nine leaf discs were separated into three samples (three discs per tube), and the titers from these three samples were used to perform statistical analysis. Leaf discs were collected from the infiltrated area, ground in 10 mM MgCl2, and serially diluted to measure bacterial numbers. The RIN4 transgenes were induced by spraying 20 μM Dex (Sigma-Aldrich) solution containing 0.005% (v/v) Silwet L-77 (Lehle) 48 h before bacterial infiltration. P values were calculated using Student’s t test. All growth assays are representative of at least three independent biological replicates.
Protein
Total protein extraction and cell fractionations were performed as described previously (Mackey et al., 2002). Briefly, total protein extracts were prepared by grinding ~2 cm2 of tissue per 0.2 mL of grinding buffer (20 mM Tris-HCl [pH 7.5]), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 5 mM DTT, and plant protease inhibitor cocktail (Sigma-Aldrich) and pelleting insoluble debris by centrifugation at 20,000 × g for 10 min at 4°C. Concentration of protein in the supernatant was determined by using the Bio-Rad protein assay reagent (Bio-Rad). Samples, typically 30 μg, were separated on SDS-PAGE gels (mini protean; Bio-Rad) and transferred to polyvinylidene fluoride membrane. All SDS-PAGE gels were 12 to 15% acrylamide except in Supplemental Figure 12 online (10%). Immunoblots were done by standard methods. Anti- RIN4 sera (Mackey et al., 2002), anti-T7 monoclonal antibody (Novagen), and anti–PR-1 sera (Kliebenstein et al., 1999) were used at dilutions of 1:5000, 1:10,000, and 1:10,000, respectively. Chemiluminescent detection and band quantification were done using the ChemiDoc XRS system (Bio-Rad).
Subcellular Fractionation
Membrane proteins were fractionated by grinding 0.1 g of tissue per 1 mL of buffer (10 mM Tris-HCl [pH 7.0], 0.33 M Suc, 1 mM EDTA, and plant protease inhibitor cocktail [Sigma-Aldrich]) and pelleting insoluble debris by centrifugation at 20,000 × g for 20 min at 4°C. The supernatant of this spin constituted the total (T) fraction. A total of 10 μL of 1 M CaCl2 was added to 500 μL of the total fraction, and the microsomal fraction was pelleted at 25,000 × g for 90 min at 4°C. The supernatant from this spin was the soluble (S) fraction. The pellet was resuspended in 30 μL of 1× SDS-PAGE loading dye, heated at 65°C for 15 min. This constituted the membrane (M) fraction.
Callose Staining and Quantification
Leaves from 4-week-old plants were syringe-infiltrated with 30 μM flg22 or distilled water and were collected after 16 h. Transgenic lines were induced by spraying 20 μM Dex containing 0.005% Silwet L-77 at 48 h before the infiltration with flg22. Five leaves for each treatment were then stained with aniline blue (Sigma-Aldrich) (Kim and Mackey, 2008). Whole leaves were cleared by submersion in lactophenol alcohol (a mixture of 1 volume of a 1:1:1:1 volume mix of glycerol, saturated phenol, lactophenol, and deionized water and 2 volumes of 95% ethanol) and incubated for 5 min at 95°C followed by an overnight incubation in fresh lactophenol alcohol at room temperature. The cleared leaves were rinsed in 50% ethanol and then in water and were then stained with 0.01% aniline blue in 150 mM phosphate buffer (pH 9.5). Stained leaves were mounted in 50% glycerol and were visualized under epifluorescent illumination by a Nikon eclipse 80i microscope. Four pictures of different areas were taken of each leaf (for 20 images per treatment), and callose deposits were counted using the ImageJ software (http://rsbweb.nih.gov/ij/).
Trypan Blue Staining
Trypan blue staining was performed as described previously (Koch and Slusarenko, 1990). Briefly, leaves were submerged in a staining solution composed of 1 part staining mix (1:1:1:1 mix of phenol, lactic acid, glycerin, and water plus 0.05% [w/v] trypan blue) and 2 parts ethanol. The leaves were incubated in the staining solution at 95°C for 3 min followed by an additional overnight incubation. Stained leaves were cleared in 15 M chloral hydrate solution and mounted in 70% glycerol.
DNA Manipulation and Generation of Transgenic Plants
The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Dayhoff matrix based method (Schwartz and Dayhoff, 1979), and branch lengths correspond to the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option) so that there were a total of 28 positions in the final data set. All phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
To generate plants expressing RIN4 derivatives under control of the native promoter, the encoding fragments (with the 1340 bp RIN4 native promoter) were subcloned from pMAC100c vectors into pBAR1 (promoterless binary vector conferring Basta resistance) and moved into GV3101 by electroporation. Transgenic plants were generated by vacuum-assisted floral dipping (Clough and Bent, 1998) of flowering rpm1 rps2 rin4 RPM1-myc plants (Boyes et al., 1998; Belkhadir et al., 2004). Transgenic progeny were selected by spraying soil-grown seedlings on 3 consecutive days with 0.04% (w/v) Basta and 0.005% Silwet L-77 solution. Independent T1 lines with single insertion loci, as determined by 3:1 segregation of Basta resistance in the T2 generation, were selected and propagated to homozygosity.
Bacteria, HR, and Conductance Measurements for Assessment of RPM1 Function
PstDC3000 carried either pVSP61 or derivatives of this plasmid containing avrRpm1 or avrB. For HR and conductance assays, leaves were infiltrated with a suspension of 5 × 107 CFU/mL bacteria in 10 mM MgCl2. For measurements of conductance, eight leaf discs (8 mm diameter) were removed immediately after infiltration (time 0) and were floated in 25 mL of water to wash away ions released during leaf disc removal. After 10 min, the wash water was replaced with 10 mL of fresh water, and the conductance in microsiemens per centimeter was measured over time.
ROS Accumulation
Four-week-old rpm1 rps2 rin4 plants expressing RIN4 derivatives and control plants were sprayed with Dex (20 μM). After 36 h, eight leaf disks (6.5 mm diameter) were excised from each plant and were floated on distilled water for 4 to 6 h. Then three leaf disks were transferred to a microfuge tube containing 100 μL of luminol solution, Immun-Star HRP Substrate (Bio-Rad), and 1 μL of horseradish peroxidase-streptavidin (Jackson Immunoresearch) and were supplemented with either 1 μL of 1 mM flg22 peptide (10 μM final concentration) or 1 μL water. Luminescence was measured every 10 s for 18 min by a Glomax 20/20 luminometer (Promega). Within each biological replicate, the data were normalized, with the average peak value from rpm1 rps2 rin4 plants exposed to flg22 set to 1.
Real-Time PCR
Four- to five-week-old plants expressing RIN4 derivatives and control plants were sprayed with Dex (20 μM). After 48 h, leaves were infiltrated with flg22 (30 μM) or water. Samples were collected 2 h after infiltration and were flash-frozen in liquid nitrogen. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) followed by DNase I (Invitrogen) treatment. The RNA quality was determined by gel electrophoresis and quantitated using a nano-drop model ND-1000 (Thermo Scientific). First-strand cDNA was synthesized from 1 μg of RNA using oligo-dT primer and AMV reverse transcriptase (Promega). Quantitative real-time PCR analyses were performed using a Bio-Rad iQ5 real-time PCR detection system with iQ SYBR green supermix (Bio-Rad). Actin was used as a control, and data were analyzed using iQ5 software (Bio-Rad). Primers used for real-time PCR are listed in Supplemental Table 1 online.
Computational Modeling
The computation models for RIN4FL and the RIN4 derivatives were built by submitting primary sequences to the I-TASSER server (http://zhang.bioinformatics.ku.edu/I-TASSER). The I-TASSER server was ranked number 1 during the Critical Assessment of Techniques for Protein Structure Prediction experiment three assessments in a row (CASP 2006, CASP 2008, and CASP 2010). The model was generated using a meta-threading approach followed by Monte Carlo simulations, with unaligned regions built by ab initio modeling (Zhang, 2008). To generate the RIN4FL model, the following six proteins were used as templates: 1m11_R (human decay-accelerating factor/coat protein), 2nud (AvrB complexed with a high-affinity RIN4 peptide), 1nkw_B (large ribosomal subunit from Deinococcus radiodurans), 2dpm_A (DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine), 1mv3_A (Myc box–dependent interacting protein 1), and 2qzu_A (putative sulfatase yidJ from Bacteroides fragilis). Visualization and optimization of the predicted model was done using the Jmol server (http://firstglance.jmol.org) and Chime (http://www.symyx.com/downloads). The PROCHECK software suite (http://www.ebi.ac.uk/thornton-srv/software/PROCHECK/) was used to generate the Ramachandran plot and to assign residues to the favored, allowed, and disallowed regions.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RIN4 homologs from other plant species (and accession numbers) are Arabidopsis lyrata (ABR46111.1), Brassica juncea (ABM30198), Medicago truncatula (ACJ83941), Lactuca saligna (GQ497776), Solanum lycopersicum (TC174419), Populus trichocarpa (XM_002316554), Vitis vinifera (CBI35706), Oryza sativa (NM_001058429), Zea mays (NP_001152021), Brachypodium distachyon (Bradi3g40950), and Physcomitrella patens (XP_001766424). Arabidopsis genes analyzed in this study are: RIN4 (AT3G25070), MYB51 (AT1G18570), FRK1 (AT2G19190), ACT2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Accumulation of ACP2 and ACP3 upon Cleavage of Native RIN4 by AvrRpt2.
Supplemental Figure 2. Residues between the NOI Domains Are Not Essential for PTI Suppression by RIN4.
Supplemental Figure 3. Membrane Association of RIN4 Derivatives Depends on the C-Terminal Cysteines.
Supplemental Figure 4. Alignment of NOI Domains.
Supplemental Figure 5. A Small N-Terminal Deletion Disrupts the PTI-Suppressing Activity of RIN4.
Supplemental Figure 6. 1Δ31 Retains the Ability to Support RPM1 Function.
Supplemental Figure 7. Alignment of the C Termini of RIN4 Homologs and NOI-Containing Proteins.
Supplemental Figure 8. Expression of RIN4 Derivatives in Plants Screened for Use in Figure 5A.
Supplemental Figure 9. Expression of RIN4 Derivatives in Plants Screened for Use in Figure 5B and Supplemental Figure 10.
Supplemental Figure 10. Flg22-Induced ROS Accumulation in Plants Expressing RIN4 Derivatives.
Supplemental Figure 11. Flg22-Induced Expression of FRK1 and MYB51 in Plants Expressing RIN4 Derivatives.
Supplemental Figure 12. Summary of Results Obtained from RIN4 Structure:Function Analysis.
Supplemental Figure 13. Structural Model of RIN4.
Supplemental Figure 14. RIN4 Is Modeled as Globular in Additional Structural Predictions.
Supplemental Figure 15. Validation of Structural Model of RIN4FL by Ramachandran Plot.
Supplemental Figure 16. Ribbon Diagrams of Predicted Structures of RIN4FL, 1Δ31, and 1Δ64.
Supplemental Table 1. List of Primers Used for Real-Time PCR.
Supplemental Data Set 1. Text File of Alignment Used to Generate Tree in Figure 3.
Acknowledgments
This work was supported by funding to D.M. from the National Science Foundation (MCB-0718882) and the Ohio Agricultural Research and Development Center. L.D.C. was supported by an Excellence in PMBB Fellowship from the Plant Molecular Biology and Biotechnology Program at Ohio State University.
AUTHOR CONTRIBUTIONS
A.J.A., L.d.C., and D.M. designed the research. A.J.A. and L.d.C. performed research. A.J.A., L.d.C., and D.M. analyzed data and wrote the article.
References
- Aiyar A., Xiang Y., Leis J. (1996). Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 57: 177–191 [DOI] [PubMed] [Google Scholar]
- Alfano J.R., Collmer A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42: 385–414 [DOI] [PubMed] [Google Scholar]
- Aoyama T., Chua N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Chisholm S.T., Dahlbeck D., Staskawicz B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Nimchuk Z., Hubert D.A., Mackey D., Dangl J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Kunkel B.N., Dahlbeck D., Brown K.L., Schmidt R., Giraudat J., Leung J., Staskawicz B.J. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Nam J., Dangl J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Boller T., Robatzek S. (2007). Flagellin signalling in plant immunity. Adv. Exp. Med. Biol. 598: 358–371 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Dahlbeck D., Krishnamurthy N., Day B., Sjolander K., Staskawicz B.J. (2005). Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl. Acad. Sci. USA 102: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.H., da Cunha L., Wu A.J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha L., Sreerekha M.V., Mackey D. (2007). Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 10: 349–357 [DOI] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Huang J., Chisholm S.T., Li D., Staskawicz B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D., Singer A.U., Wu A.J., McNulty B.C., Musselwhite L., Nimchuk Z., Sondek J., Dangl J.L. (2007). Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 3: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Geng X., Mackey D. (2011). Dose–response to and systemic movement of dexamethasone in the GVG-inducible transgene system in Arabidopsis. Methods Mol. Biol. 712: 59–68 [DOI] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Guo M., Tian F., Wamboldt Y., Alfano J.R. (2009). The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant Microbe Interact. 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P., Thilmony R., He S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim H.S., Desveaux D., Singer A.U., Patel P., Sondek J., Dangl J.L. (2005a). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102: 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.G., Mackey D. (2008). Measuring cell-wall-based defenses and their effect on bacterial growth in Arabidopsis. Methods Mol. Biol. 415: 443–452 [DOI] [PubMed] [Google Scholar]
- Kim M.G., Geng X., Lee S.Y., Mackey D. (2009). The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 57: 645–653 [DOI] [PubMed] [Google Scholar]
- Kim M.G., da Cunha L., McFall A.J., Belkhadir Y., DebRoy S., Dangl J.L., Mackey D. (2005b). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Dietrich R.A., Martin A.C., Last R.L., Dangl J.L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.T., Kunkel B.N. (2004). Mutations in the Pseudomonas syringae avrRpt2 gene that dissociate its virulence and avirulence activities lead to decreased efficiency in AvrRpt2-induced disappearance of RIN4. Mol. Plant Microbe Interact. 17: 313–321 [DOI] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.J., Coaker G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Fuglsang A.T., Palmgren M.G., Staskawicz B.J., Coaker G. (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Caldwell K.S., Wroblewski T., Wright M.E., Michelmore R.W. (2009). Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell 21: 2458–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., McFall A.J. (2006). MAMPs and MIMPs: Proposed classifications for inducers of innate immunity. Mol. Microbiol. 61: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F., III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K.A., Dangl J.L., Dietrich R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26: 403–410 [DOI] [PubMed] [Google Scholar]
- McHale L., Tan X., Koehl P., Michelmore R.W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann S., Bourdais G., Rietz S., Robatzek S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.L., MacArthur M.W., Hutchinson E.G., Thornton J.M. (1992). Stereochemical quality of protein structure coordinates. Proteins 12: 345–364 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schwartz R.M., Dayhoff M.O. (1979). Protein and nucleic acid sequence data and phylogeny. Science 205: 1038–1039 [DOI] [PubMed] [Google Scholar]
- Selote D., Kachroo A. (2010). RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 153: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Li X., Cui H., He P., Thilmony R., Chintamanani S., Zwiesler-Vollick J., Gopalan S., Tang X., Zhou J.M. (2006). RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 103: 19200–19205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Takemoto D., Jones D.A. (2005). Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol. Plant Microbe Interact. 18: 1258–1268 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H.S., Han B., Zhu T., Zou G., Katagiri F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Glazebrook J., Cohen J.D., Katagiri F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53: 763–775 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A., Kamoun S. (2008). From Guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Hou S., Wang X., Li Y., Ren D., Chen S., Tang X., Zhou J.M. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22: 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M., Subramaniam R., Elmore J., Felsensteiner C., Coaker G., Desveaux D. (2010). The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 107: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., He S.Y. (1996). The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J. Bacteriol. 178: 6399–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]