Carbon allocation to arbuscular mycorrhizal fungi is the reward that plants offer their symbiotic partners in exchange for mineral nutrients. This study identifies a monosaccharide transporter that plays a key role in this process.
Abstract
For more than 400 million years, plants have maintained a mutualistic symbiosis with arbuscular mycorrhizal (AM) fungi. This evolutionary success can be traced to the role of these fungi in providing plants with mineral nutrients, particularly phosphate. In return, photosynthates are given to the fungus, which support its obligate biotrophic lifestyle. Although the mechanisms involved in phosphate transfer have been extensively studied, less is known about the reciprocal transfer of carbon. Here, we present the high-affinity Monosaccharide Transporter2 (MST2) from Glomus sp with a broad substrate spectrum that functions at several symbiotic root locations. Plant cell wall sugars can efficiently outcompete the Glc uptake capacity of MST2, suggesting they can serve as alternative carbon sources. MST2 expression closely correlates with that of the mycorrhiza-specific Phosphate Transporter4 (PT4). Furthermore, reduction of MST2 expression using host-induced gene silencing resulted in impaired mycorrhiza formation, malformed arbuscules, and reduced PT4 expression. These findings highlight the symbiotic role of MST2 and support the hypothesis that the exchange of carbon for phosphate is tightly linked. Unexpectedly, we found that the external mycelium of AM fungi is able to take up sugars in a proton-dependent manner. These results imply that the sugar uptake system operating in this symbiosis is more complex than previously anticipated.
INTRODUCTION
Arbuscular mycorrhizal (AM) fungi are obligate biotrophs that form one of the most widespread and ancient mutualistic symbioses with plant roots (Remy et al., 1994). These fungi, which belong to the phylum Glomeromycota, are soil inhabitants whose life cycle is only completed after passage through symbiosis. The single plant-independent phase is the presymbiotic growth that occurs after spore germination, but this ceases promptly if compatible plants are not found in the surroundings (Logi et al., 1998). In the presence of a host plant, a signal exchange takes place that allows the fungus to penetrate the root cortex without elicitation of major plant defenses (Bonfante and Genre, 2010; Kloppholz et al., 2011). Once inside the plant, the fungus grows inter- and intracellularly, but never penetrates the plasma membrane. Within the inner cortical cells, the fungus develops arbuscules, the hallmark of the symbiosis (Gianinazzi-Pearson, 1996). Arbuscules are highly dichotomously branched structures in which phosphate and other nutrients are delivered to the root. Concomitantly, the fungal mycelium spreads out of the root and into the soil for the uptake of those nutrients that will be provided to the plant. Phosphate is a key element driving the symbiosis (Rausch et al., 2001; Harrison et al., 2002; Paszkowski et al., 2002; Nagy et al., 2005; Maeda et al., 2006). Because of phosphate’s low mobility and thus its availability in soil (Schachtman et al., 1998), plants have developed several mechanisms to cope with phosphate limitation. One of them is the formation of symbiosis with AM fungi (Kara–ov and Bucher, 2005). That plants are able to profit from this association is highlighted by the fact that mycorrhizal plants can rely exclusively on the symbiotic route for phosphate acquisition (Liu et al., 1998; Smith et al., 2003). The price is a carbon fee that the plant pays by transferring carbohydrates to the structures of the fungus growing within the root cortex (see Supplemental Figure 1 online). The carbon transfer from the plant to the fungus was first demonstrated by Ho and Trappe (1973), who showed that 14C accumulates in AM fungal spores derived from plants exposed to 14CO2. However, it was long debated in which form this carbon is given to the fungus and where precisely the transfer takes place (Smith and Smith, 1990). Experiments in the late 1990s using NMR and respirometric studies showed that Glc is the most likely form in which the fungus takes up sugar from the apoplast (Shachar-Hill et al., 1995; Solaiman and Saito, 1997; Pfeffer et al., 1999). These results correlated well with increased apoplastic acid invertase and Suc synthase activities in mycorrhizal roots (Wright et al., 1998; Schaarschmidt et al., 2006). From a physiological perspective, an important question is to what extent is carbon supply linked to the ability of the fungus to deliver phosphate. The answer is of major importance, because it determines the efficiency of the symbiosis and predicts plant performance. Plant phosphate status and fungal development are known to be linked, because high phosphate levels in the soil reduce fungal growth within the root (Menge et al., 1978; Bruce et al., 1994; Breuillin et al., 2010) as well as carbon transfer toward the fungus (Olsson et al., 2010). Importantly, inactivation of symbiotic plant phosphate transporters leads to fungal growth arrest (Maeda et al., 2006; Javot et al., 2007). In this regard, it has recently been shown that more cooperative fungi, in terms of plant growth responses, cost of carbon per unit of P transferred, and resource hoarding strategies, are allocated with more carbon (Kiers et al., 2011). Thus, it seems that the plant, as proposed by Fitter (2006), has mechanisms to control carbon allocation locally that depend on the phosphate homeostasis of the cell. The question is whether these changes in sugar allocation to sink tissues are perceived by the fungus and have an effect on the sugar uptake mechanisms of the fungus.
To answer these questions, the molecular characterization of fungal transporter(s) operating at the plant–fungal interface is essential. However, the identification and nature of the specific fungal transporter(s) involved has remained elusive thus far. To date, only a monosaccharide transporter from Geosiphon pyriformis, a member of the Glomeromycota phylum that forms a symbiotic relationship with the cyanobacterium Nostoc punctiforme, has been identified (Schüßler et al., 2006). However, G. pyriformis has not been found to form symbiotic relationships with higher plants. Therefore, no further investigations of sugar uptake and metabolism and its relation to phosphate nutrition have been performed on a molecular level in fungi forming the archetypical arbuscule structure.
The sequencing project of the arbuscular mycorrhizal fungus Glomus intraradices DAOM 197198, now Glomus sp DAOM 197198 (Stockinger et al., 2009), will surely allow the identification of key components of the symbiosis that so far have been elusive, because of the obligate biotrophy of the fungus. However, its completion has been delayed by assembly difficulties (Martin et al., 2008). Using a first draft of this project, we have been able to identify several partial sequences that correspond to putative sugar transporters. In this work, we present the detailed characterization of a high-affinity monosaccharide transporter that plays a major role in sugar uptake in the arbuscular mycorrhizal fungus Glomus sp DAOM 197198. In addition, we show that the extraradical mycelium of AM fungi is able to take up Glc and Xyl.
RESULTS
MST2, a Fungal Monosaccharide Transporter Highly Induced During AM Symbiosis
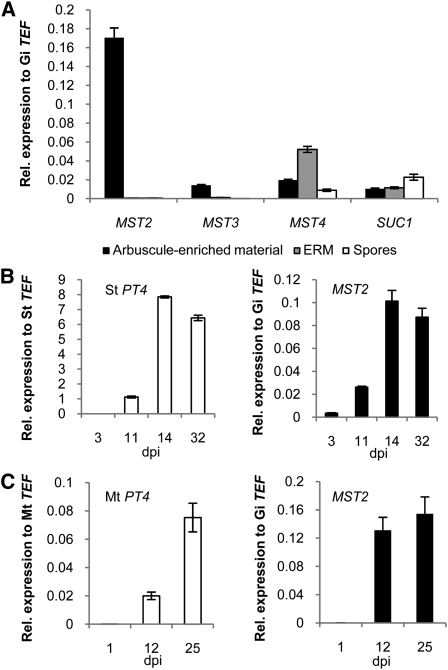
Blast searches in the first drafts of the genome sequencing project of the AM fungus Glomus sp DAOM 197198 (Martin et al., 2008) allowed us to identify four partial sequences that belong to the major facilitator superfamily (see Supplemental Table 1 online). Three of these showed high similarity to genes encoding monosaccharide transporters, and one of them had higher similarity to a Suc transporter. Expression analysis using quantitative real-time RT-PCR (qRT-PCR) during different developmental stages of the life cycle of the fungus revealed that only one of them (MST2) was highly expressed during the in planta phase and was only just detectable during the other stages (Figure 1A). This indicates that this transporter could account for the uptake of sugar from the plant. The full-length cDNA sequences of all transporters were isolated from libraries created from microdissected arbuscule-containing cortex regions or from extraradical mycelium (see Supplemental Figure 2 and Supplemental Table 1 online). Sequence analysis of the deduced proteins showed that they all contain the conserved motif cd06174, which is a characteristic of the major facilitator superfamily. In addition, MST2 and MST3 contain the motif PRK10077 (xylE motif of d-Xyl transporters, E-values 8.8 e−61 and 8.8 e−64, respectively) and SUC1 contains the motif GPH Suc superfamily (TIGR01301, E-value 6.12 e−44). Phylogenetic analysis showed that MST2 and MST3 cluster with a group that includes several Xyl transporters, whereas MST4 clusters with a group that includes the G. pyriformis monosaccharide transporter 1 (see Supplemental Figures 3 and 4 and Supplemental Data Set 1 online). It is difficult to predict whether MST4 and the transporter of G. pyriformis are orthologs, because the amino acid identity between these two proteins is only 31%. However, MST4 was the only sugar transporter that was retrieved when the G. pyriformis transporter was blasted against the Glomus sp database.
Figure 1.
Real-Time Expression Analysis of Sugar Transporters from the AM Fungus Glomus sp.
(A) Transcript accumulation of four putative Glomus sp sugar transporters in different fungal tissues: Arbuscule-enriched material from microdissected mycorrhizal potato hairy roots, ERM, and germinated spores.
(B) Time-course analysis of the expression of S. tuberosum St PT4 and Glomus sp MST2 in mycorrhizal potato hairy roots 3, 11, 14, and 32 DAI (dpi).
(C) Time-course analysis of the expression of M. truncatula Mt PT4 and MST2 in mycorrhizal roots 1, 12, and 25 DAI (dpi).
(Error bars represent sd; n = 3.)
To gain information about the expression of MST2 during symbiosis, we performed qPCR analysis with roots of potato (Solanum tuberosum) and of Medicago truncatula colonized with Glomus sp DAOM 197198. As a reference for the symbiotic status of the roots, we measured the gene expression of the mycorrhiza-specific plant phosphate transporter PT4 (Harrison et al., 2002), which is activated in arbuscule-containing cells and located at the periarbuscular membrane. PT4 is considered to be one of the best indicators of a functional arbuscular–mycorrhizal association. Inactivation of this transporter by RNA interference leads to degradation of arbuscules and to arrest of fungal development within the root (Maeda et al., 2006; Javot et al., 2007). We observed that MST2 transcript levels correlated well with those of PT4 over time. Interestingly, MST2 expression preceded that of PT4, indicating that MST2 might be expressed in other fungal structures besides arbuscules (Figures 1B and 1C). By contrast, expression of MST4 did not correlate with that of PT4 during the time course of colonization (see Supplemental Figure 5A online).
MST2 Is Expressed in Arbuscules and Intercellular Hyphae
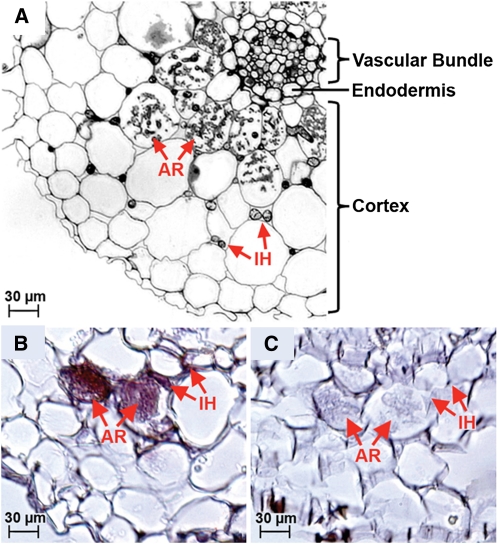
The precise location of carbon transfer from the plant to the fungus has long been a subject of debate (Fitter, 2006). The arbuscule in the inner cortical cells is perhaps the most obvious location for carbon transfer. However, the fungus develops other structures (i.e., intercellular hyphae and coils) in the outer cortical cells before forming arbuscules (Smith and Smith, 1997), suggesting that other possible sites for sugar uptake might exist. In support of this possibility, arbuscules are not always formed in some colonization types (e.g., Paris), indicating that coils or intercellular hyphae have to absorb carbon (Smith and Smith, 1990). To identify the site of sugar uptake, we performed an in situ hybridization analysis of M. truncatula roots colonized with Glomus sp DAOM 197198. Although the symbiosis of Glomus-M. truncatula results in rapid penetration and arbuscule formation, we detected expression of MST2 not only in arbuscules but also in intercellular hyphae, indicating that both structures are likely sites of sugar uptake (Figure 2).
Figure 2.
In Situ Hybridization of M. truncatula Hairy Roots Colonized with Glomus sp DAOM 197198.
(A) Overview of a cross-sectioned mycorrhizal M. truncatula root. AR, arbuscule; IH, intraradical hyphae.
(B) Detail of a cross-sectioned mycorrhizal root hybridized with the antisense probe for MST2. Positive purple staining at arbuscules and IH indicates MST2 expression.
(C) No staining is visible in sections of the same root hybridized with the MST2 sense probe.
[See online article for color version of this figure.]
MST2 Is a Potent Monosaccharide Scavenger
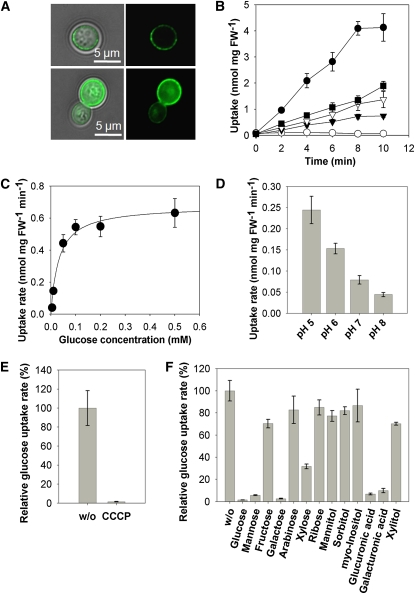
To characterize MST2 biochemically and to investigate its substrate specificity, we expressed it in the monosaccharide transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 (Wieczorke et al., 1999). We first demonstrated its membrane localization with a green fluorescent protein (GFP)-tagged version of the protein (Figure 3A). Then, using 14C-labeled sugars, we showed that MST2 is able to transport Glc, Xyl, Man, and Fru (Figure 3B) with decreasing affinity in that order. By contrast, MST2 is unable to transport disaccharides, such as Suc or maltose (see Supplemental Figures 5B and 5C online). Affinity studies showed that MST2 works as a high-affinity transporter for Glc with a Km of 33 ± 12.5 μM (se) (Figure 3C). This Km is in the range of the highest affinity reported for plant monosaccharide transporters (Büttner, 2007), including the Arabidopsis thaliana transporter STP4 (Km 15 μM) (Truernit et al., 1996; Büttner and Sauer, 2000). STP4, together with STP1, STP7, and STP13, are the most highly expressed monosaccharide transporters in Arabidopsis roots (Büttner, 2010), and with the exception of STP7, whose Km has not been determined, they are all high-affinity transporters (Km 15 to 74 μM), suggesting that the concentration of available monosaccharides at the root apoplast is very low. For MST2, a pH optimum was found at pH 5.0 (Figure 3D), a likely apoplastic pH at the plant–fungal interface (Guttenberger 2000). This optimum is comparable with that of other mycorrhiza-induced plant transporters located at the same interface, such as the M. truncatula phosphate transporter PT4 and the Lotus japonicus ammonium transporter AMT2, with pH optima of 4.25 and 4.5, respectively (Harrison et al., 2002; Guether et al., 2009). Glc transport was found to be sensitive to the presence of a protonophore, indicating the existence of a secondary H+ cotransport (Figure 3E). We showed in a previous work that AM fungi have several (H+)–ATPases, and one of them is upregulated in planta (Requena et al., 2003). We propose that the energization required for carbon uptake by MST2 could be performed by this enzyme.
Figure 3.
Functional Characterization of MST2 in Yeast.
(A) A MST2-GFP fusion is targeted to the plasma membrane of S. cerevisiae. Two examples of transformed yeast cells are shown in the top and bottom panels. In both cases, left panels are bright-field and GFP overlays, whereas right panels show GFP fluorescence alone.
(B) to (F) Sugar uptake experiments with yeast cells expressing MST2. FW, fresh weight.
(B) Uptake of different radiolabeled substrates (1 mM). Open circles, vector control; closed circles, d-Glc; closed squares, d-Xyl; open triangles, d-Man; closed triangles, d-Fru).
(C) Michaelis-Menten kinetics of Glc uptake at pH 5. (D) Uptake of Glc is optimal at acidic pH values. (E) Glc transport is sensitive to the protonophore CCCP. (F) Uptake of [14C]Glc (0.1 mM) without competitor (w/o) or with potential substrates of MST2 (100-fold molar excess). (Error bars represent se for all panels; n = 3.)
[See online article for color version of this figure.]
The versatility of MST2 as a sugar transporter was confirmed in competition assays. Plant cell wall monosaccharides, such as Xyl, glucuronic and galacturonic acid, Man or Gal were able to efficiently outcompete the uptake of Glc (Figure 3F). This is interesting, because Glc uptake by the sugar transporter identified in G. pyriformis was also shown to be outcompeted by Man (Schüßler et al., 2006). This result suggests that MST2 might transport not only Glc but also cell wall monosaccharides. This is in agreement with cytological and biochemical studies that show that the plant–fungal interface in AM symbiosis contains nonassembled primary plant cell wall components, such as nonsterified polygalacturonans, xyloglucans, β-1,4-glucans, and arabinogalactan proteins (Bonfante and Perotto, 1995; Gianinazzi-Pearson, 1996).
Xyl Triggers the Expression of MST2 in the Extraradical Mycelium
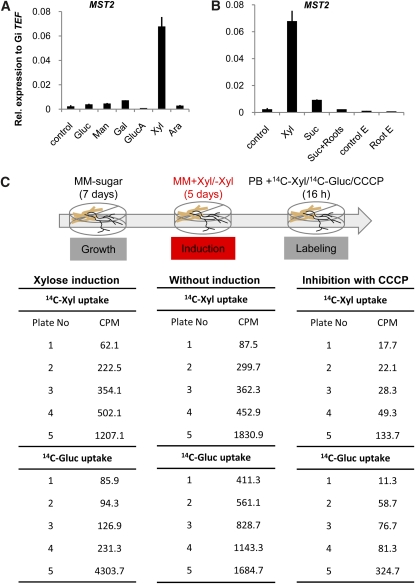
We next investigated whether any of the transported substrates of MST2 could function as a signal to induce its own expression. Thus, we tested the influence of several monosaccharides on the transcription of all of the identified sugar transporters in the extraradical mycelium (ERM). MST2 is expressed in the ERM at almost undetectable levels under routine growing conditions without sugar in the medium (Figure 1A). Interestingly, the addition of Xyl was able to specifically induce the expression of MST2 in the ERM (Figure 4A), a tissue believed thus far to be incapable of sugar uptake (Pfeffer et al., 1999). None of the other analyzed monosaccharides, including Glc, Man, Gal, Ara, or glucuronic acid, induced MST2 in the ERM. By contrast, the expression of the other sugar transporters identified was either unmodified or nonspecifically induced by Xyl (see Supplemental Figure 6 online). Because the ERM develops in the soil where other roots are present, we tested whether exposure of the mycelium to roots or their exudates could induce the expression of MST2. This was not the case, and only free Xyl was able to trigger MST2 expression (Figure 4B). Our results suggest the possibility that Xyl could be the signal that triggers MST2 expression in planta.
Figure 4.
Evidence of Sugar Uptake Via the Extraradical Mycelium of Glomus sp.
(A) Transcript accumulation of MST2 in extraradical mycelium exposed to different sugars (2%) for 5 d. Gluc, Glc; GlucA, glucuronic acid.
(B) Expression of MST2 in extraradical mycelium in response to Xyl, Suc, tomato roots (Suc + roots), tomato root exudates (Root E), or control of root exudates (Control E).
(C) Uptake of radiolabeled Xyl or Glc by extraradical mycelium. Extraradical mycelium was grown for 7 d in liquid M medium and exposed to [14C]Xyl or [14C]Glc after incubation for an additional 5 d in M medium with or without Xyl. Xyl and Glc uptake was inhibited in the presence of the protonophore CCCP. Each value represents one measurement/plate with different amounts of mycelium. The experiment was conducted twice with similar results. PB, phosphate buffer; Gluc, Glc; MM, minimal medium. (Error bars represent sd; n = 3.)
[See online article for color version of this figure.]
Evidence of Xyl Uptake and Catabolism in AM Fungi
The results above demonstrate that the ERM expresses several sugar transporters whose expression can be induced by different monosaccharides. However, studies using [13C]Glc showed that the ERM was unable to take up Glc added to the medium as a single carbon source (Pfeffer et al., 1999). We wondered whether transport of Xyl or Glc could be monitored in the ERM. To investigate this possibility, we exposed the ERM to Xyl for 6 d and then added radioactive Xyl or Glc. The results showed that the mycelium was indeed able to take up both Xyl and Glc. Furthermore, we showed that preincubation with Xyl is not essential, because the mycelium can readily take up both sugars (Figure 4C). The sugar uptake by the ERM is not simple adsorption or diffusion, because it can be blocked using a protonophore (Figure 4C). By contrast, no radioactivity was observed in the roots, suggesting that sugar imported by the ERM is retained for fungal use.
To use Xyl as a carbon source, it has to be converted to d-xylulose and then to d-xylulose-5-P, which is incorporated into the pentose phosphate pathway (see Supplemental Figure 7A online). In contrast with bacteria, fungi are not able to convert d-Xyl directly into d-xylulose. The conversion requires first the reduction of d-Xyl to d-xylitol and then the dehydrogenation to d-xylulose (Noguchi et al., 2009). Indications of the use of Xyl as a carbon source in the AM symbiosis are found in the work of Schliemann et al. (2008), who described an increased content of xylitol in M. truncatula mycorrhizal roots from 35 days after inoculation (DAI) onward, although at very low levels. We searched the genome of Glomus sp DAOM 197198 for genes related to Xyl catabolism and studied their expression during different stages of the fungal life cycle. We identified two Xyl reductases (XR1 and XR2), one xylitol dehydrogenase (XDH1) and one xylulose kinase (XK1) (see Supplemental Table 1 online). Gene expression analysis showed that both Xyl reductase genes are highly induced in planta (see Supplemental Figure 7B online). By contrast, the xylitol dehydrogenase and the xylulose kinase genes were not differentially regulated, possibly indicating that as yet unidentified isozymes might perform the next steps.
Expression of MST2 in Planta Is Dependent on the Phosphate Homeostasis of the Root
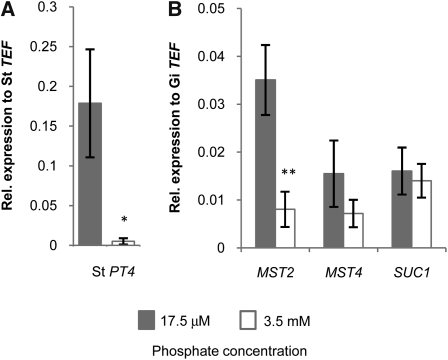
The correlation between the expression of MST2 and the symbiotic plant phosphate transporter PT4 (Figure 1) suggests that expression of the fungal sugar transporter could be regulated by phosphate homeostasis in the root. To test the hypothesis that the exchange of phosphate and carbon are interconnected, we increased the amount of available phosphate from 17.5 μM to 3.5 mM in the medium where mycorrhizal potato roots were growing. As a consequence, the roots shut off the expression of the symbiotic phosphate transporter PT4, possibly to rely on the nonsymbiotic phosphate pathway (Figure 5A). At this stage, 1 week after phosphate addition, no phenotypical differences in mycorrhizal colonization were visible, which was confirmed by the molecular quantification of the amount of fungus within the root (see Supplemental Figure 8 online). This agrees with the observations of Breuillin et al. (2010), who reported that high phosphate levels only had significant effects in reducing the colonization levels after 2 weeks, although the levels of symbiotic phosphate transporters were already decreased after 2 d. Remarkably, the expression of MST2 was downregulated in parallel to that of PT4, whereas the expression of MST4 was nonsignificantly reduced, and the expression of SUC1 was not affected (Figure 5B). Our results indicate that the expression of MST2 in planta is linked to the symbiotic phosphate delivery, even before phenotypical effects can be observed. Javot et al. (2007) proposed that symbiotic Pi delivery to the cortical cell serves as a signal for continued development of the arbuscule.
Figure 5.
Expression Levels in Mycorrhizal Potato Hairy Roots in Response to Changes in Phosphate (KH2PO4) Concentration.
Expression of St PT4 (A) and of MST2, MST4 and SUC1 (B) at the indicated phosphate concentrations (error bars represent sd; *P value < 0.05, **P value < 0.01; n = 3).
MST2 Is Critical for the Symbiosis
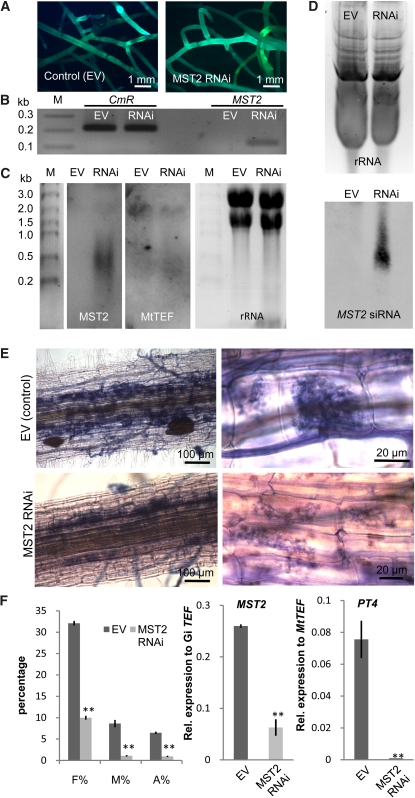
To gain evidence of the relevance of MST2 for the symbiosis, and because there is no known stable transformation system for AM fungi, we attempted its inactivation using Host-Induced Gene Silencing (HIGS) (Nowara et al. 2010). A hairpin RNAi-silencing construct targeting a 300-bp region at the N terminus of MST2 was constitutively expressed in M. truncatula hairy roots. Transformed roots (control with empty vector or MST2 RNAi) constitutively expressed the GFP cassette (Figure 6A). Expression of the hairpin construct was verified by RT-PCR using primers that target the intron forming the hairpin loop as well as with primers in the region corresponding to MST2 (Figure 6B). RNA gel blot analysis of MST2 RNAi roots showed expression of the full-length MST2 inactivation cassette, as expected, as well as a smear of smaller size (Figure 6C). By contrast, expression of the TEF1α analyzed by RNA gel blot was similar in both samples. SiRNAs were isolated and analyzed in polyacrylamide gels by RNA gel blot using a specific MST2 riboprobe. SiRNAs were only detected in MST2 RNAi roots (Figure 6D). We subsequently inoculated MST2 RNAi roots and control roots with G. intraradices and analyzed the colonization 25 DAI. MST2 RNAi roots showed much lower mycorrhization levels than control roots for all parameters analyzed, including the number of arbuscules (Figures 6E and 6F). The arbuscule morphology in MST2 RNAi roots was abnormal. Arbuscules were either not fully developed or showed premature senescence (Figure 6E). Analysis of MST2 and Mt PT4 expression in those roots correlated well with that finding. Thus, MST2 expression, using primers outside the RNAi cassette, was reduced approximately fivefold in MST2 RNAi roots, and Mt PT4 expression was almost completely abolished (Figure 6F). Taken together, these results indicate that the reduction in the expression of MST2 by HIGS compromises the normal development of arbuscules and demonstrates that this fungal sugar transporter is indispensable for a functional AM symbiosis.
Figure 6.
Host-Induced Gene Silencing of MST2.
(A) Vector control and MST2 RNAi hairy root lines of M. truncatula constitutively expressing GFP. EV, empty vector.
(B) Expression of the RNAi vector in roots was tested by PCR using primers that amplify the chloramphenicol resistance cassette (CmR) localized in the hairpin loop intron and in the 300-bp MST2 sequence chosen as target for RNAi silencing (30 cycles).
(C) Degradation of the MST2 dsRNA was shown in an RNA gel blot using a probe specific for the MST2 inverted repeats in a representative root line. The size of the MST2 RNAi construct is 2.3 kb. The integrity of the RNA was verified by hybridization of the same blot with a specific probe for TEF1α of M. truncatula (size 2.2 kb) and by ethidium bromide staining of the rRNA.
(D) The formation of MST2 siRNAs was examined by RNA gel blot of a 15% acrylamide gel with an MST2–specific riboprobe (Bottom). Equal loading was estimated by ethidium bromide staining of rRNA or predominant RNAs (Top).
(E) Mycorrhizal colonization of control and MST2 RNAi roots 25 d after inoculation with G. intraradices shown by ink staining.
(F) Estimations of mycorrhization levels in MST2 RNAi hairy roots in comparison with control roots (EV) 25 DAI. The parameters measured are frequency (F%) and intensity (M%) of mycorrhiza and arbuscule (A%) abundance. Transcript accumulation of MST2 and Mt PT4 in MST2 RNAi and control roots was measured by quantitative real-time PCR. (Error bars represent sd; **P value < 0.01; n = 3, three biological replicates with three technical replicates.)
[See online article for color version of this figure.]
DISCUSSION
This research shows that the symbiotic partner of plants Glomus sp DAOM 197198 expresses a sugar transporter (MST2) that is able to transport not only Glc but also other sugars that are normal components of the plant cell wall. Its high affinity, pH optimum, expression in arbuscules and intercellular hyphae, and broad range of substrates favorably position the fungus to efficiently compete for sugars present in the plant–fungal interface. The ability of AM fungi to feed on cell wall components was proposed long ago (Smith and Smith, 1990; Bonfante and Perotto, 1995; Gianinazzi-Pearson, 1996) and fits well with other observations: systemic activation of a plant xyloglucan endotransglucosylase/hydrolyase gene (XTH) in mycorrhizal roots (Maldonado-Mendoza et al., 2005) (see Supplemental Figure 9 online), xyloglucanase capacity of AM fungi (Rejon-Palomares et al., 1996), and presence of noncrosslinked cell wall components in the periarbuscular matrix (Bonfante and Perotto, 1995; Gianinazzi-Pearson, 1996; Ballestrini and Bonfante, 2005). Furthermore, the activation of a β-xylosidase was shown to be induced more than sevenfold in tomato mycorrhizal roots (Fiorilli et al., 2009). We now provide evidence that AM fungi posses the required machinery to catabolize Xyl in addition to Glc. Our data could explain the unexpected results that artificially induced invertase activity in roots does not alter the AM fungal colonization status (Schaarschmidt et al., 2007), suggesting that Glc and Fru are not the only carbon sources and/or that enough carbohydrates are available. The increased activities of plant invertase and Suc synthase observed in other studies might have a more important role in providing the highly metabolically active arbuscule-containing cells with an extra supply of sugars. Indeed, the activation of a plant Glc transporter in those cells has been observed (Harrison, 1996). Moreover, it has been speculated that AM fungi as mutualistic symbionts might avoid the excessive induction of invertases to keep plant defense responses low (Schaarschmidt et al., 2006; Wahl et al., 2010). In this context, the use of plant cell wall sugars as carbon source by AM fungi seems to be paradoxical. However, it is known that biotrophic fungi have developed mechanisms to subvert the plant defense response, and AM fungi are not an exception. We have recently shown that AM fungi do secrete and translocate effector proteins to the plant cell that counteract the elicitation of microbe-associated molecular pattern–triggered defense responses (Kloppholz et al., 2011). Under such conditions, a versatile sugar transporter able to efficiently transport different monosaccharides from the apoplast might be an optimal adaptation for the biotrophic modus of life. An unexpected and exciting result of our research is the finding that several of the sugar transporters identified are expressed in the ERM. Furthermore, we present evidence that the ERM is able to take up not only Glc but also Xyl, in contrast with previously published results. This finding might illuminate why these nonsaprophytic fungi benefit from growing in patches of organic matter (Hodge et al., 2001) and might open biotechnological possibilities for improving the cultivation of these obligate biotrophs.
The significance of the MST2 transporter for the functioning of the symbiosis is highlighted by the fact that its inactivation through HIGS leads to fungal growth arrest. In addition to an overall reduced degree of mycorrhization, arbuscule development is severely impaired in MST2-silenced roots, correlating with lower expression of the periarbuscular membrane-located phosphate transporter PT4. We showed that phosphate is an important factor in the regulation of the transcriptional activation of MST2 in planta, and that Xyl per se might be a signal that is able to induce MST2 expression. These results suggest the following model: The growth of the fungus within the root cortex induces a signal that increases carbon sink and promotes the availability of Xyl. Xyl itself then triggers MST2 expression. Subsequent arbuscule formation, which only occurs in inner cortical cells, induces PT4 activation shortly after MST2 induction. If the root is provided with sufficient phosphate, however, PT4 and MST2 expression are downregulated. It has been shown that high phosphate negatively regulates not only PT4 expression (Breuillin et al., 2010), but also carbon partitioning to the root (Olsson et al., 2010). Furthermore, the data of Breuillin et al. (2010) for Petunia show that high phosphate supply counteracts the AM-induced accumulation of several transcripts coding for xyloglucan endotransglucosylase/hydrolyases putative orthologs of M. truncatula XTH1 (i.e., cn5551, cn8026). This suggests that high phosphate might act by reducing the level of Xyl that induces MST2. The resulting scenario concluded from our data indicates a tight regulation of the exchange of phosphate for carbon that is mainly modulated at the arbuscular interface.
METHODS
Biological Material
Glomus sp DAOM 197198 was used to colonize roots of Medicago truncatula Jemalong A17 or hairy roots of Solanum tuberosum cv Désiré (potato) as described elsewhere (Kuhn et al., 2010). Spores and extraradical mycelium of Glomus sp DAOM 197198 were produced in the bicompartmental system as described elsewhere (Helber and Requena, 2008). To analyze the expression of fungal sugar transporters in the ERM, mycelium grown for 10 d in liquid M medium (Bécard and Fortin, 1988) without sugar were cultivated for an additional 5 d in 8 mL of fresh M medium supplemented with different sugars used in an end concentration of 2%. As a control, the ERM was cultivated in fresh liquid M medium without sugar. For the treatment with root exudates, the medium of 1-week-old ERM liquid cultures was replaced with 8 mL fresh medium (3 mL dH2O and 5 mL M medium without Suc) or with 8 mL medium supplemented with root exudates (3 mL root exudate solution and 5 mL M medium without Suc). The ERM was harvested after 1 week of incubation. Root exudates were obtained from Solanum lycopersicum cv Rio Grande hairy roots as described in Bücking et al. (2008).
Phosphate Treatment of Mycorrhizal Potato Roots
Potato hairy roots expressing the StPT3promoter-Fluorescent-Timer construct (Kara–ov et al., 2004) were mycorrhized as described under standard conditions on M medium with 17.5 μM KH2PO4. After 1 week, mycorrhizal potato roots were covered with 8 mL M medium containing 3.5 mM or 17.5 μM of KH2PO4. Roots were harvested after 1 week of incubation at 27°C in the dark.
Blast Searches
Blast searches in the genome of Glomus sp DAOM 197198 to look for sugar transporters were done using as query the Tuber borchii TbHXT1 (AAY2639) and the Pyrenophora tritici-repentis (XP_001938670) high-affinity Glc transporters.
The cDNA library of Arbuscule-Enriched Root Fragments
Arbuscule formation in potato hairy roots was monitored after the expression of the StPT3 promoter-Fluorescent-Timer construct (Kara–ov et al., 2004) after colonization with Glomus sp. Regions enriched in young arbuscules were manually dissected under the fluorescence stereomicroscope using self-made 2-mm scalpels. RNA (500 ng) was isolated and copied according to the SMART library construction protocol (Clontech).
Race Analysis and Sequence Accession
The full-length cDNA sequences of MST2, MST3, MST4, and SUC1 were obtained after rapid amplification of cDNA ends using the protocols of the SMART (Clontech) or the Gene Racer (Invitrogen) kits.
Phylogenetic Analysis
Phylogenetic analyses were conducted using Mega 4.0 software and ClustalW for the alignment and the neighbor-joining method for the construction of the phylogeny (Tamura et al., 2007). Bootstrap tests were performed using 1000 replicates. The alignment used for phylogenetic tree construction can be found in Supplemental Data Set 1 online.
Gene Expression Analyses
Total RNA from the different tissues was extracted with Trizol and quantified spectrophotometrically using the Nanodrop device. cDNA was synthesized as described elsewhere (Kuhn et al., 2010). Transcript levels of genes were determined on a Bio–Rad iCycler MyIQ using MESA Green 231qPCR MasterMix Plus (Eurogentec) in three biological replicates with three technical replicates per reaction. Plant transcript levels were normalized to the translation elongation factor 1-alpha (TEF1α) of the different organisms used (M. truncatula TC106470, DFCI gene index; S. tuberosum AJ536671; and for Glomus sp DAOM 197198: G. intraradices DQ 282611). Primers used are listed in Supplemental Table 2 online.
Localization in the Yeast Plasma Membrane
The subcellular localization of MST2 in yeast was performed with an N-terminal fusion of MST2 to GFP in strain EBY.VW4000 as described elsewhere (Wahl et al., 2010).
Transport Studies with Radiolabeled Substrates in Yeast
MST2 full-length cDNA was cloned in the vector NEV-N and transformed into the yeast strain EBY.VW4000. Sugar uptake measurements were performed as described elsewhere (Wahl et al., 2010). For uptake studies, the yeast cells were resuspended in 50 mM potassium phosphate buffer (pH 5.0). Uptake was started by the addition of 1 mmol of radiolabeled sugar ([14C]Glc, [14C]Fru, [14C]Xyl, or [14C]Man). For inhibition studies, yeast cells were preincubated for 1 min with 50 μM of the protonophore carbonyl cyanide m-chlorophenyl hydrazone, (CCCP), before the reaction commenced with the addition of [14C]Glc. Competition for Glc uptake was performed by adding 10 mM of the competitor. Yeast samples were filtered and washed two times with Milipore water before being measured in the scintillation counter (Liquid Scintillation Analyzer TRI-CARB 2100TR; Packard). Uptake of [14C]Man and [14C]Suc was tested correspondingly in the yeast mutant SEY2102. All uptake experiments were repeated with independent samples at least three times.
Sugar Uptake Experiments with Radiolabeled Substrates in Glomus sp Extraradical Mycelium
ERM of Glomus sp DAOM 197198 was cultivated as described in liquid M medium without sugar (Helber and Requena, 2008). After 7 d of incubation, the medium in the distal compartment was replaced with 8 mL of fresh M medium without sugar or supplemented with 5% Xyl. After 5 d, the medium was removed, and the mycelium was washed five times with 50 mM sodium phosphate buffer (pH 5.0). In the uptake assay, the washed mycelium was incubated for 16 h at room temperature in 1 mL of 50 mM sodium phosphate buffer (pH 5.0) containing 1 mM [14C]Xyl or [14C]Glc (specific activity 0.2 μCi/μL). For uptake inhibition, 50 μM CCCP was added to the uptake assay mixture containing 1 mM [14C]Xyl or [14C]Glc. Mycelium and root samples were washed extensively five times with Millipore water before harvest and were counted in a scintillation counter.
In Situ Hybridization of Mycorrhizal Roots
In situ hybridization was performed as described elsewhere (Schaarschmidt et al., 2006). M. truncatula roots colonized with Glomus sp DAOM 197198 were fixed with paraformaldehyde, embedded in paraplast, and hybridized using digoxigenin (DIG)-labeled probes.
Host-Induced Gene Silencing (HIGS)
For MST2 silencing via the plant host, 300 bp of the MST2 cDNA, starting from the ATG, was amplified using the primers RNAiMST2-F1_and RNAiMST2_R1 and recombined in the Gateway binary RNAi vector pK7GWIWG2D(II) (Karimi et al., 2002). The resulting MST2 RNAi vector was used to generate transgenic M. truncatula hairy roots via Agrobacterium rhizogenes transformation as described elsewhere (Kuhn et al., 2010). MST2 RNAi and control M. truncatula hairy roots containing the empty RNAi vector were mycorrhized for 25 d with Glomus sp as described above. The roots were harvested and used for RNA extraction and acid ink staining to visualize fungal structures (Kuhn et al., 2010). cDNA was obtained from mRNA using the reverse transcriptase enzyme Superscript II from Invitrogen. RT-PCR analysis was performed with primers that amplify a different region to the RNAi cassette, and PCR products were detected after EtBr staining (see Supplemental Table 2 online).
Quantification of Mycorrhizal Colonization
Quantification of mycorrhizal colonization was done according to Kloppholz et al. (2011).
RNA Analysis and Detection of siRNAs
RNA analysis was done according to the DIG Application Manual for Filter Hybridization from Roche (http://www.roche-applied-science.com/PROD_INF/MANUALS/DIG_MAN/dig_toc.htm). DIG-labeled probes for MST2 and MtTEF were PCR amplified with the primers RNAiMST2_F1 and RNAiMST2_R1 and MtTEF_F1 and MtTEF_R1 using the Roche PCR DIG Probe Synthesis Kit. The nylon membrane was hybridized overnight in high SDS hybridization buffer at 42°C. After hybridization, the membrane was washed with 2× SSC, 0.1% SDS and 0.1× SSC, 0.1% SDS at 42°C.
Isolation, hybridization, and detection of siRNAs was performed after Nicolás et al. (2003). For the specific detection of MST2 siRNAs, we used a DIG-labeled MST2 RNA probe, generated with the DIG Northern Starter Kit from Roche, which corresponds to the first 300 bp of the MST2 open-reading frame. The riboprobe was hydrolyzed to small RNA fragments and purified using G-50 Sephadex Quick Spin Columns from Roche.
Statistical Analyses
All parameters and calculated variables were tested for differences between treatments and their respective controls using the Student’s t test incorporated into Microsoft Excel 7.0 (Microsoft). Differences are described as significant when a value of P < 0.05 (marked with *) and P < 0.01 (**), respectively, was obtained. n indicates the number of biological replicates, each with three technical replicates.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers: MST2 (HM143864), MST3 (HQ848964), MST4 (HQ848965), SUC1 (HQ848966), XR1 (JK511418), XR2 (JN686742), XDH1 (JK511416), and XK1 (JK511417).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Carbon Flow in the Arbuscular Mycorrhizal Symbiosis.
Supplemental Figure 2. Microdissection of Mycorrhized Potato Roots and Arbuscule Enrichment.
Supplemental Figure 3. Phylogenetic Tree of Fungal Monosaccharide Transporter Protein Sequences.
Supplemental Figure 4. ClustalW Alignment of the Deduced Amino Acid Sequence of Glomus sp MST2, MST3, and MST4 with Amino Acid Sequences of Putative Fungal Xyl or Glc transporters.
Supplemental Figure 5. Time-Course Analysis of Expression of the Glomus sp Sugar Transporter MST4, Suc Uptake into Yeast Cells Expressing MST2, and Maltose Uptake into Yeast Cells Expressing MST2.
Supplemental Figure 6. Transcript Accumulation of Three Glomus sp DAOM 197198 Sugar Transporters in Extraradical Mycelium Exposed to Different Sugars (2%) for 5 d.
Supplemental Figure 7. Scheme of the Xyl Utilization Pathways in Fungi and Bacteria, and Expression of Genes Involved in Xyl Catabolism in Glomus sp during Different Stages of the Life Cycle.
Supplemental Figure 8. Molecular Quantification of the Fungal Colonization of Potato Roots in Response to Two Phosphate Concentrations.
Supplemental Figure 9. Transcript Accumulation of Xyloglucan Endotrans-Glucosylase/Hydrolase from S. tuberosum and from M. truncatula in Hairy Roots Colonized with Glomus sp DAOM 197198.
Supplemental Table 1. ESTs or Contig Sequences Analyzed in This Work.
Supplemental Table 2. Primer List Used in This Work.
Supplemental Data Set 1. Amino Acid Sequence Alignment in Fasta Format Used for the Phylogenetic Analysis Presented in Supplemental Figures 3 and 4 Online.
Acknowledgments
We thank the Glomus Genome Consortium (Joint Genome Institute) and the Institut National de la Recherche Agronomique GlomusDB database for access to the first drafts of the sequences from the Glomus sp DAOM 197198 genome. We are thankful for the funding support from the Leibnitz society within the “Mykopakt” project. We thank H. Slater for her comments on the manuscript.
AUTHOR CONTRIBUTIONS
N.H. and N.R. identified and isolated all genes analyzed in this article and carried out the expression analyses and inactivation experiments. N.H., S.S., and B.H. did the in situ hybridization experiments. N.H., K.W., and N.S. carried out the biochemical characterization of MST2. All authors discussed the results and constructed the figures. N.H. and N.R. planned the experiments, analyzed the data, and wrote the article.
References
- Ballestrini V., Bonfante P. (2005). The interface compartment in arbuscular mycorrhizae: A special type of plant cell wall? Plant Biosyst. 139: 8–15 [Google Scholar]
- Bécard G., Fortin J.A. (1988). Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108: 211–218 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (July 27, 2010). Mechanisms underlying beneficial plant — fungus interactions in mycorrhizal symbiosis. Nature Commun. (online), doi/10.1038/ncomms1046 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Perotto S. (1995). Strategies of arbuscular mycorrhizal fungi when infecting host plants. New Phytol. 130: 3–21 [Google Scholar]
- Breuillin F., et al. (2010). Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 64: 1002–1017 [DOI] [PubMed] [Google Scholar]
- Bruce A., Smith S.E., Tester M. (1994). The development of mycorrhizal infection in cucumber: Effects of P supply on root growth, formation of entry points and growth of infection units. New Phytol. 127: 507–514 [Google Scholar]
- Bücking H., Abubaker J., Govindarajulu M., Tala M., Pfeffer P.E., Nagahashi G., Lammers P., Shachar-Hill Y. (2008). Root exudates stimulate the uptake and metabolism of organic carbon in germinating spores of Glomus intraradices. New Phytol. 180: 684–695 [DOI] [PubMed] [Google Scholar]
- Büttner M. (2010). The Arabidopsis sugar transporter (AtSTP) family: An update. Plant Biol (Stuttg) 12(Suppl 1): 35–41 [DOI] [PubMed] [Google Scholar]
- Büttner M. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581: 2318–2324 [DOI] [PubMed] [Google Scholar]
- Büttner M., Sauer N. (2000). Monosaccharide transporters in plants: Structure, function and physiology. Biochim. Biophys. Acta 1465: 263–274 [DOI] [PubMed] [Google Scholar]
- Fitter A.H. (2006). What is the link between carbon and phosphorus fluxes in arbuscular mycorrhizas? A null hypothesis for symbiotic function. New Phytol. 172: 3–6 [DOI] [PubMed] [Google Scholar]
- Fiorilli V., Catoni M., Miozzi L., Novero M., Accotto G.P., Lanfranco L. (2009). Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 184: 975–987 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V. (1996). Plant cell responses to arbuscular mycorrhizal fungi: Getting to the roots of the symbiosis. Plant Cell 8: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M., Neuhäuser B., Balestrini R., Dynowski M., Ludewig U., Bonfante P. (2009). A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 150: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenberger M. (2000). Arbuscules of vesicular-arbuscular mycorrhizal fungi inhabit an acidic compartment within plant roots. Planta 211: 299–304 [DOI] [PubMed] [Google Scholar]
- Harrison M.J. (1996). A sugar transporter from Medicago truncatula: Altered expression pattern in roots during vesicular-arbuscular (VA) mycorrhizal associations. Plant J. 9: 491–503 [DOI] [PubMed] [Google Scholar]
- Harrison M.J., Dewbre G.R., Liu J.Y. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helber N., Requena N. (2008). Expression of the fluorescence markers DsRed and GFP fused to a nuclear localization signal in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 177: 537–548 [DOI] [PubMed] [Google Scholar]
- Ho I., Trappe J.M. (1973). Translocation of 14C from Festuca plants to their endomycorrhizal fungi. Nature 224: 30–31 [DOI] [PubMed] [Google Scholar]
- Hodge A., Campbell C.D., Fitter A.H. (2001). An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413: 297–299 [DOI] [PubMed] [Google Scholar]
- Javot H., Pumplin N., Harrison M.J. (2007). Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ. 30: 310–322 [DOI] [PubMed] [Google Scholar]
- Kara–ov V., Bucher M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10: 22–29 [DOI] [PubMed] [Google Scholar]
- Kara–ov V., Nagy R., Wegmüller S., Amrhein N., Bucher M. (2004). Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 101: 6285–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kiers E.T., et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882 [DOI] [PubMed] [Google Scholar]
- Kloppholz S., Kuhn H., Requena N. (2011). A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21: 1204–1209 [DOI] [PubMed] [Google Scholar]
- Kuhn H., Küster H., Requena N. (2010). Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula. New Phytol. 185: 716–733 [DOI] [PubMed] [Google Scholar]
- Liu H., Trieu A.T., Blaylock L.A., Harrison M.J. (1998). Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol. Plant Microbe Interact. 11: 14–22 [DOI] [PubMed] [Google Scholar]
- Logi C., Sbrana C., Giovannetti M. (1998). Cellular events involved in survival of individual arbuscular mycorrhizal symbionts growing in the absence of the host. Appl. Environ. Microbiol. 64: 3473–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda D., Ashida K., Iguchi K., Chechetka S.A., Hijikata A., Okusako Y., Deguchi Y., Izui K., Hata S. (2006). Knockdown of an arbuscular mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol. 47: 807–817 [DOI] [PubMed] [Google Scholar]
- Maldonado-Mendoza I.E., Dewbre G.R., Blaylock L., Harrison M.J. (2005). Expression of a xyloglucan endotransglucosylase/hydrolase gene, Mt-XTH1, from Medicago truncatula is induced systemically in mycorrhizal roots. Gene 345: 191–197 [DOI] [PubMed] [Google Scholar]
- Martin F., Gianinazzi-Pearson V., Hijri M., Lammers P., Requena N., Sanders I.R., Shachar-Hill Y., Shapiro H., Tuskan G.A., Young J.P.W. (2008). The long hard road to a completed Glomus intraradices genome. New Phytol. 180: 747–750 [DOI] [PubMed] [Google Scholar]
- Menge J.A., Johnson E.L.V., Platt R.G. (1978). Mycorrhizal dependency of several citrus cultivars under three nutrient regimes. New Phytol. 81: 553–559 [Google Scholar]
- Nagy R., Kara–ov V., Chague V., Kalinkevich K., Tamasloukht M., Xu G., Jakobsen I., Levy A.A., Amrhein N., Bucher M. (2005). The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 42: 236–250 [DOI] [PubMed] [Google Scholar]
- Nicolás F.E., Torres-Martínez S., Ruiz-Vázquez R.M. (2003). Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 22: 3983–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi Y., Sano M., Kanamaru K., Ko T., Takeuchi M., Kato M., Kobayashi T. (2009). Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 85: 141–154 [DOI] [PubMed] [Google Scholar]
- Nowara D., Gay A., Lacomme C., Shaw J., Ridout C., Douchkov D., Hensel G., Kumlehn J., Schweizer P. (2010). HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22: 3130–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson P.A., Rahm J., Aliasgharzad N. (2010). Carbon dynamics in mycorrhizal symbioses is linked to carbon costs and phosphorus benefits. FEMS Microbiol. Ecol. 72: 125–131 [DOI] [PubMed] [Google Scholar]
- Paszkowski U., Kroken S., Roux C., Briggs S.P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer P.E., Douds , D.D., Jr, Becard G., Shachar-Hill Y. (1999). Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol. 120: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C., Daram P., Brunner S., Jansa J., Laloi M., Leggewie G., Amrhein N., Bucher M. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–470 [DOI] [PubMed] [Google Scholar]
- Rejon-Palomares A., Garcia-Garrido J.M., Ocampo J.A., Garcia-Romera I. (1996). Presence of xyloglucan-hydrolyzing glucanases (xyloglucanases) in arbuscular mycorrhizal symbiosis. Symbiosis 21: 249–261 [Google Scholar]
- Remy W., Taylor T.N., Hass H., Kerp H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena N., Breuninger M., Franken P., Ocón A. (2003). Symbiotic status, phosphate, and sucrose regulate the expression of two plasma membrane H+-ATPase genes from the mycorrhizal fungus Glomus mosseae. Plant Physiol. 132: 1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt S., González M.C., Roitsch T., Strack D., Sonnewald U., Hause B. (2007). Regulation of arbuscular mycorrhization by carbon. The symbiotic interaction cannot be improved by increased carbon availability accomplished by root-specifically enhanced invertase activity. Plant Physiol. 143: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt S., Roitsch T., Hause B. (2006). Arbuscular mycorrhiza induces gene expression of the apoplastic invertase LIN6 in tomato (Lycopersicon esculentum) roots. J. Exp. Bot. 57: 4015–4023 [DOI] [PubMed] [Google Scholar]
- Schachtman D.P., Reid R.J., Ayling S.M. (1998). Phosphorus uptake by plants: From soil to cell. Plant Physiol. 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliemann W., Ammer C., Strack D. (2008). Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 69: 112–146 [DOI] [PubMed] [Google Scholar]
- Schüßler A., Martin H., Cohen D., Fitz M., Wipf D. (2006). Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 444: 933–936 [DOI] [PubMed] [Google Scholar]
- Shachar-Hill Y., Pfeffer P.E., Douds D., Osman S.F., Doner L.W., Ratcliffe R.G. (1995). Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol. 108: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F.A., Smith S.E. (1997). Structural diversity in (vesicular)-arbuscular mycorrhizal symbioses. New Phytol. 137: 373–388 [DOI] [PubMed] [Google Scholar]
- Smith S.E., Smith F.A. (1990). Structure and function of the interfaces in biotrophic symbioses as they relate to nutrient transport. New Phytol. 114: 1–38 [DOI] [PubMed] [Google Scholar]
- Smith S.E., Smith F.A., Jakobsen I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133: 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaiman M.D.Z., Saito M. (1997). Use of sugars by intraradical hyphae of arbuscular mycorrhizal fungi revealed by radiorespirometry. New Phytol. 136: 533–538 [DOI] [PubMed] [Google Scholar]
- Stockinger H., Walker C., Schüssler A. (2009). ‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 183: 1176–1187 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Truernit E., Schmid J., Epple P., Illig J., Sauer N. (1996). The sink-specific and stress-regulated Arabidopsis STP4 gene: Enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8: 2169–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R., Wippel K., Goos S., Kämper J., Sauer N. (2010). A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 8: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C.P., Boles E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464: 123–128 [DOI] [PubMed] [Google Scholar]
- Wright D.P., Read D.J., Scholes J.D. (1998). Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ. 21: 881–891 [Google Scholar]