This work examines the functions of the Medicago truncatula and rice GRAS-type transcription factors NSP1 and NSP2. They were found to be essential for strigolactone synthesis, possibly through direct regulation of DWARF27.
Abstract
Legume GRAS (GAI, RGA, SCR)-type transcription factors NODULATION SIGNALING PATHWAY1 (NSP1) and NSP2 are essential for rhizobium Nod factor-induced nodulation. Both proteins are considered to be Nod factor response factors regulating gene expression after symbiotic signaling. However, legume NSP1 and NSP2 can be functionally replaced by nonlegume orthologs, including rice (Oryza sativa) NSP1 and NSP2, indicating that both proteins are functionally conserved in higher plants. Here, we show that NSP1 and NSP2 are indispensable for strigolactone (SL) biosynthesis in the legume Medicago truncatula and in rice. Mutant nsp1 plants do not produce SLs, whereas in M. truncatula, NSP2 is essential for conversion of orobanchol into didehydro-orobanchol, which is the main SL produced by this species. The disturbed SL biosynthesis in nsp1 nsp2 mutant backgrounds correlates with reduced expression of DWARF27, a gene essential for SL biosynthesis. Rice and M. truncatula represent distinct phylogenetic lineages that split approximately 150 million years ago. Therefore, we conclude that regulation of SL biosynthesis by NSP1 and NSP2 is an ancestral function conserved in higher plants. NSP1 and NSP2 are single-copy genes in legumes, which implies that both proteins fulfill dual regulatory functions to control downstream targets after rhizobium-induced signaling as well as SL biosynthesis in nonsymbiotic conditions.
INTRODUCTION
Strigolactones (SLs) exuded by plant roots into the rhizosphere are well-known stimuli for symbiotic arbuscular mycorrhizal fungi of the order Glomeromycota (Akiyama et al., 2005). These ex planta signals are coopted by root-parasitic plants of the Orobanchaceae family and are essential to induce their germination (Cook et al., 1966; Bouwmeester et al., 2007). Recently, it was found that SLs, or their derivatives, also function as endogenous plant hormones controlling outgrowth of axillary shoot buds (Gomez-Roldan et al., 2008; Umehara et al., 2008). They do so in crosstalk with auxin, the most prominent plant hormone (Beveridge and Kyozuka, 2010; Xie et al., 2010; Domagalska and Leyser, 2011). Biosynthesis and subsequent secretion of SLs is highly adaptive upon availability of nutrients, mainly phosphate (Yoneyama et al., 2007; López-Ráez et al., 2008; Umehara et al., 2008; Lin et al., 2009). To understand this adaptive regulation of these novel hormones, it is important to unravel the molecular mechanisms of SL biosynthesis, transport, and signaling. Here, we show that the GRAS-type transcription factors NODULATION SIGNALING PATHWAY1 (NSP1) and NSP2, which in legumes (Fabaceae) are essential for rhizobium root nodule formation, are indispensable for SL biosynthesis under nonsymbiotic conditions.
Legumes can establish an endosymbiosis with nitrogen-fixing rhizobium bacteria. To host rhizobium, a novel lateral root organ, the root nodule, is formed in response to specific lipo-chito-oligosaccharides secreted by the bacterium. These signals, named nodulation (Nod) factors, show strong resemblance to lipo-chito-oligosaccharides produced by mycorrhizal fungi (Maillet et al., 2011). In legumes, rhizobium Nod factors can trigger cell divisions in the root cortex, resulting in the formation of a nodule primordium. Furthermore, Nod factors are essential for intracellular infection by rhizobium. The Nod factor-signaling cascade has been genetically dissected (reviewed by Kouchi et al., 2010). These studies showed that several signaling components have been recruited from the network that are also essential for endomycorrhizal symbiosis. Genes that are essential for mycorrhizal as well as rhizobium Nod factor-induced signaling form the so-called common symbiotic signaling pathway and comprise a plasma membrane receptor kinase (DMI2 in Medicago truncatula and SYMRK in Lotus japonicus); several components in the nuclear envelope, including a cation–ion channel (M. truncatula DMI1 and L. japonicus CASTOR and POLLUX); subunits of nuclear pores (L. japonicus NUP85 and NUP133); and a nuclear localized calcium calmodulin-dependent kinase (CCaMK) (M. truncatula DMI3 and L. japonicus CCaMK) (reviewed by Kouchi et al., 2010). Mycorrhizae and rhizobium induced signaling bifurcates downstream of CCaMK, possibly because of differences in the nature of the calcium signal (Kosuta et al., 2008).
Nod factor signaling also requires several transcription factors, among which are the GRAS-type proteins NSP1 and NSP2 (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006). Both of these transcription factors are essential for nearly all rhizobium-induced symbiotic responses, including Nod factor-induced early nodulin gene expression, nodule organogenesis, and nodule functioning (Catoira et al., 2000; Oldroyd and Long, 2003; Mitra et al., 2004b; Heckmann et al., 2006). Biochemical studies indicate that NSP1 and NSP2 form a protein complex that binds to a specific DNA element present in the promoter of some early nodulin genes, such as ENOD11 (Hirsch et al., 2009). This suggests that NSP1 and NSP2 can function as Nod factor-responsive transcription factors upon heterodimerization (Smit et al., 2005; Hirsch et al., 2009). NSP1 and NSP2, which are essential for rhizobium Nod factor-induced signaling, are positioned downstream of CCaMK. Although both transcription factors are not essential for mycorrhizal symbiosis, it was recently found that an NSP2-dependent signaling pathway facilitates mycorrhizal root colonization (Maillet et al., 2011). This suggests that NSP2 does not function exclusively in rhizobium Nod factor signaling, a hypothesis that is supported by the presence of orthologous NSP genes in nonlegume plant species (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006). Here, we aim to characterize this novel function because it will provide insight into how these GRAS-type transcription factors have been recruited during nodule evolution.
GRAS-type transcription factors can be grouped in at least eight different classes, which are largely conserved in higher plants (Tian et al., 2004; Lee et al., 2008). For example, potential orthologs of NSP1 (class III) and NSP2 (class VII) can be found in many higher plant species, including rice (Oryza sativa) (Os 03g29480/Os NSP1 and Os 03g15680/Os NSP2) and Arabidopsis thaliana (At3g13840 and At4g08250) (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006). Arabidopsis is unable to establish mycorrhizal symbiosis, which suggests a more generic function for both transcription factors. Notably, NSP1 and NSP2 are functionally conserved in higher plants as demonstrated in trans-complementation studies of legume nsp1 and nsp2 knockout mutants with nonlegume NSP1 and NSP2 homologs (Heckmann et al., 2006; Yokota et al., 2010). This supports the idea that NSP1 and NSP2 fulfill a conserved function in plant growth and development.
We studied the genetic network controlled by NSP1 and NSP2 under nonsymbiotic conditions and provide evidence that both transcription factors are indispensable for SL biosynthesis in legumes as well as nonlegumes. M. truncatula nsp1 mutants, nsp1 nsp2 double knockout mutants, and rice Os nsp1 Os nsp2 double knockdown lines hardly produce SLs. By analyzing the root transcriptome in M. truncatula, we found that the effect on SL production correlates with a strongly reduced DWARF27 (Mt D27) expression. Likewise, rice Os nsp1 Os nsp2 double RNA interference (RNAi) knockdown lines have reduced DWARF27 (Os D27) expression. This underlines a conserved function of NSP1 and NSP2 in regulating SL biosynthesis.
RESULTS
M. truncatula NSP1 and NSP2 Regulate Genes in the Carotenoid Biosynthetic Pathway
To identify genes that are directly or indirectly activated by NSP1 and NSP2 under nonsymbiotic conditions, we analyzed the transcriptome of M. truncatula nsp1 and nsp2 knockout mutants. NSP1 and NSP2 are expressed mainly in root and nodules (Benedito et al., 2008). Therefore, we conducted microarray studies on 7-d-old roots and compared the nsp1-1 and nsp2-2 mutants with the wild-type M. truncatula (Jemalong A17) grown on minimal medium without a nitrogen source (1.7 mM Pi). Expression values were obtained from three independent biological replicates for each of the two mutants against the wild type (National Center for Biotechnology Information Gene Expression Omnibus accession no. GSE26548). Because NSP1 and NSP2 can act as heterodimers (Hirsch et al., 2009), we searched for genes with decreased expression (at least twofold) in both mutants. In total, 42 probe sets (representing 39 genes) fulfill this criterion (see Supplemental Table 1 online). The expression of these genes was subsequently studied by quantitative (q) RT-PCR in an independent experiment that now also included the M. truncatula nsp1 nsp2 double mutant. Reduced expression in roots of both nsp mutants could be confirmed for 16 genes. These genes were also downregulated in the double mutant (Table 1).
Table 1.
Genes Downregulated in Root Tissue of M. truncatula nsp1, nsp2, and nsp1 nsp2
| Fold Change Microarray |
Fold Change qRT-PCR |
|||||
| Gene | Gene ID | nsp1 | nsp2 | nsp1 | nsp2 | nsp1 nsp2 |
| DWARF27 | – | −3.2 | −3.4 | −6.1 | −8.0 | −7.4 |
| Carotenoid isomerase | – | −2.1 | −2.2 | −2.2 | −2.5 | −3.5 |
| MAX1 | Medtr3g104560 | −2.5 | −2.3 | −1.9 | −2.0 | −1.6 |
| 9-cis-epoxycarotenoid dioxygenase 4 | – | −2.0 | −3.0 | −2.6 | −2.4 | −3.2 |
| 1-Cys peroxiredoxin | – | −3.6 | −3.7 | −2.3 | −1.9 | −9.2 |
| Abnormal gametophytes | Medtr3g014420 | −3.4 | −2.8 | −3.2 | −2.5 | −2.5 |
| Ala–tRNA ligase | Medtr4g074670 | −3.3 | −3.4 | −1.9 | −1.7 | −4.4 |
| Allyl alcohol dehydrogenase | – | −2.1 | −2.0 | −18.2 | −14.9 | −23.8 |
| C2H2-type Zinc finger protein 6 | Medtr7g082260 | −7.8 | −10.1 | −5.0 | −5.4 | −29.4 |
| Cytokinin-specific binding protein | Medtr3g055120 | −2.4 | −2.5 | −1.9 | −1.7 | −2.1 |
| Dirigent-like protein | Medtr4g122110 | −6.1 | −5.4 | −4.4 | −3.5 | −5.3 |
| Man-6-phosphate isomerase | Medtr3g104560 | −2.5 | −2.3 | −1,7 | −1.8 | −1.6 |
| Nudix hydrolase | Medtr5g078020 | −3.5 | −4.5 | −1.5 | −3.4 | −15.6 |
| Seed maturation protein LEA 4 | Medtr7g093160 | −3.2 | −3.0 | −1.2 | −1.2 | −11.1 |
| Legume-specific protein | Medtr5g061550 | −7.7 | −7.7 | −1.9 | −1.8 | −10.5 |
| Legume-specific protein | – | −4.6 | −7.8 | −1.5 | −3.0 | −2.5 |
Plants were grown in the absence of a nitrogen source. Gene expression was analyzed using microarray analysis and qRT-PCR. Gene ID is based on M. truncatula genome annotation (IMGAG MT3.5).
To obtain insight in the biological function of the downregulated genes, extended sequences were retrieved from the M. truncatula genome data (IMGAG genome annotation version MT3.5) and were compared with homologous genes in other plant species. Two genes seemed to be legume specific, whereas the remaining 14 genes have clear homologs in nonlegume species (Table 1). Four of these encode a protein that catalyzes a conversion in the carotenoid biosynthetic pathway (a carotenoid isomerase homolog) or pathways leading from carotenoids to the plant hormones SL and abscisic acid (a MAX1 homolog, a 9-cis-epoxycarotenoid dioxygenase homologous to Arabidopsis NCED4 and an iron-containing protein with high sequence similarity to rice DWARF27 [Os D27]) (Beveridge and Kyozuka, 2010; Xie et al., 2010). In particular, the expression of the DWARF27 homologous gene was strongly affected (~90% reduced expression) in roots of both nsp1 and nsp2 mutants and in the double mutant (Figure 1A). Subsequent searches in the M. truncatula genome and EST resources revealed that M. truncatula has only a single DWARF27 homologous gene that was covered by two probes on the M. truncatula GeneChip (see Supplemental Table 1 online). The encoded M. truncatula protein displays 54% identity and 70% similarity with rice D27 (Figure 2A). Subsequent phylogenetic analysis revealed that the M. truncatula protein and rice D27 belong to the same orthology group (Figure 2B; see Supplemental Data Set 1 online). Common ancestry is further supported by a largely conserved exon–intron structure of both genes (Figure 2C). Based on these findings, we named the corresponding M. truncatula gene Mt DWARF27 (Mt D27), although it must be noted that we did not provide a formal proof for orthology because functional trans-complementation experiments of the rice d27 knockout mutant have not been done.
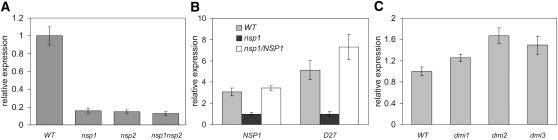
Figure 1.
M. truncatula D27 Expression Is NSP1 and NSP2 dependent.
(A) Relative expression level of D27 in nitrogen-starved roots of M. truncatula wild-type (WT), nsp1, nsp2, and nsp1 nsp2 mutant plants as determined by qRT-PCR.
(B) Expression of D27 can be rescued in the roots of nsp1 mutant complemented with pMt-NSP1:Mt-NSP1.
(C) Expression of D27 in nitrogen-starved roots of the M. truncatula common symbiotic signaling pathway mutants dmi1, dmi2, and dmi3 determined by qRT-PCR. Data are means ± sd.
Figure 2.
M. truncatula D27 Is Orthologous to Rice D27.
(A) Protein alignment of M. truncatula D27 and rice D27 (70% homology/54% identity).
(B) Maximum-likelihood phylogenetic analysis of rice D27 homologous proteins of M. truncatula (M. truncatula), soybean (Glycine max), poplar (Populus trichocarpa), cassava (Manihot esculenta), wild strawberry (Fragaria vesca), maize (Zea mays), and rice. Branch lengths are proportional to the number of amino acid substitutions per site. Branch support was obtained from 100 bootstrap repetitions.
(C) Largely conserved exon–intron structure in M. truncatula and rice D27. The lengths of exons 2 to 6 are conserved between M. truncatula D27 and rice D27 (aligned with dashed lines). Black boxes, exons; line, introns. GenBank accession number JN629088.
To confirm that downregulation of M. truncatula D27 was the result of the knockout of NSP genes and not of a background mutation present in both mutants, we complemented the M. truncatula nsp1-1 mutant with a pMt-NSP1:Mt-NSP1 construct and subsequently determined the D27 expression in transgenic roots. This revealed that the expression of D27 did indeed depend on the presence of a functional NSP1 protein (Figure 1B).
Next, we determined whether expression of D27 under nonsymbiotic conditions depends on components of the common symbiotic signaling pathway that is essential for rhizobium Nod factor signaling and mycorrhizal signaling. D27 transcript levels were quantified in roots of M. truncatula dmi1, dmi2, and dmi3 mutants. D27 expression was not reduced in these mutants (Figure 1C). Therefore, we conclude that the regulation of D27 expression in the absence of rhizobium or a mycorrhizal fungus does not depend on the common symbiotic signaling pathway but does depend on NSP1 and NSP2.
SL Biosynthesis in M. truncatula Requires NSP1 and NSP2
In rice, D27 is essential for SL biosynthesis (Lin et al., 2009). Because the expression of D27 in M. truncatula is nearly undetectable in roots of nsp1, nsp2, and nsp1 nsp2 mutants grown in the absence of a nitrogen source, we determined whether SL biosynthesis is affected in these mutants. To this end, we characterized the SLs produced by M. truncatula. It has been reported that the amount of SLs in root exudate strongly increases upon phosphate starvation (Yoneyama et al., 2007; López-Ráez et al., 2008; Umehara et al., 2008; Lin et al., 2009). Therefore, we conducted two experiments and grew plants in one-half-strength Hoagland medium under either low and normal nitrogen conditions (0.14 and 2.8 mM NH4NO3, respectively) or no and normal phosphate conditions (0.2 mM Pi). First, we determined the expression levels of NSP1, NSP2, and D27 in root systems grown under these conditions. Expression levels of NSP1 and NSP2 were not affected by the nutrient levels in the medium. By contrast, D27 already displayed a strong (>30-fold) upregulation after 1 d of phosphate starvation, whereas nitrogen starvation had little to no effect on D27 expression (Figures 3A and 3B). Next, root exudates from phosphate-starved and -nonstarved plants and from nitrogen-starved and -nonstarved plants were collected and purified using C18 column chromatography. In all root exudates, two peaks were detected that could correspond to SLs. The major peak had a retention time and transitions corresponding to didehydro-orobanchol (see Supplemental Figure 1A online). The nature of this SL was confirmed by coinjection and comparison of that of the tandem mass spectrometry (MS/MS) spectrum with the MS/MS spectrum obtained from one of the didehydro-orobanchol isomers found in tomato (Solanum lycopersicum) (López-Ráez et al., 2008) (see Supplemental Figure 1B online). The minor peak was identified as orobanchol based on the retention time and transitions (see Supplemental Figure 1C online), and this was confirmed by comparing this MS/MS spectrum with that of an orobanchol standard (see Supplemental Figure 1D online). Taken together, we conclude that M. truncatula produces two different SLs: didehydro-orobanchol and orobanchol. Although both SLs could be detected in the exudate of nonphosphate-starved plants (with or without nitrogen), amounts increased ~10-fold upon phosphate starvation (Figures 3C and 3D). This indicates that, as with many other species, SL secretion, and likely biosynthesis, by M. truncatula roots is induced by phosphate-limiting conditions, whereas the nitrogen status of the plant has no effect. This increase in SL secretion under phosphate starvation correlates with the transcriptional upregulation of D27.
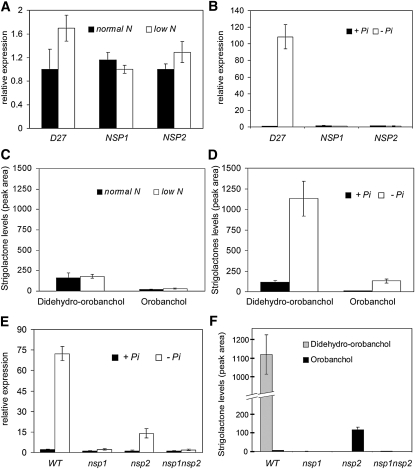
Figure 3.
SL Biosynthesis Correlates with M. truncatula D27 Expression.
(A) Relative expression levels as detected by qRT-PCR of D27, NSP1, and NSP2 in M. truncatula wild-type roots under low (0.14 mM for 24 h) and sufficient (2.8 mM) NH4NO3 growth conditions. Data are means ± sd.
(B) Relative expression levels as detected by qRT-PCR of D27, NSP1, and NSP2 in M. truncatula wild-type roots under no (for 24 h) and sufficient (0.2 mM Pi) phosphate growth conditions. Lowest expressed gene is set to 1. Data are means ± sd.
(C) Levels of the SLs didehydro-orobanchol and orobanchol in M. truncatula wild-type root exudates as detected by MRM-LC-MS/MS under low (0.14 mM for 7 d) and sufficient (2.8 mM) NH4NO3 growth conditions. Data are means ± se.
(D) Levels of the SLs didehydro-orobanchol and orobanchol in M. truncatula wild-type root exudates as detected by MRM-LC-MS/MS under no (for 7 d) and sufficient (0.2 mM Pi) phosphate conditions. Data are means ± se.
(E) Relative expression levels as detected by qRT-PCR of Mt D27 in roots of M. truncatula wild-type (WT), nsp1, nsp2, and nsp1 nsp2 mutant plants under no (for 24 h) and sufficient (0.2 mM Pi) phosphate growth conditions. Data are means ± sd.
(F) Analysis of the SLs didehydro-orobanchol and orobanchol in root exudates of phosphate-starved (0.14 mM for 7 d) M. truncatula wild-type, nsp1, nsp2, and nsp1 nsp2 mutant plants (n = 3). Data are means ± se.
Subsequently, the M. truncatula nsp1, nsp2, and nsp1 nsp2 mutants as well as an independent set of the wild-type plants were grown under phosphate starvation. Profiling of D27 expression under these growth conditions revealed that D27 expression was repressed in all three mutants. In nsp1 and nsp1 nsp2 mutant plants, D27 expression was nearly absent, whereas in roots of the nsp2 mutant, D27 expression was reduced to ~20% of wild-type levels (Figure 3E). The finding that D27 expression in phosphate-starved roots is dependent on NSP1 and NSP2 is in line with results obtained in the microarray study, although it also shows that, under phosphate limitation, transcriptional activation of D27 can occur partly independent of NSP2. Analyses of root exudates by multiple reaction monitoring–liquid chromatography (MRM-LC)-MS/MS showed that no SLs are secreted by the nsp1 mutant or the nsp1 nsp2 double mutant (Figure 3F). By contrast, in root exudates of the nsp2 mutant, orobanchol accumulated in 10-fold higher levels than those observed in exudate of the wild-type plants (Figure 2F). Interestingly, didehydro-orobanchol was absent in the root exudate of this mutant, suggesting that NSP2 controls a specific step in SL conversion. To rule out the possibility that NSP1 and NSP2 are merely affected in secretion of certain SLs and not in biosynthesis, we also investigated the SL content of root extracts. This revealed similar results as observed in root exudates (see Supplemental Figure 2 online). Taken together, these findings demonstrate that genes controlled by NSP1 and NSP2 are essential in the SL biosynthetic pathway and include D27.
NSP1/NSP2 Control SL Biosynthesis in Rice
Because NSP1 and NSP2 orthologous genes are present in nonlegume species, we investigated whether the control of SL biosynthesis is a conserved function of these proteins. The role of DWARF27 in SL biosynthesis is characterized in rice (Lin et al., 2009; Wang and Li, 2011), and rice, as a monocot, is phylogenetically distinct from M. truncatula. Therefore, we decided to focus on this species. qRT-PCR analysis on root RNA showed that transcripts of both NSP genes are present, although the level of NSP2 expression was close to the detection limit (Figure 4A). This is in line with a previous study that reported that the expression levels of rice NSP1 and NSP2 are extremely low (Yokota et al., 2010).
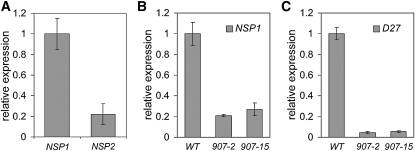
Figure 4.
Expression Analysis of NSP1, NSP2, and D27 in Rice Determined by qRT-PCR.
(A) NSP1 and NSP2 expression in roots of wild-type rice (ZH11).
(B) Expression of NSP1 in roots of the rice nsp1 nsp2 double knockdown lines 907-2 and 907-15. WT, wild type.
(C) Expression of D27 in roots of the rice nsp1 nsp2 double knockdown lines 907-2 and 907-15. Data are means ± sd.
To study whether NSP1 and NSP2 are essential for SL biosynthesis in rice, double RNAi knockdown lines were created by Agrobacterium tumefaciens–mediated transformation. In total, 14 lines were obtained, and two of these showed severe knockdown (>90%) of NSP1 in roots (Figure 4B). Because NSP2 is expressed at a very low level, the knockdown level of this gene could not be determined reliably. The expression of D27 in these knockdown lines is reduced >90% when compared with the wild type (Figure 4C), showing that in rice, as in M. truncatula, the expression of D27 relies on the presence of NSP1 and possibly NSP2.
To determine whether downregulation of rice D27 expression correlates with reduced SL biosynthesis, root exudates of nsp1 nsp2 RNAi lines were analyzed using MRM-LC-MS/MS analysis. In rice, 2′-epi-5-deoxystrigol and orobanchol are the major SLs (Umehara et al., 2008), and both could be detected in root exudates of the wild-type rice plants (Figure 5A). In the nsp1 nsp2 RNAi lines, the amounts of 2′-epi-5-deoxystrigol and orobanchol in the root exudates were approximately five- and eightfold lower, respectively, than in the wild-type plants (Figure 5A). This reduction was also visualized in a Striga hermonthica seed germination bioassay. Seeds of this parasitic plant are known to respond in a semiquantitative way to SLs (Matusova et al., 2005). The germination-inducing activity of the rice nsp1 nsp2 RNAi root exudates was four- to fivefold reduced compared with exudates from the wild-type plants (Figure 5B). Therefore, we conclude that NSP1 and possibly NSP2 are essential for SL biosynthesis in rice as well.
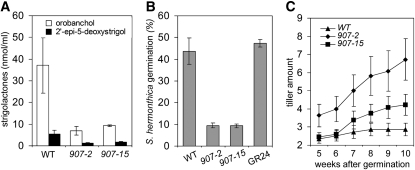
Figure 5.
Tillering Phenotype and SL Concentrations in Root Exudates of Wild-Type Rice (ZH11) and Two Independent Rice nsp1 nsp2 Knockdown Lines.
(A) Analysis of 2′-epi-5-deoxystrigol and orobanchol content in root exudates of phosphate-starved wild-type (WT) rice (ZH11) and nsp1 nsp2 knockdown lines 907-2 and 907-15 (n = 3).
(B) Germination of S. hermonthica seeds induced by root exudates of the wild type (ZH11) and nsp1 nsp2 knockdown lines 907-2 and 907-15 (n = 3). Data are means ± se
(C) Quantification of tillering in rice. Tillers of wild-type rice (ZH11) and nsp1 nsp2 double knockdown lines (907-2 and 907-15) were quantified 5 to 10 weeks after planting (n = 15). Data are means ± se.
NSP1/NSP2 Control Lateral Shoot Growth in Rice but Not in M. truncatula
In rice, a d27 knockout mutation causes increased tillering in combination with reduced plant height (Lin et al., 2009). Because the absence of NSP1–NSP2 expression in rice and M. truncatula resulted in markedly reduced SL biosynthesis in root tissue, we tested whether these plant mutants also display a more branched shoot phenotype. In rice, the amount of tillers was quantified at different time points from 5 to 10 weeks after germination. The nsp1 nsp2 RNAi lines had an increased number of tillers when compared with the wild-type rice (Figure 5C). However, these knockdown lines did not display obvious differences in plant height, as reported for the rice d27 knockout mutant (Lin et al., 2009). Therefore, although the increased tillering is in line with the phenotype of the rice d27 knockout mutant, plant height seems less critical because residual levels of D27 expression and SL biosynthesis seem to be sufficient to support normal shoot growth. Next, we studied shoot branching in the M. truncatula nsp1 nsp2 double mutant and compared it with the wild type. Wild-type M. truncatula formed three to four branches at the first four knots and well-developed secondary branches during 10 weeks of plant growth. We did not find an increased amount of branches in the nsp1 nsp2 double mutant. Therefore, we conclude that severe reduction in SL concentration in roots of the M. truncatula nsp1 nsp2 mutant plants has no obvious effect on shoot architecture.
M. truncatula nsp1 nsp2 Double Mutant Displays Reduced Mycorrhizal Infection
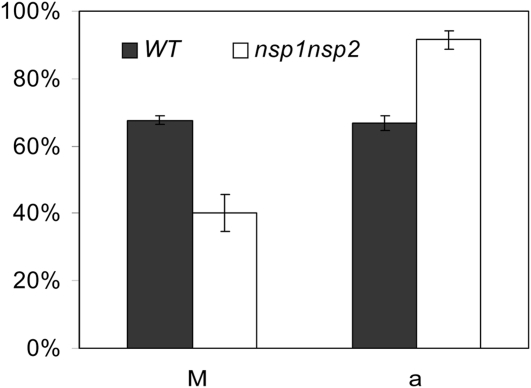
Because NSP1 and NSP2 control biosynthesis of SLs, which are important ex planta stimuli for branching of mycorrhizal hyphae, it seems probable that these GRAS-type transcription factors also have a function in the interaction of plants with arbuscular mycorrhizae. A recent study in M. truncatula indeed revealed that the nsp2 mutant displays a ~40% reduction in mycorrhizal colonization (Maillet et al., 2011). Our study revealed that the nsp2 knockout mutant still produces orobanchol. We investigated the mycorrhizal phenotype of the nsp1 nsp2 double mutant that does not produce detectable amounts of SLs. Mutant and wild-type plants were grown under low phosphate conditions and inoculated with Glomus intraradices. Six weeks after inoculation, the mycorrhizal status of the root systems was evaluated. It was found that nsp1 nsp2 can be mycorrhized effectively by G. intraradices. However, the fraction of the root system that was mycorrhized was significantly lower in the mutant than in the wild-type plants (40.1% versus 67.6%, respectively) (Figure 6). When quantifying the number of arbuscules within mycorrhized root sections, we noted that such segments of the nsp1 nsp2 mutant contained more arbuscules when compared with the wild type (91.4% versus 66.7%, respectively) (Figure 6). These data suggest that absence of SLs affects the initial infection of G. intraradices, whereas intracellular arbuscule formation is not hampered.
Figure 6.
Mycorrhizal Colonization of M. truncatula Wild Type and nsp1 nsp2 Mtnsp1 Mtnsp2 Double Mutant. (P < 0.05).
Data are means ± se. a, presence of arbuscules in the mycorrhized root segments; M, intensity of mycorrhization in total root system.
DISCUSSION
Here, we show that the GRAS-type transcriptional regulators NSP1 and NSP2 regulate SL biosynthesis in M. truncatula and rice. These two species represent distinct phylogenetic lineages that split ~150 million years ago (Moore et al., 2007; Smith et al., 2010). Therefore, we conclude that the regulation of SL biosynthesis by NSP1 and NSP2 is an ancestral function conserved in higher plants. During evolution of legumes, these two transcriptional regulators have been recruited to play an essential role in rhizobium root nodule symbiosis. NSP1 and NSP2 are single-copy genes in legumes, which implies that single proteins fulfill both functions in these species.
The absence of SL biosynthesis in nsp1 nsp2 mutant backgrounds correlates with a reduced expression of several genes that encode enzymes of the carotenoid and SL biosynthetic pathways, including D27. The precise biochemical function of the plastid-localized, iron-containing protein D27 remains to be elucidated, but it was shown to be essential for SL biosynthesis in rice (Lin et al., 2009; Wang and Li, 2011). Our study in M. truncatula revealed that the transcriptional regulation of the SL biosynthetic enzyme D27 is tightly regulated by the nutrient status of the plant. Phosphate starvation in particular triggers a dramatic upregulation of D27 transcription. Because NSP1 and NSP2 expression is not affected or is only very mildly affected by the nutrient status of the plant (Barbulova et al., 2007), we conclude that the activity of both transcription factors is controlled at the protein level, similar to their hypothesized symbiotic functioning in rhizobium Nod factor-induced signaling in legumes (Geurts et al., 2005).
We showed that M. truncatula produces two different SLs (orobanchol and didehydro-orobanchol), similar to its close relative red clover (Trifolium pratense) (Yokota et al., 1998; Xie et al., 2010). Intriguingly, M. truncatula SL biosynthesis is differentially regulated by NSP1 and NSP2. In the nsp1 mutant background, no detectable amounts of SLs are produced, but the nsp2 mutant specifically secretes orobanchol in concentrations higher than those found in the wild-type plants (Figure 7). This shows that in M. truncatula, the markedly decreased expression level of D27 in an nsp2 mutant background is still sufficient to control orobanchol biosynthesis. Because the precise biochemical pathway of different SLs has not yet been elucidated, the finding that the M. truncatula nsp2 mutant accumulates orobanchol rather than didehydro-orobanchol, as found in the wild-type M. truncatula plants, may provide a tool to identify key enzymes in the biosynthesis of different SLs. It is assumed that 5-deoxystrigol is the first genuine SL and that all other SLs are derived from it (Matusova et al., 2005; Rani et al., 2008). Subsequently, didehydro-orobanchol is derived from orobanchol in a three-step process: a homoallylic hydroxylation of orobanchol to hydroxyorobanchol, an oxidation or dehydration to oxo-orobanchol, and a migration of the methyl group leading to didehydro-orobanchol (Matusova et al., 2005; Beveridge and Kyozuka, 2010; Xie et al., 2010). The enzymes involved in these steps might be under the transcriptional regulation of NSP2 (Figure 7). A preliminary analysis of M. truncatula genes that are differentially downregulated in nsp2 mutant roots but not in the wild type or the nsp1 mutant revealed a subset of such genes (see Supplemental Table 2 online) that included several candidates that encode enzymes that could be involved in this process. Functional analysis of these genes could provide access to key enzymes in SL conversion.
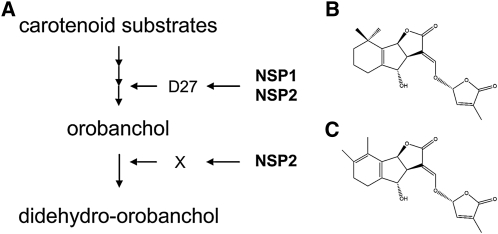
Figure 7.
Model for SL Biosynthesis and Structure of the Two Main SLs in M. truncatula.
(A) Schematic representation of the SL biosynthesis pathway in M. truncatula controlled by the GRAS-type transcription factors NSP1 and NSP2. Orobanchol is synthesized in a multistep process from carotenoid substrates and requires D27, which is under transcriptional control of NSP1–NSP2. In this step, NSP2 is partly redundant. The conversion of orobanchol in didehydro-orobanchol requires unknown enzyme X, which is under direct or indirect control of NSP2. WT, wild type.
(B) Structural formula of orobanchol.
(C) Putative structure formula of didehydro-orobanchol.
Besides D27, two different carotenoid cleavage dioxygenase (CCD) enzymes are essential for SL biosynthesis, specifically CCD7 and CCD8. However, the precise relation between both CCDs and D27 remains to be elucidated (Xie et al., 2010). M. truncatula homologs of both CCD genes are represented on the gene chip (CCD7/Medtr7g045370.1/probe ID Mtr.32038.1.S1_at and CCD8/Medtr3g109610.1/Mtr.1606.1.S1_at). Interestingly, the absence of functional NSP1 or NSP2 does not affect the expression of these M. truncatula CCD7 and CCD8 homologous genes. Assuming that both CCD genes are also essential for SL biosynthesis in M. truncatula, this suggests that key enzymes essential for this biosynthetic pathway are controlled by different transcriptional networks.
In legumes, NSP1 and NSP2 are key components in the rhizobium Nod factor-induced signaling pathway, and a knockout mutation in either of these genes impairs gene induction by Nod factors, including ENOD11 in M. truncatula (Catoira et al., 2000; Oldroyd and Long, 2003; Mitra et al., 2004a). ENOD11 is a direct target of NSP1 because this transcription factor binds to the conserved cis-regulatory binding element AATTT (Hirsch et al., 2009). This motif is named the nodulation-responsive element (NRE), and NSP1 binds to this element in an NSP2-dependent manner. ENOD11 has two NREs in in the −500-bp promoter region. Interestingly, NRE-like motifs are abundantly present in putative promoter regions of rice and M. truncatula D27 (see Supplemental Figures 3A and 3B online). In the M. truncatula D27 promoter, nine NRE-like motifs can be found in ~1000 regions: seven times in the forward orientation and two times in the reverse orientation, respectively (see Supplemental Figure 3A online). This raises the question of whether NSP1 also can bind directly to the D27 promoter. Preliminary studies using in vitro electrophoretic mobility shift assays suggest that this is the case (see Supplemental Figures 3C to 3E online), indicating that M. truncatula D27 indeed could be a direct target of NSP1. However, such studies need to be confirmed by complementary approaches.
The finding that M. truncatula D27 expression is under (direct or indirect) control of NSP1/NSP2 leads to a puzzling difference in regulation when compared with functioning of both GRAS-type transcription factors in the Nod factor signaling pathway. In the latter case, NSP1 needs to be activated upon Nod factor signaling. By contrast, none of the components of this signaling pathway, not even the kinase CCaMK directly active upstream of NSP1 and NSP2, are required for D27 expression. The finding that both NSPs can function independent of CCaMK is in line with the fact that some plant species that cannot establish a mycorrhizal symbiosis (e.g., Arabidopsis) have lost many of the components in the common symbiotic signaling pathway, including CCaMK, but have maintained NSP1 and NSP2 orthologous genes (Kaló et al., 2005; Smit et al., 2005; Zhu et al., 2006). The recruitment of a conserved transcription factor into the signaling pathway triggered by rhizobium Nod factors, as occurred in legumes, is therefore intriguing. It suggests that in the event of Nod factor signaling, the NSP-controlled transcriptional regulation is activated in a different manner. It remains to be elucidated how this difference is created at a molecular level. We hypothesize that this is achieved by modification of the NSP1 and NSP2 protein complexes upon CCaMK activity. This might be a direct modification or may involve a different component that affects the binding affinity of the transcription factor complex. However, the occurrence of NRE-like cis regulatory elements in the M. truncatula D27 promoter region suggests that both mechanisms could have parts in common.
SLs are stimuli for mycorrhizal hyphae, and SL biosynthesis knockout mutants in tomato (ort1) and pea (Pisum sativum; ccd8) display a reduction in mycorrhizal root colonization (Gomez-Roldan et al., 2008; Koltai et al., 2010). The M. truncatula nsp1 nsp2 double mutant has a similar reduction in mycorrhizal root infection (Figure 6). We also investigated arbuscule formation and showed that the nsp1 nsp2 mutant efficiently forms well-developed arbuscules. This finding suggests that SLs are not essential for arbuscule formation. Therefore, we hypothesize that SLs stimulate root colonization exclusively ex planta, in line with this are the mycorrhizae symbiotic phenotypes of nsp mutants that can be fully explained by the role of both transcription factors in SL biosynthesis.
Plant mutants affected in SL biosynthesis or signaling are known for a distinctive shoot branching phenotype. However, in M. truncatula nsp1 and nsp2, we did not observe such phenotype. This is in contrast with rice nsp knockdown lines. This suggests that M. truncatula is possibly not ideal for detecting shoot branching phenotypes, likely because it does not display strong apical dominance. Because shoot architecture can be influenced by environmental conditions, a more detailed study on bud outgrowth, stem length, and/or polar auxin transport may reveal weaker phenotypes in SL biosynthesis mutants in M. truncatula. Additionally, the D27 dependent shoot architecture in rice may be enhanced by the expression profile of the corresponding gene. In rice, D27 is transcribed in shoot bases, culms, panicles, and axillary buds (Lin et al., 2009), whereas in M. truncatula, D27 is expressed exclusively in roots (see Supplemental Figure 4 online) (Benedito et al., 2008).
During the past decade, genes encoding enzymes essential for SL biosynthesis have been elucidated. To our knowledge, we have identified the first two transcription factors, NSP1 and NSP2, which are key regulators of SL biosynthesis. SL biosynthesis is highly regulated by environmental and other conditions. Therefore, NSP1 and NSP2 will be important tools in future studies on the molecular mechanisms by which environmental sensing is translated into regulation of SL biosynthesis.
METHODS
Plant Materials, Growth Conditions, and Transformation
Medicago truncatula was grown in a growth chamber at 20°C and 16-h-day/8-h-night regime. Jemalong A17, nsp1-1 (B85) (Catoira et al., 2000; Smit et al., 2005), and nsp2-2 (0 to 4) (Oldroyd and Long, 2003; Kaló et al., 2005) were used as the wild type, nsp1, and nsp2, respectively. The nsp1 nsp2 double mutant was obtained by pollinating nsp1-1 plants with nsp2-2 pollen. nsp1 nsp2 homozygote plants were selected by PCR-based genotyping of F2 individuals.
M. truncatula plants used for gene expression analysis were grown vertically on Fåhraeus medium plates without nitrate (Fåhraeus, 1957). RNA was isolated from the Nod factor-susceptible zone of 7-d-old roots samples snap-frozen in liquid N2. Total RNA was extracted using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek), combined with Qiagen RNase-Free DNase Set for on-column DNase treatment.
Agrobacterium rhizogenes–based root transformation of M. truncatula was conducted according to Limpens et al. (2004). Plants with transgenic roots were selected based on DsRED1 fluorescence.
Mycorrhization studies were conducted with fine sand and calcined clay mixtures, which contain spores of Glomus intraradices. Six weeks after transplantation, the whole-root systems were harvested and evaluated according to Trouvelot et al. (1986).
Rice (Oryza sativa subsp japonica cv Zhonghua 11) was used as the wild-type rice. Rice plants were grown in a greenhouse at 28°C in a 16-h-day/8-h-night regime. For tillering assays, rice plants were grown on fine sand with one-half-strength full-nutrient Hoagland solution (Hoagland and Arnon, 1950) and watered once a week. Rice transformations were conducted using Agrobacterium tumefaciens strain AGL1 according to Toki et al. (2006) and subsequent hygromycin B selection.
Constructs and Plasmids
For complementation of the M. truncatula nsp1 mutant, we used the pMt-NSP1:Mt-NSP1 construct as generated by Smit et al. (2005).
Based on the principle that the RNAi functions through an ~20-nucleotide fragment, a chimeric RNAi construct was made by fusing two fragments from Os NSP1 (295 bp) and Os NSP2 (488 bp) in a single hairpin. First, fragments of OsNSP1 and Os NSP2 were PCR amplified from rice genomic DNA with primer pairs Os NSP1-F/OsNSP-mr and Os NSP-mf/Os NSP2-R, respectively (see Supplemental Table 3 online). Subsequently, both fragments were fused by overlap PCR with primer pair Os NSP1-F and Os NSP2-R (see Supplemental Table 3 online). This chimeric fragment then was subcloned into pENTRL1L2, resulting in pENTRL1L2_OsNSP1-2i. Then, this construct was recombined into pHGWIWG2 (II)-RR-R1R2 to get the binary construct pHGWIWG2 (II)-RR-OsNSP1+2i.
Affymetrix GeneChip Oligoarray Hybridization, Scanning, and Quality Control
Total RNA (5 μg) was labeled using the Affymetrix One-Cycle Target Labeling Assay kit (Affymetrix). Labeled RNA samples were hybridized on Affymetrix M. truncatula Genome arrays (product 900735) and washed, stained, and scanned on an Affymetrix GeneChip 3000 7G scanner. The detailed protocols that we used for array handling can be found in the Genechip Expression Analysis Technical Manual, section 2, chapter 2 (Affymetrix; product 701028, revision 5). Packages from the Bioconductor project (Gentleman et al., 2004) were used to analyze the array data. Various advanced quality metrics, diagnostic plots, pseudoimages, and classification methods were used to determine the quality of the arrays before statistical analysis (Heber and Sick, 2006, Brettschneider et al. 2008). In brief, the library “affy” was used to determine for each array the average background signal, percentage present calls, scale factor, and RNA degradation plots essentially as described by Alvord et al. (2007). All arrays passed the guidelines recommended by Affymetrix, as described in the Affymetrix Microarray Suite Users Guide, Version 5.0. In addition, the library “AffyPLM” was used to fit probe-level linear models that provide parameter estimates for probes and arrays on a probe-by-probe basis (Bolstad et al., 2005). Two-dimensional pseudoimages of the arrays based on probe-level quantities, namely the weights and residuals computed by fitPLM, were inspected for stains, scratches, and other artifacts. Even if present, all artifacts covered less than 5% of the area of the array. Moreover, numerical quality was assessed based on two distributions computed at the probe set level, the normalized unscaled se, and relative log expression (RLE). No outlier arrays were observed for all these quality control parameters; that is, all arrays were reasonably centered around the median normalized unscaled se of 1, and boxplots for RLE had a small spread and were centered at an RLE of 0. Array data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE26548.
Expression estimates were obtained by GeneChip Robust Multiarray Averaging analysis, using the empirical Bayes approach to adjust for background (Wu et al., 2004) in the Bioconductor library “gcrma.” Differentially expressed probe sets were identified using linear models, applying moderated t statistics that implemented empirical Bayes regularization of se values (library “limma”). The moderated Student’s t test statistic has the same interpretation as an ordinary Student’s t test statistic, except that the se values have been moderated across genes (i.e., shrunk to a common value) using a Bayesian model (Smyth, 2004). To adjust for both the degree of independence of variance relative to the degree of identity and the relationship between variance and signal intensity, the moderated t statistic was extended by a Bayesian hierarchical model to define an intensity-based moderated t statistic (Sartor et al., 2006). Intensity-based moderated t statistics improve the efficiency of the Empirical Bayes moderated t statistics and thereby achieve greater power while correctly estimating the true proportion of false positives. P values were corrected for multiple testing using a false discovery rate method proposed by Storey and Tibshirani (2003). Probe sets that satisfied the criterion of false discovery rate < 5% (q value < 0.05) were considered to be significantly regulated.
qRT-PCR Analysis
Total RNA (1 μg) was reverse transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad) following the supplier’s manual. Real-time PCR was set up in a 20-μL reaction system using the Eurogentec qPCR Core kit and the iQ5 Real-time PCR detecting system according to the manuals supplied by the manufacturers. Gene-specific primers were designed with Primer-3-Plus software (Untergasser et al., 2007) and are listed in Supplemental Table 3 online. For M. truncatula, ubiquitin (UBQ) and polypyrimidine tract binding protein were used as internal control, whereas for rice, UBQ was used (Lin et al., 2009).
Phylogenetic Analysis
Maximum-likelihood phylogenetic analysis was performed on rice D27 homologous proteins of M. truncatula, soybean (Glycine max), poplar (Populus trichocarpa), cassava (Manihot esculenta), wild strawberry (Fragaria vesca), maize (Zea mays), and rice . Protein sequences were aligned using Geneious Pro software package 5.4.6 (Biomatters) by applying Blosum62 matrix, a gap open penalty of 12, and a gap extension penalty of 3. Phylogenetic trees were reconstructed by maximum likelihood and 1000 bootstrap repetitions to evaluate statistical support of the branches. Rice Os08g0114100 was used as an outgroup because it is the closest paralog of rice D27.
Characterization and Quantification of SLs
For SL analysis, 15 1-week-old M. truncatula plants were transplanted (16 h of light, 23°C, 60% relative humidity) into an X-stream 20 aeroponic system (Nutriculture) operating with 5 L of modified one-half-strength Hoagland solution (Hoagland and Arnon, 1950; López-Ráez et al., 2008). Four weeks after transplanting, phosphate starvation was introduced by replacing the nutrient solution with one-half-strength Hoagland solution without phosphate. Twenty-four hours before exudate collection, the nutrient solution was refreshed to remove all accumulated SLs. For SL analysis in rice, plants were grown as previously described by Jamil et al. (2010).
M. truncatula exudates were purified and concentrated as previously described by López-Ráez et al. (2010) with some modifications. Five liters of root exudate was loaded onto a preequilibrated C18 column (Grace Pure C18-Fast 5000 mg/20 mL) per sample. Subsequently, columns were washed with 50 mL of deionized water and 50 mL of 30% acetone/water. SLs were eluted with 50 mL of 60% acetone/water. Rice exudates were purified and concentrated as previously described by Jamil et al. (2010). All exudates were collected within 3 h and stored at −20°C before further use. SLs were extracted from M. truncatula root material as previously described for tomato (Solanum lycopersicum) by López-Ráez et al. (2010).
Analysis of SLs was performed by comparing retention times and mass transitions with those of available SL standards (sorgolactone, strigol, 2′-epistrigol, orobanchol, 2′-epiorobanchol, 5-deoxystrigol, 2′-epi-5-deoxystrigol, solanacol, and orobanchyl acetate, sorgomol, 7-oxoorobanchol, 7-oxoorobanchyl acetate) using ultra-performance LC coupled to MS/MS, as previously described by Kohlen et al. (2011). Didehydro-orobanchol MS/MS fragmentation spectra of M. truncatula and tomato root exudates were obtained as previously described López-Ráez et al. (2008).
Root exudates obtained from different rice lines were assessed for germination stimulatory activity by germination bioassays with Striga hermonthica as previously described by Jamil et al. (2010).
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assay experiments were performed mainly as described previously (Kaufmann et al., 2005). The coding sequence of M. truncatula NSP1 was PCR amplified from a pool of cDNA using the following primers with NcoI (forward) and ClaI (reverse) restriction sites (see Supplemental Table 3 online). The amplified fragment was cloned into the pSPUTK, an in vitro transcription/translation vector (Stratagene). The protein was produced by in vitro transcription/translation with the TNT SP6 High-Yield Wheat Germ Protein Expression System following the manufacturer’s instructions (Promega). Promoter fragments of M. truncatula D27 and ENOD11 were PCR amplified from M. truncatula genomic DNA and cloned via TA-cloning into the pGEM-T vector (see Supplemental Table 3 online). Biotin-labeled pGEM-T–specific primers, directly flanking the cloning site, were used for generation of the DNA probes by PCR. The Mt NSP1 protein (2 μL of the in vitro reaction) was incubated with 40 fmol of biotin-labeled DNA probes in binding solution (1 mM EDTA, pH 8.0, 0.25 mg/mL BSA, 7 mM HEPES, pH 7.3, 0.7 mM DTT, 60 μg/mL salmon sperm DNA, 1.3 mM spermidine, 2.5% 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, and 8% glycerol) for 1 h on ice. DNA–protein complexes were analyzed on a 5% native PAGE (37.5:1 acrylamide:bisacrylamide). After electrophoresis, the gel was blotted to Amersham Hybond-N+ membrane, and the signal was detected using the chemiluminescent nucleic acid detection module (Pierce Chemical Co.) in a Genius:BOX gel documentation system (Westburg).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: M. truncatula nsp1 and nsp2 microarray data, GSE26548; and M. truncatula D27, JN629088. The following genes were used in qRT-PCR experiments: M. truncatula UBQ, BT053109; M. truncatula polypyrimidine tract binding protein, CT963079; M. truncatula NSP1, AJ972478; M. truncatula NSP2, AJ832138; rice NSP1, AC135559; rice NSP2, AC135206; rice D27, FJ641055; and rice UBQ, AC103891.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MRM-LC-MS/MS Analysis of M. truncatula Root Exudates.
Supplemental Figure 2. Amount of Didehydro-Orobanchol and Orobanchol in M. truncatula Root Extracts of the Wild Type (A17), nsp1, nsp2, and nsp1 nsp2 Mutants (n = 6).
Supplemental Figure 3. Putative NRE-Like cis Regulatory Binding Elements for NSP1 in the Promoter Region of M. truncatula D27 and Rice D27.
Supplemental Figure 4. Expression Analysis of D27 in M. truncatula Shoot, Stem, and Root Tissue Determined by qRT-PCR.
Supplemental Table 1. M. truncatula Probe Sets Representing Genes Downregulated in Root Tissue of the M. truncatula nsp1 and nsp2 Knockout Mutant.
Supplemental Table 2. M. truncatula Probe Sets Representing Genes Downregulated in Root Tissue of M. truncatula nsp2 Mutant versus the Wild Type and nsp1 Mutant Plants.
Supplemental Table 3. Primers Used in This Study.
Supplemental Data Set 1. D27 Homologs Coding and Protein Sequences of Different Species Used for Phylogenetic Reconstruction as Shown in Figure 2B.
Acknowledgments
This research was funded by the Chinese Academy of Sciences (CAS) and the Royal Netherlands Academy of Arts and Sciences (KNAW) (CAS-KNAW Joint PhD Training Programme 06PhD12 to W.L., W.C.Y., T.B., and R.G.), the Dutch Science Organization NWO (VIDI 864.06.007 to R.G., Dutch Russian Research Cooperation 047.018.001 to S.I., E.L., R.G., and T.B., VICI 865.06.002 to H.J.B., and Equipment 834.08.001 to H.J.B.), EC Marie Curie Research Training Network Program MRTN-CT-2006-035546 NODPERCEPTION (to A.L. and R.G.), and Pakistan Higher Education Commission (to M.J.). H.J.B. is cofinanced by the Centre for BioSystems Genomics, which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research.
AUTHOR CONTRIBUTIONS
R.G., T.B., and H.J.B. designed the research. W.L., W.K., M.J., A.L., R.O.d.C., S.I., M.H., E.L., G.J.E.J.H., and T.C. performed the research. W.K., M.J., C.S., K.K., W.-C.Y., G.J.E.J.H., T.C., and H.J.B. contributed analytic tools. W.L., W.K., M.J., G.J.E.J.H., T.C., H.J.B., T.B., and R.G. analyzed data, and W.L., W.K., H.J.B., T.B., and R.G. wrote the article.
References
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alvord W.G., Roayaei J.A., Quiñones O.A., Schneider K.T. (2007). A microarray analysis for differential gene expression in the soybean genome using Bioconductor and R. Brief. Bioinform. 8: 415–431 [DOI] [PubMed] [Google Scholar]
- Barbulova A., Rogato A., D’Apuzzo E., Omrane S., Chiurazzi M. (2007). Differential effects of combined N sources on early steps of the Nod factor–dependent transduction pathway in Lotus japonicus. Mol. Plant-Mircobe Interact. 20: 994–1003 [DOI] [PubMed] [Google Scholar]
- Beveridge C.A., Kyozuka J. (2010). New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bolstad B., Collin F., Brettschneider J., Simpson K., Cope L., Irizarry R., Speed T.P. (2005). Quality assessment of Affymetrix GeneChip data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor Book Series: Statistics for Biology and Health, Gentleman R., Vincent J.C., Wolfgang H., Irizarry R.A., Dudoit S., (New York: Springer; ) pp. 33–47 [Google Scholar]
- Bouwmeester H.J., Roux C., Lopez-Raez J.A., Bécard G. (2007). Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12: 224–230 [DOI] [PubMed] [Google Scholar]
- Brettschneider J., Collin F., Bolstad B.M., Speed T.P. (2008). Quality assessment for short oligonucleotide microarray data. Technometrics 50: 241–264 [Google Scholar]
- Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C.E., Whichard L.P., Turner B., Wall M.E., Egley G.H. (1966). Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Domagalska M.A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Bio. 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Fåhraeus G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Heber S., Sick B. (2006). Quality assessment of Affymetrix GeneChip data. OMICS 10: 358–368 [DOI] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland A., Arnon D. (1950). The water-culture method for growing plants without soil. California Agricultural Experimental Station Circular, No. 347 (Berkeley, CA: University of California; ), pp. 1–32 [Google Scholar]
- Jamil M., Charnikhova T., Verstappen F., Bouwmeester H. (2010). Carotenoid inhibitors reduce strigolactone production and Striga hermonthica infection in rice. Arch. Biochem. Biophys. 504: 123–131 [DOI] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Anfang N., Saedler H., Theissen G. (2005). Mutant analysis, protein-protein interactions and subcellular localization of the Arabidopsis B sister (ABS) protein. Mol. Genet. Genomics 274: 103–118 [DOI] [PubMed] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M.A., Beguerie S., Verstappen F., Leyser O., Bouwmeester H., Ruyter-Spira C. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H., LekKala S.P., Bhattacharya C., Mayzlish-Gati E., Resnick N., Wininger S., Dor E., Yoneyama K., Yoneyama K., Hershenhorn J., Joel D.M., Kapulnik Y. (2010). A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 61: 1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E.D. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Imaizumi-Anraku H., Hayashi M., Hakoyama T., Nakagawa T., Umehara Y., Suganuma N., Kawaguchi M. (2010). How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., et al. (2008). Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 67: 659–670 [DOI] [PubMed] [Google Scholar]
- Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., Yan C., Jiang B., Su Z., Li J., Wang Y. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez J.A., Charnikhova T., Gómez-Roldán V., Matusova R., Kohlen W., De Vos R., Verstappen F., Puech-Pages V., Bécard G., Mulder P., Bouwmeester H. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178: 863–874 [DOI] [PubMed] [Google Scholar]
- López-Ráez J.A., Kohlen W., Charnikhova T., Mulder P., Undas A.K., Sergeant M.J., Verstappen F., Bugg T.D.H., Thompson A.J., Ruyter-Spira C., Bouwmeester H. (2010). Does abscisic acid affect strigolactone biosynthesis? New Phytol. 187: 343–354 [DOI] [PubMed] [Google Scholar]
- Maillet F., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Matusova R., Rani K., Verstappen F.W.A., Franssen M.C.R., Beale M.H., Bouwmeester H.J. (2005). The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139: 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E.D., Long S.R. (2004a). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Shaw S.L., Long S.R. (2004b). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Bell C.D., Soltis P.S., Soltis D.E. (2007). Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA 104: 19363–19368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Miwa H., Imaizumi-Anraku H., Kouchi H., Downie J.A., Kawaguchi M., Kawasaki S. (2006). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 13: 255–265 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Long S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani K., Zwanenburg B., Sugimoto Y., Yoneyama K., Bouwmeester H.J. (2008). Biosynthetic considerations could assist the structure elucidation of host plant produced rhizosphere signalling compounds (strigolactones) for arbuscular mycorrhizal fungi and parasitic plants. Plant Physiol. Biochem. 46: 617–626 [DOI] [PubMed] [Google Scholar]
- Sartor M.A., Tomlinson C.R., Wesselkamper S.C., Sivaganesan S., Leikauf G.D., Medvedovic M. (2006). Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics 7: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith S.A., Beaulieu J.M., Donoghue M.J. (2010). An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc. Natl. Acad. Sci. USA 107: 5897–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3 [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S., Li J., Chen M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54: 519–532 [DOI] [PubMed] [Google Scholar]
- Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., Tanaka H. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Trouvelot A., Kough J., Gianinazzi-Pearson V. (1986). Measuring the rate of VA mycorrhization of root systems. In Physiological and Genetical Aspects of Mycorrhizae, Gianinazzi-Pearson V., Gianinazzi S., (Paris: INRA Press; ), pp. 217–221 [Google Scholar]
- Umehara M., Hanada A., Magome H., Takeda-Kamiya N., Yamaguchi S. (2010). Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 51: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35: W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J. (2011). Branching in rice. Curr. Opin. Plant Biol. 14: 94–99 [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917 [Google Scholar]
- Xie X., Yoneyama K., Yoneyama K. (2010). The strigolactone story. Annu. Rev. Phytopathol. 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Yokota K., Soyano T., Kouchi H., Hayashi M. (2010). Function of GRAS proteins in root nodule symbiosis is retained in homologs of a non-legume, rice. Plant Cell Physiol. 51: 1436–1442 [DOI] [PubMed] [Google Scholar]
- Yokota T., Sakai H., Okuno K., Yoneyama K., Takeuchi Y. (1998). Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry 49: 1967–1973 [Google Scholar]
- Yoneyama K., Yoneyama K., Takeuchi Y., Sekimoto H. (2007). Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Zhu H., Riely B.K., Burns N.J., Ané J.M. (2006). Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172: 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]