Abstract
The bypass of AP sites in yeast requires the Rev1 protein in addition to the Pol ζ translesion synthesis DNA polymerase. Although Rev1 was originally characterized biochemically as a dCMP transferase during AP-site bypass, the relevance of this activity in vivo is unclear. The current study uses highly sensitive frameshift- and nonsense-reversion assays to monitor the bypass of AP sites created when uracil is excised from chromosomal DNA. In the frameshift-reversion assay, an unselected base substitution frequently accompanies the selected mutation, allowing the relative incorporation of each of the four dNMPs opposite endogenously created AP sites to be inferred. Results with this assay suggest that dCMP is the most frequent dNMP inserted opposite uracil-derived AP sites and demonstrate that dCMP insertion absolutely requires the catalytic activity of Rev1. In the complementary nonsense-reversion assay, dCMP insertion likewise depended on the dCMP transferase activity of Rev1. Because dAMP insertion opposite uracil-derived AP sites does not revert the nonsense allele and hence could not be detected, it also was possible to detect low levels of dGMP or dTMP insertion upon loss of Rev1 catalytic activity. These results demonstrate that the catalytic activity of Rev1 is biologically relevant and is required specifically for dCMP insertion during the bypass of endogenous AP sites.
Keywords: Polζ, Rev1, AP sites, mutagenesis, translesion synthesis
1. Introduction
Apurinic/apyrimidinic (AP) sites are the most common endogenous DNA lesion [1] and are formed by hydrolysis of the glycosidic bond between the base and deoxyribose sugar of a nucleoside. Base loss can occur spontaneously or through enzymatic removal of damaged bases by specialized DNA N-glycosylases of the base excision repair (BER) pathway [2]. Because an unrepaired AP site is a potent block to replicative DNA polymerases, bypass is critical for maintaining genetic integrity and completing genome duplication. Translesion synthesis (TLS) is one of two general tolerance/bypass pathways that circumvent the replication blocks caused by AP sites, as well as a variety of other DNA lesions (for recent reviews, see [3, 4]). TLS can be divided into two steps: an insertion step in which a nucleotide is incorporated opposite the lesion and an extension step in which the unpaired primer-template terminus is extended. While the overall efficiency of TLS is determined by the extension step, whether or not the bypass event is mutagenic depends on the nucleotide inserted opposite a non-instructive AP site. Under conditions where AP-site repair is compromised, mutagenesis increases in a TLS-dependent manner [5].
The yeast Saccharomyces cerevisiae contains three, highly conserved TLS polymerases that potentially can participate in AP-site bypass: Pol η, Pol ζ and Rev1. Pol η is a Y-family DNA polymerase whose loss results in a variant form of the human cancer-predisposition syndrome Xeroderma Pigmentosum, which is characterized by extreme sensitivity to UV light [6, 7]. In yeast, Pol η is encoded by the RAD30 gene and its absence is associated with enhanced UV-induced sensitivity and mutagenesis [8]. Pol ζ is a B-family DNA polymerase comprised of two subunits: the Rev3 catalytic and Rev7 accessory proteins (reviewed in [9]). Pol ζ is required for most induced, as well as a substantial fraction of spontaneous, mutagenesis in yeast and is essential in mammalian cells [10, 11]. While it is capable of independently bypassing lesions in vitro, the primary role of Pol ζ in vivo is thought to reflect its unique ability to extend an unpaired primer-template terminus [12, 13]. Finally, Rev1 is a Y-family DNA polymerase that is required for Pol ζ-dependent mutagenesis. It was initially described biochemically as a deoxycytidyl (dCMP) transferase, specifically inserting cytosine opposite template lesions [14]. In addition to the catalytic activity, an N-terminal BRCT domain is important for DNA binding [15] and a C-terminal scaffoloding domain interacts with Rev3 and Rev7 [16, 17].
In contrast to the general biological significance of the BRCT and C-terminal domains of Rev1, the relevance of the dCMP transferase activity in vivo appears to be lesion-specific. This activity, for example, is not required for survival or Rev1-dependent mutagenesis following UV irradiation, but is important for surviving 4-nitroquinoline-1-oxide (4-NQO)-induced damage [18]. With regard to AP-site bypass, there are conflicting data concerning the relevance of the Rev1 dCMP transferase activity. Early experiments examined genomic mutations induced by the base-alkylating agent methyl methanesulfonate (MMS), which generates AP sites primarily at purines. Most MMS-induced mutations were GC > TA transversions, a mutation pattern inconsistent with dCMP insertion opposite AP sites and shown not to require Rev1 catalytic activity [13]. While these data were used to argue for a dAMP insertion bias during Rev1-dependent bypass of guanine-derived AP sites, it should be noted that dCMP insertion would not have been mutagenic and hence could not have been detected in these experiments (see [19]. A study of mutagenesis associated with expression of T- or C-specific glycosylases reported Rev1-dependent mutation patterns consistent with dCMP insertion opposite AP sites [5], as did a study examining the mutagenic consequence of uracil-derived AP sites [20]. Neither of these studies, however, could have detected non-mutagenic dAMP insertion opposite thymine-derived AP sites and neither examined the relevance of the protein’s catalytic activity. As an alternative to studying the bypass of physiologically produced AP sites, oligonucleotides or gapped plasmids containing a single, defined AP site have been used in transformation-based studies. These analyses have reported preferential insertion of dCMP opposite an engineered AP site, [19, 21–24], and have implicated the catalytic activity of Rev1 during bypass [24].
We previously described very sensitive frameshift- and nonsense-reversion assays that monitor the bypass of AP sites produced when uracil is excised from highly transcribed DNA [25, 26]. Because uracil specifically replaces thymine in these assays, the base substitution pattern at AT base pairs provides a read-out of nucleotides inserted opposite thymine-derived AP sites. In contrast to previous assays, where non-mutagenic AP-site bypass via dAMP insertion could not be detected, the frequent occurrence of non-selected base substitutions in the frameshift-reversion assay allows the relative insertion efficiencies of all four dNMPs to be inferred. The data reported here suggest that dCMP is the predominant nucleotide inserted opposite uracil-derived AP sites in this system and confirm that dCMP insertion, but not that of alternative dNMPs, requires the catalytic activity of Rev1. Accompanying shifts in mutation spectra suggest that back-up activities compensate for dCMP transferase loss by inserting primarily dAMP. When considered together with the nonsense-reversion assay, results indicate that the insertion preference opposite AP sites in yeast is dCMP > dAMP ≫ dGMP ~ dTMP. Finally, we extend the importance of the BRCT DNA-binding and C-terminal scaffold domains of Rev1 to include the bypass of AP sites.
2. Materials and Methods
2.1. Media and growth conditions
All growth of yeast strains was at 30°C. Cells were grown non-selectively in YEP medium (1% yeast extract, 2% peptone; 1.5% agar for plates) supplemented with 2% dextrose (YEPD) or 2% each of glycerol and ethanol (YEPGE). It should be noted that under these growth conditions, pTET (whose repression requires doxycycline) is maximally activated. Selective growth was on synthetic, 2% dextrose (SD) medium supplemented with all but the one relevant amino acid or base. The presence of a hygromycin-resistance marker was selected by plating transformants on YEPD plates supplemented with 300 μg/ml hygromycin.
2.2. Strain constructions
All yeast strains were derived from YPH45 (MATα ura3-52 ade2-101oc trp1Δ1). Wild-type (WT) strains containing the pTET-lys2ΔA746 [27] or pTET-lys2-TAA [26] allele near ARS306 on Chromosome III were previously described. The rev1-BRCT (rev1-G192A,G193A), rev1-AA (rev1-D467A,E468A) or rev1-CDEL (rev1-Y914Stop,L915Stop) allele was introduced into these strains by two-step replacement using plasmids pSR836, pSR838 or pSR986, respectively. To construct these plasmids, REV1 sequences were subcloned into the URA3-marked integrating vector pRS306 [28]. The desired sequence changes were then introduced using the Quik-Change Site-Directed Mutagenesis kit (Stratagene; see Table S1 for mutagenic oligonucleotides used). Deletion strains were derived by one-step gene disruption using PCR-generated cassettes containing an appropriate selectable marker flanked by ~60 bp of homology bordering the coding region of the relevant gene [29]. When appropriate, the marker gene was subsequently deleted using a Cre/loxP-mediated procedure [30]. A complete list of strains is provided in Table S2.
2.3. Mutation rates and spectra
For mutation rate determinations, 1-ml YEPGE cultures were inoculated with 250,000 cells from an overnight culture grown in the same medium. After 3 days growth, appropriate dilutions were plated on SD-Lys and YEPD plates to determine the number of Lys+ revertants and the total number of cells, respectively, in each culture. Mutation rates were calculated using the method of the median and the corresponding 95% confidence intervals were calculated as described previously [31]. Ten to 24 cultures were used for each rate determination.
To isolate independent Lys+ revertants, 1-ml YEPGE cultures were started from independent colonies. Following 2–3 days growth, an appropriate fraction of each culture was plated on SD-Lys medium. A single revertant from each culture was purified and genomic DNA was prepared using an enzymatic lysis method modified for a 96-well format. The relevant region of LYS2 was amplified using primers LYSWINDF (5′-GCCTCATGATAGTTTTTCTAACAAATACG) and LYSWINDR (5′-CCCATCACACATACCATCAAATCCAC), and sequenced by the Duke University DNA Analysis Facility.
3. Results
3.1. AP sites in the pTET- lys2ΔA746 assay are produced by Ung1 and require Pol ζ for bypass
The lys2ΔA746 allele contains a 1-bp deletion in a nonessential region of the LYS2 coding sequence and reverts by any net +1 frameshift mutation that restores the correct reading frame of the gene [32]. The Pol ζ TLS polymerase generates a distinctive mutation signature in this assay: “complex” 1-bp insertions that additionally contain a nearby, unselected base substitution. Such complex events are proportionally enhanced in repair-defective backgrounds, indicating that most are produced during the Pol ζ-dependent bypass of unrepaired lesions [33]. Because changing the identity of the base pair most often mutated at complex hotspots eliminates the selected frameshift, we have proposed that complex mutations are produced by a misincorporation-slippage mechanism [32]. In this mechanism, misinsertion opposite a discrete lesion precedes and may facilitate slippage in a short homopolymer run that follows, thereby generating the selected frameshift mutation. A key feature of misincorporation-slippage is that the position of the base substitution at complex-mutation hotspots marks the site of the initiating lesion. Because the base substitution is not the selected event, no functional constraints are placed on the dNMP inserted opposite the initiating lesion.
When fused to the heterologous pTET promoter, high transcription of the lys2ΔA746 allele elevates its reversion rate 10–20 fold [27]. This effect was weakly enhanced upon loss of Rad14, a protein required for nucleotide excision repair (NER; [34]; Apn1, the major AP endonuclease in yeast [35]; or the Ntg1 and Ntg2 lyases, which also nick the DNA backbone at AP sites [36]. In the apn1 mutant, transcription-associated mutagenesis was accompanied by the appearance of a novel complex-mutation hotspot, with the associated base substitutions occurring at AT base pairs. Upon additional deletion of RAD14 or NTG1/NTG2 from the apn1 background, very strong synergism was observed, indicating functional overlap in removing potentially mutagenic AP lesions. Mutagenesis in these mutants was completely dependent on the Ung1 DNA glycosylase, which creates AP sites by excising uracil from the DNA backbone, and on Rev3, the catalytic subunit of Pol ζ (Table S3). Furthermore mutagenesis was suppressed by overproduction of the Dut1 dUTPase [25, 26]. These genetic data, together with the occurrence of the frameshift-associated base substitutions at AT base pairs, provided compelling evidence that complex mutations reflect the Pol ζ-dependent bypass of AP sites that are generated by the excision of uracil, which is specifically incorporated in place of thymine. Because we know how (uracil excision) and where (the thymine of AT base pairs) AP sites are generated in the pTET-lys2ΔA746 system, the unselected base substitutions at complex hotspots provide an unbiased read-out of dNMPs inserted opposite the initiating lesion. In the current study, we use this system, plus a pTET-based nonsense-reversion assay, to explore the roles of Rev1 during AP-site bypass in apn1 single, apn1 rad14 double, and apn1 ntg1 ntg2 triple mutant strains; all corresponding data are presented in Table S3. Because of the much stronger mutagenesis in the double and triple mutants, however, only these data are described in the sections that follow. Hereafter, we refer to the apn1 ntg1 ntg2 triple and apn1 rad14 double mutants as the BER- and BER/NER-defective backgrounds, respectively.
3.2. AP site bypass yields simple as well as complex frameshifts at a common hotspot
In the BER- and BER/NER-defective strains, the 1-bp insertion at the major complex-mutation hotspot occurs within a run of six AT base pairs (Figure 1; [25, 26]). Using the convention of describing sequences in terms of the coding/nontranscribed strand of LYS2, this position is referred to as the 6A hotspot. Although our previous analyses focused exclusively on complex mutations at this hotspot, the rates of “simple” frameshifts that expand the 6A run to a 7A run were also elevated under high-transcription conditions. These simple events are likewise dependent on Ung1 and Rev3 with loss of either protein causing Lys+ rate decreases in the BER- or BER/NER-defective backgrounds, respectively (Figure 2 and Table S3).
Figure 1.
Effect of rev1-AA allele on pTET-lys2ΔA746 reversion spectra. The 6A hotspot is underlined; complex insertions and simple insertions are indicated by “cins” and “+,” respectively. In A and B, cins that have an associated T > G base substitution are highlighted in gray; in C and D, cins that have an associated A > C base substitution are highlighted. When an insertion occurs outside a run, the base inserted is indicated. Only the first half of the reversion window is shown, as >95% of reversion events were in this region. N, number of revertants sequenced. The apn1 ntg1 ntg2 and apn1 rad14 spectra were published previously [25, 26]
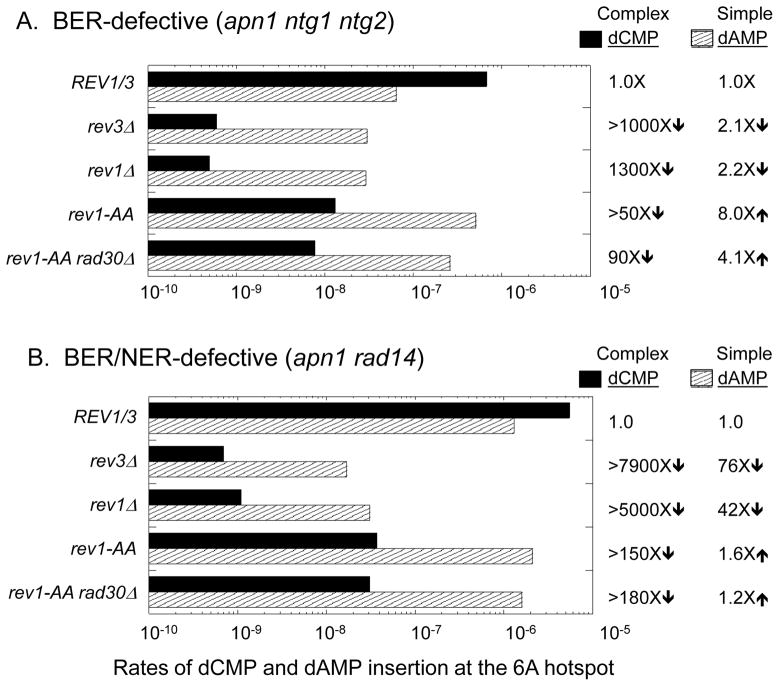
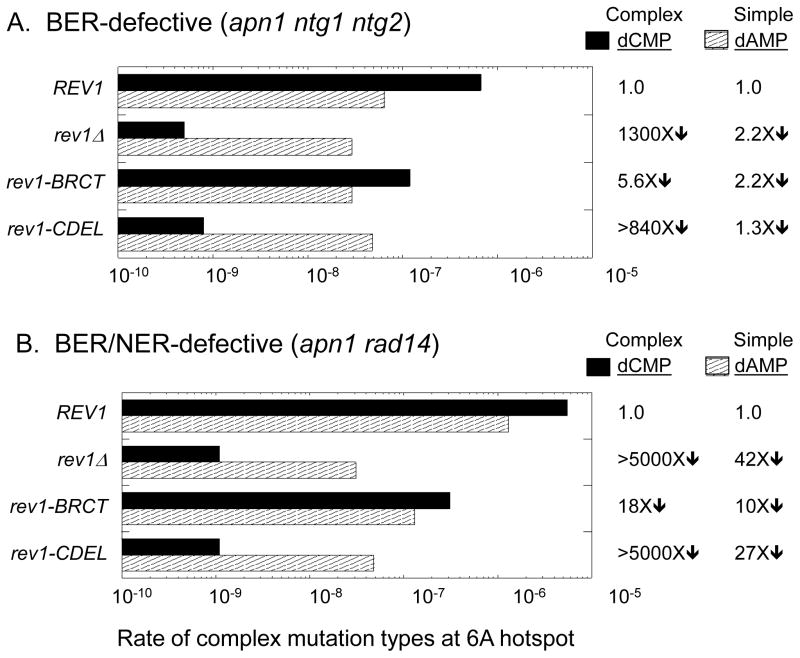
Figure 2.
The catalytic activity of Rev1 is required for dCMP insertion opposite AP sites in the pTET-lys2ΔA746 assay. Striped and solid bars correspond to the rates of simple and complex frameshifts (dAMP and dCMP insertion opposite uracil-derived AP sites, respectively) at the 6A hotspot. Changes in simple and complex rates in TLS-polymerase mutants relative to the REV1/3 background are indicated to the right of the graphs. When none of the relevant event was observed in the corresponding spectrum, the rate was calculated assuming one event and “>” precedes the rate change. The total Lys+ rate and the number of mutants sequenced in each genetic background are in Table S3.
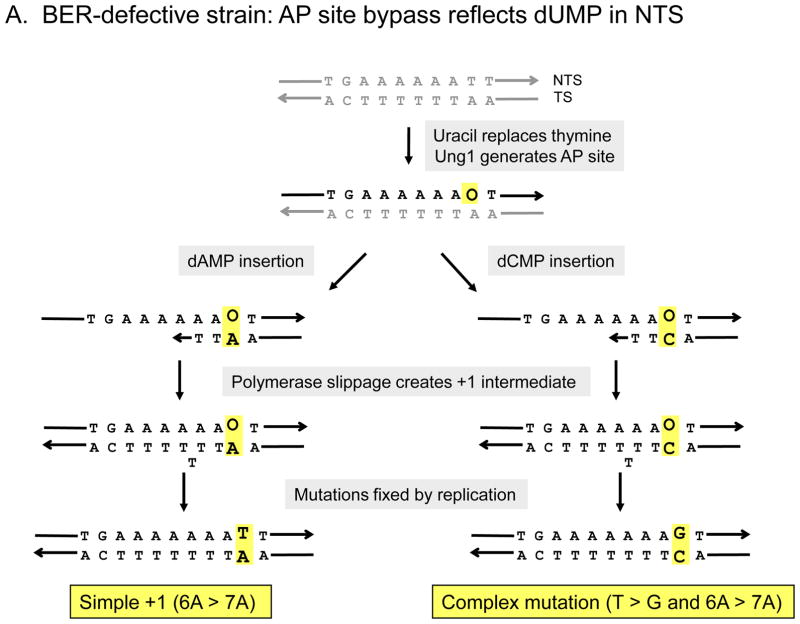
Because the simple and complex events at the 6A hotspot have common genetic requirements, the most parsimonious explanation is that both are derived via a common AP-site bypass mechanism. As illustrated in Figure 3, each can be generated by the misincorporation-slippage mechanism, with the simple versus complex mutation outcome being determined by the specific dNMP inserted opposite the AP site. For complex events in a BER-defective background, we previously argued that an Ung1-dependent AP site arises when uracil replaces one of the thymines immediately 3′ of the 6A run. Insertion of dCMP opposite the AP site, followed by slippage in the 6A run, gives rise to a complex mutation in which a T to G transversion accompanies the selected 6A > 7A frameshift (right side of Figure 3A; see [25]). Insertion of dAMP instead of dCMP opposite the same AP site does not produce a base substitution, but it will, if followed by slippage in the 6A run, result in a simple 6A >7A mutation (left side of Figure 3A).
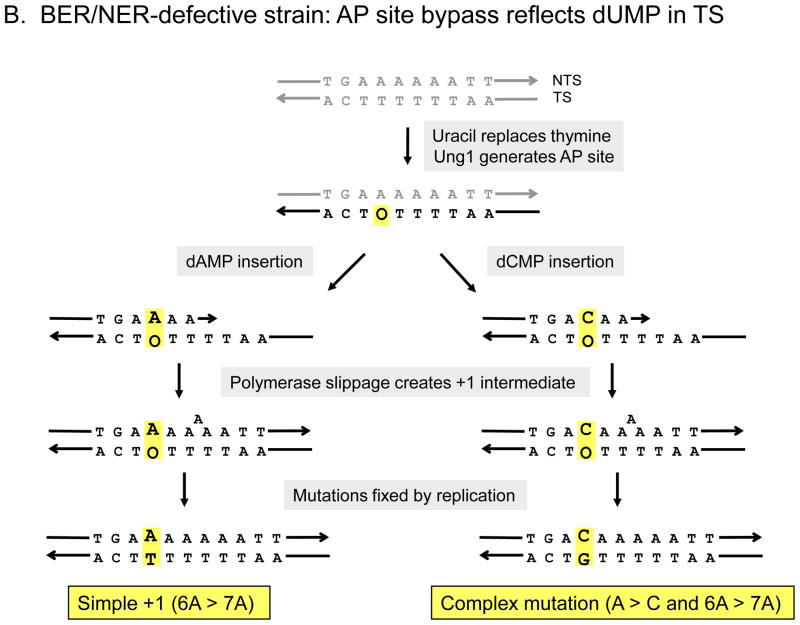
Figure 3.
Mutagenic bypass of uracil-derived AP sites in the pTET-lys2-ΔA746 assay. In (A) the uracil-derived AP site is on the NTS of LYS2, as occurs in the apn1 ntg1 ntg2 background. Insertion of dCMP or dAMP opposite the AP site followed by slippage during extension through the remainder of the 6T run, results in a complex frameshift with an A > C change or a simple +1 frameshift, respectively. In (B), the uracil-derived AP site is on the TS of LYS2, as occurs in the apn1 rad14 background. Insertion of dCMP or dAMP opposite the AP site, followed by slippage in the 6A run that follows, results in a complex frameshift with a T > G change or a simple +1 frameshift, respectively.
In the BER/NER-defective strain, we proposed that uracil is substituted for one of the thymines opposite the 6A run [26]. When the resulting Ung1-generated AP site is bypassed, insertion of dCMP or dAMP opposite the AP site, followed by slippage, results in either a complex frameshift characterized by an A to C transversion or a simple 6A > 7A frameshift, respectively (Figure 3B). Additional genetic analyses have shown that the accumulation of uracil-derived AP sites on complementary strands of the reporter in BER- versus BER/NER-defective strains reflects transcription-coupled NER of lesions located on the transcribed/noncoding strand [26]. In either the BER- or BER/NER-defective background, it is important to note that the rate of complex events at the 6A run exceeds that of simple events (10- and 4-fold, respectively; Figure 2). This suggests that dCMP insertion opposite the relevant AP sites normally occurs much more frequently than dAMP insertion. An alternative possibility is that dCMP insertion is associated with more slippage than is dAMP insertion, but we think this is unlikely. Finally, complex mutations predicted by dTMP or dGMP insertion at the 6A hotspot are very rare, suggesting that these two dNMPs are infrequently inserted relative to dCMP or dAMP.
3.3. Rev1 is essential for AP-site bypass, and its catalytic activity is required for dCMP insertion in the pTET-lys2ΔA746 assay
The effect of REV1 deletion on reversion of the pTET-lys2ΔA746 allele in the BER- or BER/NER-defective background was indistinguishable from that of REV3 deletion, with either deletion reducing the Lys+ rate to the level observed in the WT strain (Table S3). In the BER- and BER/NER-defective backgrounds, complex events at the 6A hotspot were reduced over 100-fold, demonstrating that dCMP insertion is absolutely dependent on Rev1 as well as on Pol ζ (Figure 2). Simple events at the 6A hotspot were not as dramatically affected by Rev1 or Rev3 loss. The smaller effects on simple events are presumably because they occur frequently in the WT background via slippage of replicative DNA polymerases during genome duplication [32].
The relevance of Rev1 dCMP transferase activity during AP-site bypass was assessed using the rev1-AA allele, in which two amino acid residues essential for catalytic activity were changed to alanine (see Materials and Methods). Although this allele did not significantly reduce the overall reversion rate of pTET-lys2ΔA746 allele in the BER-defective background (Table S3), it dramatically changed the patterns and rates of mutations at the 6A hotspot (Figures 1 and 2). The complex events reflecting dCMP incorporation were eliminated from the spectrum and were replaced by a large and compensatory (8-fold) increase in the rate of the simple 1-bp insertions, indicating the presence of a back-up, AP-site bypass activity that specifically inserts dAMP. The loss of dCMP insertion in the rev1-AA mutant clearly demonstrates the biological relevance of Rev1 catalytic activity during AP-site bypass. The large increase in dAMP insertion in the rev1-AA mutant is particularly significant, as it indicates that dCMP insertion opposite AP sites in this system normally exceeds dAMP insertion by 5–10 fold.
In the BER/NER-defective background, introduction of the rev1-AA allele was associated with a modest, ~ 3-fold reduction in the overall Lys+ rate. Importantly, the rate reduction was accompanied by the disappearance of complex mutations from the corresponding reversion spectrum, confirming an absolute requirement of Rev1 catalytic activity for dCMP insertion opposite the uracil-derived AP sites (Figures 1 and 2). The loss of dCMP insertion in the rev1-AA mutant was furthermore associated with a 1.6-fold increase in the rate of the simple 1-bp insertions diagnostic of dAMP insertion, again suggesting that dCMP is the primary dNMP inserted opposite endogenous AP sites. We note that, in contrast to what was observed in the BER-defective background, dAMP insertion did not completely compensate for the loss of dCMP insertion in the BER/NER-defective background. We suggest either that the back-up insertion polymerase is relatively inefficient in this context, or that it fails to generate the required frameshift mutation. Finally, of the very small number of complex mutations that persisted at the 6A hotspot in the BER- and BER/NER-defective backgrounds (Table S4), most can be explained by dTMP or dGMP insertion opposite the AP site.
3.4. Loss of Rev1 catalytic activity eliminates most reversion of the pTET-lys2-TAA nonsense allele
The lys2-TAA allele contains an engineered, in-frame TAA stop codon, the reversion of which occurs via a base substitution at one of the three nucleotides of the TAA codon [37]. The TAA stop codon monitors AP sites in a locally different sequence context, which is ~100 bp downstream of the 6A hotspot in the pTET-lys2ΔA746 reversion assay. We previously demonstrated that reversion of the highly-transcribed pTET-lys2-TAA allele, like that of the pTET-lys2ΔA746 allele, results from the Pol ζ-dependent bypass of uracil-derived AP sites in BER- and BER/NER-defective backgrounds (Table S5; [26]). T > G or A > C mutations are produced if dCMP is inserted opposite the AP site; insertion of dTMP leads to T > A or A > T transversions; and insertion of dGMP results in T > C and A > G transitions, with A > G mutations escaping detection (Figure S1). In contrast to the frameshift-reversion assay, which was also able to infer dAMP insertion opposite uracil-derived AP sites, similar dAMP insertion in the base substitution assay would regenerate the stop codon and hence cannot be detected.
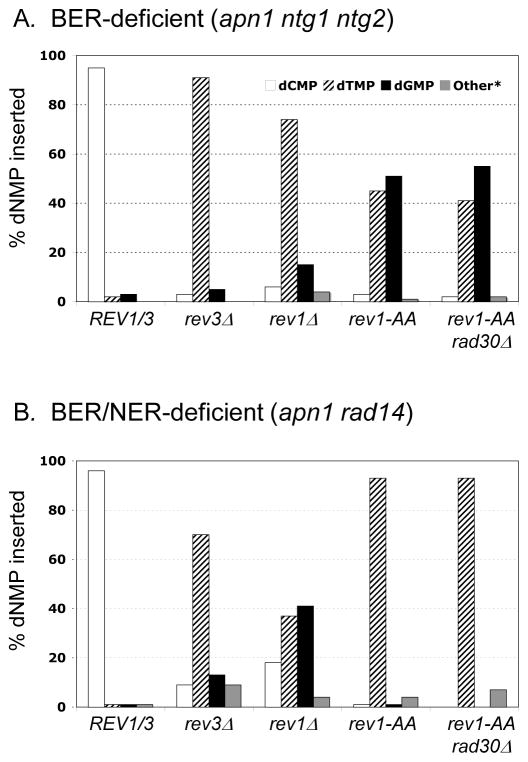
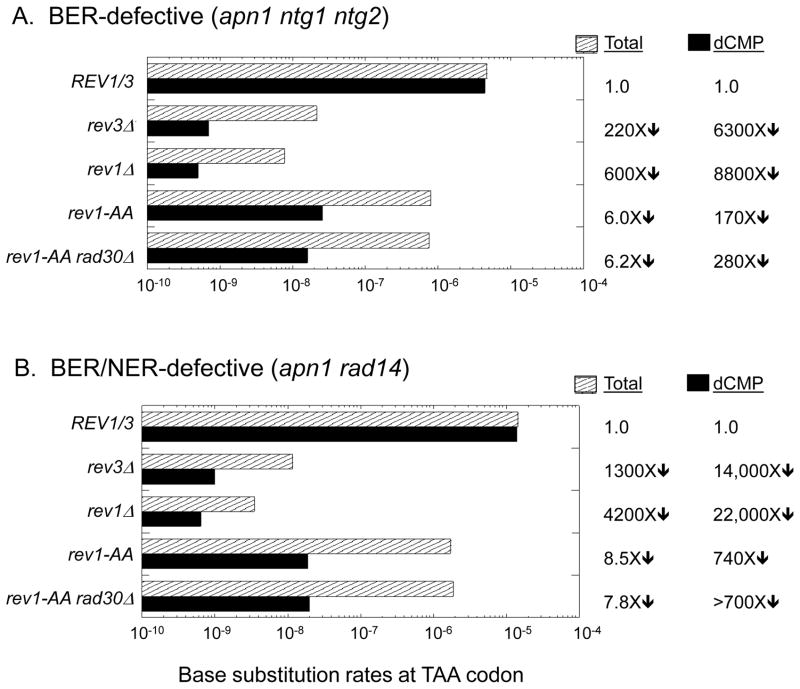
In a WT strain background, all possible base changes at the stop codon were recovered at comparable frequencies among pTET-lys2-TAA revertants (Table S6 and [26]. The reversion rate of the pTET-lys2-TAA allele was elevated at least several hundred fold in BER- and BER/NER-defective backgrounds and there was an accompanying shift in the reversion spectra, with > 95% of revertants containing the T > G or A > C change diagnostic of dCMP insertion (Figure 4; [26]). Deletion of the REV3 or REV1 gene led to dramatic (1000-fold) decreases in the reversion rate of pTET-lys2-TAA allele in the BER- and BER/NER-defective backgrounds (Figure 5). The reversion rate was consistently ~3-fold lower in the absence of Rev1 than in the absence of Rev3, however, suggesting that there may be a very inefficient, Pol ζ-independent AP-site bypass mechanism that requires Rev1. In addition to the large Lys+ rate reductions in the rev3Δ and rev1Δ mutants, the percentages of T > G and A > C transversion mutations were reduced to < 20% of the corresponding spectra (Figure 4).
Figure 4.
Proportions of dNMPs inserted opposite AP sites in the pTET-lys2-TAA assay. The numbers of each mutant type are in Table S6. “Other” mutations include deletions and multiple base changes within a single revertant.
Figure 5.
Base substitution rates in the pTET-lys2-TAA reversion assay. The total Lys+ rate and the rate of dCMP insertion are indicated by striped and solid bars, respectively. Changes in these rates in TLS-polymerase mutants relative to the REV1/3 background are indicated to the right of the graphs. When none of the relevant event was observed in the corresponding spectrum, the rate was calculated assuming one event and “>” precedes the rate change. The rates and the number of mutants sequenced in each genetic background are in Table S5.
When the rev1-AA mutation was introduced into the BER- and BER/NER-defective backgrounds, the reversion rate of the lys2-TAA allele decreased 6- to 8-fold (Figure 5). Although loss of Rev1 catalytic activity still allowed some mutagenic bypass, the T > G and A > C mutation classes were almost completely eliminated (Figures 4–5). In the BER/NER-defective background, there was an accompanying shift in the mutation spectrum to ~90 % TA > AT transversions. In the BER-defective background, however, TA > CG transitions indicative of dGMP insertion opposite the corresponding AP sites were as abundant as TA > AT transversions (Figure 4). We suggest that the apparent lack of dGMP insertion in the BER/NER-defective strain can be explained if, as we argued previously, AP sites in this background accumulate primarily on the transcribed strand of the reporter (see Figure 3B; [26]). The A > G class, which reflects dGMP insertion opposite AP sites on the transcribed strain, cannot be detected in the nonsense reversion assay because it regenerates a stop codon. The reciprocal T > C class, which can be detected, reflects the bypass of AP sites on the nontranscribed strand, which do not accumulate in the BER/NER-defective background. Consistent with this interpretation, it should be noted that the A > T mutations, which reflect dTMP insertion opposite AP sites on the transcribed strand, were ~ 10-fold more frequent than T > A mutations in the NER/BER-defective background (Table S6).
3.5. Pol η is not required for dNMP insertion opposite AP sites in the absence of Rev1 catalytic activity
The bypass of AP sites was completely dependent on the presence of Rev1 in both the frameshift- and nonsense-reversion assays, but significant bypass persisted when only the catalytic site was mutated (rev1-AA strains). This was most evident in the pTET-lys2ΔA746 reversion assay, where dAMP insertion could be detected as simple +1 events at the 6A hotspot. We considered the possibility that insertion of dAMP might be catalyzed by the remaining TLS polymerase, Pol η, in the rev1-AA mutants. While additional deletion of RAD30 gene did not lead to any significant changes in the pTET-lys2ΔA746 reversion rates or spectra in the BER/NER-defective background, there was a small (35%), but significant, decrease in the Lys+ rate in the BER-defective background (Figure 2 and Table S3). In the case of the base-substitution assay, additional deletion of the RAD30 gene in the rev1-AA strains did not result in any significant changes in mutation rates or spectra (Figures 4 and 6). Although the data fail to convincingly implicate Pol η in AP-site bypass, it is possible that a minor activity is obscured by one of the replicative DNA polymerases.
Figure 6.
Importance of the Rev1 BRCT and C-terminal domains during AP-site bypass in the pTET-lys2ΔA746 assay. Striped and solid bars correspond to the rates of simple and complex frameshifts at the 6A hotspot. Changes in simple and complex rates in TLS-polymerase mutants relative to the REV1/3 background are indicated to the right of the graphs. When none of the relevant event was observed in the corresponding spectrum, the rate was calculated assuming one event and “>” precedes the rate change. The total Lys+ rate and the number of mutants sequenced in each genetic background are in Table S3.
3.6. Mutation of Rev1 BRCT domain reduces AP-site bypass in the pTET-lys2ΔA746 assay
The original rev1-1 allele (G193R), which has a mutation within the N-terminal BRCT domain of Rev1, did not affect dCMP transferase activity, but eliminated or reduced bypass of a 6–4 photoproduct or AP site, respectively, engineered into a gapped plasmid [38]. Transformation with an oligonucleotide containing an AP site into a strain with an N-terminally deleted form of Rev1 also greatly reduced, but did not completely eliminate bypass [24]. There were, however, conflicting data regarding the nucleotides inserted opposite the AP site. Preferential insertion of dCMP persisted during the gap-filling reaction [38], whereas there was a shift to dTMP or dGMP insertion following oligonucleotide transformation [24].
To examine the effect of a BRCT domain mutation on the bypass of AP sites created in a chromosomal context, we introduced a rev1-BRCT allele into strains containing the pTET-lys2ΔA746 allele. In the BER-defective background, both the Lys+ rate and the rate of complex mutations decreased 5-fold when the rev1-BRCT mutation was introduced, rates which were significantly higher than those obtained in the rev1Δ background (Figure 6). The decrease in complex insertions at the 6A hotspot revealed a novel class of complex deletions that comprised ~20% of the corresponding spectrum (see Table S4). This new class contained the T > G base-substitution signature diagnostic of dCMP insertion during AP-site bypass, but instead of an accompanying 1-bp insertion contained a 2-bp deletion that reduced the 6A run to a 4A run. Although we cannot discern whether the rate of complex deletions remained the same or increased upon introduction of the rev1-BRCT allele, their appearance nevertheless suggests that the BRCT domain of Rev1 regulates slippage during Pol ζ-dependent bypass.
In the BER/NER-defective background, introduction of the rev1-BRCT mutation resulted in similar (10–20 fold) decreases in overall reversion rate as well as in the rates of simple and complex events at the 6A hotspot (Figure 6). In contrast to the BER-defective mutant, however, the novel complex deletions at the 6A run were not evident in the BER/NER-defective background. It is possible that their absence reflects a greater difficult in generating the requisite deletion intermediate when the 6A run is effectively shortened by interruption with an AP site.
3.7. Deletion of the C-terminal domain of Rev1 eliminates AP-site bypass in the pTET-lys2ΔA746 assay
The C-terminal domain of yeast Rev1 mediates physical interaction with Rev3, Rev7 and Rad30 [17, 39, 40]. Mutations in this region lead to enhanced UV sensitivity and to a significant reduction in UV-induced mutagenesis, demonstrating its significance in the bypass of UV-induced lesions [16, 17, 40]. In order to determine whether the C-terminal domain of Rev1 is similarly important in the bypass of AP sites, we introduced a mutant rev1 allele (rev1-CDEL) that eliminates the C-terminal 72 amino acids of the protein into the BER- and BER/NER-defective strains. In both backgrounds, the rev1-CDEL allele resulted in pTET-lys2ΔA746 reversion rates and spectra that were indistinguishable from those associated with the rev1Δ allele (Figure 6).
4. Discussion
In previous experiments carried out in other labs as well as in the work presented here, the Rev1 Y-family DNA polymerase is required for Pol ζ-dependent lesion bypass, with its loss resulting in a reversionless phenotype identical to that associated with loss of Rev3 (reviewed in [3]). Deciphering the precise function of Rev1 in lesion bypass has been complicated by the fact that, in addition to its ability to catalyze dCMP insertion opposite lesions in vitro, it has a DNA-binding BRCT domain and an essential C-terminal scaffolding domain. Whereas the latter two domains are assumed to be universally important, the relevance of the dCMP transferase activity is likely lesion specific [18]. Here, we focus specifically on the functions of Rev1 during the bypass of endogenously generated AP sites, which is thought to be a multi-polymerase process requiring different polymerases for the insertion and extension steps [13]. Accumulated evidence indicates that Pol ζ is required for the extension step, but which polymerase is primarily responsible for the insertion step has remained controversial. The controversy revolves around whether dCMP is the predominant nucleotide inserted, which would support a catalytic role for Rev1 during AP-site bypass, or whether dAMP is the preferred nucleotide, which would indicate a primary role for a different DNA polymerase during the insertion step.
In the current study, highly sensitive pTET-lys2 frameshift- and nonsense-reversion assays were used as tools to generate AP sites. The uniqueness of these assays is that the source of the AP sites has been defined: uracil specifically replaces thymine in highly transcribed DNA, and it is the subsequent excision of uracil by Ung1 that creates the AP sites [25, 26]. Deletion of the C-terminal 72 amino acids of Rev1 resulted in mutation rates and spectra that were indistinguishable from those associated with the rev1Δ allele, indicating no AP-site bypass. This is in agreement with previous results demonstrating a requirement for the Rev1 C-terminal domain in Pol ζ-mediated UV survival and mutagenesis [16]. Similar to the yeast protein, the analogous C-terminal domain of human Rev1 also mediates interaction with human Rev7 [41], and is required for DNA-damage tolerance in the avian DT40 cell line [42]. In contrast to the null phenotype associated with the rev1-CDEL allele, mutation of the BRCT domain reduced, but did not eliminate AP-site bypass in the pTET-lys2ΔA746 assay. While this is consistent with previous transformation-based assays using engineered AP sites [23, 24, 38, 43], the associated changes in frameshift-reversion spectrum suggest that the DNA-binding activity of the BRCT domain affects primer-template slippage during lesion bypass.
In BER (apn1 ntg1 ntg2)- and BER/NER (apn1 rad14)-defective backgrounds the pTET-lys2ΔA746 assay selects for net +1 frameshift mutations that arise during the bypass of uracil-derived AP sites that accumulate when uracil replaces thymine. In particular, complex frameshifts at a 6A hotspot result from dCMP, dGMP or dTMP insertion, while non-mutagenic dAMP insertion yields simple +1 frameshifts within the 6A run (see Figure 3). Because the base substitution is not the selected event, we believe that this specific assay allows an unbiased assessment of the frequency with which each dNMP is inserted opposite AP sites, as well as the relevance of Rev1 catalytic activity to the insertion specificity. While it could be argued that concerted misincorporation-slippage during AP-site bypass is rare and, therefore, does not provide an appropriate read-out of insertion specificity, it should be noted that the overall Lys+ rates in the frameshift-reversion assay were within 7-fold of those obtained in the nonsense-reversion assay. This suggests that, if there is a mononucleotide run appropriately positioned near an AP site, slippage subsequent to the insertion step is a frequent event.
In the presence of Rev1, both complex and simple frameshifts were common in the pTET-lys2ΔA746 assay, with almost all complex events containing the T > G or A > C transversion diagnostic of dCMP insertion opposite the AP site. Although introduction of the catalytically inactive rev1-AA allele had only minor effects on the total rate of Lys+ revertants, there were very striking and distinctive changes in mutation types at the 6A hotspot. The complex events that reflect dCMP insertion opposite AP sites were eliminated in the rev1-AA mutants and there was a compensatory increase in the simple frameshifts predicted to result when dAMP is inserted opposite the initiating AP site. These data demonstrate that insertion of dCMP absolutely depends on the catalytic activity of Rev1 and suggest that dCMP, rather than dAMP, is the most frequently inserted dNMP during AP-site bypass in this system.
Analogous experiments were done with BER- and BER/NER-defective strains containing the pTET-lys2-TAA nonsense allele, a system that only allows detection of dCMP, dGMP or dTMP insertion opposite uracil-derived AP sites (dAMP insertion does change the stop codon). Consistent with Rev1-catalyzed dCMP insertion opposite AP sites, the reversion rate of the pTET-lys2-TAA allele was reduced 5–10 fold upon introduction of the rev1-AA allele. Reversion spectra confirmed that dCMP insertion is strongly preferred to that of dGMP or dTMP, and that dCMP insertion absolutely requires the catalytic activity of Rev1. Because compensatory dAMP insertion is not detected in the pTET-lys2-TAA reversion assay, we also were able to observe what would normally be very minor insertion of dGMP or dTMP opposite AP sites. Our data suggest that the nucleotide selectivity during AP site bypass in yeast follows the pattern of dCMP > dAMP ≫ dGMP ~ dTMP.
The role of Rev1 in inserting the nucleotides other than dCMP is presumably structural, and the identity of the DNA polymerase that steps in to accomplish insertion opposite AP sites in rev1-AA strains is not known. While Pol η can accomplish the insertion step in vitro [44], its loss in the rev1-AA background had very little, if any, effect on reversion rates or spectra in either assay used here. Among the three yeast replicative DNA polymerases, both Pol δ and Pol α have been reported to insert nucleotides opposite AP sites in biochemical assays [13, 45]; Pol ε has not been examined. Given that Pol δ has a strong preference for dAMP insertion opposite AP sites in vitro [13], and the requirement for its noncatalytic, Pol32 subunit for Pol ζ-dependent mutagenesis in vivo [5, 19, 46–49], this replicative polymerase may be the most likely candidate.
Finally, it should be noted that both of our reporters were located close to the strong ARS306 origin of replication on Chromosome III. Because AP sites accumulate on complementary strands in BER- versus BER/NER-defective backgrounds [26], our systems have the potential to detect replication-related differences in Rev1 activity during AP-site bypass. AP sites remain mainly on the nontranscribed strand in BER-defective strains, which corresponds to the lagging-strand template for a fork originating from the ARS306 [27]. Conversely, in BER/NER-defective strains, AP lesions persist on the transcribed strand, which is the leading-strand template during replication. Although there were generally no differences in the mutagenic effects associated with various rev1 alleles tested in these two strain backgrounds, there was a difference in the compensatory insertion of dAMP in the rev1-AA mutants. In the BER-defective background, there was complete compensation so that there was no associated drop in the overall reversion rate; in the BER/NER-defective background, there was little compensation, resulting in a 3-fold drop in Lys+ rate. One interpretation of this difference is that the dAMP-inserting polymerase has better access to AP sites encountered during lagging-strand than during leading-strand synthesis. Given that Pol δ is responsible for most lagging-strand synthesis in yeast [50], this would be consistent with it being the major polymerase responsible for dAMP insertion opposite AP sites.
By demonstrating the significance of Rev1 dCMP transferase activity during AP-site bypass, our data further support a model in which engagement of the catalytic activity of this protein depends on the specific lesion being bypassed [18, 51]. The dCMP insertion specificity of Rev1 would make biological sense if most spontaneous AP sites occur at guanines, as this would serve to limit AP-associated mutagenesis. Available data suggest, however, that direct incorporation of uracil is the major source of endogenous AP sites in yeast DNA [52], which would result in the TA > GC mutations of the type documented here. It still remains possible, however, that the most biologically relevant lesion, especially outside a laboratory environment, is a damaged form of guanine, which may or may not lead to AP site formation. If this were the case, then the dCMP transferase activity of Rev1 would provide an error-free bypass mechanism.
In higher eukaryotes, the TLS of uracil-derived AP sites is a critical step in the somatic hypermutation of immunoglobulin genes [4, 53, 54]. In mammalian cells, Rev1 appears to play an important role in this process, with its loss affecting the spectrum of mutations [55]. Similar shifts in mutation spectra have been observed in mouse or chicken cells expressing catalytically inactive Rev1 [56, 57]. Finally, the dCMP transferase activity of Rev1 documented here might be especially relevant when the cellular ratio of dUTP to dTTP is perturbed. Inhibitors of thymidylate synthase and anti-folates, for example, are commonly used chemotherapeutics that elevate uracil in DNA [58]. The presence and/or catalytic activity of Rev1 might have implications for cell survival and especially mutagenesis following treatment with such drugs.
Supplementary Material
Mutagenic AP-site bypass in the pTET-lys2-TAA assay. The substitution of uracil for thymine leads to a mutagenic AP site at the thymine of the stop codon, or opposite either of the two adenines. The specific base changes and corresponding amino acid substitutions resulting from the insertion of each dNMP are illustrated.
AP-site bypass in yeast requires Pol ζ and Rev1
Rev1 catalytic activity is biologically relevant during AP-site bypass
Insertion specificity opposite AP sites is dCMP > dAMP ≫ dGMP ~ dTMP
Acknowledgments
This work was supported by the National Institutes of Health (grant numbers GM038464 and GM093197).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Loeb LA. Apurinic sites as mutagenic intermediates. Cell. 1985;40:483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- 2.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 3.Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auerbach PA, Demple B. Roles of Rev1, Pol ζ, Pol32 and Pol η in the bypass of chromosomal abasic sites in Saccharomyces cerevisiae. Mutagenesis. 2010;25:63–69. doi: 10.1093/mutage/gep045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 7.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 8.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 10.Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 11.Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 13.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 15.de Groote FH, Jansen JG, Masuda Y, Shah DM, Kamiya K, de Wind N, Siegal G. The Rev1 translesion synthesis polymerase has multiple distinct DNA binding modes. DNA Repair (Amst) 2011;10:915–925. doi: 10.1016/j.dnarep.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 16.D’Souza S, Waters LS, Walker GC. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair (Amst) 2008;7:1455–1470. doi: 10.1016/j.dnarep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acharya N, Johnson RE, Prakash S, Prakash L. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26:9555–9563. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiltrout ME, Walker GC. The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics. 2011;187:21–35. doi: 10.1534/genetics.110.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol δ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillet M, Van Der Kemp PA, Boiteux S. dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:2056–2066. doi: 10.1093/nar/gkl139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs PE, Lawrence CW. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J Mol Biol. 1995;251:229–236. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka C, Kobayashi K, Kawaguchi N, Kunitomi N, Moriyama K, Hata Y, Iwai S, Loakes D, Noskov VN, Pavlov Y, Negishi K. Use of yeast transformation by oligonucleotides to study DNA lesion bypass in vivo. Mutat Res. 2002;502:53–60. doi: 10.1016/s0027-5107(02)00023-4. [DOI] [PubMed] [Google Scholar]
- 23.Kow YW, Bao G, Minesinger B, Jinks-Robertson S, Siede W, Jiang YL, Greenberg MM. Mutagenic effects of abasic and oxidized abasic lesions in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:6196–6202. doi: 10.1093/nar/gki926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otsuka C, Kunitomi N, Iwai S, Loakes D, Negishi K. Roles of the polymerase and BRCT domains of Rev1 protein in translesion DNA synthesis in yeast in vivo. Mutat Res. 2005;578:79–87. doi: 10.1016/j.mrfmmm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Kim N, Jinks-Robertson S. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature. 2009;459:1150–1153. doi: 10.1038/nature08033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spell RM, Jinks-Robertson S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods in molecular biology (Clifton, NJ. 2004;262:3–12. doi: 10.1385/1-59259-761-0:003. [DOI] [PubMed] [Google Scholar]
- 32.Harfe BD, Jinks-Robertson S. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4766–4773. doi: 10.1128/mcb.19.7.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harfe BD, Jinks-Robertson S. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 34.Bankmann M, Prakash L, Prakash S. Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature. 1992;355:555–558. doi: 10.1038/355555a0. [DOI] [PubMed] [Google Scholar]
- 35.Popoff SC, Spira AS, Johnson AW, Demple B. The yeast structural gene (APN1) for the major apurinic endonuclease: homology to E. coli endonuclease IV. Proc Natl Acad Sci USA. 1985;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meadows KL, Song B, Doetsch PW. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res. 2003;31:5560–5567. doi: 10.1093/nar/gkg749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdulovic AL, Minesinger BK, Jinks-Robertson S. The effect of sequence context on spontaneous Polζ-dependent mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:2082–2093. doi: 10.1093/nar/gkn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 39.Acharya N, Haracska L, Prakash S, Prakash L. Complex formation of yeast Rev1 with DNA polymerase η. Mol Cell Biol. 2007;27:8401–8408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Souza S, Walker GC. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol Cell Biol. 2006;26:8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 42.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otsuka C, Sanadai S, Hata Y, Okuto H, Noskov VN, Loakes D, Negishi K. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase η. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase η in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci USA. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minesinger BK, Jinks-Robertson S. Roles of RAD6 epistasis group members in spontaneous polζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2005;169:1939–1955. doi: 10.1534/genetics.104.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 49.Huang ME, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 50.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Wang J, Zhang Y, Wang Z. The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis. Nucleic Acids Res. 2010;38:5036–5046. doi: 10.1093/nar/gkq225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 54.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA--general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 55.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang JY. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- 57.Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 58.Berger SH, Pittman DL, Wyatt MD. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem Pharmacol. 2008;76:697–706. doi: 10.1016/j.bcp.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutagenic AP-site bypass in the pTET-lys2-TAA assay. The substitution of uracil for thymine leads to a mutagenic AP site at the thymine of the stop codon, or opposite either of the two adenines. The specific base changes and corresponding amino acid substitutions resulting from the insertion of each dNMP are illustrated.