Abstract
To provide insight as to how cells receive information from their external surroundings, synthetic hydrogels have emerged as systems for assaying cell function in well-defined microenvironments where single cues can be introduced and subsequent effects individually elucidated. However, as the field seeks to answer more complex biological questions, advanced material systems are needed that allow dynamic alteration of the 3D cellular environment with orthogonal reactions that enable multiple levels of control of biochemical and biomechanical signals. Here, we sought to synthesize one such 3D culture system using cytocompatible and wavelength-specific photochemical reactions to create hydrogels that allow orthogonal and dynamic control of the material properties through independent spatiotemporally-regulated photocleavage of crosslinks and photoconjugation of pendant functionalities. Results demonstrate the versatile nature of the chemistry to create programmable niches to study and direct cell function by modifying the local hydrogel environment.
Since its conception by Sharpless in 20011, the concept of click chemistry has been rapidly adopted in many disciplines, perhaps most notably in material science, with annual publication numbers continuing to increase exponentially2–5. The click philosophy idealizes reactions that enable researchers to link covalently two reactants in a straightforward, modular, high-yielding manner. The copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) is frequently billed as the quintessential example in meeting these criteria. Though each of the click reactions has a variety of desirable properties, their true benefit lies in the orthogonality of these reactions with respect to many common reactive groups (e.g., amines, alcohols, acids)6. Reaction orthogonality enables independent control over multiple functional groups in a single system and opens the door for the synthesis of materials with ever-increasing complexity for an ever-expanding list of applications.
There is also a growing interest in chemical reactions that can be performed in the presence of live cells6. These reactions must proceed mildly in aqueous medium and a defined chemical environment (5% CO2 and atmospheric O2 levels), under regulated pH (7.4), temperature (37°C), osmolarity (~300 mOsM), and involve non-toxic reactive moieties and (by)products. The requirement of reactions that proceed under physiological conditions is a stringent constraint and presents severe limitations on reaction selection. In particular, there are even fewer chemistries that proceed in a specific manner, while limiting side reactions with the plethora of functional groups that are found in biological systems. These elite reactions are considered bio-orthogonal and are a necessity for probing chemically and directing biological function.
In many instances, the ideal cytocompatible reaction would not only be selected by its bio-orthogonality, but also by its capacity to be controlled in both time and space. In this regard, photochemical reactions are widely regarded for their spatiotemporal control, where the reaction of interest is defined by when and where the light is delivered to the system7. Photolithographic techniques, where masked light is projected directly onto a sample, enable photoreactions to be confined to specific regions within a sample as defined by a 2D mask pattern, while focused laser light (either single- or multi-photon) provides full 3D control over where a specific reaction occurs within the volume of a material. While the effects of attenuation and scattering must be carefully taken into consideration, light-based chemistries have become a powerful tool for material synthesis and spatial modification, owing to their ease of implementation and readily available inexpensive light sources, and have become indispensible in formation and subsequent modification of biomaterials.
To date, biocompatible light-based chemistries have enabled control in space and time of either the gel degradation or gel chemistry of synthetic cell culture systems. By introducing chemical functionalities in user-defined patterns within the material, cell spreading and migration have been explicitly controlled in 3D8–13. Alternatively, cell outgrowth and stem cell fate have been directed by altering the gel’s structural properties14–18. Nevertheless, independent control over both the material’s physical and chemical makeup in 3D, along with in time, allow dynamic tailoring of a cell’s microenvironment, and has not been demonstrated. Such 4D control of material properties would be tremendously advantageous in a number of biomaterial applications, including 3D cell culture, stem cell expansion, cancer metastasis, and tissue regeneration. Of further importance and novelty, the ability for the experimenter to control gel properties at any point in space and time enables opportunities for unique experiments, such as the ability to introduce dynamically a cell ligand or allow cell-cell interactions at specified locations. These programmable cell culture niches facilitate the ability to perform newfound experiments and answer questions about the dynamic exchange of information between a cell and its niche. In this work, we present one such system where multiple wavelengths of light are utilized to control independently the functionality and architecture of a hydrogel network formed via a copper-free alkyne-azide reaction (Fig. 1). Each of the reactions is cytocompatible, and both photoconjugation and photocleavage reactions were used to spatiotemporally regulate materials properties, including the presentation of integrin-binding motifs and network erosion through cleavage of crosslinking moieties. This platform allows gel parameters to be tuned in real-time, and results demonstrate how spatiotemporal regulation of material properties can be used to direct the function of embedded cells.
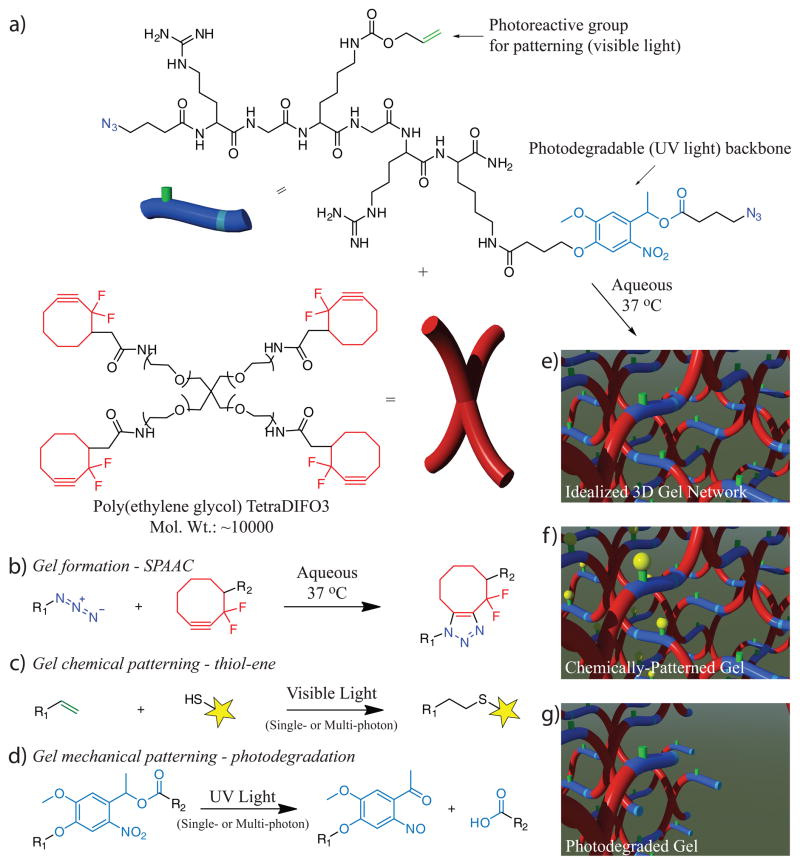
Figure 1. Synthesis, photocoupling, and photodegradation for tuning chemical and physical properties of click-based hydrogels.
(a) Click-functionalized macromolecular precursors (i.e., PEG-tetraDIFO3 and bis(azide)-functionalized polypeptides) form a 3D ideal hydrogel structure via a step-growth polymerization mechanism by the (b) SPAAC reaction. (c) In the presence of visible light (λ = 490 – 650 nm or 860 nm), thiol-containing biomolecules are covalently affixed to pendant vinyl functionalities throughout the hydrogel network via the thiol-ene reaction. (d) A nitrobenzyl ether moiety within the backbone of the polymer network undergoes photocleavage in the presence of single or multi-photon UV light (λ = 365 nm or 740 nm) that results in photodegradation of the network. Schematics of the formed SPAAC-based idealized gel, network post thiol-ene functionalization, and material post photodegradation are found in (eg), respectively.
Results and Discussion
A four-arm poly(ethylene glycol) (PEG) tetracyclooctyne (Mn ~ 10,000 Da) was reacted with a bis(azide) di-functionalized polypeptide (Azide-RGK(alloc)GRK(PLazide)-NH2) via a copper-free, strain promoted azide-alkyne cycloaddition (SPAAC) reaction between terminal difluorinated cyclooctyne (DIFO3) and azide (–N3) moieties with 1:1 stoichiometry at 10 wt% total macromer concentration to form an idealized 3D network with minimal local defects. The ring strain and electronegative fluorine substituents of DIFO3 enable the SPAAC reaction to proceed rapidly, without a catalyst, and in the presence of cells19. Network gelation occurs ~2 min after mixing as estimated by the crossover point of G′ and G″ (Supplementary Fig. S1). By including a synthetic polypeptide in the gel formulation, the precise chemical makeup of the material is tailored readily by choice of the amino acid sequence, allowing one to tailor the biofunctionality (e.g., enzymatic degradability, integrin binding ligands, protein affinity binding sites) and introduce bioorthogonal reactive moieties (e.g., vinyl groups, azides). Ultimately, the timescale and mechanism of the SPAAC reaction permits high viability (>95%) during encapsulation of both established cell lines, as well as primary cell types (Supplementary Fig. S11)8.
Biochemical Control via Thiol-ene Photoconjugation
Incorporated into the synthetic peptide is the commercially-available lysine(allyloxycarbonyl) (alloc) amino acid, whose alloc protecting group is stable to standard solid phase peptide synthesis methods and contains a vinyl functionality that is readily photocoupled to thiol-containing compounds, such as cysteine, via the thiol-ene reaction. Although a number of reactions can be controlled with light, including the CuAAC by a photogenerated copper(I) catalyst20, the radical-mediated thiol-ene addition has emerged as a versatile click reaction that can be photochemically initiated21,22. This reaction, which involves the catalytic propagation of a thiyl radical across an olefin (–C=C) and subsequent chain transfer from the resulting carbon radical to a thiol (–SH), has gained recent interest as an approach to functionalize systems with biomolecules8,23,24, control dendrimer formation25, as well as synthesize other complex materials26,27. The propagating thiyl radical is readily generated in the presence of both cleavage-type, as well as hydrogen-abstracting photoinitiators28, enabling thiol-containing molecules to be physically linked to vinyl-functionalized moieties over a variety of light conditions including in the visible range. This reaction can be employed with peptides, thiolated full proteins, and small molecules that are individually capable of diffusing throughout the hydrogel (Supplementary Figs. S2 & S3), though peptides that contain free thiols in their bioactive domain may exhibit reduced biological effect upon thiol-ene coupling. The reaction is regarded as cytocompatible (Supplementary Fig. S11), as well as bioorthogonal, facilitating its use in biological systems21 and enables materials to be functionalized dynamically with specific molecules of interest at any given location and time.
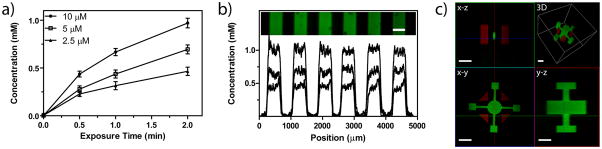
After gel formation, fluorescently-labeled thiol-containing biomolecules were swollen into the network along with a small amount of eosin Y photoinitiator (2.5 – 10 μM), which was followed by visible light irradiation (λ = 490 – 650 nm) at low intensities (10 mW cm−2) and short durations (0.5 – 2 min). The extent of photocoupling was visualized and quantified using confocal microscopy and was controlled by the photoinitiator concentration and exposure time (Fig. 2a). Specifically, patterning concentrations between 0 and 1 mM were obtained with short light exposures of only a few minutes. By irradiating through a photomask, patterning was confined to specific locations throughout the gel, as demonstrated by the transfer of a 400 μm wide line pattern through the depth of the sample (Fig. 2b, Supplementary Fig. S4). Additionally, multi-photon initiation techniques (λ = 860 nm) were used to create elaborate, user-defined, 3D biochemical patterns within the hydrogel (Fig. 2c). Here, a 300 × 400 × 400μm interconnected 3D structure composed of multiple shapes and two distinct peptides was created. The resolution that we achieve with multi-photon-based patterning is ~1 μm in the x–y plane and ~3–5 μm in the z plane, which are values typical of multi-photon imaging methods and represent a limitation of the optics and not the chemistry. The photocoupling process can be repeated many times over, with each cycle requiring on the time scale of a few hours for introduction and removal of the signal via diffusion (depending on the gel dimensions). Thus, multiple signals can be incorporated with micron-scale patterning resolution on time and size scales that are relevant for many cell culture experiments.
Figure 2. Biochemical patterning within preformed click hydrogels using visible light.
Upon swelling thiol-containing biomolecules into preformed gels, pendant functionalities are affixed to the hydrogel backbone via the thiol-ene reaction upon exposure to visible light (λ = 490 – 650 nm). (a) The final patterned concentration of a fluorescent RGD peptide (AF488-AhxRGDSC-NH2) depends on the amount of photoinitiator present (2.5, 5, and 10 mM Eosin Y), as well as the exposure time to visible light (0 – 2 min, 10 mW cm−2). (b) The network functionalization with pendant fluorescently-labeled peptides is confined to user-defined regions within preformed gels using photolithography (0.5, 1, or 2 min exposure with increased patterning concentration for increased exposure time, 10 mW cm−2, 10 μM eosin Y). (c) The photocoupling reaction is controlled in 3D by rastering the focal point of multi-photon laser light (λ = 860 nm) over defined volumes within the gel, affording micron-scale resolution in all spatial dimensions. Additionally, the patterning process can be repeated many times to introduce multiple biochemical cues within the same network, as demonstrated by the red- and green- labeled patterned peptides within the same gel. Images represent confocal projections and 3D renderings. Scale bar = 400 μm for (b) and 100 μm for (c).
Biophysical Control via Photodegradation
The utility of a peptide linker enables desired sequences, as well as desired functionalities, to be precisely incorporated in a modular fashion. In addition to the pendant alloc vinyl functionality, the peptide includes a photodegradable nitrobenzyl ether moiety (PLazide) within its backbone, enabling photocleavage of the crosslinks upon exposure to UV light (either λ = 365 nm for single-photon of λ = 740 nm for multi-photon14,29). Specifically, the irreversible photocleavage of an o-nitrobenzyl ether moiety into nitroso- and acid-terminated byproducts permits a previously-intact chemical linkage to be cleaved photolytically. The photolabile group degrades under cytocompatible irradiation conditions, including 365 nm light29, and has been used for the uncaging of proteins30, to cleave peptides from a solid support31, as well as to control cell adhesion9,32. The functionality has also been incorporated into materials to produce networks that are capable of degrading in the presence of light14,33,34, allowing the effects of physical material cues on cell function to be probed35–37.
Based on kinetic nuclear magnetic resonance (NMR) as well as photorheometry studies, the photoscission of the PLazide moiety (see Supplementary Figs. S5) was found to follow a first order degradation with a rate constant (k) that can be expressed as:
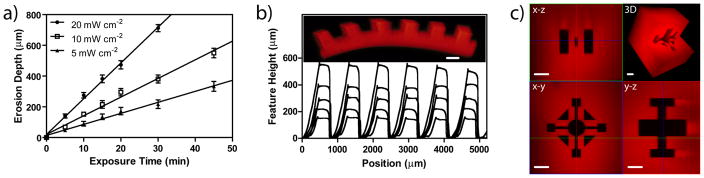
where φ is the quantum yield (determined to be 0.020), ε is the molar absorptivity of the sample (4780 M−1 cm−1 for PLazide at λ = 365 nm), I is the intensity of light, NA is Avogadro’s number, h is the Planck constant, and ν is the frequency of the associated electromagnetic wave (see Supplementary Fig. S6, S7, & S8). For typical exposure conditions (λ = 365 nm, 10 mW cm−2), k was determined to be 2.9 × 10−3 sec−1, and correlates well with other photodegradable moieties14,31. Physical channels were eroded downward from the surface of optically-thick samples, with the depth of photodegradation directly related to the total light intensity (5, 10 and 20 mW cm−2), as well as the exposure time of UV light (0 – 45 min) (Fig. 3a, Supplementary Fig. S9), and the total light dosage delivered to the material (See Supplementary Fig. S10). As with the photocoupling reaction, the photocleavage reaction was confined to regions of interest within the sample using photolithographic processes to create channels of varying depth (~150 – 600 μm) (Fig. 3b), as well as multi-photon patterning approaches to erode precisely defined 3D regions of interest with user-defined shapes and connectivity (Fig. 3c, λ = 740 nm). Each process affords a high level of patterning fidelity, similar to that by the photocoupling reaction.
Figure 3. Biophysical patterning within preformed click hydrogels using UV light.
In the presence of UV light (λ = 365 nm), photolabile functionalities within the hydrogel crosslinks undergo an irreversible cleavage, thereby decreasing the total network connectivity, resulting in local material degradation and removal of the fluorescent hydrogel material. (a) In optically-thick samples, the depth of photodegradation is directly related to the incident light intensity (5, 10 and 20 mW cm−2), as well as the exposure time to visible light (0 – 45 min). (b) Using mask-based photolithographic techniques, network degradation was confined to user-defined regions within fluorescently-labeled gels (10, 15, 20, 30, and 45 min, feature height increasing with exposure time), as measured by profilometry. (c) The photodegradation reaction is controlled in 3D with micron-scale resolution in all dimensions using focused multi-photon laser light (λ = 740 nm). Images represent confocal projections and 3D renderings. Scale bar = 400 μm for (b) and 100 μm for (c).
Orthogonal Photoreactions for Advanced 3D Cell Culture
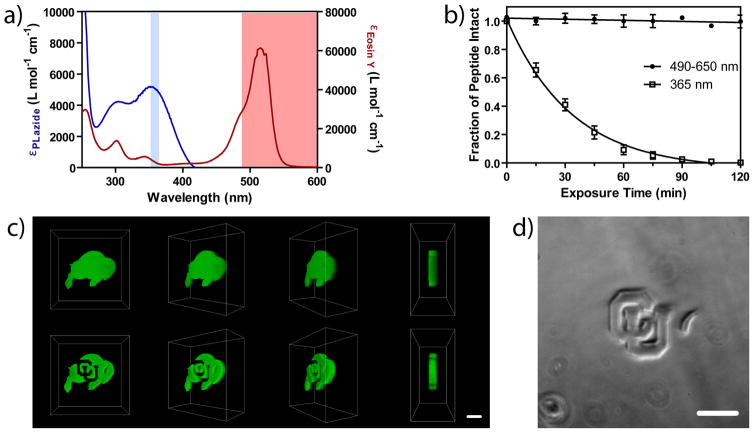
The utility of the photocoupling and photocleavage reactions ultimately stems from their ability to be performed orthogonally, such that both network mechanical and chemical makeup are controlled independently. The peak absorbance for the photolabile group and the visible photoinitiator was found to be ~350 and ~520 nm, respectively, with relatively little overlap of the absorbance spectra (Fig. 4a). As eosin Y also has a low absorbance at λ = 365 nm, photocoupling during photodegradation was readily prevented by performing degradation only in the absence of photoinitiator. Alternatively, photocoupling was initiated first with visible light (λ = 490 – 650 nm) and photodegradation was commenced with subsequent UV irradiation. The orthogonality of these reactions was confirmed by solution NMR studies where photocleavage was quantified under both visible and UV light initiation conditions using model compounds (Fig. 4b). To illustrate orthogonality of the reactions within the same material system, a buffalo logo was first photocoupled within the 3D network using visible light, and user-defined letters (CU) were eroded within the fluorescent logo at a later time (Fig. 4c). Photodegradation was confirmed to be confined only to the areas of interest by brightfield microscopy (Fig. 4d), as well as with the disappearance of the fluorescently-labeled reporter peptide, indicating that cues can be spatially coupled and subsequently removed with orthogonal reactions.
Figure 4. Orthogonality of photocoupling and photodegration reactions.
(a) The peak absorbance for the photoinitiator (red) and the photolabile group (blue) is well separated (~520 and ~350 nm, respectively), thus enabling photocoupling and photodegradation reactions to be performed independently from one another using different light sources (illustrated with colored bars). (b) NMR studies indicate that the photodegradable moiety cleaves readily in the presence of UV light (λ = 365 nm, 10 mW cm−2), but remains intact when exposed to the visible light used to initiate the photocoupling reaction (λ = 490 – 650 nm, 10 mW cm−2). (c) Multi-photon visible light was first used to couple a fluorescently-labeled peptide within the center of the hydrogel in a user-defined 3D pattern (top, buffalo), and the network was subsequently degraded locally with multi-photon UV light, thereby removing the peptide from selected regions (bottom, CU and horn). (d) Brightfield microscopy confirms that photocleavage is confined only to user-defined locations within the gel, and that the photocoupling light conditions do not give rise to undesired degradation. Images in (c) represent 3D renderings of confocal z-stacks. Scale bars = 100 μm.
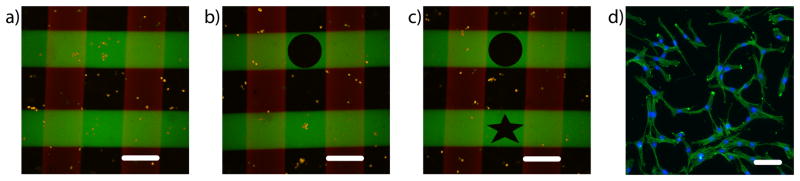
To provide a demonstration of the potential utility of these two photoreactions for advanced 3D cell culture, Figure 5 presents an approach where one might assay the specific effects of a variety of biomolecular cues on cell function within an otherwise uniform gel culture platform. Here, human mesenchymal stem cells (hMSCs) were encapsulated in gels synthesized via SPAAC chemistry, and their cellular microenvironment was patterned via the thiol-ene photocoupling reaction with perpendicular lines of width = 200 μm of integrin-binding peptide ligands, RGD and PHSRN, at ~1 mM each. These peptide sequences are both derived from sequences found in fibronectin and are known to elicit some degree of synergy on cell adhesion38. The patterning created a repeating array of four distinct biochemical culture conditions (no cue, RGD alone, PHSRN alone, or both RGD and PHSRN) within the same gel (Fig. 5a). At a later time point, the photodegradation reaction was exploited to capture cells in spatially-defined regions of interest by exposing the gel to a given condition of light to induce erosion and liberate cells from their 3D culture environment (Fig. 5b). This process can be repeated many times over at different time points and locations to collect cells that have been exposed to either the same or different biochemical conditions, demonstrating full spatiotemporal control over cell subpopulation sampling (Fig. 5c). The released cells are readily collected, subsequently plated, expanded, and available for additional biological assays, including those that may be more difficult to perform on encapsulated cells. hMSCs remained viable (>95%) throughout the entire process (Supplementary Fig. S11). Here, we plated the released hMSCs and visualized their cytoskeletal organization using a fluorescent phalloidin, which stains for F-actin (Fig. 5d).
Figure 5. Culture and recovery of hMSCs from hydrogel microenvironments.
CellTracker Orange-labeled human mesenchymal stem cells (hMSCs) were encapsulated within the click hydrogel formulation at 5 × 106 cells mL−1. (a) 24 hours post encapsulation, perpendicular 200 μm wide lines of ~1 mM RGD and PHSRN were patterned throughout the hydrogel via thiol-ene photocoupling to create an array of four distinct biochemical conditions (no cue, RGD, PHSRN, RGD and PHSRN). (b) 4 hours later, channels of user-defined shape (cylindrical) were eroded down from the surface of the hydrogel to capture entrapped cells exposed to a specific cue. (c) This process was repeated 1 hour later to release entrapped cells within a different location of the material and a different shape (star-shaped cylinder). (d) The released cells were isolated by centrifugation, cultured in a 96-well plate for 48 hours, and their cytoskeleton was visualized with a fluorescent phalloidin. (a–c) RGD is shown in green, PHSRN red, and hMSCs orange. Images represent single confocal slices within the 3D gel. (d) F-actin is shown in green, nuclei blue. Image represents inverted fluorescence micrograph. Scale bars = 200 μm in (a–c), 50 μm in (d).
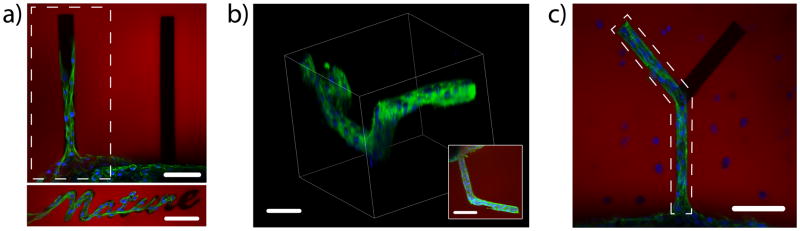
To further demonstrate how these reactions can be used to manipulate cellular functions in a spatiotemporally-regulated manner, a cell-laden (3T3 fibroblasts) fibrin clot was encapsulated within the click hydrogel formulation. After 2 hours, physical channels were eroded radially from the spherical clot via multi-photon photodegradation of the network to direct collective cell migration. Additionally, only specific regions of the gel were functionalized with RGD via the thiol-ene photocoupling reaction. Cells were found to leave the clot and migrate into the patterned hydrogel channels, but only when eroded migration channels were present and their surfaces decorated with the RGD adhesive ligand (Fig. 6a). Using two-photon patterning techniques, cell outgrowth was explicitly directed in all three spatial dimensions (Fig. 6b). This directed outgrowth can be performed in the presence of other encapsulated cells or with combinations of cell types. For example, hMSCs were encapsulated in the gel surrounding the 3T3-fibroblast-laden clot, and the fibroblasts were directed into the surrounding hMSC microenvironment in a manner controlled by changes in the local gel environment. The patterned 3T3 fibroblasts were found to create complex structures in the presence of encapsulated hMSCs (Fig. 6c). These photoreactions are included to demonstrate how one might engineer complex, multicellular structures, ultimately expanding the potential for engineering tissue constructs with spatially varying cellularity in advanced bioreactors or culture systems.
Figure 6. Directed 3D cell motility within patterned hydrogels.
(a) A fibrin clot containing 3T3 fibroblasts was encapsulated within the click hydrogel formulation. Chemical channels of RGD, a cell-adhesive fibronectin motif, as well as physical channels of user-defined shape were created radially out of the roughly spherical clot. The combination of having physical space to spread as well as chemical moieties to bind to were found to be required for collective cell migration. By day 10, cells were found to migrate only down the physical channel that was functionalized with RGD. (b) By creating 3D functionalized channels, cell outgrowth was controlled in all three spatial dimensions, with the image inset illustrating a top-down projection. (c) The outgrowth of 3T3 fibroblast cells was controlled in the presence of encapsulated human mesenchymal stem cells (hMSCs) and confined to branched photodegraded channels that were functionalized with RGD. The regions of RGD-functionalization are depicted by the dashed polygons in (a) and (c). The hydrogel is shown in red, F-actin green, and cell nuclei blue. Scale bars = 100 μm.
As presented, this work utilizes two novel photoreaction schemes to combine and exploit features of previously mutually exclusive technologies. Namely, the physical and chemical properties of the network can be controlled independently with orthogonal light-based chemistries, allowing for real-time manipulation of cell function within a simplified synthetic microenvironment. These reactions are performed dynamically with full spatiotemporal control, enabling full user-direction over these programmable cell niches. The cytocompatibility of the reaction processes should enable newfound opportunities for experiments to test basic hypotheses about critical events regulating cell-materials interactions at multiple time and size scales and, with this knowledge, improve strategies for stem cell culture, biomaterial design, 3D cell culture assays and tissue regeneration.
Methods
Synthesis of Click-Functionalized Macromolecular Precursors
Synthesis of PEG-tetraDIFO3
DIFO319,39 (121 mg, 0.6 mmol, See Supplementary Fig. S12) and 2-(1H-7-Azabenzotriazol-1-yl)--1,1,3,3-tetramethyl uronium hexafluorophosphate Methanaminium (HATU, 225 mg, 0.6 mmol, Anaspec) were dissolved in minimal dimethylformamide (DMF, 5 mL) with N,N-diisopropylethylamine (DIEA, 210μL, 1.2 mmol) and reacted for 5 min at RT. This solution was then added to 4-arm PEG tetraamine (Mn ~ 10,000 Da, 1 g, 0.4 mmol NH2, JenKem) and stirred overnight, concentrated, dissolved in dH2O, dialyzed (MWCO ~ 2 kDa, SpectraPor), filtered, and lyophilized to yield a white powder (1.03 g, 96%). Functionalization was confirmed to be >95% by 1H-NMR.
Synthesis of Bis(azide)-functionalized photodegradable peptide crosslinker
The allyl-ester containing peptide H-RGK(alloc)GRK(dde)-NH2 was synthesized (Protein Technologies Tribute peptide synthesizer) through Fmoc solid-phase methodology and HATU activation. 4-azidobutanoic acid (See Supplementary Fig. S13) was coupled to the N-terminal amine with HATU, the 1-(4,4-dimethyl-2,6-dioxacyclohexylidene)ethyl (Dde) group was removed with 2% hydrazine monohydrate (Sigma) in DMF (3 × 10 min), and 4-(4-(1-(4-azidobutanoyloxy)ethyl)-2-methoxy-5-nitrophenoxy)butanoic acid (PLazide, see Supplementary Fig. S13) was coupled to the ε-amino group of the C-terminal lysine. Resin was treated with trifluoroacetic acid/triisopropylsilane/water (95:2.5:2.5) for 2 h and precipitated in and washed with (2x) ice-cold diethyl ether. The crude peptide was purified using semipreparative reversed-phase high-performance liquid chromatography (RP-HPLC) (Waters Delta Prep 4000) using a 70 min linear gradient (5 – 95% of acetonitrile and 0.1% trifluoroacetic acid) and lyophilized to give the product (Azide-RGK(alloc)GRK(PLazide)-NH2) as a fluffy, yellow solid. Peptide purity was confirmed with analytical RP-HPLC and matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (Applied Biosystems DE Voyage) using a-cyano-4-hydroxycinnamic acid matrix (Sigma): Calculated ([M+H]+ 1288.4); observed ([M+H]+ 1288.1) (See Supplementary Fig. S14).
Synthesis of fluorescently-labeled adhesive ligand
H-AhxRGDSC-NH2 (0.5 mmol) was synthesized and modified with Alexa FluorR 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester (2 mg, Invitrogen) in DMF overnight at room temp. The peptide was cleaved from resin, precipitated, and lyophilized to give a yellow solid (denoted AF488-AhxRGDSC-NH2). AF633-AhxPHSRNC-NH2 was synthesized in a similar manner.
Gel Formation
Hydrogels were created by mixing 10 wt% total macromer photodegradable, photocouplable click gel formulation in media in between an azide-functionalized (See Supplementary Fig. S15) and Rain-XR-treated glass slides spaced at a known distance (typically 500 μm), and reacted for 30 min at 37 °C. The slides were separated, and the gel remained covalently attached to the azide-functionalized slide.
Biochemical Patterning
Hydrogels were swollen in phenol red-free media containing 3 mg mL−1 patterning agent AF488-AhxRGDSC-NH2 and eosin Y (10 μM) for one hour. For photolithographic-based experiments, gels were exposed to collimated visible light (λ = 490 – 650 nm), achieved with an Acticure (EXFO) high pressure mercury lamp equipped with an internal bandpass filter (350 – 650 nm) and an external 490 nm longpass filter (Edmund Optics), through a patterned chrome photomask. Alternatively, 3D patterning was obtained via two-photon techniques where subvolumes within the hydrogel were selectively exposed to pulsed laser light (λ = 860 nm, power = 350 mW/μm2, scan speed = 1.27 μsec/μm2) at 1 μm z-plane increments on a 710 LSM NLO confocal microscope stage (Carl Zeiss) equipped with a 20x/0.8 Plan-Apochromat objective (NA = 1.0). Unreacted patterning agent and initiator were swollen into fresh media as the sample was gently agitated on an orbital shaker (2 hours), yielding the final patterned hydrogel. In both the photolithographic and multi-photon patterning techniques, photocoupling of the peptide was confined to volumes exposed to light within the material and was visualized by fluorescent confocal microscopy.
Biophysical Patterning
Gels containing 0.125 mM Alexa FluorR 594 azide (Invitrogen) were patterned using photolithographic techniques, where hydrogels were exposed to collimated UV light (λ = 365 nm) from an Omnicure S1000 (EXFO) high pressure mercury lamp equipped with an internal bandpass filter (365 nm). 3D patterning was obtained via two-photon techniques where regions of interest (x–y control) within the hydrogel were selectively exposed to pulsed laser light (λ = 740 nm, power = 670 mW/μm2, scan speed = 1.27 μsec/μm2). Photodegraded monomer was swollen into fresh media, yielding the final patterned hydrogel.
Cell Culture
NIH 3T3s (mouse) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% fetal bovine serum (Invitrogen), 1% penicillin/streptomycin (Gibco), 0.2% fungizone, and 0.4% gentamicin. hMSCs were cultured in low-glucose DMEM with 10% fetal bovine serum, 1% penicillin/streptomycin, 0.2% fungizone, and 0.4% gentamicin. All cells were maintained in 5% CO2 at 37 °C. Cells were used between passages P4 and P6.
Fibrin Clot Encapsulation
Cells were suspended at 10 × 106 cells mL−1 in a fibrinogen solution (10 mg mL−1 in PBS, Sigma) containing thrombin (5 U mL−1, Sigma) and reacted for 30 min at 37 °C. The formed cell-laden clots were suspended in a 10 wt% total macromer photodegradable, photocouplable click gel formulation sandwiched between azide-functionalized (See Supplementary Fig. S10) and Rain-XR-treated glass slides spaced at 1 mm, and reacted for an additional 30 min at 37 °C. The slides were separated, and the gel remained covalently attached to the azide-functionalized slide. After 2 hours in media, physical channels were patterned into the network with two-photon patterning (λ = 740 nm). The media was then supplemented with Ac-RGDSC-NH2 (3 mg mL−1) and eosin Y (10 μM), equilibrated for 1 hr, and selected regions within the gel were biochemically decorated with RGD (λ = 860 nm). On day 10, the hydrogels were fixed in formalin for 1.5 h, followed by cell permeabilization with 0.5% Triton® X-100 (Fisher) in PBS for 2 h. The samples were blocked with 3% bovine serum albumin (BSA, Sigma) in PBS for 1 h and rinsed with PBS. F-actin was visualized using Alexa Fluor® 488 Phalloidin Conjugate (5 U/mL, Invitrogen), while nuclei were stained with DAPI (500 nM, Invitrogen), each for 2 h. The samples were washed with PBS prior to confocal visualization.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. A. Kloxin and M. Tibbitt for their useful discussions on photopatterning, Dr. Chien-Chi Lin for advice with cell outgrowth experiments, Drs. A. Aimetti and P. Hume for communication on general experimental design, as well as Dr. C. Kloxin for insightful feedback on the written manuscript. Fellowship assistance to C.A.D. was awarded by the US Department of Education’s Graduate Assistantships in Areas of National Need. This work was made possible by financial support from the National Science Foundation (DMR 1006711) and the Howard Hughes Medical Institute.
Footnotes
Author contribution
C.A.D. and K.S.A. developed the material concept, C.A.D. and K.S.A. designed the experiments, C.A.D. carried out the experiments, and C.A.D. and K.S.A. composed the manuscript.
Competing financial interests
The authors declare that they have no competing financial interests.
References
- 1.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 3.Moses JE, Moorhouse AD. The growing applications of click chemistry. Chem Soc Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 4.Hawker CJ, Wooley KL. The convergence of synthetic organic and polymer chemistries. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- 5.Barner-Kowollik C, et al. “Clicking” polymers or just efficient linking: What is the difference? Angew Chem Int Ed Engl. 2011;50:60–62. doi: 10.1002/anie.201003707. [DOI] [PubMed] [Google Scholar]
- 6.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: Fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman CN, Kloxin CJ. Toward an enhanced understanding and implementation of photopolymerization reactions. Aiche J. 2008;54:2775–2795. [Google Scholar]
- 8.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 10.Aizawa Y, Wylie R, Shoichet M. Endothelial cell guidance in 3d patterned scaffolds. Adv Mater. 2010;22:4831. doi: 10.1002/adma.201001855. [DOI] [PubMed] [Google Scholar]
- 11.Lee SH, Moon JJ, West JL. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3d cell migration. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann JC, West JL. Three-dimensional photolithographic patterning of multiple bioactive ligands in poly(ethylene glycol) hydrogels. Soft Matter. 2010;6:5056–5063. [Google Scholar]
- 13.Seidlits SK, Schmidt CE, Shear JB. High-resolution patterning of hydrogels in three dimensions using direct-write photofabrication for cell guidance. Adv Funct Mater. 2009;19:3543–3551. [Google Scholar]
- 14.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khetan S, Katz JS, Burdick JA. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter. 2009;5:1601–1606. [Google Scholar]
- 16.Khetan S, Burdick JA. Patterning network structure to spatially control cellular remodeling and stem cell fate within 3-dimensional hydrogels. Biomaterials. 2010;31:8228–8234. doi: 10.1016/j.biomaterials.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 17.Sarig-Nadir O, Livnat N, Zajdman R, Shoham S, Seliktar D. Laser photoablation of guidance microchannels into hydrogels directs cell growth in three dimensions. Biophys J. 2009;96:4743–4752. doi: 10.1016/j.bpj.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilina O, Bakker GJ, Vasaturo A, Hofmann RM, Friedl P. Two-photon laser-generated microtracks in 3d collagen lattices: Principles of mmp-dependent and -independent collective cancer cell invasion. Phys Biol. 2011;8 doi: 10.1088/1478-3975/8/1/015010. [DOI] [PubMed] [Google Scholar]
- 19.Codelli JA, Baskin JM, Agard NJ, Berozzi CR. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J Am Chem Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzima BJ, et al. Spatial and temporal control of the alkyne-azide cycloaddition by photoinitiated cu(ii) reduction. Nat Chem. 2011;3:258–261. doi: 10.1038/nchem.980. [DOI] [PubMed] [Google Scholar]
- 21.Hoyle CE, Bowman CN. Thiol-ene click chemistry. Angew Chem Int Ed. 2010;49:1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 22.Dondoni A. The emergence of thiol-ene coupling as a click process for materials and bioorganic chemistry. Angew Chem Int Ed. 2008;47:8995–8997. doi: 10.1002/anie.200802516. [DOI] [PubMed] [Google Scholar]
- 23.Polizzotti BD, Fairbanks BD, Anseth KS. Three-dimensional biochemical patterning of click-based composite hydrogels via thiolene photopolymerization. Biomacromolecules. 2008;9:1084–1087. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 24.DeForest CA, Sims EA, Anseth KS. Peptide-functionalized click hydrogels with independently tunable mechanics and chemical functionality for 3d cell culture. Chem Mater. 2010;22:4783–4790. doi: 10.1021/cm101391y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killops KL, Campos LM, Hawker CJ. Robust, efficient, and orthogonal synthesis of dendrimers via thiol-ene “click” chemistry. J Am Chem Soc. 2008;130:5062–5064. doi: 10.1021/ja8006325. [DOI] [PubMed] [Google Scholar]
- 26.Fairbanks BD, et al. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Adv Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta N, et al. A versatile approach to high-throughput microarrays using thiol-ene chemistry. Nat Chem. 2010;2:138–145. doi: 10.1038/nchem.478. [DOI] [PubMed] [Google Scholar]
- 28.Uygun M, Tasdelen MA, Yagci Y. Influence of type of initiation on thiol-ene “click” chemistry. Macromol Chem Phys. 2010;211:103–110. [Google Scholar]
- 29.Alvarez M, et al. Single-photon and two-photon induced photocleavage for monolayers of an alkyltriethoxysilane with a photoprotected carboxylic ester. Adv Mater. 2008;20:4563–4567. [Google Scholar]
- 30.Deiters A. Principles and applications of the photochemical control of cellular processes. Chembiochem. 2010;11:47–53. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes CP. Model studies for new o-nitrobenzyl photolabile linkers: Substituent effects on the rates of photochemical cleavage. J Org Chem. 1997;62:2370–2380. doi: 10.1021/jo961602x. [DOI] [PubMed] [Google Scholar]
- 32.Ohmuro-Matsuyama Y, Tatsu Y. Photocontrolled cell adhesion on a surface functionalized with a caged arginine-glycine-aspartate peptide. Angew Chem Int Ed. 2008;47:7527–7529. doi: 10.1002/anie.200802731. [DOI] [PubMed] [Google Scholar]
- 33.Johnson JA, Baskin JM, Bertozzi CR, Koberstein JT, Turro NJ. Copper-free click chemistry for the in situ crosslinking of photodegradable star polymers. Chem Commun. 2008:3064–3066. doi: 10.1039/b803043j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong DY, Griffin DR, Reed J, Kasko AM. Photodegradable hydrogels to generate positive and negative features over multiple length scales. Macromolecules. 2010;43:2824–2831. [Google Scholar]
- 35.Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat Protoc. 2010;5:1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloxin AM, Benton JA, Anseth KS. In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials. 2010;31:1–8. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frey MT, Wang YL. A photo-modulatable material for probing cellular responses to substrate rigidity. Soft Matter. 2009;5:1918–1924. doi: 10.1039/b818104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redick SD, Settles DL, Briscoe G, Erickson HP. Defining fibronectin’s cell adhesion synergy site by site-directed mutagenesis. J Cell Biol. 2000;149:521–527. doi: 10.1083/jcb.149.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims EA, DeForest CA, Anseth KS. A mild, large-scale synthesis of 1,3-cyclooctanedione: Expanding access to difluorinated cyclooctyne for copper-free click chemistry. Tetrahedron Lett. 2011;52:1871–1873. doi: 10.1016/j.tetlet.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.