Abstract
The molecular mechanisms of major mental illnesses, such as schizophrenia and bipolar disorder, are unclear. To address this fundamental question, many groups have studied molecular expression profiles in postmortem brains and other tissues from patients compared with those from normal controls. Development of unbiased high-throughput approaches, such as microarray, RNA-seq, and proteomics, have supported and facilitated this endeavor. In addition to genes directly involved in neuron/glia signaling, especially those encoding for synaptic proteins, genes for metabolic cascades are differentially expressed in the brains of patients with schizophrenia and bipolar disorder, compared with those from normal controls in DNA microarray studies. Here we propose the importance and usefulness of genetic mouse models in which such differentially expressed molecules are modulated. These animal models allow us to dissect the mechanisms of how such molecular changes in patient brains may play a role in neuronal circuitries and overall behavioral phenotypes. We also point out that models in which the metabolic genes are modified are obviously untested from mental illness viewpoints, suggesting the potential to re-address these models with behavioral assays and neurochemical assessments.
Introduction
Schizophrenia (SCZ) and bipolar disorder (BPD) are major mental illnesses with onset in adulthood. Although the clinical manifestation and disease course are substantially different from each other, these two disorders share genetic risks at several loci and have some similarities in the patterns of cognitive impairment (Berrettini, 2004; Craddock et al., 2006; Hill et al., 2008).
To address molecular mechanisms underlying mental illnesses, many groups have studied molecular expression profiles in postmortem brains and other tissues from patients compared with those from normal controls. Since 2000, in addition to candidate molecule approaches, an unbiased methodology for expression study with microarrays has been frequently employed (Horvath et al., 2011; Mirnics et al., 2006; Mirnics et al., 2000). More recently, RNA-seq has been introduced as a revolutionary tool for transcriptomics (Twine et al., 2011; Wang et al., 2009). At the protein level, several types of proteomic approaches are also utilized (English et al., 2011; Wittmann-Liebold et al., 2006). A major concern in these molecular profile studies is that this approach may not be able to address brain circuitry-dependent mechanisms.
To complement this limitation, rodents (rats and mice) are used to model mental diseases (Chen et al., 2006; Cryan and Holmes, 2005; Nestler and Hyman, 2010). These models have been characterized primarily by behavioral changes: for example, hyperlocomotion, social interaction deficits, impaired prepulse inhibition, and working memory deficits in rodents are used as indicators of behavioral deficits possibly corresponding to positive symptoms, negative symptoms, disturbance in information processing, and cognitive deficits in SCZ patients, respectively (Arguello and Gogos, 2006). In addition, mild but significant abnormalities in anatomy and histology are used as translatable characteristics between mice and humans, including enlarged ventricles, dendritic changes, and interneuron deficits (Jaaro-Peled et al., 2010). Nonetheless, due to the uncertain etiology of mental illnesses, the construct and etiological validity of these models is always debated. The models that modulate genes whose expression is altered in patient tissues may reflect disease pathophysiologies, although there is no guarantee that the model really mimics disease etiologies. Such animal models may complement the difficulty to study brain circuitry-dependent mechanisms at functional levels using expression studies of patient cells and tissues. For example, transient overexpression of dopamine D2 receptor specifically in the striatum can cause persistent functional abnormalities in the prefrontal cortex, affecting some endophenotypes relevant to schizophrenia (Kellendonk et al., 2006), while transient knockdown of DISC1 specifically in the cells of pyramidal neuron lineage in the developing cortex affects postnatal mesocortical dopaminergic maturation and causes functional deficits in mesolimbic dopaminergic neurons, leading to neurochemical and behavioral deficits relevant to SCZ in adulthood (Niwa et al., 2010). These two papers may support the idea of how gene manipulations in mice can provide us with clues to our understanding of circuitry-based mechanisms of mental illnesses. Specific targets found in human gene expression studies can be a source for genetic mouse models that could further enhance such insight into the mechanisms underlying mental illnesses.
In this review, we first summarize DNA microarray studies in postmortem brains from patients with SCZ and BPD. From the many genes in the literature we discuss those that have been found to change in more than one study. In the second part, we review mouse models built by modulating genes that have been identified in human brain gene expression studies. Finally, we discuss a promising research strategy for better understanding major mental illnesses by combining human molecular profiling and studies with mouse models.
Human gene expression profiles of SCZ and BPD in postmortem brains
Most DNA microarray analysis studies have used postmortem brains of patients with SCZ, BPD, and related mental illnesses. However, there are three major limitations in studies with postmortem brains, which may possibly generate inconsistency among the results:
Autopsied brains may be contaminated with long-term medications, drug abuse, and smoking especially in the cases of SCZ). Therefore, differences from control brains may not directly reflect the primary pathological changes, but may be mere secondary effects of the confounding factors (Chan et al., 2010; Halim et al., 2008; Mexal et al., 2006). Moreover, influences of aging and age-associated co-morbid conditions cannot be ignored.
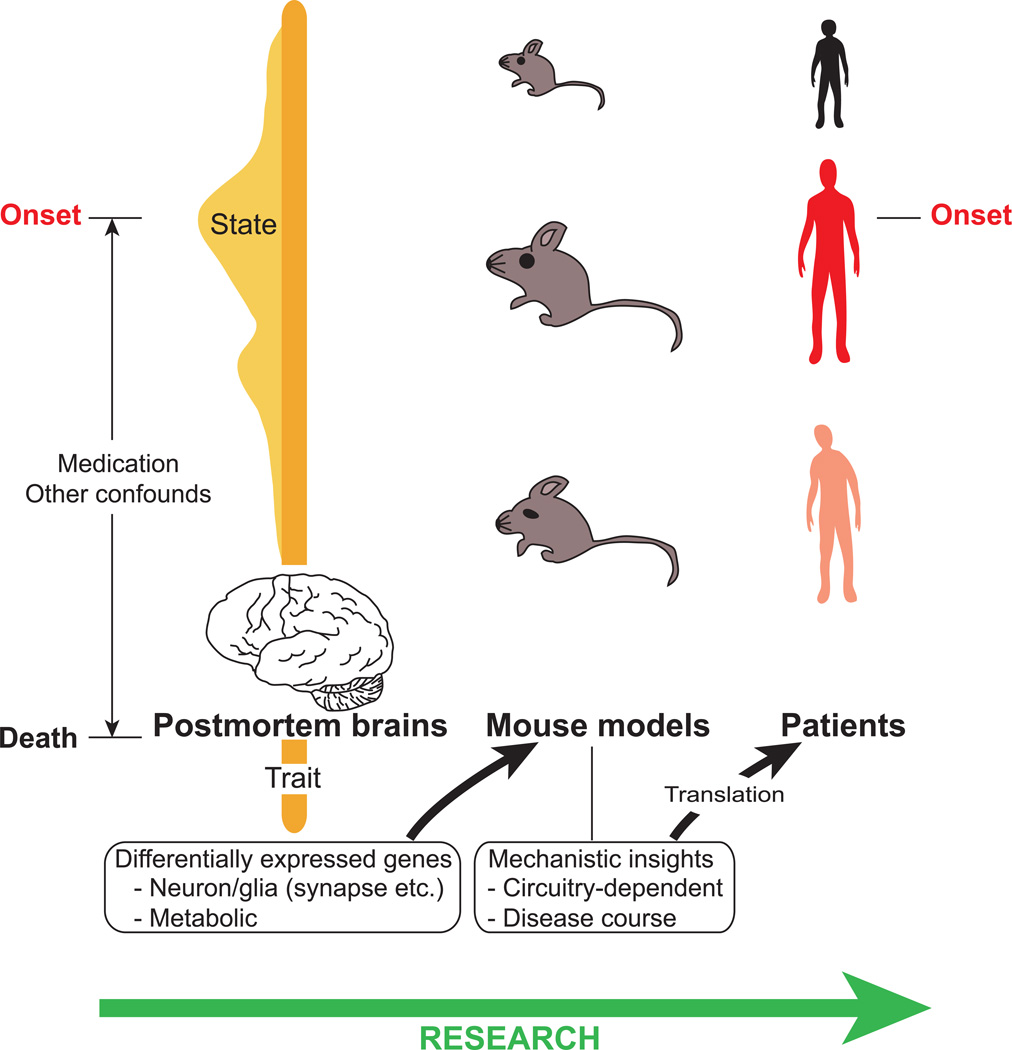
Critical molecular changes associated with pathogenic processes and disease onset may not be apparent at the time of death, and therefore in autopsied brains. In other words, we may be able to detect “trait” (relatively stable core features of the disease) changes but possibly fail to detect critical “state” (features dynamically changed in association with clinical manifestations, such as those during manic or depressive stage or only during psychotic episode) markers associated with early stages of the disease (Figure 1) (Endler and Kocovski, 2001; Extein and Bowers, 1979; Friedman et al., 2006). To fully understand the pathologic processes of mental illnesses, both trait and state markers are important.
The brain homogenates used for DNA microarray and other simple biochemical approaches are a mixture of different cell-types (neurons, various glial cells, and endothelial cells), and unique composition occurs in each different brain region.
Fig. 1.
From gene expression profiling studies of postmortem brains to mouse models for mental illnesses. Thus far, gene array studies of postmortem brains of patients with schizophrenia and bipolar disorder have revealed numerous differentially expressed genes, such as those involved in neuronal or glial signaling and metabolic genes. Postmortem brains may represent the core “trait” pathophysiological changes, but may not fully reflect the “state” changes associated with the disease onset. Additionally, the gene expression changes may be influenced by several confounds, such as long-term medication. We suggest using mouse models for the differentially expressed genes to study circuitry-dependent mechanisms and the time course of the disease, which are difficult to address in human studies. We hope that mechanistic insights obtained by this approach will help in finding new avenues for translation.
Nonetheless, we believe that the molecular information from patient brains is still very important for dissecting SCZ and BPD, as they are “brain” disorders. Furthermore, several efforts have been made in order to overcome these limitations (Lewis, 2002; McCullumsmith and Meador-Woodruff, 2010). Thus, here we searched published literature for DNA microarray studies of postmortem brains from patients with SCZ and/or BPD and summarized the genes that were differentially expressed in more than one study when compared to controls. These genes were then categorized according to their functional properties using DAVID Functional Annotation Bioinformatics Microarray Analysis (Huang da et al., 2009). The genes were clustered into four major groups as follows: synaptic genes (Table 1), genes associated specifically with neuronal and/or glial functions (Table 2), metabolic genes (Table 3), and genes with general functions (signaling, protein degradation, cytoskeletal) (Table 4).
Table 1.
Synaptic genes differentially expressed in postmortem brains of SCZ and BPD patients and the corresponding genetic mouse models.
| Gene | References | Mouse models | Positive findings | References |
|---|---|---|---|---|
| Schizophrenia: decreased | ||||
| Gamma-aminobutyric acid (GABA) A receptor, alpha 5 (GABRA5) |
(Duncan et al., 2010) | Gabra5 KO | Altered synaptic transmission, enhanced learning and memory | (Collinson et al., 2002) |
| Glutamate receptor, metabotropic 3 (GRM3) | (Hemby et al., 2002) | mGlu3 KO (exon 3) mGlu3 KO (exon 2) |
Disrupted anxiolytic activity of mGLu2/3 agonist in the EPM Disrupted neuroprotection against excitotoxicity in mixed culture |
(Linden et al., 2005) (Corti et al., 2007)* |
| Dopamine supersensitive striata | (Seeman et al., 2009)* | |||
| Neurexin 3 (NRXN3) | (Mexal et al., 2005) (Chu et al., 2009) |
Nrxn 3a KO | Impaired survival, abnormal NMDA-R function | (Missler et al., 2003), (Kattenstroth et al., 2004) |
| Synapsin II (SYN2) | (Mirnics et al., 2000) (Arion et al., 2007) |
Syn2 KO | Seizures, decreased post-tetanic potentiation and severe synaptic depression upon repetitive stimulation |
(Rosahl et al., 1995), |
| (Arion et al., 2007) | PPI deficits, social interaction deficit | (Dyck et al., 2007)* | ||
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (YWHAH) |
(Vawter et al., 2002) (Chu et al., 2009) |
Uch-L1 KO | Retarded axon formation Impaired memory and LTP |
(Ferreira et al., 1998) (Sakurai et al., 2008) |
| Schizophrenia and bipolar: decreased | ||||
| Glutamate decarboxylase 1 (brain, 67kDa) (GAD1) | (Vawter et al., 2002) (Hashimoto et al., 2008) (Duncan et al., 2010) (Konradi et al., 2004) |
Gad67 KO Gad1 KO Floxed Gad1 KO |
Lethal cleft palate Deficient perisomatic GABAergic innervation in the visual cortex |
(Asada et al., 1997) (Condie et al., 1997) (Chattopadhyaya et al., 2007) |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) |
(Chu et al., 2009) (Konradi et al., 2004) |
|||
| V-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) (ERBB3) |
(Hakak et al., 2001) (Tkachev et al., 2003) (Aston et al., 2004) |
|||
| Schizophrenia and bipolar: increased | ||||
| Monoamine oxidase A (MAOA) | (Shao and Vawter, 2008) | |||
| Bipolar: increased | ||||
| Gamma-aminobutyric acid (GABA) A receptor, alpha 5 (GABRA5) |
(Choudary et al., 2005) | Gabra5 H10R knockin | Facilitated trace fear conditioning | (Crestani et al., 2002) |
| Glutamate receptor, metabotropic 3 (GRM3) | (Choudary et al., 2005) | |||
Representative mouse models characterized from mental illness viewpoints
Table 2.
Other neuronal and glial genes differentially expressed in postmortem brains of SCZ and BPD patients and the corresponding genetic mouse models.
| Subgroup | Gene | References | Mouse models | Positive findings | References |
|---|---|---|---|---|---|
| Schizophrenia: decreased | |||||
| Myelin proteins |
2',3'-cyclic nucleotide 3' phosphodiesterase (CNP) | (Hakak et al., 2001) (McCullumsmith et al., 2007) (Dracheva et al., 2006) |
Cnp KO | Compromised axonal survival | (Lappe-Siefke et al., 2003) |
| Neurogenesis | Neural precursor cell expressed, developmentally down-regulated 8 (NEDD8) |
(Bowden et al., 2008) (Altar et al., 2005) |
|||
| Neuron development |
Sema domain, seven thrombospondin repeats (type 1 and type 1- like), transmembrane domain and short cytoplasmic domain, (semaphorin) 5A (SEMA5A) |
(Altar et al., 2005) | |||
| Ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (UCHL1) |
(Vawter et al., 2001) (Altar et al., 2005) |
||||
| Schizophrenia: increased | |||||
| Neuron development |
Sema domain, seven thrombospondin repeats (type 1 and type 1- like), transmembrane domain and short cytoplasmic domain, (semaphorin) 5A (SEMA5A) |
(Bowden et al., 2008) | |||
| Schizophrenia and bipolar: decreased | |||||
| Myelin proteins |
Claudin 11 (CLDN11) | (Dracheva et al., 2006) (Tkachev et al., 2003) |
OSP/claudin-11 KO | Tight junctions missing in CNS myelin, slowed conduction, hindlimb weakness |
(Gow et al., 1999) |
| Myelin associated glycoprotein (MAG) | (Hakak et al., 2001) (Aston et al., 2004) (Dracheva et al., 2006) (McCullumsmith et al., 2007) (Tkachev et al., 2003) |
MAG KO | Motor deficits, hyperactive, axon degeneration |
(Pan et al., 2005) | |
| SRY (sex determining region Y)-box 10 (SOX10) | (Dracheva et al., 2006) (Tkachev et al., 2003) (Iwamoto et al., 2005) |
SOX10 KO | Die ~birth, no peripheral glia, no terminal differentiation of myelin forming oligodendrocytes |
(Britsch et al., 2001), (Stolt et al., 2002) |
|
| Transferrin (TF) | (McCullumsmith et al., 2007) (Hakak et al., 2001) (Arion et al., 2007) (Tkachev et al., 2003) |
||||
| Neuropeptides | Neuropeptide Y (NPY) | (Kuromitsu et al., 2001) | NPY KO | Anxiogenic in the open field, hypoalgesic |
(Bannon et al., 2000) |
| Somatostatin (SST) | (Hashimoto et al., 2008) (Hashimoto et al., 2008) (Bowden et al., 2008) (Konradi et al., 2004) |
NPY Y2 receptor KO Sst KO |
Hyperlocomotion in a novel open field Impaired motor learning |
(Karl et al., 2010) (Zeyda et al., 2001) |
|
Table 3.
Metabolic genes differentially expressed in postmortem brains of SCZ and BPD patients and the corresponding genetic mouse models.
| Subgroup | Gene | References | Mouse models | Positive findings | References |
|---|---|---|---|---|---|
| Schizophrenia: Decreased | |||||
| Mitochondria and energy metabolism |
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) (NDUFS4) Phosphoglycerate mutase 1 (brain) (PGAM1) |
(Altar et al., 2005) (Arion et al., 2007) (Altar et al., 2005) (Bowden et al., 2008) |
Conditional NDUFS4 KO | Fatal encephalomyopathy | (Kruse et al., 2008) |
| Schizophrenia: Increased | |||||
| Aminoglycan metabolic process |
Chitinase 3-like 1 (cartilage glycoprotein-39) (CHI3L1) | (Arion et al., 2007) (Chung et al., 2003) |
|||
| Schizophrenia and bip olar: Decreased | |||||
| Mitochondria and energy metabolism |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (ATP6V1A) |
(Chu et al., 2009) (Konradi et al., 2004) |
|||
| Amino acid metabolism |
Glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (GOT1) |
(Arion et al., 2007) (Konradi et al., 2004) |
|||
| Bipolar: Decreased | |||||
| Mitochondria and energy metabolism |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) (ATP5G3) ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 (ATP5C1) Mitochondrial ribosomal protein L3 (MRPL3) Ubiquinol-cytochrome c reductase core protein II (UQCRC2) |
(Sun et al., 2006) (Konradi et al., 2004) (Sun et al., 2006) (Konradi et al., 2004) (Konradi et al., 2004) (Sun et al., 2006) |
|||
| Bipolar: increased | |||||
| Mitochondria and energy metabolism |
Mitochondrial ribosomal protein L3 (MRPL3) Ubiquinol-cytochrome c reductase core protein II (UQCRC2) |
(Iwamoto et al., 2005) (Iwamoto et al., 2005) |
|||
Table 4.
Genes with general functions differentially expressed in postmortem brains of SCZ and BPD patients and the corresponding genetic mouse models.
| Subgroup | Gene | References | Mouse models | Positive findings | References |
|---|---|---|---|---|---|
| Schizophrenia: Decreased | |||||
| Signal transduction | Phosphodiesterase 1A, calmodulin-dependent (PDE1A) | (Aston et al., 2004) | |||
| Regulator of G-protein signaling 4 (RGS4) | (Mirnics et al., 2001) (Arion et al., 2007) (Bowden et al., 2008) |
Floxed Rgs4 KO | Lower body weight, mildly impaire balance/motor coordination |
(Grillet et al., 2005)* | |
| Ubiquitin/proteasome /protein degradation |
Proteasome (prosome, macropain) subunit, alpha type, 1 (PSMA1) |
(Vawter et al., 2001) (Altar et al., 2005) |
|||
| Schizophrenia and bipolar: Decreased | |||||
| Ubiquitin/proteasome /protein degradation |
Proteasome (prosome, macropain) 26S subunit, ATPase, 6 (PSMC6) |
(Altar et al., 2005) (Konradi et al., 2004) |
|||
| Schizophrenia and bipolar: Increased | |||||
| Cytoskeleton | Heat shock 27kDa protein 1 (HSPB1) | (Arion et al., 2007) (Iwamoto et al., 2004) |
Tg overexpressing human HSP27 |
Protects against ischemia/reperfusion injury |
(Hollander et al., 2004) |
| Bipolar: Increased | |||||
| Signal transduction | Phosphodiesterase 1A, calmodulin-dependent (PDE1A) | (Iwamoto et al., 2004) | |||
Representative mouse models characterized from mental illness viewpoints
Although multiple brain regions have been examined in gene expression studies of SCZ and BPD, the majority of research has focused on the prefrontal cortex for the microarray studies. Therefore, many genes listed in the tables are data from the prefrontal cortex. Other brain regions analyzed include the anterior cingulate cortex (McCullumsmith et al., 2007), the hippocampus (Altar et al., 2005; Konradi et al., 2004; Mexal et al., 2005), and the temporal cortex (Aston et al., 2004; Hemby et al., 2002). There is one study focusing on the altered thalamic gene expression in both SCZ and BPD (Chu et al., 2009). Of note, although we focus on DNA microarray studies in this review article, altered levels of BDNF and TrkB in SCZ brains have been reported via in situ hybridization and Western blotting (Hashimoto et al., 2005; Weickert et al., 2003).
Category 1: synaptic genes (Table 1)
The number of molecules differentially expressed is the largest for the category of “synaptic genes” (in particular, genes involving GABAergic and glutamatergic synapses) in postmortem SCZ and BPD brains, which is consistent with a proposal that SCZ is a synaptic disorder (Mirnics et al., 2001a).
Most of the dysregulated genes in this synapse group are from studies of the prefrontal cortex areas. Among them, the expression of V-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) (ERBB3) is reduced in postmortem tissues from the prefrontal cortex (Hakak et al., 2001; Tkachev et al., 2003) and the temporal cortex (Aston et al., 2004) in SCZ, as well as the prefrontal cortex (Tkachev et al., 2003) in BPD. Also, the expression of Synapsin II (SYN2) has been shown to be decreased based on two different microarray studies in the prefrontal cortex in SCZ (Arion et al., 2007; Mirnics et al., 2000). Moreover, the expression of Monoamine oxidase A (MAOA) is increased in the prefrontal cortex in both SCZ and BPD (Shao and Vawter, 2008). Those genes whose expression change has not been reported in the prefrontal cortex but other brain regions are Neurexin 3 (NRXN3) (Chu et al., 2009; Mexal et al., 2005), Tyrosine 3-onooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) (Chu et al., 2009; Konradi et al., 2004), and Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (YWHAH) (Chu et al., 2009; Vawter et al., 2001). Expression of these three genes is reduced in the temporal cortex, the hippocampus, or the thalamus in SCZ or both SCZ and BPD. Very interestingly, they are also genetically associated with SCZ or BPD (Grover et al., 2009; Jia et al., 2004; Novak et al., 2009; Toyooka et al., 1999).
Abnormalities in glutamatergic neurotransmission and associated signaling (Tamminga, 1998), as well as GABAergic deficits (Benes, 2000) have been consistently reported in SCZ. Several genes listed in Table 1 are glutamate- or GABA-related. Among them, the expression of Gamma-aminobutyric acid A receptor, alpha 5 (GABRA5) and Glutamate receptor, metabotropic 3 (GRM3 i.e. mGluR3) is increased in the prefrontal cortex and anterior cingulate cortex in BPD (Choudary et al., 2005), whereas their expression is downregulated in the prefrontal cortex (Duncan et al., 2010) and the entorhinal cortex in SCZ (Hemby et al., 2002). Several groups have consistently reported that the expression of Glutamate decarboxylase 1 (GAD1) is downregulated in both SCZ and BPD (Duncan et al., 2010; Hashimoto et al., 2008; Konradi et al., 2004; Vawter et al., 2002). Genetic associations of these glutamatergic and GABAergic molecules with SCZ and BPD have also been reported (Cherlyn et al., 2010).
In parallel to the expression data and genetic information described above, synaptic disturbances have been reported in many other studies with human subjects (e.g., decrease in synaptic density in SCZ brains and disturbed cortical synchrony as a sign of GABAergic interneuron deficits) (Lewis et al., 2003; Uhlhaas and Singer, 2010). Thus, the molecules that are altered in expression in the patient brains can be key probes in studying the mechanistic roles of synaptic disturbances in the pathophysiologies of SCZ and BPD.
Category 2: genes associated specifically with neuronal and/or glial functions (Table 2)
This category includes genes that are involved not directly in synaptic roles but in functions otherwise associated with neuronal and glial cells. In contrast to the dominance of expression changes in the synaptic genes (Category 1 above) in the prefrontal cortex, the expression changes of this second category are from various brain regions, including the prefrontal cortex, the temporal cortex, and the hippocampus. Although we do not know whether these differences imply the disease pathophysiology or are a mere bias of the brain regions studied, it might be worth mentioning.
In this category, many myelin-related genes are downregulated in SCZ and BPD (Dracheva et al., 2006; Hakak et al., 2001; McCullumsmith et al., 2007; Tkachev et al., 2003). Considering a recent excitement on the detection and characterization of white matter pathology in SCZ and BPD (Connor et al., 2009; Regenold et al., 2007), these expression data may be very important. In SCZ, their downregulation is reported in multiple brain areas (Bowden et al., 2008; Dracheva et al., 2006; Hakak et al., 2001; McCullumsmith et al., 2007; Tkachev et al., 2003). Among them, alteration in the expression of Myelin associated glycoprotein (MAG) is most frequently reported in at least three distinct brain areas in SCZ (Aston et al., 2004; Dracheva et al., 2006; Hakak et al., 2001; McCullumsmith et al., 2007) and the prefrontal cortex in BPD (Tkachev et al., 2003). The expression of 2',3'-cyclic nucleotide 3' phosphodiesterase (CNP) is decreased in the prefrontal cortex (Hakak et al., 2001), the anterior cingulate cortex (Dracheva et al., 2006; McCullumsmith et al., 2007), and the hippocampus (Dracheva et al., 2006) in SCZ. The expression of SRY (Sex determining region Y)-box 10 (SOX10) is reduced in multiple brain regions of SCZ (Dracheva et al., 2006; Iwamoto et al., 2005b) and in the prefrontal cortex of BPD (Tkachev et al., 2003). The expression of Claudin 11 (CLDN11) and Transferrin (TF) is decreased in the prefrontal cortex in both diseases (Arion et al., 2007; Hakak et al., 2001; Tkachev et al., 2003) and in the anterior cingulate cortex in SCZ (Dracheva et al., 2006; McCullumsmith et al., 2007).
In addition to myelin-related genes, the expression of some neuropeptides is also downregulated in SCZ and BPD. The expression of Somatostatin (SST) is reduced in the prefrontal (Hashimoto et al., 2008) and temporal cortices in SCZ (Bowden et al., 2008) as well as in the hippocampus in BPD (Konradi et al., 2004). Neuropeptide Y (NPY) expression is decreased in the prefrontal cortex in both SCZ and BPD (Hashimoto et al., 2008; Kuromitsu et al., 2001).
In addition to genes associated with myelination and neuropeptides, the following molecules may be of scientific interest in this category: expression of Neural precursor cell expressed, developmentally downregulated 8 (NEDD8) is decreased in the temporal cortex (Bowden et al., 2008) and in the hippocampus (Altar et al., 2005) in SCZ. Ubiquitin carboxyl-terminal esterase L1 (Ubiquitin thiolesterase) (UCHL1) is downregulated in several brain regions of SCZ as well (Altar et al., 2005; Vawter et al., 2001). These observations may give us a hint to consider roles for misfolded proteins and endoplasmic reticulum (ER) stress in major mental illnesses (Hayashi et al., 2009; Ottis et al., 2011). The expression of Sema domain, seven thrombospondin repeats, transmembrane domain and short cytoplasmic domain, 5A (SEMA5A) is increased in the temporal cortex (Bowden et al., 2008) while reduced in the hippocampus (Altar et al., 2005) in SCZ.
Category 3: metabolic genes (Table 3)
Recent studies have suggested intrinsic abnormalities in metabolic cascades in SCZ (Holmes et al., 2006; Khaitovich et al., 2008; Kirkpatrick et al., 2009; Prabakaran et al., 2004). Metabolic disturbances are observed in drug-naïve SCZ patients (Thakore, 2004): for example, metabolic profiling of cerebrospinal fluid of drug-naïve patients with first-onset SCZ shows alterations in glucoregulatory processes (Holmes et al., 2006) and individuals with non-affective psychosis display increased prevalence of abnormal glucose tolerance prior to antipsychotic treatment (Fernandez-Egea et al., 2009). Thus, it is now very important to address molecular mechanisms underlying such changes. Interestingly, through this categorization of the past publications of gene expression in SCZ and BPD brains, we came across many genes differentially expressed in these disorders, forming a category of “metabolic genes.”
Many genes in this category are downregulated in SCZ and BPD. Among them, the expression of NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) (NDUFS4) and Phosphoglycerate mutase 1 (brain) (PGAM1) is reduced in the hippocampus of SCZ (Altar et al., 2005). The expression of ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) (ATP5G3), ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 (ATP5C1), ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (ATP6V1A), and Glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (GOT1) is decreased in the hippocampus of BPD (Konradi et al., 2004). These genes are also downregulated in other brain regions, such as the prefrontal cortex, the superior temporal gyrus, and the thalamus (Arion et al., 2007; Bowden et al., 2008; Chu et al., 2009; Sun et al., 2006). In contrast, the expression of Chitinase 3-like 1 (cartilage glycoprotein-39) (CHI3L1) in SCZ is increased both in the hippocampus (Chung et al., 2003) and the prefrontal cortex (Arion et al., 2007).
The expression of Mitochondrial ribosomal protein L3 (MRPL3) in BPD is increased in the prefrontal cortex (Iwamoto et al., 2005a), whereas it is reduced in the hippocampus (Konradi et al., 2004). Ubiquinol-cytochrome c reductase core protein II (UQCRC2) has been reported with opposite expression changes in the prefrontal cortex of BPD in two different studies (Iwamoto et al., 2005a; Sun et al., 2006), which is speculated to be due to a medication effect (Iwamoto et al., 2005a).
Category 4: genes with general functions (Table 4)
Although the genes categorized in this group have functions not only in the central nervous system but also in many organs, their roles in cellular signaling in neurons and glia are also well known. For example, the sub-category of signal transduction includes Regulator of G-protein signaling 4 (RGS4), and Phosphodiesterase 1A, calmodulin-dependent (PDE1A), which are known as key players in modulating the signals from dopamine and glutamate receptors (Levitt et al., 2006; Menniti et al., 2006). Downregulation of RGS4 in multiple brain regions of SCZ has been consistently reported in several studies (Arion et al., 2007; Bowden et al., 2008; Mirnics et al., 2001b) and expression of PDE1A is increased in the temporal cortex in SCZ (Aston et al., 2004) whereas its expression is increased in the prefrontal cortex in BPD (Iwamoto et al., 2004).
Some genes involved in protein degradation cascade were already classified in the group of “genes associated specifically with neuronal and/or glial functions” (see category 2 above and Table 2), but several of these genes are also listed in the ubiquitin/proteasome/protein degradation subgroup of this category, including Proteasome subunit, alpha type, 1 (PSMA1) (Altar et al., 2005; Vawter et al., 2001).
Finally, genetic susceptibility factors for SCZ and its interacting proteins have indicated the involvement of cytoskeletal disturbances in major mental illnesses (Kamiya et al., 2008). Consistent with this notion, some genes in the cytoskeleton subgroup are differentially expressed between patients and controls, and most of the reports are from the studies of the prefrontal cortex (Arion et al., 2007; Iwamoto et al., 2004).
Animal models for genes that are differentially expressed in patient brains
Here we summarize mouse models in which the differentially expressed genes detected in patients are targeted. For the sake of simplicity we looked for publications of knockout (KO) models for genes that are downregulated in patients and those of transgenic overexpression models for genes that are upregulated in patients, respectively. Of note, in addition to mimicking the condition of a loss of function, KO mice are useful to study physiological role for the target gene. The information for unpublished KO models has been found at the website of International Knockout Mice Consortium (http://www.knockoutmouse.org/). Compared with abundant KO models, we came across only a few transgenic models in which genes upregulated in patients are overexpressed. Among all the publications on mouse models with disease relevance in human expression studies, none of them referred to the relevance for BPD and only three mouse models did for SCZ. This gap may imply that many mouse geneticists at present generate models that are potentially useful in studies of mental disorders, without acknowledging such merits and potentials towards translation (Figure 1).
Mouse models based on the genes in categories 1, 2, and 4 (Tables 1, 2, and 4)
Although KO mice have been generated for almost all neuroscience-related genes that are differentially expressed in patients with SCZ and/or BPD, only a few have been characterized from the viewpoint of mental illnesses. Basic neuroscience research has focused mainly on electrophysiology of the cerebellum and the hippocampus, because they are structurally relatively simple and anatomically well defined. Although the hippocampus is relevant to mental illnesses, attention to the circuitry between the hippocampus and the prefrontal cortex or the striatum may be expected for better understanding of major mental illnesses. Furthermore, behavioral changes in the animals have not been well studied yet under the light of endophenotypes for mental illnesses. This is partly because many of these homozygotes become embryonic lethal. Here we introduce three KO mice as representative models, in which genes included in these categories are genetically depleted and characterized from mental illness viewpoints. These include KO mice for Grm3 (mGluR3) (Table 1), syn2 (Table 1), and rgs4 (Table 4).
Grm3/mGluR3 KO mice: Glutamate receptor, metabotropic 3 (mGluR3) is downregulated in the entorhinal cortex layer II stellate neurons of SCZ patients (Hemby et al., 2002). Since mGlu2/3 agonists have recently attracted attention as a potential new type of antipsychotics (Cartmell et al., 2000; Patil et al., 2007), the first paper on Grm3/ mGluR3 KO mice focused on dissecting the effect of an mGlu2/3 agonist on mGlu2 versus mGlu3 receptors (Linden et al., 2005). The question of a possible interaction between mGlu2/3 and dopamine D2 receptors has also been addressed (Seeman et al., 2009). The striatum of mGlu3 KO is supersensitive to dopamine as demonstrated by its higher proportion of D2 receptors in the high-affinity state and its supersensitivity to a dopamine receptor agonist. These results suggest that mGlu3 receptors may normally regulate D2 receptors by reducing the proportion of high affinity D2 receptors in membranes. By using cultures of cortical neurons and astrocytes from the KO mice, requirement of astrocytic mGlu3 receptors activation for neuroprotection has been reported (Corti et al., 2007).
Syn2 (synapsin II) KO mice: Synapsins are a major component of synaptic vesicles and important for neurotransmitter release (Cesca et al., 2010), and a decrease in the expression of Syn2 in SCZ is shown in two different microarray studies in the prefrontal cortex (Arion et al., 2007; Mirnics et al., 2000). Syn2 KO mice are viable and fertile, but accompany seizures (Rosahl et al., 1995). Syn2 KO CA1 pyramidal neurons in hippocampal slices display decreased post-tetanic potential and severe synaptic depression upon repetitive stimulation of physiological frequencies (Rosahl et al., 1995). Dyck et al. (Dyck et al., 2007) aimed to confirm a role of Syn2 in the pathophysiology of SCZ and conducted behavioral assays, showing that syn2 KO had prepulse inhibition (PPI) and social interaction deficits.
Rgs4 KO mice: RGS4 has been repeatedly shown to be decreased in the SCZ postmortem brain in the prefrontal cortex (Mirnics et al., 2001b), dorsolateral prefrontal cortex (Arion et al., 2007) and superior temporal gyrus (Bowden et al., 2008). Rgs4 KO mice have lower body weight but seem healthy otherwise (Grillet et al., 2005). Phenotypes relevant to SCZ have been explored in this model, but thus far the only abnormalities detected are poorer performance in the rotarod test and reduction in the pain sensitivity, and no robust abnormalities have been observed in the open field, tail suspension, fear conditioning, and Y-maze tests (Grillet et al., 2005).
Mouse models based on the genes in category 3 (Table 3)
Although many genes associated with metabolic cascades are differentially expressed in SCZ and BPD brains compared with those from normal controls (Table 3), psychiatry-oriented studies with KO mice deficient in such metabolic genes have rarely been performed. However, considering the high prevalence of metabolic disturbances in SCZ (McEvoy et al., 2005) and BPD (Taylor and MacQueen, 2006), these models will be very useful and provide us with valuable information especially when their behavioral and neurochemical changes are examined in the light of endophenotypes relevant to major mental illnesses. The high incidence of metabolic abnormalities is accounted at least in part by poor life style and side effects of second generation antipsychotics (Correll et al., 2008); however, recent studies have supported a notion that mental illness-associated intrinsic abnormalities include metabolic disturbances, which may be further augmented by these exogenous factors. Mediators of metabolic signaling, such as insulin and leptin, are known to modulate cognitive functions (Benedict et al., 2011; Gerozissis, 2003; Harvey, 2007; Paz-Filho et al., 2008; Shanley et al., 2001). Animal models will be useful to test mechanistic details in the crosstalk of intrinsic susceptibility and exogenous factors.
Future perspectives
Here we propose a research strategy in which molecular profiling in patient tissues is combined with characterization of genetic animal models (Figure 1). As etiological and construct validity is still debated in modeling major mental illnesses, this approach to focus on the molecules associated with pathophysiology may be a promising alternative.
Through the overview of the past publications, we now realize that there are several technical barriers and missing points to enhance this research strategy:
Many CNS-relevant genes differentially expressed in SCZ and BPD are involved in neurodevelopment, and conventional homozygous KO mice are lethal or have too severe phenotypes to allow us to conduct behavioral assays. Thus, heterozygotes or conditional KOs may be useful for more subtle and psychiatry-relevant phenotypes, including cognitive deficits (Morozov et al., 2003). In addition, a relatively fast way to generate a rodent model for a candidate gene without lethality is to introduce an inhibitory (such as RNAi) or overexpression construct into a specific brain region and activate it at a specific time point. Stereotaxic viral infection enables excellent spatiotemporal control of the construct expression postnatally (Seshadri and Hayashi-Takagi, 2009) and in utero gene transfer enables gene manipulation of the developing brain (Kamiya, 2009; Niwa et al., 2010). A novel technique to generate mouse models is bacterial artificial chromosome (BAC)-driven microRNA-mediated silencing of gene expression (Garbett et al., 2010). In summary, by considering non-lethal and spatial/temporal-specific modulation of expression for a target molecule, the effects of CNS-relevant genes differentially expressed in SCZ and BPD are to be tested not only from basic neuroscience, but also from translational psychiatry viewpoints. Furthermore, such genetic models can also be utilized in testing interactions of genetic and environmental factors relevant to mental illnesses (Ayhan et al., 2009; Oliver, 2011; Sawa et al., 2004). Good collaboration between basic neuroscience and psychiatry would advance the field.
Genetic mouse models in which genes involved in metabolic signaling are modulated are another underused resource in biological psychiatry. In the light of a concept that SCZ (and BPD) are systemic disorders (Kirkpatrick, 2009), these metabolic models are to be re-examined with behavioral assays and neurochemical assessments. An interdisciplinary approach will enhance the research of mental illnesses.
Finally, the premise of this research strategy in using mouse models is heavily dependent on molecular profiling studies in human tissues. Although the autopsied brain is still a very useful resource, we also need to acknowledge its limitations in patients with mental illnesses, such as secondary effects of long-term medication, drug abuse, and smoking. Thus, use of biopsied cells and tissues with neuronal relevance and induced CNS-like cells will be considered as alternative templates for molecular profiling in the future (Brennand et al., 2011; Rioux et al., 2005; Tajinda et al., 2010).
Acknowledgements
We thank Yukiko L. Lema for organizing the manuscript and preparing the figure. We also thank Dr. Minae Niwa and Ms. Ashley Wilson for critical reading. Grant supports: from MH-084018 Silvo O. Conte center (A.S.), MH-069853 (A.S.), MH-085226 (A.S.), MH-088753 (A.S.), Stanley (A.S.), RUSK (A.S.), S-R foundations (A.S.), NARSAD (H.J-P., A.S.) and Maryland Stem Cell Research Fund (A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altar CA, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Arguello PA, Gogos JA. Modeling madness in mice: one piece at a time. Neuron. 2006;52:179–196. doi: 10.1016/j.neuron.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Arion D, et al. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, et al. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- Ayhan Y, et al. Animal models of gene-environment interactions in schizophrenia. Behav Brain Res. 2009;204:274–281. doi: 10.1016/j.bbr.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C, et al. Intranasal insulin as a therapeutic option in the treatment of cognitive impairments. Exp Gerontol. 2011;46:112–115. doi: 10.1016/j.exger.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Benes FM. Emerging principles of altered neural circuitry in schizophrenia. Brain Res Brain Res Rev. 2000;31:251–269. doi: 10.1016/s0165-0173(99)00041-7. [DOI] [PubMed] [Google Scholar]
- Berrettini W. Bipolar disorder and schizophrenia: convergent molecular data. Neuromolecular Med. 2004;5:109–117. doi: 10.1385/NMM:5:1:109. [DOI] [PubMed] [Google Scholar]
- Bowden NA, et al. Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genomics. 2008;9:199. doi: 10.1186/1471-2164-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmell J, et al. Attenuation of specific PCP-evoked behaviors by the potent mGlu2/3 receptor agonist, LY379268 and comparison with the atypical antipsychotic, clozapine. Psychopharmacology (Berl) 2000;148:423–429. doi: 10.1007/s002130050072. [DOI] [PubMed] [Google Scholar]
- Cesca F, et al. The synapsins: key actors of synapse function and plasticity. Prog Neurobiol. 2010;91:313–348. doi: 10.1016/j.pneurobio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Chan MK, et al. Evidence for disease and antipsychotic medication effects in post-mortembrain from schizophrenia patients. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.100. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. Genetic mouse models of schizophrenia: from hypothesis-based to susceptibility gene-based models. Biol Psychiatry. 2006;59:1180–1188. doi: 10.1016/j.biopsych.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Cherlyn SY, et al. Genetic association studies of glutamate, GABA and related genes in schizophrenia and bipolar disorder: a decade of advance. Neurosci Biobehav Rev. 2010;34:958–977. doi: 10.1016/j.neubiorev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Choudary PV, et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci U S A. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu TT, et al. Thalamic transcriptome screening in three psychiatric states. J Hum Genet. 2009;54:665–675. doi: 10.1038/jhg.2009.93. [DOI] [PubMed] [Google Scholar]
- Chung C, et al. Schizophrenia hippocampus has elevated expression of chondrex glycoprotein gene. Synapse. 2003;50:29–34. doi: 10.1002/syn.10228. [DOI] [PubMed] [Google Scholar]
- Connor CM, et al. Cingulate white matter neurons in schizophrenia and bipolar disorder. Biol Psychiatry. 2009;66:486–493. doi: 10.1016/j.biopsych.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll CU, et al. Equally increased risk for metabolic syndrome in patients with bipolar disorder and schizophrenia treated with second-generation antipsychotics. Bipolar Disord. 2008;10:788–797. doi: 10.1111/j.1399-5618.2008.00625.x. [DOI] [PubMed] [Google Scholar]
- Corti C, et al. The use of knock-out mice unravels distinct roles for mGlu2 and mGlu3 metabotropic glutamate receptors in mechanisms of neurodegeneration/neuroprotection. J Neurosci. 2007;27:8297–8308. doi: 10.1523/JNEUROSCI.1889-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, et al. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Holmes A. The ascent of mouse: advances in modelling human depression and anxiety. Nat Rev Drug Discov. 2005;4:775–790. doi: 10.1038/nrd1825. [DOI] [PubMed] [Google Scholar]
- Dracheva S, et al. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol Dis. 2006;21:531–540. doi: 10.1016/j.nbd.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Duncan CE, et al. Prefrontal GABA(A) receptor alpha-subunit expression in normal postnatal human development and schizophrenia. J Psychiatr Res. 2010;44:673–681. doi: 10.1016/j.jpsychires.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Dyck BA, et al. Synapsin II knockout mice show sensorimotor gating and behavioural abnormalities similar to those in the phencyclidine-induced preclinical animal model of schizophrenia. Schizophr Res. 2007;97:292–293. doi: 10.1016/j.schres.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Endler NS, Kocovski NL. State and trait anxiety revisited. J Anxiety Disord. 2001;15:231–245. doi: 10.1016/s0887-6185(01)00060-3. [DOI] [PubMed] [Google Scholar]
- English JA, et al. The neuroproteomics of schizophrenia. Biol Psychiatry. 2011;69:163–172. doi: 10.1016/j.biopsych.2010.06.031. [DOI] [PubMed] [Google Scholar]
- Extein I, Bowers MB., Jr State and trait in psychiatric practice. Am J Psychiatry. 1979;136:690–693. doi: 10.1176/ajp.136.5.690. [DOI] [PubMed] [Google Scholar]
- Fernandez-Egea E, et al. Metabolic profile of antipsychotic-naive individuals with non-affective psychosis. Br J Psychiatry. 2009;194:434–438. doi: 10.1192/bjp.bp.108.052605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JN, et al. Bipolar disorder: imaging state versus trait. J Neuropsychiatry Clin Neurosci. 2006;18:296–301. doi: 10.1176/jnp.2006.18.3.296. [DOI] [PubMed] [Google Scholar]
- Garbett KA, et al. Novel animal models for studying complex brain disorders: BAC-driven miRNA-mediated in vivo silencing of gene expression. Mol Psychiatry. 2010;15:987–995. doi: 10.1038/mp.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerozissis K. Brain insulin: regulation, mechanisms of action and functions. Cell Mol Neurobiol. 2003;23:1–25. doi: 10.1023/A:1022598900246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N, et al. Generation and characterization of Rgs4 mutant mice. Mol Cell Biol. 2005;25:4221–4228. doi: 10.1128/MCB.25.10.4221-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet D, et al. Family-based association of YWHAH in psychotic bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:977–983. doi: 10.1002/ajmg.b.30927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim ND, et al. Increased lactate levels and reduced pH in postmortem brains of schizophrenics: medication confounds. J Neurosci Methods. 2008;169:208–213. doi: 10.1016/j.jneumeth.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey J. Leptin regulation of neuronal excitability and cognitive function. Curr Opin Pharmacol. 2007;7:643–647. doi: 10.1016/j.coph.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, et al. Aberrant endoplasmic reticulum stress response in lymphoblastoid cells from patients with bipolar disorder. Int J Neuropsychopharmacol. 2009;12:33–43. doi: 10.1017/S1461145708009358. [DOI] [PubMed] [Google Scholar]
- Hemby SE, et al. Gene expression profile for schizophrenia: discrete neuron transcription patterns in the entorhinal cortex. Arch Gen Psychiatry. 2002;59:631–640. doi: 10.1001/archpsyc.59.7.631. [DOI] [PubMed] [Google Scholar]
- Hill SK, et al. Neurocognitive allied phenotypes for schizophrenia and bipolar disorder. Schizophr Bull. 2008;34:743–759. doi: 10.1093/schbul/sbn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, et al. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030327. e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, et al. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry. 2011;69:157–162. doi: 10.1016/j.biopsych.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, et al. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005a;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, et al. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005b;25:5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, et al. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Jaaro-Peled H, et al. Review of pathological hallmarks of schizophrenia: comparison of genetic models with patients and nongenetic models. Schizophr Bull. 2010;36:301–313. doi: 10.1093/schbul/sbp133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, et al. An association study between polymorphisms in three genes of 14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) family and paranoid schizophrenia in northern Chinese population. Eur Psychiatry. 2004;19:377–379. doi: 10.1016/j.eurpsy.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Kamiya A. Animal models for schizophrenia via in utero gene transfer: understanding roles for genetic susceptibility factors in brain development. Prog Brain Res. 2009;179:9–15. doi: 10.1016/S0079-6123(09)17902-5. [DOI] [PubMed] [Google Scholar]
- Kamiya A, et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch Gen Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C, et al. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick B. Schizophrenia as a systemic disease. Schizophr Bull. 2009;35:381–382. doi: 10.1093/schbul/sbn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick B, et al. Differences in glucose tolerance between deficit and nondeficit schizophrenia. Schizophr Res. 2009;107:122–127. doi: 10.1016/j.schres.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, et al. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Kuromitsu J, et al. Reduced neuropeptide Y mRNA levels in the frontal cortex of people with schizophrenia and bipolar disorder. Brain Res Gene Expr Patterns. 2001;1:17–21. doi: 10.1016/s1567-133x(01)00003-5. [DOI] [PubMed] [Google Scholar]
- Levitt P, et al. Making the case for a candidate vulnerability gene in schizophrenia: Convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2006;60:534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Lewis DA. The human brain revisited: opportunities and challenges in postmortem studies of psychiatric disorders. Neuropsychopharmacology. 2002;26:143–154. doi: 10.1016/S0893-133X(01)00393-1. [DOI] [PubMed] [Google Scholar]
- Lewis DA, et al. Altered cortical glutamate neurotransmission in schizophrenia: evidence from morphological studies of pyramidal neurons. Ann N Y Acad Sci. 2003;1003:102–112. doi: 10.1196/annals.1300.007. [DOI] [PubMed] [Google Scholar]
- Linden AM, et al. Anxiolytic-like activity of the mGLU2/3 receptor agonist LY354740 in the elevated plus maze test is disrupted in metabotropic glutamate receptor 2 and 3 knock-out mice. Psychopharmacology (Berl) 2005;179:284–291. doi: 10.1007/s00213-004-2098-x. [DOI] [PubMed] [Google Scholar]
- McCullumsmith RE, et al. Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophr Res. 2007;90:15–27. doi: 10.1016/j.schres.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullumsmith RE, Meador-Woodruff JH. Novel approaches to the study of postmortem brain in psychiatric illness: old limitations and new challenges. Biol Psychiatry. 2010;69:127–133. doi: 10.1016/j.biopsych.2010.09.035. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, et al. Prevalence of the metabolic syndrome in patients with schizophrenia: baseline results from the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) schizophrenia trial and comparison with national estimates from NHANES III. Schizophr Res. 2005;80:19–32. doi: 10.1016/j.schres.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Menniti FS, et al. Phosphodiesterases in the CNS: targets for drug development. Nat Rev Drug Discov. 2006;5:660–670. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- Mexal S, et al. Brain pH has a significant impact on human postmortem hippocampal gene expression profiles. Brain Res. 2006;1106:1–11. doi: 10.1016/j.brainres.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Mexal S, et al. Differential modulation of gene expression in the NMDA postsynaptic density of schizophrenic and control smokers. Brain Res Mol Brain Res. 2005;139:317–332. doi: 10.1016/j.molbrainres.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Mirnics K, et al. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Mirnics K, et al. Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci. 2001a;24:479–486. doi: 10.1016/s0166-2236(00)01862-2. [DOI] [PubMed] [Google Scholar]
- Mirnics K, et al. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Mirnics K, et al. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001b;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Morozov A, et al. Using conditional mutagenesis to study the brain. Biol Psychiatry. 2003;54:1125–1133. doi: 10.1016/s0006-3223(03)00467-0. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65:480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak G, et al. Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: a preliminary study. World J Biol Psychiatry. 2009;10:929–935. doi: 10.1080/15622970903079499. [DOI] [PubMed] [Google Scholar]
- Oliver PL. Challenges of analysing gene-environment interactions in mouse models of schizophrenia. Scientific World Journal. 2011;11:1411–1420. doi: 10.1100/tsw.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottis P, et al. Convergence of Two Independent Mental Disease Genes on the Protein Level: Recruitment of Dysbindin to Cell-Invasive Disrupted-In-Schizophrenia 1 Aggresomes. Biol Psychiatry. 2011 doi: 10.1016/j.biopsych.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Patil ST, et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: a randomized Phase 2 clinical trial. Nat Med. 2007;13:1102–1107. doi: 10.1038/nm1632. [DOI] [PubMed] [Google Scholar]
- Paz-Filho GJ, et al. Leptin replacement improves cognitive development. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003098. e3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001511. 643. [DOI] [PubMed] [Google Scholar]
- Regenold WT, et al. Myelin staining of deep white matter in the dorsolateral prefrontal cortex in schizophrenia, bipolar disorder, and unipolar major depression. Psychiatry Res. 2007;151:179–188. doi: 10.1016/j.psychres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Rioux L, et al. Characterization of olfactory bulb glomeruli in schizophrenia. Schizophr Res. 2005;77:229–239. doi: 10.1016/j.schres.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Rosahl TW, et al. Essential functions of synapsins I and II in synaptic vesicle regulation. Nature. 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- Sawa A, et al. Neuron-glia interactions clarify genetic-environmental links in mental illness. Trends Neurosci. 2004;27:294–297. doi: 10.1016/j.tins.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Seeman P, et al. Glutamate receptor mGlu2 and mGlu3 knockout striata are dopamine supersensitive, with elevated D2(High) receptors and marked supersensitivity to the dopamine agonist (+)PHNO. Synapse. 2009;63:247–251. doi: 10.1002/syn.20607. [DOI] [PubMed] [Google Scholar]
- Seshadri AJ, Hayashi-Takagi A. Gene manipulation with stereotaxic viral infection for psychiatric research: spatiotemporal components for schizophrenia. Prog Brain Res. 2009;179:17–27. doi: 10.1016/S0079-6123(09)17903-7. [DOI] [PubMed] [Google Scholar]
- Shanley LJ, et al. Leptin enhances NMDA receptor function and modulates hippocampal synaptic plasticity. J Neurosci. 2001;21:RC186. doi: 10.1523/JNEUROSCI.21-24-j0001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, et al. Downregulation in components of the mitochondrial electron transport chain in the postmortem frontal cortex of subjects with bipolar disorder. J Psychiatry Neurosci. 2006;31:189–196. [PMC free article] [PubMed] [Google Scholar]
- Tajinda K, et al. Neuronal biomarkers from patients with mental illnesses: a novel method through nasal biopsy combined with laser-captured microdissection. Mol Psychiatry. 2010;15:231–232. doi: 10.1038/mp.2009.73. [DOI] [PubMed] [Google Scholar]
- Tamminga CA. Schizophrenia and glutamatergic transmission. Crit Rev Neurobiol. 1998;12:21–36. doi: 10.1615/critrevneurobiol.v12.i1-2.20. [DOI] [PubMed] [Google Scholar]
- Taylor V, MacQueen G. Associations between bipolar disorder and metabolic syndrome: A review. J Clin Psychiatry. 2006;67:1034–1041. doi: 10.4088/jcp.v67n0704. [DOI] [PubMed] [Google Scholar]
- Thakore JH. Metabolic disturbance in first-episode schizophrenia. Br J Psychiatry Suppl. 2004;47:S76–S79. doi: 10.1192/bjp.184.47.s76. [DOI] [PubMed] [Google Scholar]
- Tkachev D, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Toyooka K, et al. 14-3-3 protein eta chain gene (YWHAH) polymorphism and its genetic association with schizophrenia. Am J Med Genet. 1999;88:164–167. [PubMed] [Google Scholar]
- Twine NA, et al. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer's disease. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016266. e16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11:100–113. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- Vawter MP, et al. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55:641–650. doi: 10.1016/s0361-9230(01)00522-6. [DOI] [PubMed] [Google Scholar]
- Vawter MP, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, et al. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- Wittmann-Liebold B, et al. Two-dimensional gel electrophoresis as tool for proteomics studies in combination with protein identification by mass spectrometry. Proteomics. 2006;6:4688–4703. doi: 10.1002/pmic.200500874. [DOI] [PubMed] [Google Scholar]