Abstract
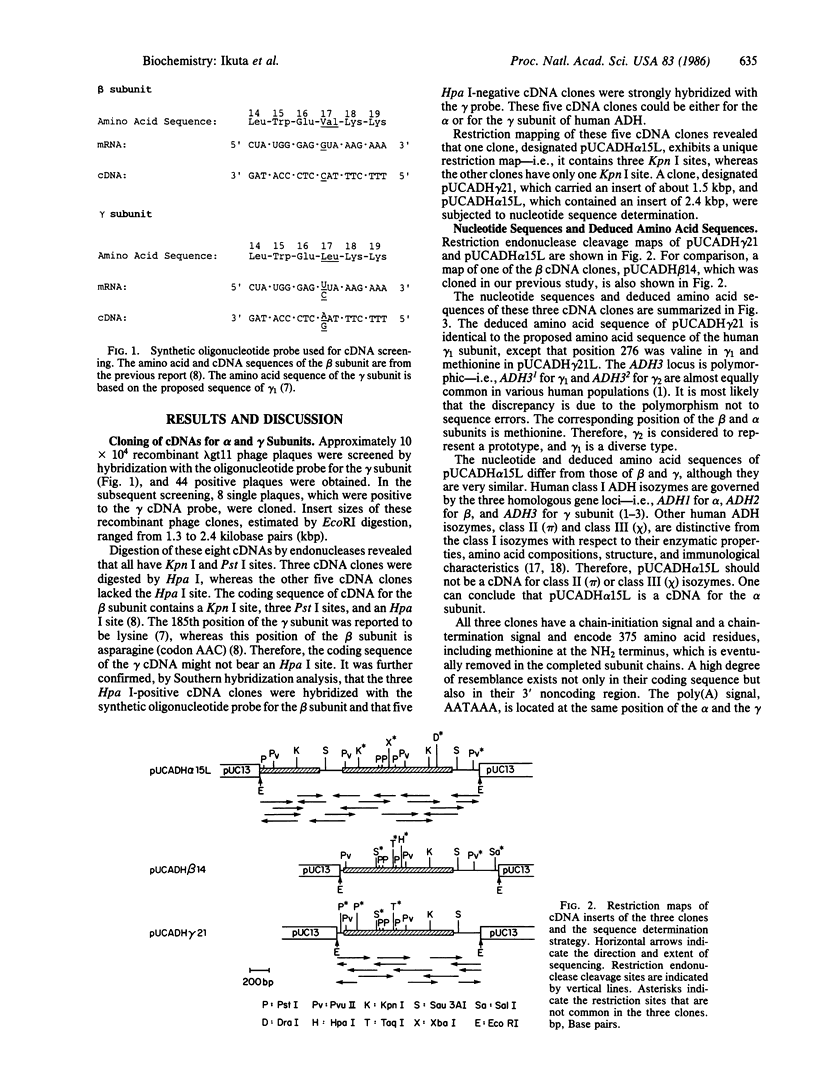
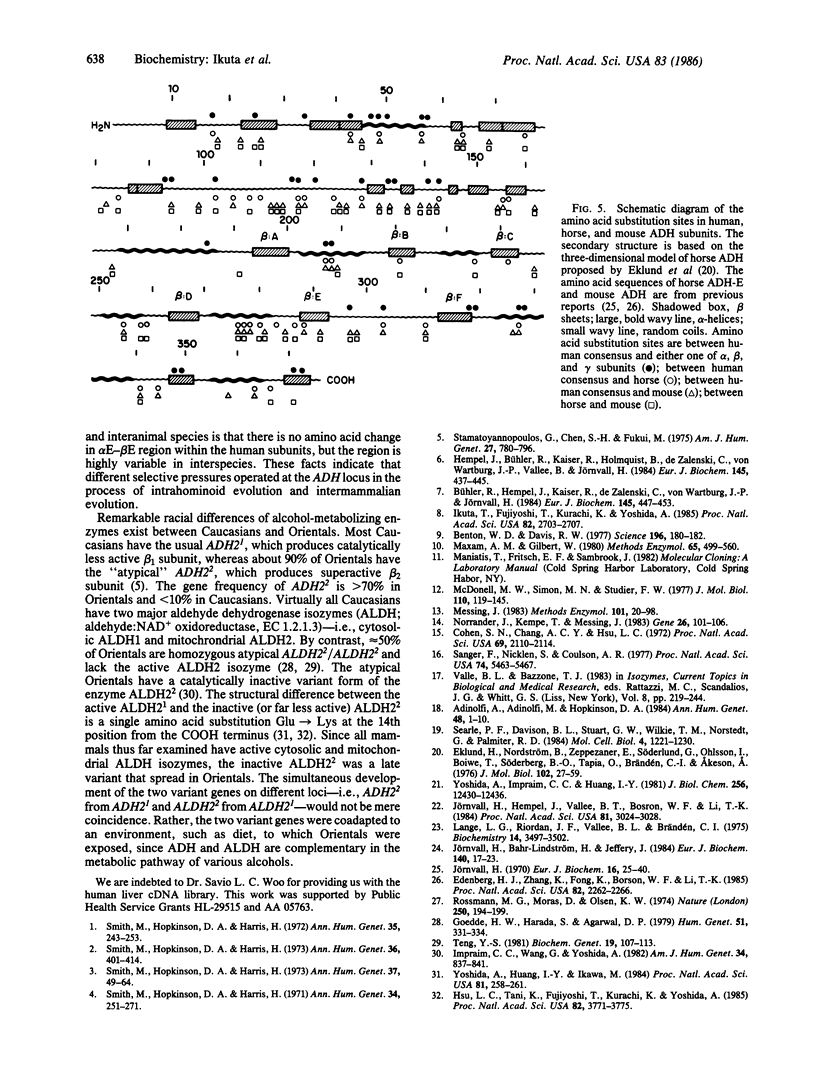
Class I human alcohol dehydrogenase (ADH; alcohol:NAD+ oxidoreductase, EC 1.1.1.1) consists of several homo- and heterodimers of alpha, beta, and gamma subunits that are governed by the ADH1, ADH2, and ADH3 loci. We previously cloned a full length of cDNA for the beta subunit, and the complete sequence of 374 amino acid residues was established. cDNAs for the alpha and gamma subunits were cloned and characterized. A human liver cDNA library, constructed in phage lambda gt11, was screened by using a synthetic oligonucleotide probe that was matched to the gamma but not to the beta sequence. Clone pUCADH gamma 21 and clone pUCADH alpha 15L differed from beta cDNA with respect to restriction sites and hybridization with the nucleotide probe. Clone pUCADH gamma 21 contained an insertion of 1.5 kilobase pairs (kbp) and encodes 374 amino acid residues compatible with the reported amino acid sequence of the gamma subunit. Clone pUCADH alpha 15L contained an insertion of 2.4 kbp and included nucleotide sequences that encode 374 amino acid residues for another subunit, the alpha subunit. In addition, this clone contained the sequences that encode the COOH-terminal part of the beta subunit at its extended 5' region. The amino acid sequences and coding regions of the cDNAs of the three subunits are very similar (approximately 93-95% identity). A high degree of resemblance is observed also in their 3' noncoding regions. However, distinctive differences exist in the vicinity of the Zn-binding cysteine residue at position 46--i.e., Cys-Gly-Thr in the alpha, Cys-Arg-Thr in the wild-type beta 1, Cys-His-Thr in the Oriental-type beta 2, and Cys-Arg-Ser in the gamma, reflecting the differences in their kinetic properties. Based on the cDNA sequences and the deduced amino acid sequences of the three subunits, their structural and evolutionary relationships are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adinolfi A., Adinolfi M., Hopkinson D. A. Immunological and biochemical characterization of the human alcohol dehydrogenase chi-ADH isozyme. Ann Hum Genet. 1984 Jan;48(Pt 1):1–10. doi: 10.1111/j.1469-1809.1984.tb00828.x. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bühler R., Hempel J., Kaiser R., de Zalenski C., von Wartburg J. P., Jörnvall H. Human liver alcohol dehydrogenase. 2. The primary structure of the gamma 1 protein chain. Eur J Biochem. 1984 Dec 17;145(3):447–453. doi: 10.1111/j.1432-1033.1984.tb08575.x. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg H. J., Zhang K., Fong K., Bosron W. F., Li T. K. Cloning and sequencing of cDNA encoding the complete mouse liver alcohol dehydrogenase. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2262–2266. doi: 10.1073/pnas.82.8.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Goedde H. W., Harada S., Agarwal D. P. Racial differences in alcohol sensitivity: a new hypothesis. Hum Genet. 1979 Oct 2;51(3):331–334. doi: 10.1007/BF00283404. [DOI] [PubMed] [Google Scholar]
- Hempel J., Bühler R., Kaiser R., Holmquist B., de Zalenski C., von Wartburg J. P., Vallee B., Jörnvall H. Human liver alcohol dehydrogenase. 1. The primary structure of the beta 1 beta 1 isoenzyme. Eur J Biochem. 1984 Dec 17;145(3):437–445. doi: 10.1111/j.1432-1033.1984.tb08573.x. [DOI] [PubMed] [Google Scholar]
- Hsu L. C., Tani K., Fujiyoshi T., Kurachi K., Yoshida A. Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3771–3775. doi: 10.1073/pnas.82.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T., Fujiyoshi T., Kurachi K., Yoshida A. Molecular cloning of a full-length cDNA for human alcohol dehydrogenase. Proc Natl Acad Sci U S A. 1985 May;82(9):2703–2707. doi: 10.1073/pnas.82.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impraim C., Wang G., Yoshida A. Structural mutation in a major human aldehyde dehydrogenase gene results in loss of enzyme activity. Am J Hum Genet. 1982 Nov;34(6):837–841. [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H., Hempel J., Vallee B. L., Bosron W. F., Li T. K. Human liver alcohol dehydrogenase: amino acid substitution in the beta 2 beta 2 Oriental isozyme explains functional properties, establishes an active site structure, and parallels mutational exchanges in the yeast enzyme. Proc Natl Acad Sci U S A. 1984 May;81(10):3024–3028. doi: 10.1073/pnas.81.10.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H. Horse liver alcohol dehydrogenase. The primary structure of the protein chain of the ethanol-active isoenzyme. Eur J Biochem. 1970 Sep;16(1):25–40. doi: 10.1111/j.1432-1033.1970.tb01049.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., von Bahr-Lindström H., Jeffery J. Extensive variations and basic features in the alcohol dehydrogenase-sorbitol dehydrogenase family. Eur J Biochem. 1984 Apr 2;140(1):17–23. doi: 10.1111/j.1432-1033.1984.tb08061.x. [DOI] [PubMed] [Google Scholar]
- Lange L. G., 3rd, Riordan J. F., Vallee B. L., Brändén C. I. The role of arginyl residues in directing carboxymethylation of horse liver alcohol dehydrogenase. Biochemistry. 1975 Jul 29;14(15):3497–3502. doi: 10.1021/bi00686a032. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle P. F., Davison B. L., Stuart G. W., Wilkie T. M., Norstedt G., Palmiter R. D. Regulation, linkage, and sequence of mouse metallothionein I and II genes. Mol Cell Biol. 1984 Jul;4(7):1221–1230. doi: 10.1128/mcb.4.7.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Hopkinson D. A., Harris H. Alcohol dehydrogenase isozymes in adult human stomach and liver: evidence for activity of the ADH 3 locus. Ann Hum Genet. 1972 Mar;35(3):243–253. doi: 10.1111/j.1469-1809.1957.tb01398.x. [DOI] [PubMed] [Google Scholar]
- Smith M., Hopkinson D. A., Harris H. Developmental changes and polymorphism in human alcohol dehydrogenase. Ann Hum Genet. 1971 Feb;34(3):251–271. doi: 10.1111/j.1469-1809.1971.tb00238.x. [DOI] [PubMed] [Google Scholar]
- Smith M., Hopkinson D. A., Harris H. Studies on the properties of the human alcohol dehydrogenase isozymes determined by the different loci ADH1, ADH2, ADH3. Ann Hum Genet. 1973 Jul;37(1):49–67. doi: 10.1111/j.1469-1809.1973.tb01814.x. [DOI] [PubMed] [Google Scholar]
- Smith M., Hopkinson D. A., Harris H. Studies on the subunit structure and molecular size of the human alcohol dehydrogenase isozymes determined by the different loci, ADH1, ADH2, and ADH3. Ann Hum Genet. 1973 Apr;36(4):401–414. doi: 10.1111/j.1469-1809.1973.tb00604.x. [DOI] [PubMed] [Google Scholar]
- Teng Y. S. Human liver aldehyde dehydrogenase in Chinese and Asiatic Indians: gene deletion and its possible implications in alcohol metabolism. Biochem Genet. 1981 Feb;19(1-2):107–114. doi: 10.1007/BF00486141. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Huang I. Y., Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A. 1984 Jan;81(1):258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Impraim C. C., Huang I. Y. Enzymatic and structural differences between usual and atypical human liver alcohol dehydrogenases. J Biol Chem. 1981 Dec 10;256(23):12430–12436. [PubMed] [Google Scholar]



