Abstract
The cellular DNA damage response (DDR) involves changes in the functional and structural properties of a number of nuclear proteins, resulting in a coordinated control of gene expression and DNA repair. This response includes functional interactions of the DNA repair, transcription, and RNA processing machineries. Following DNA damage, cellular levels of polyadenylated transcripts are transiently decreased and normal recovery depends on transcription-coupled repair (TCR). In addition, DNA damage has gene-specific effects regulating the mRNA levels of factors involved in the DDR itself at different times after the damage. The 3′-end processing machinery, which is important in the regulation of mRNA stability, is involved in these general and gene-specific responses to DNA damage. The role of 3′-end processing in DDR supports the idea that the steady-state levels of different mRNAs change upon DNA-damaging conditions as a result of regulation of not only their biosynthesis but also their turnover. Here, we review the mechanistic connections between 3′-end processing and DDR, and discuss the implications of deregulation of this important step of mRNA maturation in the cellular recovery after DNA-damaging treatment. The relevance of these functional connections is illustrated by the increasing number of reports on this relatively unexplored field.
The cellular response to DNA damage is a protective mechanism against disease. This cellular response could be either in a survival mode, where DNA repair occurs and gene expression is controlled along with cell-cycle arrest, or in a death mode, where apoptosis is induced. After DNA damage, the steady-state levels of different mRNAs are regulated (Figure 1), reflecting changes in both their biosynthesis and their turnover. Although the mechanisms involved in this response are still unresolved, it has been shown that 3′-end processing plays an important role. In this review, we discuss the role of 3′-end formation and 3′-processing factors in the DNA damage response (DDR). All eukaryotic mRNAs possess poly(A) tails at the 3′-end, with the exception of some histone mRNAs. The poly(A) tail is synthesized in the nucleus by a two-step reaction involving endonucleolytic cleavage, which specifies the 3′-end of the mRNA, and subsequent synthesis of a 200 adenosine residues tail. The specificity and efficiency of 3′-end processing is determined by the binding of multiprotein complexes to specific elements at the 3′-untranslated region (3′-UTR) of the pre-mRNA. The poly(A) tails are critical for other aspects of mRNA metabolism associated to the 3′-UTR, such as mRNA subcellular localization, translation, and mRNA decay. For better understanding of basic aspects of 3′-end formation and its regulation, we suggest reading the review by Mandel and coworkers,1 which covers the topic. Here, we also review the role of 3′-end processing in the decrease of total mRNA levels and in the regulation of the levels of specific mRNAs after DNA damage treatment (Figures 2 and 3). This review provides a broader understanding of the interplay between different DDRs and mRNA 3′ processing pathways in gene regulation following DNA damage.
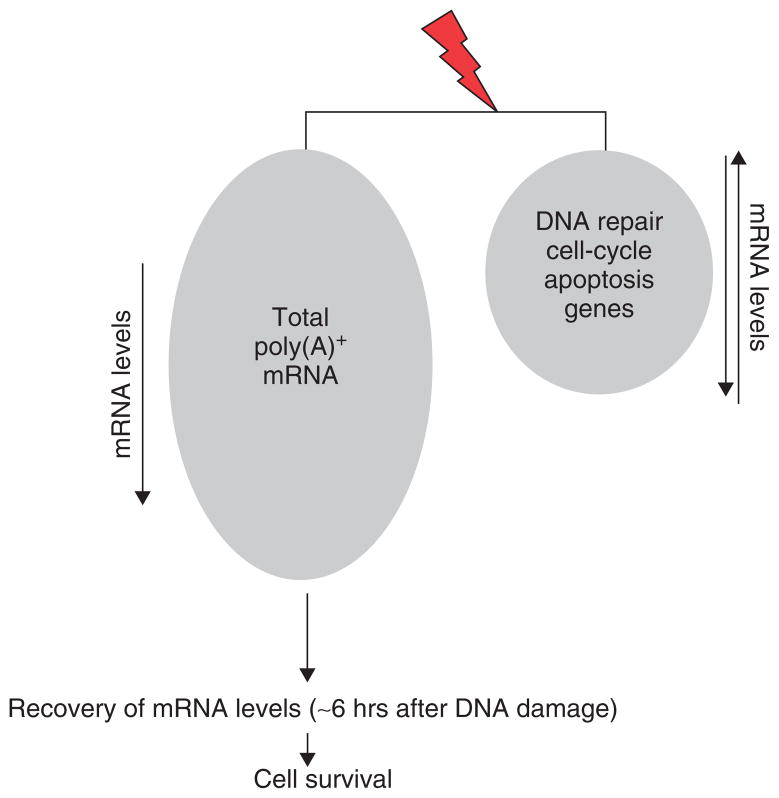
FIGURE 1.
General and gene-specific effects of DNA-damaging conditions on mRNA levels. The level of poly(A)+ mRNAs of genes not involved in the DNA damage response (DDR) decreases after DNA damage.2–6 Full recovery of mRNA levels within 6 h after the exposure correlates with the cell survival. The level of poly(A)+ mRNAs of genes involved in the DDR is down- or up-regulated at different time points after DNA damage.7–10.
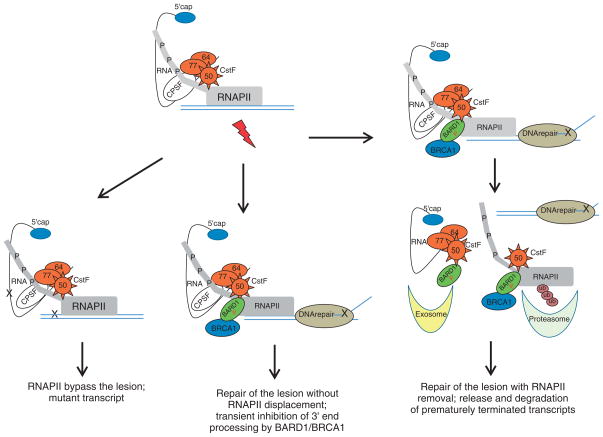
FIGURE 2.
Model for transcriptional–3′-end processing alternatives during the DNA damage response (DDR). The DNA damage-induced lesions may affect the elongating RNA polymerase II–cleavage stimulation factor (RNAP II–CstF) holoenzyme in different ways. The lesion is bypassed, generating a mutant transcript. Alternatively, RNAP II stalled at sites of DNA damage can reengage in a transcriptional process. In that scenario, the BRCA1–BARD1–CstF complex reassures the correct 3′-end processing of the nascent RNA. For some other lesions, RNAP II–CstF holoenzyme is stalled, causing premature termination or transient arrest of elongation process. BRCA1–BARD1-containing complexes are recruited to sites of DNA repair, resulting in RNAP II ubiquitination and inhibition of transcription by degradation of RNAP II. The nascent RNA product is released and its 3′-end processing is inhibited by both the CstF–BARD1 interaction and RNAP II degradation. This process facilitates repair by allowing access of the repair machinery to the DNA damage site, and at the same time prevents the formation of aberrantly processed mRNAs, which are eliminated by exosome-mediated degradation in a nuclear surveillance pathway.
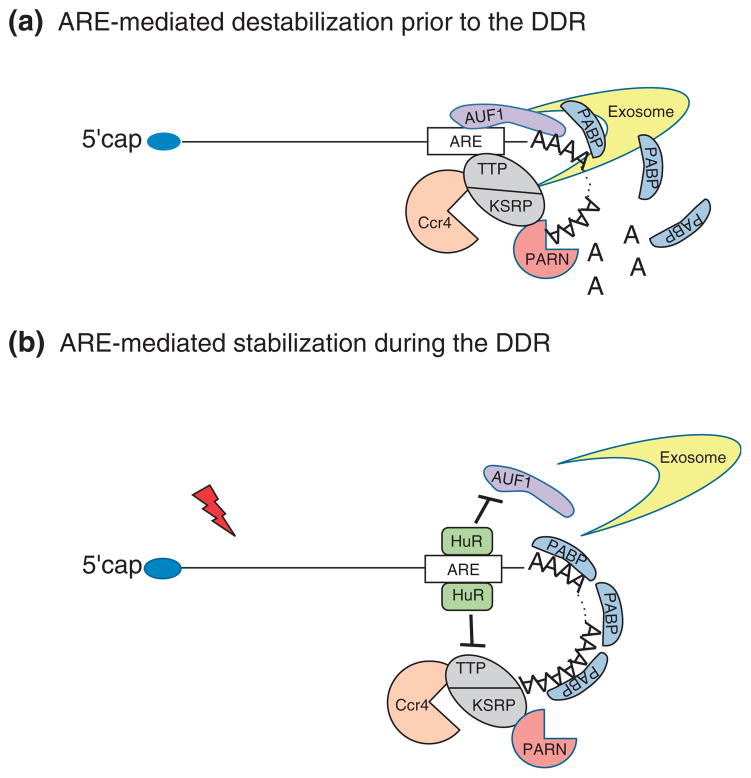
FIGURE 3.
Schematic diagram of the involvement of 3′-end processing in the mRNA surveillance pathway of genes involved in DNA damage response (DDR). (A) AU-rich elements (ARE)-mediated destabilization prior to DNA damage. Genes involved in the early DDR are rapidly degraded in non-treated cells. The ARE-binding proteins (ARE-BP) AUF1 compete with poly(A) binding protein (PABP) for binding to the poly(A) tail, destabilizing and, possibly, exposing it to deadenylases, such as poly(A)-specific ribonuclease (PARN). Alternatively, other ARE-BP, such as tristetraprolin (TTP) and KH-type splicing regulatory protein (KSRP), recruit the deadenylases Ccr4 and PARN to ARE-mRNAs and initiate deadenylation-dependent degradation of those transcripts via the exosome. (B) ARE-mediated stabilization during the DDR. Following DNA damage, genes involved in DDR are up-regulated. In that scenario, the cellular levels of the ARE-BP HuR are up-regulated and HuR competes with AUF1 for binding to the same ARE region. This competition stabilizes the association of the PABP to the poly(A) tail. Moreover, HuR also competes with the other ARE-BP, such as TTP and KRSP, preventing the recruitment of deadenylases to the ARE-mRNA and the exosomal degradation.
DECREASE OF TOTAL CELLULAR mRNA LEVELS AFTER DNA DAMAGE
Multiple strategies have evolved to minimize the genotoxic consequences of endogenous and environmental agents that damage the DNA. One example of this is the conserved nucleotide excision repair (NER) pathway that removes mainly helix-distorting lesions, including ultraviolet (UV)-induced cyclobutane pyrimidine dimers and 6–4 photoproducts, as well as bulky chemical adducts. The efficiency of NER is not equal in all parts of the genome, and different kinds of lesions are removed from the DNA with different efficiencies. For instance, global genome repair eliminates lesions throughout the genome, and transcription-coupled repair (TCR) selectively removes DNA lesions from the transcribed strands of active genes.11 Accumulating evidence suggests that the blockage of the elongating RNA polymerase II (RNAP II) at sites of DNA damage is an early event that initiates TCR.11 It has been shown that RNAP II stalled at sites of DNA damage could respond in different alternative ways depending on the nature of the lesion. For example, RNAP II could continue transcription and generate a mutant RNA; alternatively the transcriptional rate of RNAP II could be affected by the DNA repair process without leading to mutagenesis of the transcript. The stalled RNAP II could also be ubiquitinated and degraded releasing the nascent RNA product, and these prematurely terminated transcripts should be eliminated to avoid the formation of potentially deleterious proteins.
Following UV treatment, but not ionizing radiation, cellular levels of poly(A)+ mRNA are transiently decreased (Figure 1) and normal recovery depends on TCR.2 Supporting these studies, the mRNA levels of several housekeeping genes also transiently decreased after UV treatment.3–6 Although the mechanism involved in this response is still unresolved, it has been suggested that the UV-induced inhibition of transcription is responsible for the decrease in the mRNA levels. This may very well be a significant part of the mechanism; however, those early studies have not considered that the steady-state levels of cellular mRNAs change as a result of regulation of not only their biosynthesis but also their turnover. Indeed, RNA 3′-end processing, which is important in the regulation of mRNA stability, is strongly but transiently inhibited upon DNA-damaging conditions.12
The cleavage stimulation factor (CstF) is involved in this general effect of DNA damage on 3′-end processing (Figure 2).12 CstF is one of the essential polyadenylation factors and consists of three protein subunits called CstF-77, CstF-64, and CstF-50. Both the CstF-50 and CstF-77 subunits interact specifically with the carboxy-terminal domain (CTD) of RNAP II’s largest subunit, most likely facilitating the RNAP II-mediated activation of 3′-end processing.13 CstF-50 can also interact with the BRCA1-associated RING domain protein (BARD1) to inhibit 3′ processing. Together, BARD1 and BRCA1 have been involved in both the repair of DNA lesions and in the regulation of cell-cycle checkpoints in DDR.14 3′-End processing is inhibited after UV-induced DNA damage as a result of both the formation of the BRCA1–BARD1–CstF complex12 and the proteasomal-mediated degradation of RNAP II.15 Interestingly, DNA damage-induced BARD1 phosphorylation in an ATM-dependent manner is critical for these functions of BARD1 in 3′-end processing.16 As CstF-50 can interact with the tumor suppressor BARD1 to inhibit 3′ processing and with RNAP II to activate 3′ processing, it was proposed that CstF-50 plays a coordinating role in the nuclear DDR. Supporting this idea, depletion of CstF enhances sensitivity to UV treatment and reduces ability to ubiquitinate RNAP II and repair DNA;6 this depletion also leads to cell-cycle arrest and results in apoptosis.17
The UV-induced inhibition of 3′ processing might be mechanistically more complex than initially thought and other factors might also be involved. For example, a number of factors involved in DNA repair, such as the DNA-activated protein kinase complex (DNA-PKcs–Ku70–Ku86) and poly(ADP-ribose) polymerase 1, have been identified by proteomic analysis as part of the human 3′ processing complex.18 Presently, their functional roles in 3′-end processing have not been determined. Another example is the role of the antiproliferative factor BTG2 as a general activator of mRNA deadenylation.19 This BTG2 function requires a direct interaction between BTG2 and Pop2–Caf1 and Ccr4 deadenylases. As BTG2 is involved in p53-dependent and p53-independent DDR, it may represent an alternative mechanism of global regulation of gene expression after specific stresses.
Other mRNA metabolic pathways associated to the 3′-UTR might influence the role of 3′-end processing in DDR. For example, incorrectly processed and defective transcripts are generated during DDR. The exosome acts as a surveillance apparatus that eliminates those mRNAs by 3′-end RNA degradation, ensuring that only high-quality mRNAs are engaged in protein synthesis. Most of the exosome-activating signals come from the 3′-UTR and the poly(A) tail, and RNA degradation is promoted not only by different deadenylases but also by the nuclear Trf4/Air2/Mtr4p polyadenylation (TRAMP) complex.20 The role of RNA surveillance under DNA-damaging conditions is poorly understood. Interestingly, the only exclusive nuclear components of the exosome, Rrp6 and Lrp1, participate not only in general mRNA degradation but also in specific mRNA degradation upon UV treatment.21 Rrp6 has exonucleolytic activity on aberrantly 3′-end processed transcripts and retains transcripts at sites of transcription where they are presumed to be degraded. Lrp1, which is a binding partner of Rrp6, is involved in DDR and requires Rrp6 for proper localization on target mRNAs under both DNA-damaging and non-damaging conditions. Further functional studies are required to fully understand the role of the exosome in DDR.
The maintenance of genome integrity is essential for the regulation of cell proliferation and differentiation—processes that are primarily regulated by cyclin-dependent kinases (CDKs). There is increasing evidence that the catalytic activities of CDKs play critical roles in the DDR.22 In this context, another potential link between mRNA 3′ processing and DDR is suggested by the observation that poly(A) polymerase (PAP) is phosphorylated and inhibited in a cyclin-dependent manner.23 It has been suggested that PAP repression might result in reductions of poly(A)+ RNA and/or protein synthesis, both of which are observed in DDR.
REGULATION OF SPECIFIC mRNAs INVOLVED IN DDR AFTER DNA DAMAGE
One of the paradigms of the general effect of DNA damage on gene expression is that transcripts of genes involved in the DDR are expressed before the DNA repair process begins. This suggests that compensatory mechanisms should be used on genes involved in the DDR to allow transcription and RNA 3′ processing, providing the cells with a way to react to DNA-damaging conditions. For example, the activation of the p53 pathway after DNA damage treatment depends on gene-specific requirements for transcription elongation and RNAP II phosphorylation.24 Importantly, RNAP II CTD phosphorylation at Ser 2, which occurs mainly on elongating polymerase and is therefore more abundant at the 3′ region of the genes, is not required for the transcription of p53 target genes and for mRNA 3′ cleavage–polyadenylation.24 However, the elongation factor P-TEFb and RNAP II phosphorylation at Ser 2 are required for transcription and mRNA 3′ processing of immediate early response genes, such as c-fos and junB,25 indicating that different compensatory mechanisms might exist for different cellular stress-response genes.
It has been shown that 3′-end processing is tightly associated to mRNA decay mechanisms, which are also involved in the regulation of the expression of genes involved in the DDR. For instance, AU-rich elements (ARE) can decrease mRNA stability under non-stress conditions and can increase mRNA stability under DNA-damaging conditions, such as after treatment with UV, hydrogen peroxide, methyl methanesulfonate, and cyclopentenone PGA2.7–10 This tight regulation is mediated by modifications in the 3′-end processing of these ARE-containing mRNAs. Approximately 8% of human mRNAs contain AREs, including mRNAs encoding cytokines, growth factors, oncogenes, and cell-cycle regulators.26 As many proto-oncogenes, such as c-fos, c-myc, and c-jun, play roles not only in DDR but also in proliferation and differentiation of normal cells, they must be tightly regulated to prevent malignant transformation. After UV treatment, ARE-containing mRNAs, such as c-fos, kin17, c-jun, IκB, and c-myc, are transiently stabilized.7 For example, c-fos mRNA expression increased by 45 min to 1 h after UV treatment and then dramatically decreased 2 h after the treatment. ARE-binding proteins (ARE-BP) can regulate the poly(A) tail length either by inhibiting or activating deadenylation resulting in changes in the stability of ARE-containing mRNAs under different cellular conditions.26 For example, the expression level of the growth arrest and DNA damage-inducible gene GADD45α is down-regulated under regular cellular conditions by the destabilizing effect of the ARE-BP AUF1.27 Interestingly, AUF1 can compete with the poly(A) binding protein (PABP) for binding the poly(A) destabilizing the mRNA, recruiting the exosome, and labeling the mRNA for degradation.28 Moreover, upon genotoxic stress, AUF1 dissociates from the GADD45α mRNA, which results in the up-regulation of that gene. Another example comes from the UV-mediated stabilization of the mRNA of the apoptotic signal transduction factor RhoB by the ARE-BP HuR,29 which can block the recruitment of deadenylases or the exosome to the ARE-mRNAs.
A major regulation of the ARE-mRNA levels seems to come from the antagonistic behavior of different ARE-BP (Figure 3). For instance, the degradation of ARE-containing mRNAs in untreated cells involves the recruitment of the exosome via ARE-BP, such as AUF1, tristetraprolin (TTP), and KH-type splicing regulatory protein (KSRP20). KSRP and TTP can also recruit the deadenylases poly(A)-specific ribonuclease (PARN) and Ccr4, a component of the Ccr4–Caf1–Not complex, to the nascent target in order to initiate the deadenylation that precedes degradation. Upon DNA-damaging conditions, ARE-BP HuR binds AREs, resulting in the dissociation of AUF1, TTP, and KSRP from ARE-containing mRNAs and in the up-regulation of genes involved in DDR. For example, p21, an CDK inhibitor, is destabilized by AUF1 under regular cellular conditions.26. However, following UV damage, HuR interaction with p21 stabilizes the transcript via binding to its ARE.9 Interestingly, the Xenopus homolog of HuR, elrA, has been shown to play a role in polyadenylation of maternal mRNAs in fertilized eggs.30 Other ARE-mRNAs, such as c-fos, granulocyte macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and cyclin D1, are destabilized in the presence of AUF1 and stabilized by HuR under oxidative stress mediated by UV light, H2O2, or arsenite.26
In yeast, the deadenylases poly(A) nuclease (PAN) and Ccr4–Caf1 complex are also involved in DDR. Both deadenylases can interact with Dun1 kinase, which is involved in DNA damage-dependent cell-cycle arrest in the G2/M and in the transcriptional induction of repair genes.31,32 Dun1 and PAN complex act together in regulating mRNA levels of the DNA repair gene RAD5, and dun1pan2 and dun1pan3 double mutants are synthetically lethal in the presence of replication blocks.31 As part of this DDR, Dun1 and PAN are involved in posttranscriptional mechanisms involving poly(A) tail length control, representing additional checkpoint targets in the regulation of gene expression. Mutants in the mRNA deadenylase catalytic subunit Ccr4 have been found to be sensitive to DNA damage treatment,33 and the Ccr4–Not complex genetically interacts with different checkpoint pathways.32 Both transcriptional and 3′-end processing functions of Ccr4–Not contribute to the DDR affecting gene expression in a complex manner. Interestingly, Ccr4–Not and Paf1, which have mRNA 3′-end processing functions, are involved in TCR.34
Finally, the Arabidopsis thaliana ortholog of the 30 kDa subunit of the cleavage and polyadenylation specificity factor (CPSF30) has been implicated in the regulation of oxidative signaling.35 A mutation in CPSF30 results in higher expression levels of genes encoding thioredoxins and glutaredoxins, which are reactive oxygen species-associated or -induced genes. Interestingly, the disruption of CPSF30 does not have a dramatic effect on poly(A) tail length but on the poly(A) site choice, a result expected of a gene that encodes a processing endonuclease that is involved in the step that precedes poly(A) addition.1 These studies suggest that 3′-end processing can modulate stress tolerance responses induced by oxidative stress signals.
CONCLUSION
Understanding the mechanisms and consequences of 3′-end regulation in the DDR constitutes a major challenge in this growing, but mainly, unexplored field. The slow progress in this research topic is in part due to the lack of systematic studies connecting these pathways. Most of the studies have shown mechanistic connections between changes in the steady-state levels of mRNAs during the DDR and transcriptional regulation, but have not considered the important effect of mRNA processing on mRNA levels. As 3′-end processing can influence mRNA stability, mRNA nuclear export, and translational efficiency, it is essential for proper control of gene expression in different cellular conditions, especially in the DDR. The data reviewed here indicate that 3′-end processing has a role in the DNA damage-induced decrease of cellular mRNA levels and in the regulation of mRNA levels of genes involved in the DDR. Although the first steps have been taken to understand the role of 3′-end processing in the DDR, we still do not know how the coupling between these two pathways occurs. What 3′-end processing and DNA repair factors are involved in this mechanism? Is the exosome involved in the degradation of DNA damage-induced erroneously processed and mutant mRNAs? How are those defective mRNAs detected? How is RNAP II involved in the connection of these two pathways? As the medical relevance of deregulation of the DDR has been extensively analyzed over the years and growing evidence shows the important role of 3′-end mRNA processing in disease,36 what is the medical relevance of the connections between both pathways? The development of novel approaches for studying 3′-end processing in the DDR will be essential to address some of these basic questions.
Acknowledgments
We apologize to those authors whose work was not cited because of space limitations. The authors are supported by National Institute of General Medical Sciences grant SC1GM083806 to F.E.K.
References
- 1.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derheimer FA, Chang CW, Ljungman M. Transcription inhibition: a potential strategy for cancer therapeutics. Eur J Cancer. 2005;41:2569–2576. doi: 10.1016/j.ejca.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- 4.Akeo K, Funayama T, Hamada N, Akeo Y, Hiramitsu T, Kobayashi Y. The effects of ultraviolet irradiation and hypoxia on expression of glutathione peroxidase and glyceraldehyde 3-phosphate dehydrogenase in the cultured retinal pigment epithelium. Tissue Cult Res Commun. 2007;26:149–157. [Google Scholar]
- 5.Maccoux LJ, Clements DN, Salway F, Day PJ. Identification of new reference genes for the normalisation of canine osteoarthritic joint tissue transcripts from microarray data. BMC Mol Biol. 2007;8:62–72. doi: 10.1186/1471-2199-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirkin N, Fonseca D, Mohammed S, Cevher MA, Manley JL, Kleiman FE. The 3′ processing factor CstF functions in the DNA repair response. Nucleic Acids Res. 2008;36:1792–1804. doi: 10.1093/nar/gkn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, Angulo JF, Herrlich P. UV-induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol. 2000;20:3616–3625. doi: 10.1128/mcb.20.10.3616-3625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollig F, Winzen R, Kracht M, Ghebremedhin B, Ritter B, Wilhelm A, Resch K, Holtmann H. Evidence for general stabilization of mRNAs in response to UV light. Eur J Biochem. 2002;269:5830–5839. doi: 10.1046/j.1432-1033.2002.03300.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gowrishankar G, Winzen R, Bolling F, Ghebremedhin B, Redich N, Ritter B, Resch K, Kracht M, Holtmann H. Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol Chem. 2005;12:1287–1293. doi: 10.1515/BC.2005.146. [DOI] [PubMed] [Google Scholar]
- 11.Tornaletti S. DNA repair in mammalian cells: transcription-coupled DNA repair: directing your effort where it’s most needed. Cell Mol Life Sci. 2009;66:1010–1020. doi: 10.1007/s00018-009-8738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 13.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 2008;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 14.Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 15.Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HS, Li H, Cevher M, Parmelee A, Fonseca D, Kleiman FE, Lee SB. DNA damage-induced BARD1 phosphorylation is critical for the inhibition of messenger RNA processing by BRCA1/BARD1 complex. Cancer Res. 2006;66:4561–4565. doi: 10.1158/0008-5472.CAN-05-3629. [DOI] [PubMed] [Google Scholar]
- 17.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauxion F, Faux C, Séraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 21.Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yata K, Esashi F. Dual role of CDKs in DNA repair: to be, or not to be. DNA Repair. 2009;8:6–18. doi: 10.1016/j.dnarep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 24.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita T, Ryser S, Piuz I, Schlegel W. Up-regulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Mol Cell Biol. 2008;28:1630–1643. doi: 10.1128/MCB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Sagliocco F, Laloo B, Cosson B, Laborde L, Castroviejo M, Rosenbaum J, Ripoche J, Grosset C. The ARE-associated factor AUF1 binds poly(A) in vitro in competition with PABP. Biochem J. 2006;400:337–347. doi: 10.1042/BJ20060328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westmark CJ, Bartleson VB, Malter JS. RhoB mRNA is stabilized by HuR after UV light. Oncogene. 2005;24:502–511. doi: 10.1038/sj.onc.1208224. [DOI] [PubMed] [Google Scholar]
- 30.Wu L, Good PJ, Richter JD. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammet A, Pike BL, Heierhorst J. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J Biol Chem. 2002;277:22469–22474. doi: 10.1074/jbc.M202473200. [DOI] [PubMed] [Google Scholar]
- 32.Traven A, Hammet A, Tenis N, Denis CL, Heierhorst J. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics. 2005;169:65–75. doi: 10.1534/genetics.104.030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westmoreland TJ, Marks JR, Olson JA, Jr, Thompson EM, Resnick MA, Bennett CB. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryotic Cell. 2004;3:430–446. doi: 10.1128/EC.3.2.430-446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaillard H, Wellinger RE, Aguilera A. Methods to study transcription-coupled repair in chromatin. Methods Mol Biol. 2009;523:141–159. doi: 10.1007/978-1-59745-190-1_10. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS One. 2008;3:e2410. doi: 10.1371/journal.pone.0002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]