Summary
Molecular analysis of a Drosophila minichromosome, Dp(1;f)1187, revealed a relationship between position-effect variegation and the copy number reductions of heterochromatic sequences that occur in polytene cells. Heterochromatin adjacent to a defined junction with euchromatin underpolytenized at least 60-fold. Lesser reductions were observed in euchromatic sequences up to 103 kb from the breakpoint. The copy number changes behaved in all respects like the expression of yellow, a gene located within the affected region. Both copy number and yellow expression displayed a cell-by-cell mosaic pattern of reduction, and adding a Y chromosome, a known suppressor of variegation, increased both substantially. We discuss the possibility that changes in replication alter copy number locally and also propose an alternative model of position-effect variegation based on the somatic elimination of heterochromatic sequences.
Introduction
Chromosome rearrangements that juxtapose euchromatic and heterochromatic regions in plants, mammals, and Dipterans frequently inhibit the function of nearby genes on a cell-by-cell basis (reviewed in Lewis, 1950; Spofford, 1976; Eissenberg, 1989). Genetic and cytological studies in Drosophila melanogaster demonstrated distinctive and perplexing general features of such “position-effect variegation.” Inhibition frequently involves a chromosome region containing many genes, polytene chromosome bands, and, by inference, hundreds of DNA kilobases. In any given cell, genes lying closer to the breakpoint are always affected if a more distal gene has been inhibited, suggesting that some influence “spreads” along the chromosome from heterochromatin into euchromatin. Indeed, variable effects on chromosome morphology near the breakpoint are readily visualized in polytene chromosomes. Variegated phenotypic effects are sensitive to large changes in the quantity of heterochromatin anywhere in the genome, as well as to the dosage of a small number of unlinked modifier genes. Finally, maternal or paternal effects are sometimes observed, where the parental source of the rearrangement influences the amount of variegation within progeny.
The highly condensed structure of heterochromatin is widely believed to underlie position-effect variegation (see Tartof et al., 1984). Compacted heterochromatin may cause abnormal folding of any adjacent euchromatin, reducing or eliminating the transcription of genes within the affected region. The length of the condensed region is proposed to vary in different cells because the formation and spreading of the inhibitory folding pattern require the local binding of proteins whose supply is limited relative to their multiple target sites throughout the heterochromatin (Locke et al., 1988). Enhancer and suppressor genes would encode the chromosomal proteins required for condensation, while suppressive heterochromatin would contain multiple sites that compete with the breakpoint region for the binding of the compaction-stimulating proteins.
Alternatively, changes at the level of the DNA itself could underlie position-effect variegation. Although the nature of the spreading effect rules out chromosome loss, a role for other changes in “chromosome metabolism” was proposed as early as 1936 (Schultz, 1936, 1956). Most Drosophila tissues are composed of cells with polytene nuclei that form as a result of altered cell cycles in which multiple rounds of replication occur without cytokinesis. Heterochromatin becomes underrepresented as much as 1000-fold in polytene nuclei (Rudkin, 1989; Gall et al., 1971); specific heterochromatic sequences achieve different copy numbers through poorly understood processes that appear to be developmentally regulated (reviewed in Spradling and Orr-Weaver, 1987). If changes in polytenization could spread into euchromatin across an abnormal junction with heterochromatin, reduced copy number might inhibit gene function without altering transcriptional regulation.
Many previous studies have failed to detect significant changes in polytenization near variegating rearrangements. Although local changes in the appearance of variegating polytene chromosomes are easily seen (Prokofyeva-Belgovskaya, 1939; Schultz and Caspersson, 1939) only small changes in DNA copy number, at most 2- to 3-fold, have been reported (Cowell and Hartmann-Goldstein, 1980; Kornher and Kaufman, 1986). After correcting for changes in chromatin structure, Henikoff (1981) detected no differences in the polytenization of a variegating heat shock puff. Rushlow, Bender, and Chovnick (1984) found no change in rosy (ry) locus copy number in polytene larval fat body resulting from a variegating inversion, ryps11136, broken 4 kb from the 5’ end of the rosy locus. They concluded that altered transcription rather than polytenization was most likely responsible for reducing rosy expression up to 7-fold.
We reexamined the question of altered polytenization in the course of molecular studies on the Drosophila minichromosome Dp(1;f)1187 (Krivshenko and Cooper, 1954 [cited in Lindsley and Grell, 1968]). Genes on this drastically shortened X chromosome such as yellow (y) reside adjacent to centromeric heterochromatin and display variegated expression. Our studies revealed a strong, long-range position effect on the polytenization of Dp(1;f)1187 euchromatin. We concluded that reduced copy number probably contributes to gene dysfunction in Dp(1;f)1187, and that alterations at the level of DNA metabolism cause some variegated position effects.
Results
Structural Analysis of Dp(1;f)1187 by Pulsed-Field Gel Electrophoresis
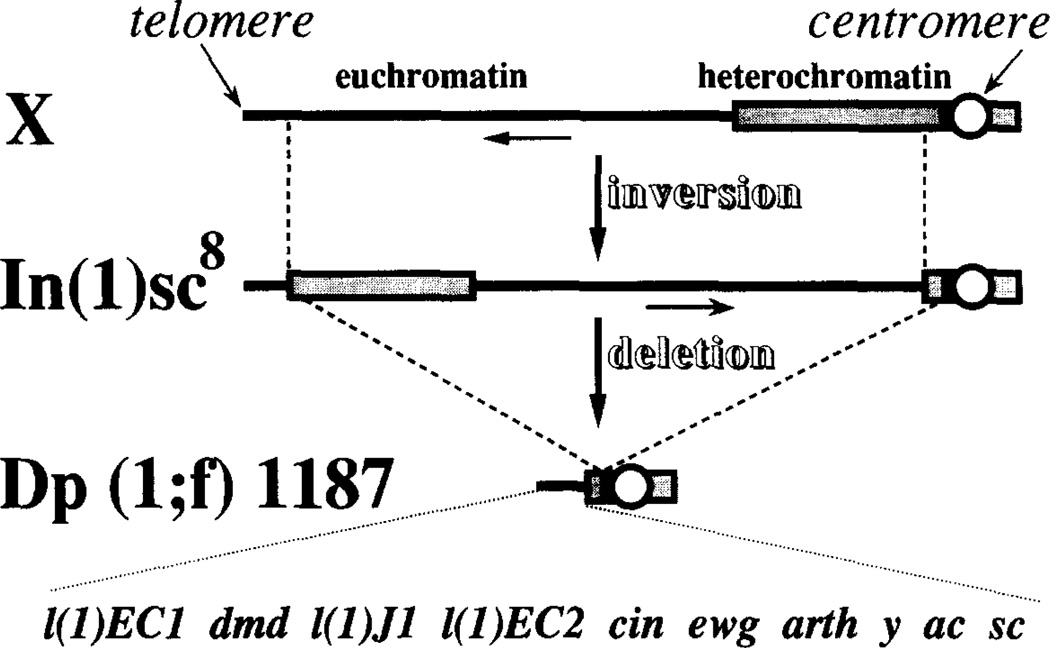
Dp(1;f)1187 (hereafter referred to as Dp1187) is the smallest of a series of free X chromosome duplications (minichromosomes) generated by deleting all but the proximal and distal sequences within ln(1)sc8, an X chromosome containing a large inversion (Krivshenko and Cooper, 1954 [cited in Lindsley and Grell, 1968]). The sc8 inversion in the progenitor chromosome joins a large block of basal heterochromatin with sequences from the distally located scute (sc) locus (Campuzano et al., 1985). Since the deletion producing Dp1187 resulted from breaks in both the proximal and distal heterochromatin, the minichromosome retains wild-type alleles for all nine of the known complementation groups distal to scute (Figure 1). In Dp1187 these genes are still juxtaposed with heterochromatin at the original distal sc8 junction; however, the exact location of the heterochromatic breakpoints is unknown.
Figure 1. Origin and Genetic Structure of Dp1187.
The origin of the ln(1)sc8 progenitor chromosome from a normal X chromosome and its subsequent derivitization to produce Dp1187 is diagrammed schematically (not drawn to scale). See text and Lindsley and Grell (1968) for further details. A genetic map showing the ten known complementation groups from the X tip that remain on Dp1187 is drawn below. Heterochromatin is shown as a stippled box and euchromatin as a solid line.
To initiate a molecular analysis of this minichromosome, we prepared DNA from brains and imaginal discs of larvae containing or lacking Dp1187 and analyzed it using pulse-oriented electrophoresis (Schwartz and Koval, 1989) a modification of pulsed-field gel electrophoresis. Bands corresponding to the minichromosome were identified by hybridizing Southern blots with a unique sequence probe immediately adjacent to the sc8 breakpoint, and sized by comparison to yeast chromosomes and multimerized λ DNA (Figure 2). Full-length Dp1187 DNA comigrated with molecules about 1400 kb in length; therefore, the minichromosome is ~1/25th the size of the normal X. A restriction map was constructed using enzymes that cut DNA infrequently. These studies revealed that the distal X euchromatin comprises about l/3 (320–400 kb) of the chromosome, and that the heterochromatin proximal to the sc8 breakpoint accounts for the remaining ~2/3 (1000 kb). Additional mapping confirmed that Dp1187 appears identical to ln(1)sc8 within the first 90 kb of distal heterochromatin (data not shown). More detailed structural studies of Dp1187 will be presented elsewhere (G. H. K. and A. C. S., unpublished data).
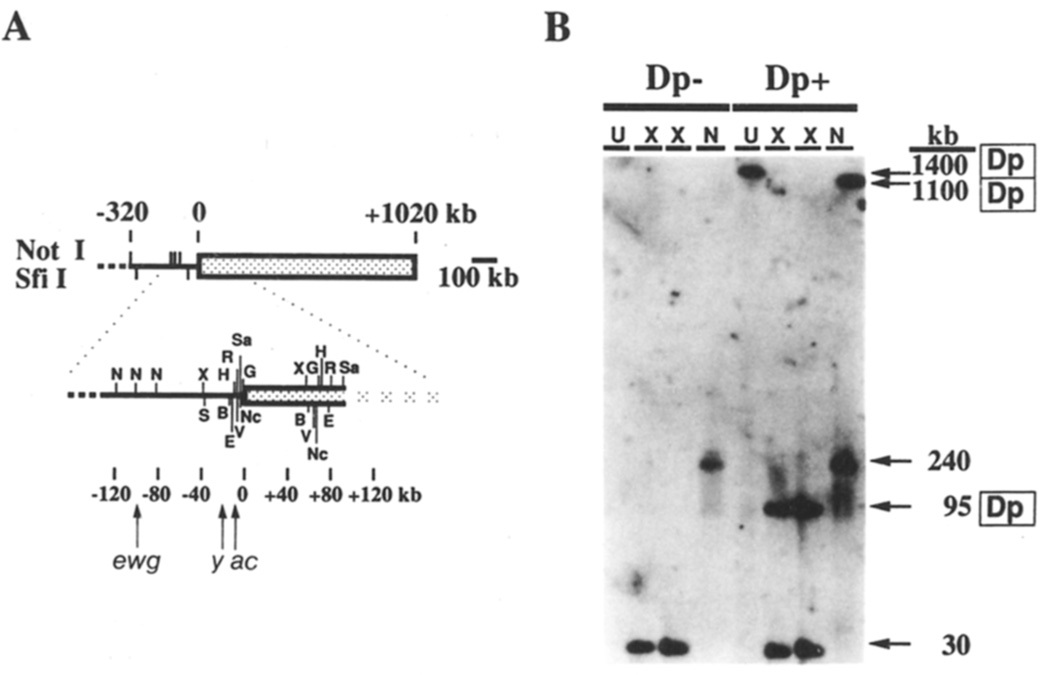
Figure 2. Molecular Structure of Dp1187.
(A) The restriction map of Dp1187 was compiled using maps and cloned DNAs from the first 130 kb of euchromatin and by Southern blot analysis similar to that shown in (B). The solid line is euchromatin and the stippled box is heterochromatin. Sites of the rare-cutting enzymes Notl (N) and Sfil (S) are indicated near the top. Additional sites may be present if the chromosome extends beyond position −320 (dashed line). Only the restriction sites flanking the sc8 breakpoint (coordinate 0) are shown for the following enzymes: X (XhoI), H (HindIII), E (EagI), B (BamHI), R (EcoRI), V (EcoRV), NC (NcoI), G (BgIII), and Sa (SaII). The locations of the erect wing (ewg, coordinates −92 to −100 kb), yellow (y, −20 to −25 kb), and achaete (ac, −10 to −13 kb) genes are indicated (Campuzano et al., 1985; Fleming et al., 1989).
(B) DNA inserts were prepared from imaginal discs and brains of sibling third instar larvae (X/O males) either lacking (Dp−) or containing (Dp+) one copy of Dp1187. Inserts were subjected to pulse-oriented gel electrophoresis either uncut (U) or following digestion with XhoI (X) or Notl (N), and the DNA was transferred and hybridized with a probe from the scute region (pBSscXR3.7). Electrophoresis conditions were 120 V, 104 s pulse at 8.5°C for 3.5 days.
Dp1187 Shows Position-Effect Variegation
On the original ln(1)sc8chromosome, at least two genes located distal to the SC8 breakpoint, achaete (ac) and yellow, display position-effect variegation (Noujdin, 1936). To learn whether these position effects had been altered by the rearrangements that produced Dp1187 we examined the ability of the duplication to complement X-linked mutant alleles. Because all of the genes on Dp1187 are duplicated on the X chromosome, their function can be analyzed without the viability effects that complicate studies of many variegating rearrangements, including ln(1)sc8. The yellow gene, which lies 20–25 kb distal to the sc8 breakpoint, provides a sensitive cell-autonomous assay for variegation. yellow expression is necessary for wild-type pigmentation of the larval and adult cuticle, bristles, and other ectodermal structures (Lindsley and Grell, 1968). The anterior edge of each adult wing contains ~80 triple-row bristles whose pigmentation is dependent on yellow (Figures 3a and 3b). Since each triple-row bristle develops from a single polytene epidermal cell, bristle pigmentation allows the expression of yellow to be monitored within individual cells.
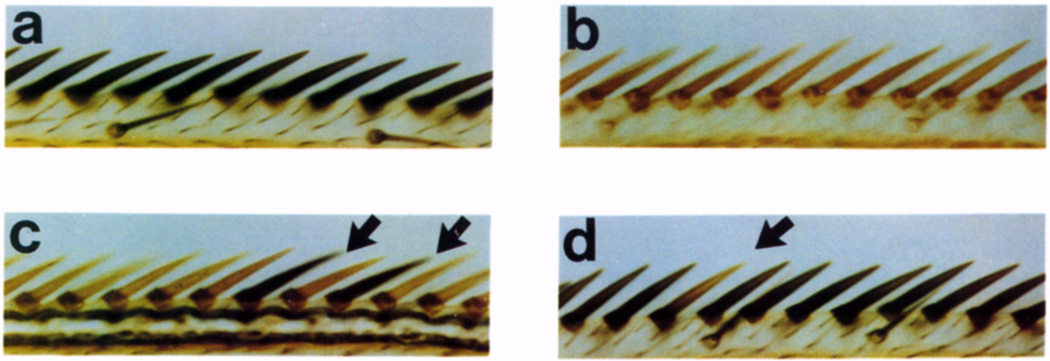
Figure 3. Position-Effect Variegation of yellow+ in Wing Triple-Row Bristles.
A segment of the triple row at the anterior margin of the wing is shown for the following males.
(a) Wild type.
(b) yellow mutant (y1).
(c) Dp1187 without Y chromosome (X,y/O; Dp1187 y+; ry506). Arrows indicate y+ bristles.
(d) Dp1187 with Y chromosome (X,y/Y; Dp1187 y+; ry506). Arrow points to a y bristle. Magnification, ×738.
These experiments revealed that the yellow gene on Dp1187 displays position-effect variegation that is similar to that reported for ln(1)sc8. We examined flies in the presence of various doses of the large, heterochromatic Y chromosome, since its presence strongly suppresses position-effect variegation. Duplication-bearing males that lacked a Y chromosome variegated extensively for yellow; 84% of the triple-row bristles lacked black pigmentation (Table 1; Figure 3c). Bristles and cuticle present in other regions also variegated for yellow (data not shown). In contrast, only about 5% of the triple-row bristles were unpigmented in adult males containing a Y chromosome (Figure 3d). Addition of a second Y chromosome suppressed the inactivation of yellow even further; now nearly 100% of the triple-row bristles were normally pigmented. When Dp1187 was inherited maternally, males displayed less variegation than when the duplication was received from the father (42% versus 84% unpigmented, respectively).
Table 1.
yellow Variegation in Dp1187
| Percent of Triple-Row Bristles | ||||
|---|---|---|---|---|
| Genotype |
yellow+ (wild-type) |
yellow+/− (intermediate) |
yellow− (mutant) |
N |
| X,y/O;Dp,y+ | 15 | 1.2 | 84 | 1466 |
| X,y/O;Dp,y+(maternal Dp) | 55 | 3.1 | 42 | 1394 |
| X,y/Y;Dp,y+ | 94 | 0.9 | 5 | 1308 |
| XY,y/Y;Dp,y+ | 99.6 | 0.4 | 0 | 1376 |
The source of the duplication was paternal in all cases except where indicated. N, the total number of bristles scored.
Dp1187 Heterochromatin Adjacent to the sc8 Breakpoint Is Underrepresented in Salivary-Gland Polytene Nuclei
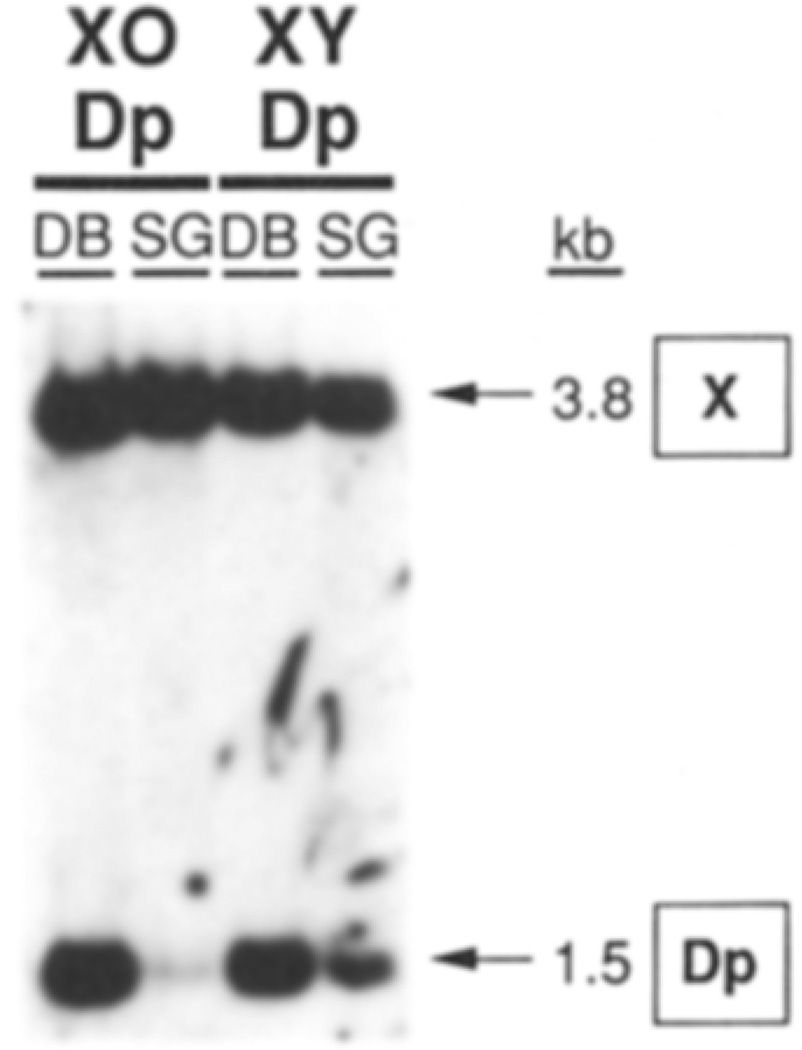
The same methods used to construct the molecular map of Dp1187 allowed us to investigate the polytenization of the specific heterochromatic region adjacent to the variegating genes. High molecular weight DNA was isolated from the polytene larval salivary gland nuclei and the predominantly diploid imaginal discs and brains (see Experimental Procedures). Pulsed-field Southern hybridization analysis, using a single-copy probe adjacent to the sc8 junction, allowed the copy number of sequences extending 60 kb into Dp1187 heterochromatin to be monitored in these cell types. As shown in Figure 4, the 85 kb Dp1187 EcoRI fragment that spans the sc8 breakpoint is severely (>60-fold) underrepresented in the polytene DNA in comparison to diploid DNA. The probe sequences present on the unrearranged X chromosome polytenize normally (represented by the 7.5 kb EcoRI fragment). We concluded that Dp1187 heterochromatin adjacent to the breakpoint is underrepresented in salivary gland cells.
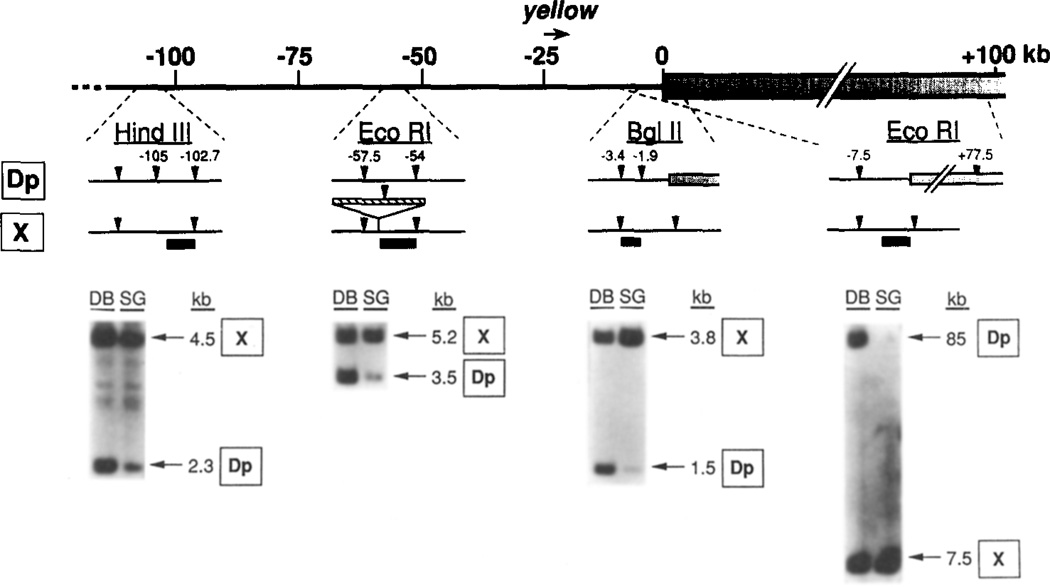
Figure 4. Underpolytenization of Dp1187 at Sites Near the sc8 Breakpoint.
A map of the region of Dp1187 flanking the sc8 breakpoint is shown at the top using the kb coordinates of Figure 2. Four regions where probes were used to determine DNA copy numbers are shown, along with local restriction site polymorphisms that were used to distinguish bands derived from the normal X(X) and Dp1187 (Dp). Below each map are corresponding lanes from Southern blots containing DNA from the predominantly diploid imaginal discs and brain cells (DB) or from highly polytene salivary gland cells (SG) isolated from the same larvae. The probe used for each blot is indicated by a heavy line on the map above along with the enzyme used; the origin and size in kb of the labeled fragments are also given (arrows at the right). The rightmost blot was from a pulse-oriented electrophoresis experiment; electrophoresis conditions were 120 V, SO s pulse at 8.5°C for 2 days. The other blots utilized standard electrophoresis. All DNAs were isolated from X/O;Dp1187 male larvae.
Underpolytenization Spreads Far into the Euchromatin of Dp1187
We studied the polytenization of euchromatic sequences within Dp1187 using similar methods, except that pulsed-field gels were not required. The representation of single-copy, euchromatic sequences spanning 105 kb from the sc8 breakpoint was studied by quantitative Southern hybridization analysis. Restriction enzyme polymorphisms were identified at three sites, 1.9 kb, 54 kb, and 103 kb from the sc8 breakpoint (Figure 4) and used to distinguish the polytenization of Dp1187 sequences from homologous sequences located on the tip of the X chromosome. The yellow locus lies within this region, 20–25 kb from the breakpoint.
Underpolytenization of Dp1187 heterochromatin extended across the rearrangement breakpoint and affected the juxtaposed euchromatin for at least 105 kb. Euchromatic sequences located 1.9 kb from the heterochromatin-euchromatin junction were severely underrepresented in X/O;Dp1187 males, the genotype that displayed high levels of position-effect variegation for the y+ gene. Densitometric measurements indicated that the 1.5 kb Dp1187-specific BgIII fragment was 39-fold underrepresented in polytene DNA relative to diploid DNA (Table 2). The copy number of these same sequences did not change in polytene DNA when present on the unrearranged X chromosome (3.8 kb BgIII fragment). A similar analysis demonstrated that sequences located 54 and 103 kb from the heterochromatin-euchromatin junction in Dp1187 were 8- and 2.4-fold underrepresented, respectively. Thus, the amount of underpolytenization decreased monotonically with increasing distance from the heterochromatin. Since duplication DNA was still 2.4-fold underrepresented 103 kb from the breakpoint (the distalmost region for which polymorphisms were available), altered copy number may extend even further than was demonstrated in these experiments.
Table 2.
Underpolytenization of Dp1187
| Amount of Underpolytenization ata | |||
|---|---|---|---|
| Genotype | −1.9b | −54b | −103b |
| X/O;Dp1187 | 39 | 8.0 | 2.4 |
| X/Y;Dp1187 | 4.7 | 2.0 | 1.0 |
Values indicate the amount of underpolytenization in salivary gland nuclei (fold difference in amount of DNA) when the sequences are present on Dp1187, in comparison with their polytenization on the un-rearranged X chromosome. These values were derived by densitometry of the quantitative Sourthern hybridization analyses reported in Figures 4 and 5. The raw differences in DNA content in the polytene nuclei (SG lanes, Figures 4 and 5) were normalized to the representation of each sequence in the predominantly diploid imaginal disc and brain cells (DB lanes, Figures 4 and 5).
Distance in kb from the heterochromatic–euchromatic junction (coordinate 0) to the first restriction site in the euchromatic polymorphisms used to distinguish Dp1187 sequences from those on the X chromosome (see Figures 4).
The Inhibition of Dp1187 Euchromatic Polytenization Responds to a Known Modifier of Position-Effect Variegation
If the underrepresentation of euchromatic sequences was due to a variegated position effect, it should respond to modifiers such as the Y chromosome. In X/O;Dp1187 male salivary gland DNA, sequences located 1.9 kb from the breakpoint were 39-fold underrepresented but were only 4.7-fold reduced in X/Y;Dp1187 males (Figure 5 and Table 2). A Y chromosome also reduced the underpolytenization observed in X/O males 54 kb and 103 kb from the breakpoint (Table 2). Thus, in the presence of a Y chromosome, the underpolytenization of Dp1187 euchromatin extended for a shorter distance from the sc8 breakpoint and was less severe at any given position.
Figure 5. Underpolytenization Responds to a Modifier of Position-Effect Variegation.
A Southern blot similar to those in Figure 4 is shown. DNA was prepared from disc and brains (DB) or salivary glands (SG) of male larvae containing Dp1187 and either a normal Y chromosome (XY) or lacking a Y chromosome (XO). DNAs were digested with BgIII and probed with the −1.9 kb region probe shown in Figure 4. The location and size of fragments deriving from the normal X and from Dp1187 are indicated.
The Copy Number of the Variegating Dp1187 Region Varies between Individual Salivary Gland Cells
The underpolytenization of Dp1187 euchromatin observed in the quantitative Southern analysis could occur to a uniform extent in all salivary gland nuclei, or it could differ from cell to cell. To determine whether the inhibition of euchromatic polytenization in Dp1187 is a variegating position effect, we performed in situ hybridization to squashes of salivary gland polytene chromosomes. The amount of polytenization at the same three positions on Dp1187 (1.9 kb, 54 kb, and 103 kb from the breakpoint) was compared with the polytenization of homologous X chromosome sequences by determining the ratio of grains in the chromocenter, where Dp1187 resides, and at the tip of the X. 35S-labeled RNA probes and appropriate exposure times were used so that sequences on the normal X showed an average of 25–40 grains per site. This experimental procedure allowed up to about a 10-fold sequence under-representation to be detected (40 grains versus 4 grains). However, copy numbers more than 10-fold below the normal X could not be reliably distinguished due to background labeling.
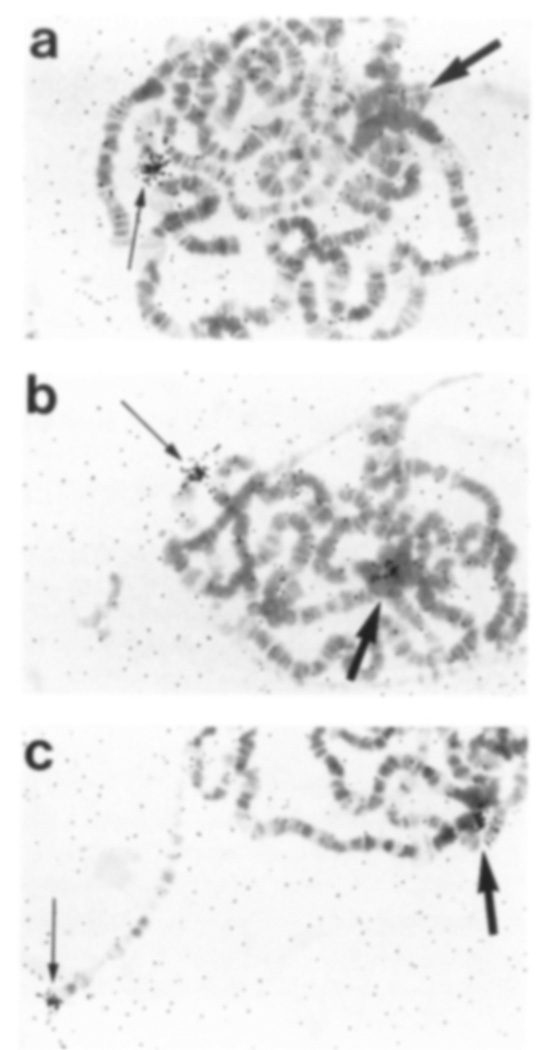
These experiments revealed that Dp1187, but not the normal X, was labeled in an unusually variable manner between cells, and at levels that depended on the location of the probe. Cell-to-cell variation in labeling over Dp1187 was particularly evident with the −54 kb probe. In the absence of a Y chromosome, no specific labeling over Dp1187 was present (Figure 6a) in 60% of cells (Figure 7), whereas at least 26 grains (average = 37 grains) were always present at the distal X site. In contrast, many other cells within the same gland distinctly labeled Dp1187, but at a lower level than at the complementary site on the normal chromosome (Figure 6b). About 10% of the cells were labeled equally at the distal end of the normal X and on Dp1187 (Figure 6c). These results suggested that the copy number of the Dp1187 regions studied varied extensively from cell to cell in the salivary gland.
Figure 6. Cell-to-Cell Variation of Dp1187 Labeling In Situ.
In situ hybridization was carried out to salivary gland polytene chromosomes from Dp1187-bearing male larvae containing or lacking a Y chromosome. A sample of the results is shown from a preparation hybridized with the −54 kb probe shown in Figure 4. Labeling of Dp1187 (large arrow) was not detected in (a), was intermediate in (b), and was approximately equal in (c) to labeling of the X tip (small arrow). All three nuclei were from a single individual of the genotype X/O;Dp1187 Magnification, ×467.
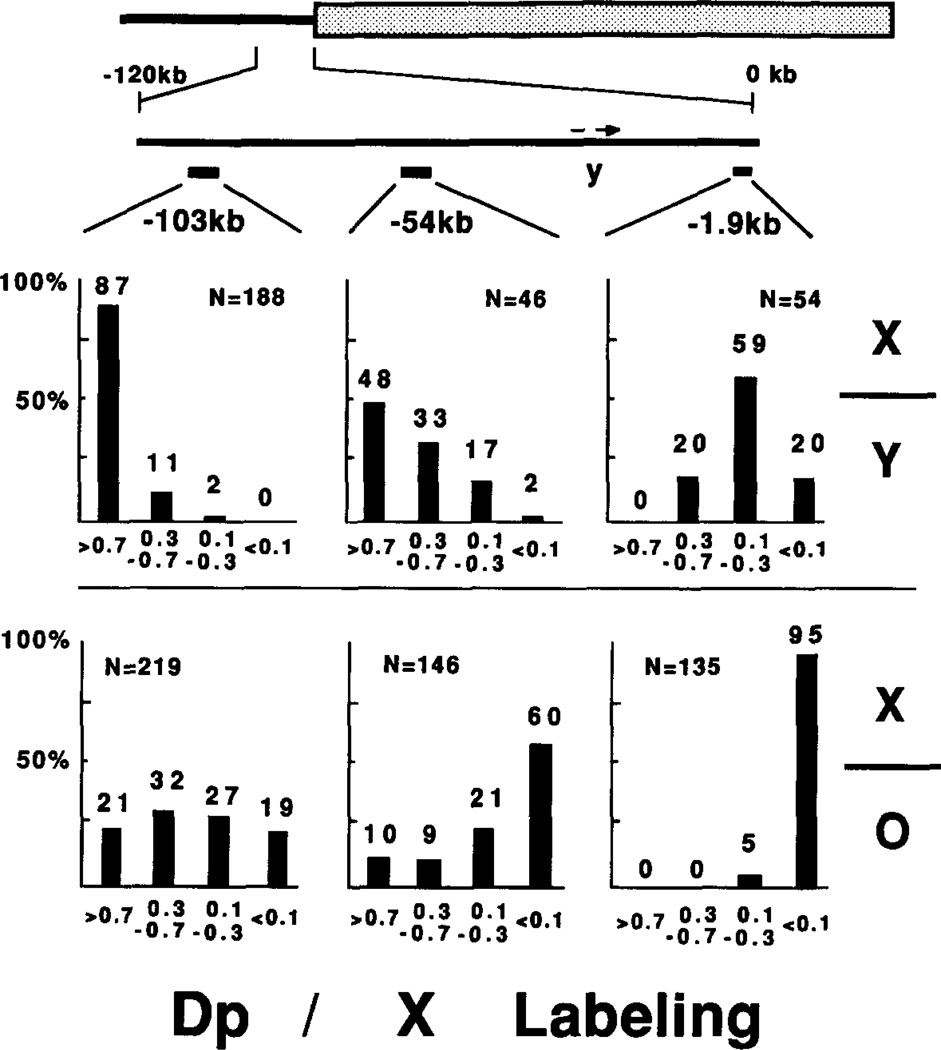
Figure 7. Dp1187 Labeling In Situ Depends on Probe Location and the Presence of a Y Chromosome.
Data from grain counts of polytene chromosomes labeled by one of the three euchromatic probes was tabulated. Each of the three probes was hybridized to chromosomes from Dp1187-bearing male larvae containing (X/Y) or lacking (X/O) a Y chromosome. In each case the ratio of grains associated with Dp1187 to those associated with the normal X was calculated after subtracting for background. The ratios (r) were grouped into four classes: 1.0>r>0.7, 0.7>r>0.3, 0.3>r>0.1, and 0.1>r. The percent of nuclei falling in each class is plotted for the six combinations and displayed at the top of the bars. N, the total number of nuclei.
The degree of underlabeling on Dp1187 depended on both the distance of the probe from the sc8 breakpoint and the presence of a Y chromosome (Figure 7). For each probe we calculated the ratio of grains on Dp1187 to those on the normal X, after subtracting background, and plotted the results in four classes. In the absence of a Y chromosome, virtually all nuclei (95%) severely underpolytenized Dp1187 sequences 1.9 kb from the breakpoint, but with increasing distance from the heterochromatin junction, only 60% (54 kb) or 19% (103 kb) severely underrepresented the duplication. Addition of a Y chromosome to the genotype shifted the distribution of nuclei toward complete polytenization in all three regions. Dp1187 polytenization therefore varied from cell to cell and responded to a modifier as expected for a variegating position effect.
If condensed chromatin was less accessible for in situ hybridization, reduced grain counts over Dp1187 could reflect increased compaction rather than lowered copy number. Henikoff (1981) detected 2-to 3-fold differences in the efficiency of in situ hybridization between puffed and unpuffed heat shock sequences. However, efficiency differences between transcriptionally inactive chromosome regions (such as the regions studied here) have not been reported. Moreover, the correspondence between the reduced hybridization in situ and the decreased copy number within purified salivary gland DNA eliminated the possibility that differences in hybridization efficiency were primarily responsible for the decreased labeling of Dp1187 in situ. The distribution of nuclei in Figure 7 predicted average copy numbers quite close to those observed by Southern blotting (Table 2). For example, in the absence of a Y the fraction of nuclei observed in each class predicted that Dp1187 was underrepresented 100-fold, 5.9-fold, and 2.6-fold (at −1.9 kb, −54 kb, and −103 kb, respectively) compared with observed values of 39-fold, 8-fold, and 2.4-fold. In X/Y males the predicted values were 4.5-fold, 1.6-fold, and 1.2-fold, while observed were 4.7-fold, 2-fold, and equal representation. As expected due to the insensitivity to low-copy number values, the in situ hybridization data predict slightly higher average copy numbers than observed in the quantitative Southern analysis. If compaction rather than underpolytenization had contributed significantly to the reduced signals on Dp1187, lower rather than higher values should have been predicted.
Discussion
A Variegated Position Effect Reduces the Copy Number of Some Euchromatic Dp1187 Sequences
The possibility that position-effect variegation locally alters the DNA content of affected polytene chromosome bands has been a source of continuing controversy. Although small reductions in DNA content were reported previously in some cases (Schultz, 1941; Ananiev and Gvozdev, 1974; Cowell and Hartmann-Goldstein, 1980; Kornher and Kaufman, 1986) other workers detected no evidence for changes in copy number (Cole and Sutton, 1941; Henikoff, 1961; Rushlow et al., 1984). Most measurements were made at an unknown distance from the inducing breakpoint. The magnitude of the expected effect was usually uncertain, since changes in the expression of genes in the affected region were unquantitated. Furthermore, the polytenization level of the heterochromatin adjacent to the breakpoint was never monitored. Although the apparent discrepancies may have reflected the fact that different rearrangements and genetic backgrounds were studied, many workers have concluded that differential polytenization does not contribute significantly to the genetic defects in variegating rearrangements, and that changes in chromosome morphology result primarily from altered condensation (reviewed by Eissenberg, 1989).
Our experiments revealed that in at least one case, the sc8 rearrangement retained on Dp1187, extensive underpolytenization does occur within the chromosome region containing variegating genes such as yellow. Because sequences in the salivary gland were underrepresented by as much as 39-fold, these observations cannot be ascribed to the difficulties of quantitating small copy number differences on Southern blots or to systematic changes in hybridization efficiency in situ. Dp1187 underpolytenization exhibited all the properties classically associated with position-effect variegation. The effect on copy number spread from the sc8 breakpoint more than 100 kb into adjacent euchromatin. More significantly, the observed copy number alterations in Dp1187 closely paralleled the pattern of yellow variegation. Reduced polytenization and yellow inhibition occurred in a cell-by-cell pattern. Addition of a Y chromosome dramatically suppressed underpolytenization and yellow variegation. Thus, there may be two classes of variegating rearrangements: those like Dp1187 that reduce DNA copy number, and those like the ryps11136 rearrangement studied by Rushlow et al. (1984) that leave average copy number unchanged. Additional studies will be required to determine the relative frequency of these types of variegation.
Does Reduced Copy Number Cause Variegated yellow Expression in Dp1187?
Because we measured copy number changes in the salivary gland but scored the expression of yellow in the triple-row bristles (and other cuticular structures), our experiments do not prove that yellow variegation was caused by changes in gene copy number. However, we strongly suspect that this was the case in at least some of the affected tissues. Bristles are produced by large polytene pupal trichogen cells that contain banded polytene chromosomes with reduced amounts of centromeric heterochromatin (Ribbert, 1979; Beckingham and Rubacha, 1984). It was not possible to correlate directly the copy number of functional yellow genes with the level of yellow expression in individual trichogen cells, because these cells constitute a small fraction of the developing epidermis and degenerate during late pupation prior to the completion of pigmentation. However, black pigmentation is reduced or abolished when yellow mRNA levels are reduced more than 10-fold (Geyer et al., 1990), the approximate magnitude of the copy number decrease that we observed in salivary glands.
The ploidy of other cells showing variegated yellow expression in flies bearing Dp1187 is unknown, such as the pupal epidermal cells that secrete the adult cuticle. If the expression of yellow is affected in diploid cells, then such effects could not be explained by alterations specific to the polytene cell cycle. Similarly, there is uncertainty as to the ploidy of cells affected by many other variegating rearrangements, such as the adult eye pigment cells.
Copy Number Variegation and the Compaction Model
Our observation that polytenization is strongly affected within a large chromosome region undergoing position-effect variegation does not necessarily contradict the major conclusions of the compaction model. Considerable evidence supports the idea that the chromatin structure of regions brought adjacent to heterochromatin can be dramatically changed, not the least of which is direct observation in the polytene chromosomes. Reduced copy number alone without any change in chromatin structure would be expected to produce a banded constriction but not to disorganize banding as observed near heterochromatic-euchromatic breakpoints. However, there is presently no way to distinguish whether the change in chromatin structure causes copy number reduction, or whether an independent process that produces sequence underrepresentation also distorts chromatin structure.
Position-effect variegation is sometimes assumed to be an all or nothing process at the level of single cells. However, cells expressing phenotypes equivalent to hypomorphic alleles of variegating loci have sometimes been observed. Dp1187 males lacking a Y chromosome contain triple-row bristles, structures produced by single cells, with intermediate levels of pigmentation (Table 1). The observation that gene copy number varies widely among individual polytene cells provides a simple and direct explanation for the quantitative variation caused by some variegating rearrangements. Alternatively, sometimes compaction would have to only reduce rather than eliminate gene expression, or fail to involve all DNA strands within a polytene chromosome.
Copy Number Variegation and the Control of Replication
We considered two basic models to account for our observations: altered replication and somatic elimination. Satellite DNA, ribosomal DNA, and other heterochromatic sequences all decrease in copy number relative to euchromatic sequences during polytenization. These changes have generally been thought to result from decreased replication of heterochromatin relative to euchromatin during polytene cell growth, although direct proof is lacking (reviewed in Spradling and Orr-Weaver, 1987). According to the underreplication model, our results suggest that mechanisms controlling chromosome replication are subject to position effect. This idea has long been considered and several previous studies detected changes in the [3H]thymidine incorporation of polytene bands near variegating rearrangements (Ananiev and Gvozdev, 1974; Wargent et al., 1974). The time of replication appeared to be prolonged in such bands. Perhaps significantly, increased incorporation preceded detectable alterations in DNA content and chromatin morphology.
Limited information on how replication is controlled at the molecular level permits only speculation about how regulatory mechanisms could be modified to produce such changes. Replicons in late third instar salivary glands average 60–192 kb in size and forks elongate about 0.21–2.7 kb/min (Steinemann, 1981; Lakhotia and Sinha, 1983). Even if active origins are rare in heterochromatin, replication of sequences distal to the breakpoint in Dp1187 should still be able to proceed from euchromatic control elements. Given the long duration of S phase, it is unclear why forks do not finish replicating the euchromatin, and possibly even proceed into the adjacent heterochromatin. Because a gradual change in average copy number was observed, forks must terminate or arrest at multiple sites along the affected euchromatin. Perhaps initiation is inhibited at nearby origins, forcing replication to proceed long distances. Alternatively, initiation at euchromatic origins that have been juxtaposed near heterochromatic control sequences may be delayed until late in S phase. Perhaps fork movement is retarded by abnormally condensed euchromatin. Whatever the cause, the underreplication model predicts that forks start too late or move too slowly to complete euchromatin replication before S phase ends.
It is difficult to explain all aspects of position-effect variegation through underreplication in polytene cells. Some variegated phenotypic effects probably occur in diploid cells. If rearrangement near heterochromatin forced genes to replicate late in S, then it is possible this could lead to reduced expression in diploid cells as well as cause underreplication in polytene cells. Susceptibility to variegation is certainly heritable in diploid ceils. For example, the yellow mosaicism in the cuticle associated with ln(1)sc8 is not random but has been reported to exhibit local correlations suggestive of a clonal origin at about the time of hatching (Noujdin, 1936) or during the late second instar (Gsell, 1971). Both times are prior to the cessation of histoblast and imaginal disc cell division and the onset of differentiation. Furthermore, yellow variegation shows a parental source effect on pigmentation (Noujdin, 1944) and on cytological heterochromatization of the X tip (Prokofyeva-Belgovskaya, 1947). Like these authors, we observed that the variegation of the yellow gene in Dp1187 was more extreme when the duplication was inherited paternally rather than maternally (Table 1). If sequence underrepresentation is related to these changes, it becomes necessary to argue that signals controlling underreplication can be set and inherited in dividing diploid ceils many generations prior to their actual execution.
Copy Number Variegation and Somatic Elimination
An alternative model for position-effect variegation, the somatic elimination model, postulates that a process operates during development to degrade some heterochromatic sequences, rather than simply failing to replicate them. A wide variety of organisms, including many lower Dipterans, eliminate certain DNA sequences from somatic cells during development (reviewed in Pimpinelli and Goday, 1989). Heterochromatic regions contain many families of transposable elements not located in euchromatin that provide one mechanism for such a process (Young et al., 1983; Traverse and Pardue, 1989). Some transposons are known to excise somatically (McClintock, 1951; Emmons and Yesner, 1984; Bryan et al., 1987). High levels of experimentally induced somatic P element excision are not necessarily lethal even when occurring in euchromatin (Laski et al., 1986; Engels et al., 1990), so excisions within heterochromatin might normally have no phenotypic effect. Indeed, McClintock (1950, 1951) postulated a common basis for position-effect variegation and unstable mutations in maize.
According to the somatic elimination model, position-effect variegation would result when juxtaposed euchromatic sequences lying near excising transposons are eliminated by imperfect repair of breaks or by transposon-associated deletions. The spreading effect would result from exonucleases moving over long distances from broken ends, or from multiple cycles of local transposition and imprecise excision within a cell lineage. The developmental timing of variegation would correspond to the time of transposon activation within different tissues. Repressor proteins produced by transposons spread throughout the Y chromosome and other heterochromatic regions would account for the suppression associated with heterochromatic DNA itself. As in the case of su(Hw) (Spana et al., 1988) modifiers of variegation would encode proteins that bind to and modulate the activity of the relevant transposons. The recent demonstration that a suppressor of variegation, Suvar(3)7, encodes a zinc finger protein is consistent with this role (Reuter et al., 1990).
The somatic elimination model provides a quite different rationale than underreplication for many perplexing aspects of position-effect variegation. Gene loss could occur in diploid as well as polytene cells, giving rise to the observed clonal patches of inactivation. Parental source effects would result from inheriting different amounts of transposon repressor proteins in the sperm and egg. If somatic elimination is a common property of hetetochromatin, it might provide insight into a wide variety of other biological phenomena. For example, excisions would produce the observed reduction in heterochromatic DNAs during polytenization. Random ligation of free ends present on different DNA strands would tangle together the heterochromatic regions of all the chromosomes, producing the disorganized banding and physical interconnections observed in the polytene chromocenter.
The changes we observed in the copy number of sequences within the 100 kb region upstream of the breakpoint in Dp1187 will allow a clear test of these differing models. Two-dimensional gel electrophoresis (Brewer and Fangman, 1987) can be used to visualize amplifying chorion genes containing forks approximately every 15 kb (Delidakis and Kafatos, 1989; Heck and Spradling, 1990). The underreplication model predicts that replication forks should densely populate the underrepresented region of Dp1187 Since the copy number within a 103 kb region changed about 20-fold (corresponding to log220 = 4.3 forks), a fork must occur on average every 103/4.3 = 24 kb, a frequency that should be detectable in two-dimensional gel analyses of salivary gland DNA. In contrast to underreplication, somatic elimination predicts that novel sequence joints rather than replication forks will be present at high levels in the underrepresented region of Dp1187. We are currently analyzing the structure and replication of Dp1187 in salivary gland cells and testing several other predictions of the models in order to distinguish them.
Experimental Procedures
Drosophila Strains and Culture
Flies were grown on standard corn meal/agar media (see Ashburner, 1990) at 22°C. All strains and mutations are as described in Lindsley and Grell (1968).
yellow Variegation
To quantitate yellow variegation, wings were removed from adults, mounted in water or permount, and examined in bright field using a Zeiss 63× Planapochromat lens. Males with different sex chromosome constitutions were obtained from the following crosses:
X/O;Dp1187:
X,y/X,y × X̅Y̅ Df(1)259, I(1)J1 y w/O;Dp1187, y+
X/Y;Dp1187:
X,y/X,y × X,y/Y;Dp1187, y+
XY/Y;Dp1187:
X̅X̅, yv/Y × X̅Y̅ Df(1)259, I(1)J1 y w/O;Dp1187; y+
X/O;Dp1187 (Dp1187 inherited maternally):
X̅X̅, yv/O; Dp1187, y+ × X,y/Y
Probes
Restriction fragments were subcloned from λ or cosmid clones into the appropriate sites of the Bluescript KS+ polylinker (Stratagene). pBSscXR3.7 contains a 3.7 kb XbaI-EcoRI fragment that adjoins the sc8 breakpoint (subcloned from λsc101, Campuzano et al., 1985). The −1.9 kb probe was from pBSscBP0.8, a 0.8 kb BgIII-PstI fragment from λsc101; the −54 kb probe corresponds to pBSTG3R3.5, which contains a 3.5 kb EcoRI fragment subcloned from λTG-3 (Fleming et al., 1989); and the −103 kb probe was derived from pBS6.1HR1.75, whose 1.75 kb HindIII-EcoRI insert was obtained from a cosmid walk generated in our laboratory (cos 6.1).
Preparation of DNA in Agarose Inserts
Unless stated otherwise, standard procedures and buffers were those described in Sambrook et al. (1989). High molecular weight DNA was prepared from male late third instar larval tissues according to the following methods, modified from the agarose insert protocols of Schwartz and Cantor (1984). Mouthhooks and ventral denticle belts were scored for yellow or yellow+ phenotype on a white background to select animals that lacked or contained Dp1187, respectively. X/O and X/Y males were produced by the same crosses outlined for scoring yellow variegation (see above).
Imaginal Discs and Brains
Twenty sets of discs and brains were collected and stored on ice in a small ground glass homogenizer (Kimble # 716-275-9097) filled with Ringer solution (see Ashburner, 1990). Following a 5 s microfuge spin to collect the tissue, the Ringer solution was removed, 40 µl of insert buffer A + Triton (0.1 M NaCl, 0.03 M Tris [pH, 8.0], 0.05 M EDTA, 7.7 mM β-mercaptoethanol, 0.5% Triton X-100) was added, and the tissue was homogenized. The mixture was heated to 37°C–42°C for 5 min, and 120 µl of 1.33% InCert agarose (FMC Corp.) in 0.125 M EDTA [pH, 7.5] was added, gently mixed, and then carefully pipetted into insert molds (~80 µl per insert). The agarose was melted and equilibrated at 50°C for 10 min or more prior to mixing with the homogenate.
Salivary Glands
Our method is modified from the salivary gland nuclear isolation procedure of Endow and Glover (1979). Salivary glands were dissected in Ringer solution from the same larvae used to prepare discs and brains, taking care to remove the fat body. Glands were periodically transferred to an Eppendorf tube containing Ringer solution and stored on ice. When ~40 gland pairs had accumulated, they were collected in the bottom of the tube by centrifuging briefly in a microfuge. Ringer solution was removed and 400 µl of Endow buffer A (60 mM KCI, 15 mM NaCl. 15 mM Tris, 0.15 mM spermine, 0.5 mM spermidine [pH, 7.4]) + 2% Triton + 0.25 M sucrose was added. Glands were drawn through a 200 µl pipetteman tip ~15 times until the tissue dispersed. Nuclei were spun out at 8000 × g for 5 min in a microfuge spin using a horizontal rotor. Following removal of the supernatant, nuclei were gently resuspended in 400 µl of Endow buffer A + 1% Triton + 0.25 M sucrose and spun for 5 min at 8008 × g, the supernatant was removed, and the pellet was resuspended in 40 µI of insert buffer A + 0.5% Triton (see above). Agarose was added and the mixture was pipetted into molds as described for discs and brains.
Lysis and Restriction Digestion
Immediately after hardening, inserts were pushed out of the mold (using a bent glass Pasteur pipette) into a 50 ml plastic screw cap tube containing at least 10 ml of NDS (0.5 M EDTA, 0.01 M Tris [pH, 9.5], 1% sarkosyl) to which proteinase K was added at 1–2 mg/ml. The tube was then incubated at 50°C overnight, with occasional mixing by inversion. Inserts could be stored in this solution at 4°C indefinitely. Prior to restriction, proteinase K was removed with at least two 4 hr washes of TE containing PMSF (1 µl of 0.1 M PMSF in isopropanol added per ml of TE), at least 3 ml per insert. Inserts were then gently shaken three to four times in TE alone at room temperature for at least 2 hr per wash. Inserts were cut in half with a glass cover slip, placed in screw cap microfuge tubes, and digested overnight with restriction enzyme at the appropriate temperature with periodic mixing by inversion. Approximately 350 µl of the recommended buffer was supplemented with acetylated BSA (100 µg/ml), 0.2% Triton X-100 (Boehringer-Mannheim), 50 U of enzyme, and 10 µg/ml DNAase-free (boiled) RNAase A. After digestion, buffer was replaced twice with 500 µl of 0.025 M NaCl, 0.010 M Tris–HCI [pH 7.5] and equilibrated for 30 min each.
Electrophoresis
Inserts containing restricted DNA were electrophoresed on conventional 0.7% agarose gels or by pulse-oriented electrophoresis (Schwartz and Koval, 1989). For pulse gels, agarose inserts were loaded directly into the wells of a 1% HGT agarose (FMC Corp.) gel filled with Tris-borate-EDTA buffer (TBE) and electrophoresed in 0.5× TBE. For conventional electrophoresis, buffer was removed, and inserts were melted for 2–5 min at 65°C, loaded immediately into dry wells, and electrophoresed in 1× TBE. The molecular weights for fragments identified in the pulse-oriented electrophoresis analyses were determined by comparison to λ multimers (Schwartz and Cantor, 1984) and to yeast chromosomal markers from strain YPH149 (90,220,280,360,445,555,610,690,760, 800,830,920,960, 1010, 1100, rDNA+1100, and 1600 kb; Heiter et al., unpublished data).
Southern Hybridization Analysis
Electrophoresed DNA was transferred to GeneScreen Plus (New England Nuclear) according to the manufacturer’s directions. After pre-hybridization for 2–4 hr at 65°C in 1 M NaCl, 1% SDS, 100 mg/ml denatured salmon sperm DNA, and 10% dextran sulfate, blots were hybridized at 65°C overnight in the same buffer plus 1–5 × 106 cpm/ml [32P]CTP labeled probes. A total of 50 µl of buffer was used per square cm of membrane. All probes were generated by random priming of gel-purified restriction fragments. Blots were washed with shaking three times for 5 min in 2× SSPE at room temperature, twice for 30 min in 2× SSPE, 1% SDS at 65°C, then twice for 30 min in 0.1 × SSPE at room temperature, wrapped in Saran wrap (Dow), and exposed for various times at −80°C using Kodak XAR-5 film and Cronex screens (Dupont). Densitometric measurements were performed using a Molecular Dynamics densitometer. Representation of the Dp1187-specific bands was quantitated relative to the X-specific band in polytene salivary gland DNA and normalized to their relative representation in diploid imaginal disc and brain DNA. For all blots, the polytenization of the unrearranged X-linked probe sequences was shown to be the same as the third chromosome rosy+ gene by stripping the blots and reprobing with rosy sequences (data not shown).
In Situ Hybridization to Sallvary Gland Polytene Chromosomes
In situ hybridization to salivary gland polytene chromosomes was carried out as described in Karpen et al. (1988), with the following modifications. RNA probes were prepared by transcription with T7 RNA poly-merase using undiluted [35S]UTP and [35S]CTP. Prior to hybridization overnight at 42°C, slides were pretreated with acetic anhydride, treated with RNAase, and denatured (Pardue, 1986). Slides were washed three to four times for 15 min in 2× SSC at 52°c and three times for 15 min in 0.1× SSC at 52°C. Grains were counted over all nuclei of the major ploidy class (determined by chromosome width) on a slide. For each nucleus, grains located at the X tip and over the region of the chromocenter showing a distinct cluster of grains were compared. Background was subtracted from the scores at the X tip by counting grains on an equal area at the tip of another chromosome, and from the chromocenter scores by counting an equal area on the euchromatic base of chromosome 3R. When Dp1187 was not distinctly labeled, an area of the expected size within the chromocenter that contained the most grains was counted and assumed to represent the duplication. This procedure probably resulted in a slight exaggeration in the labeling of Dp1187 measured in cells containing low copy numbers. In some cases, cells were placed into the categories plotted in Figure 7 by estimating the Dp1187 to X signal ratio without recording grain counts. Statistical analyses were carried out on the labeling values after correcting for background, which was small (for example: X tip = 37.5 ± 5.95 [SD], background = 1.1 ± 1.2). The variances of the X tip and Dp1187 labeling were compared using Bartiett’s test. Significant differences were found in both X/O and X/Y cells with the −54 kb probe and in X/O cells at −103 kb; in the first case it was necessary to correct for the large differences in the means. The −1.9 kb probe labeled Dp1187 so weakly in X/O cells that it was not clear whether significant variegation had occurred. Five percent of cells were distinctly labeled in the chromocenter, but only at very low levels. It was also unclear whether unusual variation in Dp1187 labeling occurred in X/Y cells with the −1.9 kb or −103 kb probe. Because yellow variegation was also infrequent in these genotypes, a similar level of Dp1187 copy number variation would probably not have been detected. Average copy number values were calculated from the histograms of Figure 7 by multiplying the fraction of cells in the three highest classes by the average values for those classes, i.e., 0.2, 0.5, and 0.85. If cells from the lowest class, where Dp1187 was labeled less than one-tenth as much as the X-tip, are not discounted, then slightly higher values would be predicted.
Acknowledgments
We thank Drs. J. Modollel, R. Fleming, K. White, and D. Lindsley for providing strains and clones. We specially thank Dr. David Schwartz for sharing, prior to publication, materials and advice that allowed us to initiate analysis of Dp1187 on pulsed-field and pulse-orientated gels. This work was supported by the Howard Hughes Medical Institute, the Carnegie Institution of Washington, and NIH Grant GM27875. G. K. was supported by a National Research Service Award of the Public Health Service.
References
- Ananiev EV, Gvozdev VA. Changed pattern of transcription and replication in polytene chromosomes of Drosophila resulting from eu-heterochromatin rearrangement. Chromosoma. 1974;45:173–191. doi: 10.1007/BF00362310. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1990. p. 1331. [Google Scholar]
- Beckingham K, Rubacha A. Different chromatin states of the intron− and type 1 intron+ rRNA genes of Calliphora erythrocephela. Chromosoma. 1984;90:311–316. doi: 10.1007/BF00287040. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Bryan GJ, Jacobson JW, Hartl DL. Heritable somatic excision of a Drosophila transposon. Science. 1987;235:1636–1638. doi: 10.1126/science.3029874. [DOI] [PubMed] [Google Scholar]
- Campuzano S, Carramolino L, Cabrera CV, Ruíz-Gómez M, Villares R, Boronat A, Modolell J. Molecular genetics of the achaete–scute gene complex of D. melanogaster. Call. 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Cole PA, Sutton E. The absorption of ultraviolet radiation by bands of the salivary gland polytene chromsomes of Drosophila melanogaster; Cold Spring Harbor Symp. Quant. Biol; 1941. pp. 66–70. [Google Scholar]
- Cowell JK, Hartmann-Goldstein IJ. Modification of the DNA content in translocated regions of Drosophila polytene chromosomes. Chromosoma. 1980;81:55–64. doi: 10.1007/BF00292422. [DOI] [PubMed] [Google Scholar]
- Delidakis C, Kafatos FC. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989;8:891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC. Position-effect variegation in Drosophila: to-wards a genetics of chromatin assembly. Bioassays. 1989;11:14–17. doi: 10.1002/bies.950110105. [DOI] [PubMed] [Google Scholar]
- Emmons SW, Yesner L. High-frequency excision of transposable element Tc1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984;36:599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- Endow SA, Glover DM. Differential replication of ribosomal gene repeats in polytene nuclei of Drosophila. Cell. 1979;17:597–607. doi: 10.1016/0092-8674(79)90267-8. [DOI] [PubMed] [Google Scholar]
- Engels W, Benz WK, Preston CR, Grahm PL, Phillis RW, Robertson HM. Somatic effects of P element activity in Drosophila melanogaster pupal lethality. Genetics. 1990 doi: 10.1093/genetics/117.4.745. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming RJ, DeSimone SM, White K. Molecular isolation and analysis of the erect wing locus in Drosophila melanogester. Mol. Cell. Biol. 1989;9:719–725. doi: 10.1128/mcb.9.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Cohen EH, Polan ML. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33:319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Geyer PK, Green MM, Corces VG. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 1990;9:2247–2256. doi: 10.1002/j.1460-2075.1990.tb07395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsell R. Untersuchungen zur Stabilkät einer yellow Postion-seffekt-Variegation in Imaginalscheiben-Kulturen von Drosophila melanogaster. Mol. Gen. Genat. 1971;110:218–237. doi: 10.1007/BF00337835. [DOI] [PubMed] [Google Scholar]
- Heck M, Spradling AC. Multiple origins are used during Drosophila chorion gene amplification. J. Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation and chromosome structure of a heat shock puff in Drosophila. Chromosoma. 1981;83:381–393. doi: 10.1007/BF00327360. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Kornher JS, Kaufman SA. Variegated expression of the sgs-4 locus in Drosophila melanogaster. Chromosoma. 1986;94:205–216. doi: 10.1007/BF00288495. [DOI] [PubMed] [Google Scholar]
- Lakhotia SC, Sinha P. Replication in Drosophila chromosomes. X. Two kinds of active replications in salivary gland polylene nuclei and their relation to chromosomal replication patterns. Chromosoma. 1983;88:265–276. doi: 10.1007/BF00292903. [DOI] [PubMed] [Google Scholar]
- Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- Lewis EB. The phenomenon of position-effect. Adv. Genet. 1950;3:73–115. doi: 10.1016/s0065-2660(08)60083-8. [DOI] [PubMed] [Google Scholar]
- Lindsley D, Grell EH. Genetic variations of Drosophila melanogaster. Carnegie Inst. Wash. Publ. 1968;627:316. [Google Scholar]
- Locke J, Kotarski MA, Tartof KD. Dosage-dependent modifiers of position-effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988;120:181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression; Cold Spring Harbor Symp. Quant. Biol; 1951. pp. 13–47. [DOI] [PubMed] [Google Scholar]
- Noujdin IJ. Genetic analysis of certain problems of the physiology of the development of Drosophila melanogaster. Biol. Zh. 1936;4:571–624. [Google Scholar]
- Noujdin IJ. The regularities of heterochromatin influence on mosaicism. Zh. Obsh. Biol. 1944;5:357–388. [Google Scholar]
- Pardue ML. In situ hybridization. In: Roberts DB, editor. Drosophila: A Practical Approach. Oxford: IRL Press; 1986. pp. 111–138. [Google Scholar]
- Pimpinelli S, Goday C. Unusual kinetochores and chromatin diminution in Parascaris. Trends Genet. 1989;5:310–315. doi: 10.1016/0168-9525(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Prokofyeva-Belgovskaya AA. Cytological mechanism of mosaicism and of chromosome rearrangement. C.R. (Dolk) Acad. Sci. URSS N. S. 1939;22:270–273. [Google Scholar]
- Prokofyeva-Belgovskaya AA. Heterochromatinization as a change of the chromosome cycle. J. Genet. 1947;48:80–98. doi: 10.1007/BF02986099. [DOI] [PubMed] [Google Scholar]
- Reuter G, Giarre M, Parah J, Gausz J, Spierer A, Spierer P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990;344:219–225. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- Ribbert D. Chromomeres and puffing in experimentally induced polytene chromosomes of Calliphora erythrocephala. Chromosoma. 1979;74:269–298. doi: 10.1007/BF01190743. [DOI] [PubMed] [Google Scholar]
- Rudkin GT. Non-replicating DNA in Drosophila. Genetics. 1969;61:227–238. [PubMed] [Google Scholar]
- Rushlow CA, Bender W, Chovnick A. Studies on the mechanism of heterochromatic position effect at the rosy locus of Drosophila melanogaster. Genetics. 1984;108:603–615. doi: 10.1093/genetics/108.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Second edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schultz J. Variegation in Drosophila and the inert heterochromatic regions. Proc Natl. Acad. Sci. USA. 1936;22:27–33. doi: 10.1073/pnas.22.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J. The evidence of the nucleoprotein nature of the gene; Cold Spring Harbor Symp. Quant. Biol; 1941. pp. 55–65. [Google Scholar]
- Schultz J. The relationship of heterochromatic chromosome regions to the nucleic acids of the cell; Cold Spring Harbor Symp. Quant. Biol; 1956. pp. 307–328. [DOI] [PubMed] [Google Scholar]
- Schultz J, Caspersson T. Heterochromatic regions and the nucleic acid metabolism of the chromosomes. Archiv. exp. Zellforsch. 1939;22:650–654. [Google Scholar]
- Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Koval M. Conformational dynamics of individual DNA molecules during gel electrophoresis. Nature. 1989;338:520–522. doi: 10.1038/338520a0. [DOI] [PubMed] [Google Scholar]
- Spana C, Harrison DA, Corces VG. The Drosophila melanogaster supressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988;2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Spofford JB. Position-effect variegation in Drosophila. In: Ashburner M, Novitiski E, editors. The Genetics and Biology of Drosophila. London: Academic Press; 1976. pp. 955–1018. [Google Scholar]
- Spradling AC, Orr-Weaver T. Regulation of DNA replication during Drosophila development. Annu. Rev. Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- Steinemann M. Chromosomal replication in Drosophila virilis III. Organization of active origins in highly polytene salivary gland cells. Chromosoma. 1981;82:289–307. doi: 10.1007/BF00286112. [DOI] [PubMed] [Google Scholar]
- Tartof KD, Hobbs C, Jones M. A structural basis for variegating position effects. Cell. 1984;37:869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]
- Traverse KL, Pardue ML. Studies of He-T DNA sequences in the pericentric regions of Drosophila chromosomes. Chromosoma. 1989;97:261–271. doi: 10.1007/BF00371965. [DOI] [PubMed] [Google Scholar]
- Young BS, Pession A, Traverse KL, French C, Pardue ML. Telomere regions in Drosophila share complex DNA sequences with pericentric heterochromatin. Cell. 1983;34:85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
- Wargent JM, Hartmann-Goldstein IJ. Replication behavior and morphology of a rearranged chromosome region in Drosophila. Chromosomes Today. 1974;5:109–116. [Google Scholar]