Abstract
Centromere function requires the coordination of many processes including kinetochore assembly, sister chromatid cohesion, spindle attachment and chromosome movement. Here we show that CID, the Drosophila homologue of the CENP-A centromere-specific H3-like proteins, colocalizes with molecular-genetically defined functional centromeres in minichromosomes. Injection of CID antibodies into early embryos, as well as RNA interference in tissue-culture cells, showed that CID is required for several mitotic processes. Deconvolution fluorescence microscopy showed that CID chromatin is physically separate from proteins involved in sister cohesion (MEI-S332), centric condensation (PROD), kinetochore function (ROD, ZW10 and BUB1) and heterochromatin structure (HP1). CID localization is unaffected by mutations in mei-S332, Su(var)2–5 (HP1), prod or polo. Furthermore, the localization of POLO, CENP-meta, ROD, BUB1 and MEI-S332, but not PROD or HP1, depends on the presence of functional CID. We conclude that the centromere and flanking heterochromatin are physically and functionally separable protein domains that are required for different inheritance functions, and that CID is required for normal kinetochore formation and function, as well as cell-cycle progression.
The centromere is cytologically visible as the primary constriction on metaphase chromosomes, and is genetically defined as a locus required for accurate chromosome inheritance1. In multicellular eukaryotes, centromeres are usually located in centric heterochromatin—a region that is rich in repeated DNA. The centromere is associated intimately with the kinetochore— the specialized protein–DNA structure responsible for attachment to spindle microtubules, prometaphase congression, and anaphase initiation and poleward movement. Defects in centromere/kinetochore function are associated with deviations from the normal chromosome complement (aneuploidy), and are linked to the cause of birth defects and cancer progression1.
An enigmatic component of the centromere/kinetochore is CENP-A, which was originally identified as one of the antigens recognized by antisera from calcinosis, Raynaud syndrome, oesophageal dysmotility, scleroderma and telangiectasia (CREST) patients, and was later found to be related to histone H3 (ref. 2). CENP-A homologues have been found in Saccharomyces cerevisiae3, Schizosaccharomyces pombe4, Caenorhabditis elegans5 and Drosophila melanogaster6. CENP-A has been proposed to be responsible for establishing a specialized chromatin structure at the centromere and may provide an epigenetic mark for centromere identity7–9. Localization of CENP-A at the electron microscopy level has not been published, but human CENP-A has been grossly localized to the inner kinetochore by light microscopy7. Furthermore, CENP-A seems to be important for initiating centromere formation and recruiting other centromere components, because mouse CENP-C, an inner kinetochore protein, is mislocalized in a CENPA knockout8.
Although CENP-A seems to be important for centromere identity (the mechanisms responsible for choosing a particular site for kinetochore formation) and kinetochore formation, very little is known about the roles of CENP-A chromatin and the flanking heterochromatin in centromere/kinetochore function. What cell-cycle and mitotic functions does CENP-A mediate? What are the spatial and functional relationships between CENP-A chromatin, outer kinetochore proteins and proteins that localize to the flanking heterochromatin?
Here we use a series of minichromosome deletion derivatives to show that localization of the Drosophila CENP-A homologue (CID; for centromere identifier6) is correlated with centromeric DNA and function, and that CID chromatin can be acquired by normally non-centromeric DNA (neocentromeres). We examined the effect of CID inactivation on cell-cycle progression, mitosis and the localization of kinetochore and centromere region proteins by using double-stranded (ds) RNA interference (RNAi) in Kc tissue-culture cells and time-lapse microscopy of early embryos injected with CID antibodies.
We used deconvolution fluorescence microscopy to examine the physical distribution of CID and proteins involved in centric condensation (proliferation disruptor; PROD10), centric sister chromatid cohesion (MEI-S332; ref. 11), outer kinetochore function (POLO kinase12) and heterochromatin structure (heterochromatin protein 1; HP1; ref. 13). Finally, we used genetic and cytological analyses to determine whether CID localization depends on the presence of these proteins, and vice versa. Our results elucidate the many roles of CENP-A in kinetochore formation and mitosis, as well as the physical and functional relationships between centromere activity and other inheritance functions encoded by the flanking heterochromatin.
Results
CID localization to the inner kinetochore correlates with centromeric DNA and function
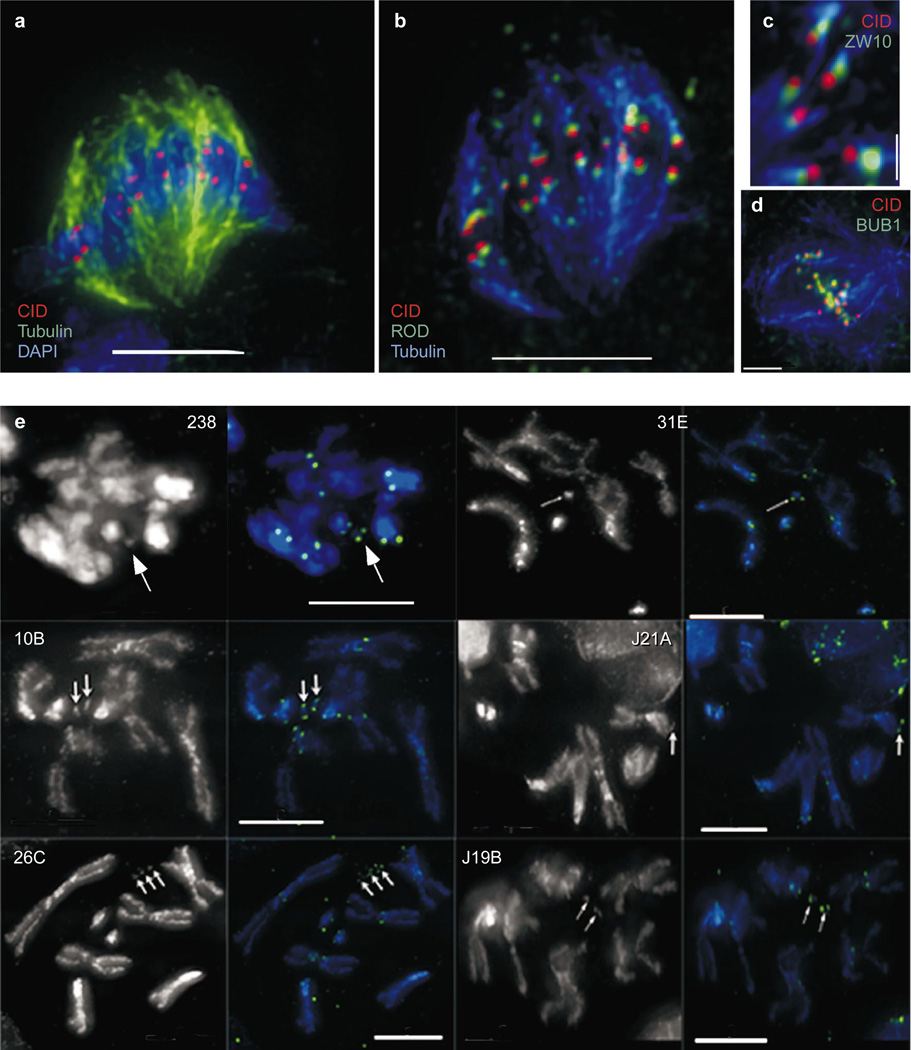
Immunolocalization experiments in mammals have shown that it is possible to distinguish between the inner and outer kinetochore by fluorescence microscopy7. To determine whether CID is located in the inner kinetochore, we simultaneously localized CID, spindle microtubules and many transient kinetochore proteins including BUB1, zeste-white 10 (ZW10) and rough deal (ROD)14–16. Each of these proteins would be expected to localize to the outer kinetochore plate or the fibrous corona, similar to other transient kinetochore proteins localized in mammals (for example, BUB1, CENP-E, Dynein)17–19.
Simultaneous detection of CID with outer kinetochore proteins showed that CID is consistently separated from ZW10, ROD (Fig. 1a–c) and POLO kinase (data not shown), and was located closer to the chromosome and further from the kinetochore microtubules than these proteins (Fig. 1a–c). CID was also offset from BUB1 (a component of the spindle assembly checkpoint) at unattached kinetochores, but CID and BUB1 showed significant overlap (Fig. 1d). This result is consistent with studies in mammals, which suggest that BUB1 may be located at both the inner and outer kinetochore plates19. Our results show that CID is located in or near the inner plate of the kinetochore in Drosophila and is likely to be associated closely with centromeric DNA.
Figure 1. CID is localized to the inner kinetochore and the functional centromere.
CID was simultaneously localized with BUB1, ZW10, ROD and spindle microtubules in mitotic figures from Kc cells. a, CID is localized in paired spots along the spindle equator at metaphase. b, CID is localized closer to the chromosomes and further from kinetochore microtubules than ROD; the same cell as in a is shown. c, High-magnification view showing that CID is located further from kinetochore microtubules than ZW10. d, CID and BUB1 are offset but show significant colocalization at unattached kinetochores. e, Indirect immunofluorescence with anti- CID antibody was performed in larval neuroblasts from animals carrying one or more copies of each of the indicated derivatives. CID (green) is present on 100% of all derivatives (arrows) in staining intensities comparable to endogenous chromosomes. In all spreads minichromosomes are present as paired sister chromatids, and CID staining appears as double dots, as observed for endogenous chromosomes. See Supplementary Information for Dp derivative structures and transmission rates to progeny. Scale bars, 2 µm (d); 1 µm (c); and 5 µm (a, b, e).
Previous work has shown that the outer kinetochore proteins ZW10 and Dynein are present on fully functional Drosophila minichromosomes (that is, 100% transmission through mitosis and meiosis), as well as structurally acentric minichromosomes that lack detectable centromeric sequence (neocentromeres)15,20. To determine the relationship of CID-containing chromatin to the functional centromere, we examined the localization of CID protein on a series of minichromosome derivatives of decreasing size and meiotic transmission efficiency (Supplementary Information Fig. 1).
CID was present on all minichromosome derivatives that contain a fully functional centromere (γ238, 31E, 10B and J21A; Fig. 1e and see Supplementary Information), indicating that CID colocalizes with the molecular-genetically defined centromere21,22. CID also was present on all of the neocentromeric derivatives that lack centric heterochromatin, including the normal minichromosome centromere (26C, J19B; Fig. 1e and Supplementary Information), consistent with the localization pattern of outer kinetochore components ZW10 and Dynein15,20.We conclude that CID localization is correlated with centromere function, regardless of the composition of the underlying DNA.
Inhibiting CID function results in interphase arrest and aberrant chromosome segregation
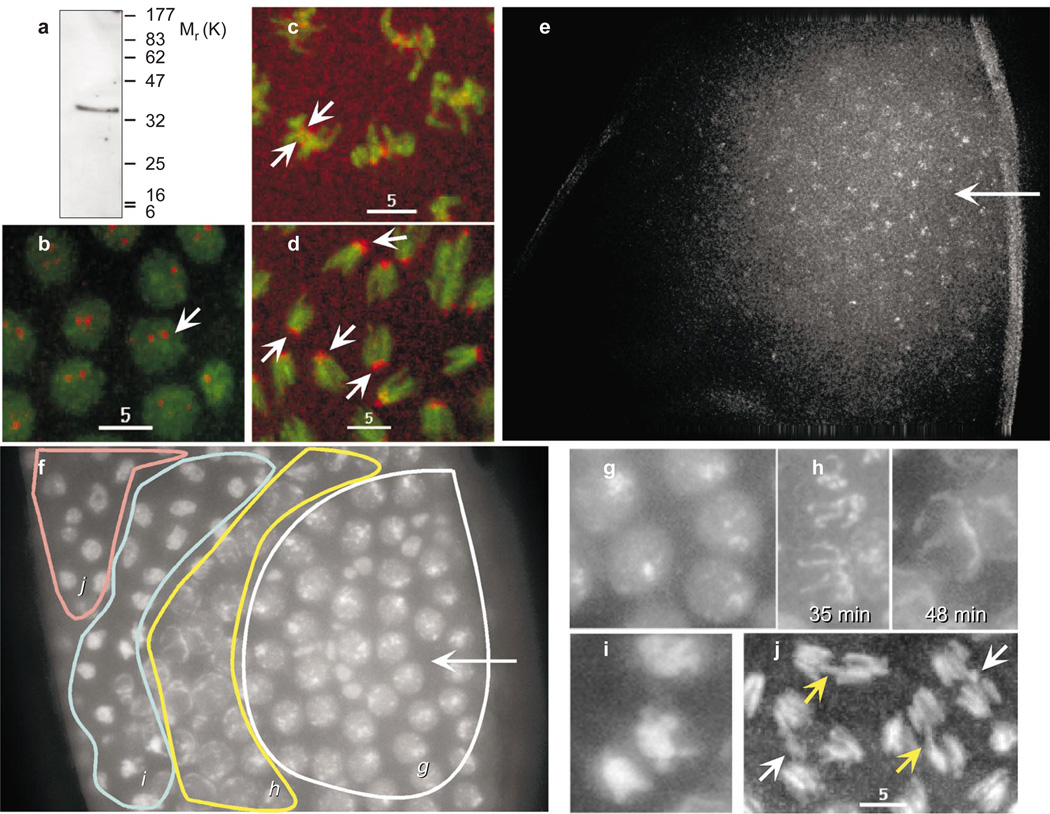
The presence of CID at the functional minichromosome centromere shows that CID localization is correlated with centromere activity. To test the role of CID in mitosis directly, we injected affinity-purified chicken anti-CID antibodies into early embryos that express histone H2A/green fluorescent protein (GFP)23 and observed chromosome behaviour using time-lapse microscopy. Rhodamine-labelled anti-CID antibodies bound specifically to centromeres in all stages of the cell cycle (Fig. 2b–d), and showed no nonspecific crossreactivity with histone H3 either in vivo or by western blot (Fig. 2a).We also observed that injected antibody bound centromeres in a gradient in which more antibody bound close to the site of injection (Fig. 2e).
Figure 2. Affinity-purified chicken anti-CID binds centromeres at all stages of the cell cycle in vivo, and induces several mitotic and cell-cycle defects.
a, Western blot shows that affinity-purified chicken anti-CID antibody recognizes a single protein of relative molecular mass (Mr) ~32,000 (~32K) in total nuclear protein prepared from embryos, consistent with the predicted size of CID and with no cross-reactivity to histone H3 (16K). b–d, Rhodamine-labelled chicken anti-CID (red) binds the centromeres (arrows) of all chromosomes (green) in vivo in all stages of the cell cycle. e, Injection of chicken anti-CID results in a gradient of antibody binding in the embryo, centred at the injection site (arrow). f, Antibody injection results in a gradient of phenotypes in the embryo. Italic lettering refers to regions that contain the phenotypes represented by the higher magnification images in g–j. g, Interphase arrest phenotype prevalent nearest the site of antibody injection. h, Chromosomes that have begun condensation, reversed condensation and arrested (compare 35 min to 48 min). i, Chromosomes arrested in metaphase. j, Chromosomes exhibiting anaphase defects, such as lagging chromosomes (yellow arrow) and chromosomes left at the metaphase plate (white arrow). See Supplementary Information for movies. Scale bars, 5 µm.
Injection of CID antibodies into early embryos resulted in a range of phenotypes affecting both cell-cycle progression and mitotic chromosome segregation (Fig. 2f–j; and Table 1). The phenotypic series is consistent with a gradient of CID inhibition mirroring the gradient of antibody concentration (Fig. 2f). Nuclei closest to the site of injection arrested in interphase (13%, Fig. 2g), whereas nuclei further from the site of injection delayed entering mitosis and exhibited different mitotic defects: specifically, entry into prophase condensation followed by a loss of condensation (3.6%; Fig. 2h); metaphase arrest (15%; Fig. 2i); and various anaphase chromosome segregation defects (failure to move toward the poles at anaphase onset, unequal chromosome segregation, failure to maintain spindle contact and karyokinesis defects at telophase, 20%; Fig. 2j). Uninjected embryos, embryos injected with heat-killed antibody, and embryos injected with 10 mg ml−1 bovine serum albumin displayed few mitotic defects, showing that these abnormalities resulted from inhibition of CID function (Table 1; see movies in Supplementary Information).
Table 1.
Quantification of cell-cycle and mitotic defects observed after anti-CID injection
| Anti-CID (n = 534) | Control (n = 481) | |
|---|---|---|
| Phenotype | (% total) | |
| Interphase arrest (G) | 13.11 | 0 |
| Condense and arrest (H) | 3.56 | 0 |
| Metaphase arrest (I) | 15.36 | 0.21 |
| Anaphase defects (J) | 20.41 | 2.29 |
G–H in parentheses refers to areas indicated in Fig. 2.
n, Number of nuclei.
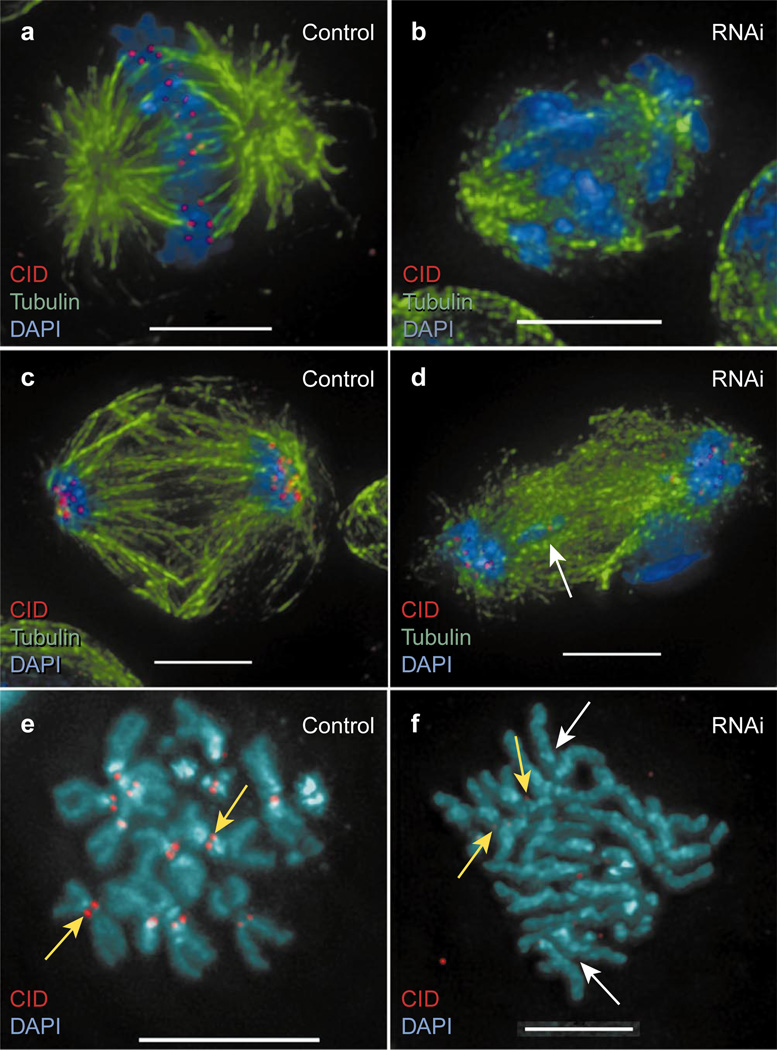
We also disrupted CID function in Drosophila Kc tissue-culture cells using RNAi. Cells were treated with dsRNA corresponding to the whole CID transcript; cells were then fixed and monitored for levels of CID protein and aberrant chromosome behaviour every 24 h after adding dsRNA. Cells in a given treated population displayed a variable penetrance of CID inhibition (compare Fig. 3b, d, f), which resulted in different phenotypes.
Figure 3. CID RNAi results in several mitotic phenotypes in tissue culture cells.
Kc cells were treated with dsRNA from the full CID transcript and observed for mitotic defects by comparison with control cells. a, Untreated control cell showing chromosome alignment at the metaphase plate and kinetochore microtubule attachment. b, RNAi cell exhibiting chromosome misalignment, failure to capture spindle microtubules, and spindle disorganization. Note the absence of CID staining. c, Untreated control anaphase showing all chromosomes present near the poles and a well-organized spindle. d, RNAi anaphase showing a lagging chromosome (white arrow) and spindle disorganization. Note the greatly reduced amount of CID staining compared with controls. e, Untreated control metaphase spread showing prominent CID staining at the primary constrictions of all chromosomes (yellow arrow). f, RNAi metaphase spread showing precocious sister chromatid separation (white arrows). Faint CID staining is visible at a reduced constriction on two autosomes that retain sister chromatid cohesion (yellow arrow). Scale bars, 5 µm (a–d, f); 10 µm (e).
Mitotic defects in Kc cells were consistent with those observed after antibody injection into embryos, including aberrant prometaphase congression (Fig. 3a, b), precocious sister chromatid separation (Fig. 3f, white arrow), kinetochore microtubule capture (Fig. 3a, b) and anaphase segregation (Fig. 3c, d). Cells treated with dsRNA were no longer dividing 8–10 d after RNAi, suggesting that interphase arrest had resulted from complete CID disruption; however, without live analysis it is difficult to differentiate between interphase arrest and a terminal phenotype resulting from massive chromosome segregation defects.
The results of CID disruption by both antibody injection into embryos and RNAi in tissue-culture cells show that CID is directly or indirectly required for many aspects of kinetochore-mediated chromosome movement as well as cell-cycle progression.
The centromere and flanking heterochromatin comprise many structurally separable domains
Centromeres in most higher eukaryotes are embedded in centric heterochromatin, suggesting that both the structure and function of heterochromatin are required for centromere function.What are the structural relationships between centromeric chromatin, defined by CID, and chromosomal proteins previously localized to the centromere region? We addressed this question using immunolocalization of three proteins and CID on mitotic chromosomes from S2 and Kc tissue-culture cells.
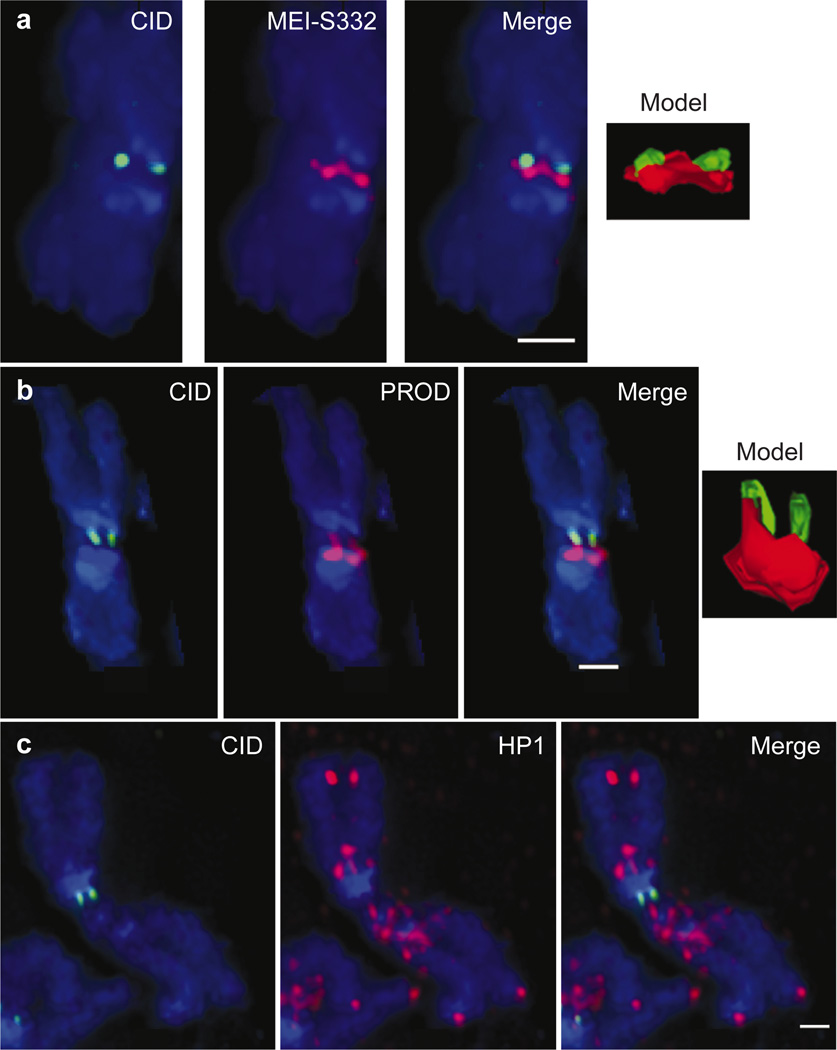
MEI-S332 is required for sister chromatid cohesion during metaphase I of meiosis, and is present in the centromeric regions of meiotic and mitotic chromosomes11. Simultaneous localization of CID with MEI-S332 showed that CID antibodies yielded typical double-dot staining, whereas MEI-S332 was localized in two concentrated foci joined by a bridge of staining that connected the sister chromatids (Fig. 4a).
Figure 4. The centromere region comprises several, spatially separable domains.
CID was simultaneously localized with MEI-S332, HP1 and PROD on metaphase chromosomes of S2 tissue culture cells. a, MEI-S332 (red) is offset from CID (green) to one side of the chromosome and seems to form a bridge between the paired sister chromatids. b, PROD (red) is displaced towards the arms, always in the same direction as MEI-S332. c, HP1 (red) is present near but not in centromere chromatin. In all panels, DNA was counterstained with DAPI (blue). All models were created directly from the raw data. All measurement bars are 1 µM.
Although MEI-S332 has been described as centromeric24 and possibly located to the inner kinetochore25, the higher resolution localization of MEI-S332 presented here showed consistent offset of antibody staining to one side of the kinetochore and along the chromosome axis on all chromosomes. The offset localization was always to the same side of the kinetochore on each chromosome type. This was especially evident on the X chromosome, in which MEI-S332 was always located on the proximal long arm side of CID, and for chromosomes 2 and 3, on the basis of colocalization with the sequence-specific satellite binding protein PROD26 (see below). We conclude that MEI-S332 is located near but not in the CID chromatin, providing a physical basis for the previous observation that kinetochore function and MEI-S332-mediated cohesion can be separated using minichromosome derivatives25.
proliferation disrupter (prod) mutant larval neuroblasts display hypo-condensation of the centromere region and metaphase/anaphase arrest10. Consistent with the decondensation phenotype, the PROD protein was localized to the centromeric region of chromosomes 2 and 3 in mitosis, suggesting that it may be involved in kinetochore function on these chromosomes. Simultaneous detection of CID and PROD on mitotic chromosomes showed, however, that PROD stains a more expansive portion of the chromosome than CID (Fig. 4b), and is offset from the kinetochore in the same manner as MEI-S332. In fact, PROD and MEI-S332 are both localized to the same side of the kinetochore on chromosomes 2 and 3 (data not shown).
HP1 mutants show dominant suppression of heterochromatin-induced position-effect variegation (PEV), and recessive telomere fusions and chromosome segregation defects13,27. Human and mouse homologues of HP1 localize to the centromere region28, and S. pombe Swi6, another chromodomain protein, is localized to the centromere and required for proper chromosome transmission29. Simultaneous localization of CID with HP1 revealed that HP1 is not present in centromeric chromatin in either interphase or metaphase. In metaphase chromosomes, HP1 is located throughout the pericentric heterochromatin, and is near but not in CID chromatin (Fig. 4c).
We conclude that PROD and HP1 are located in the pericentric heterochromatin and not in the centromeric chromatin. These results suggest that, although the centromere is embedded in large blocks of heterochromatin, centromeric chromatin is spatially separable from canonical centric heterochromatin.
CID localization is unaffected by mutations in proteins involved in heterochromatin structure, cohesion and kinetochore function
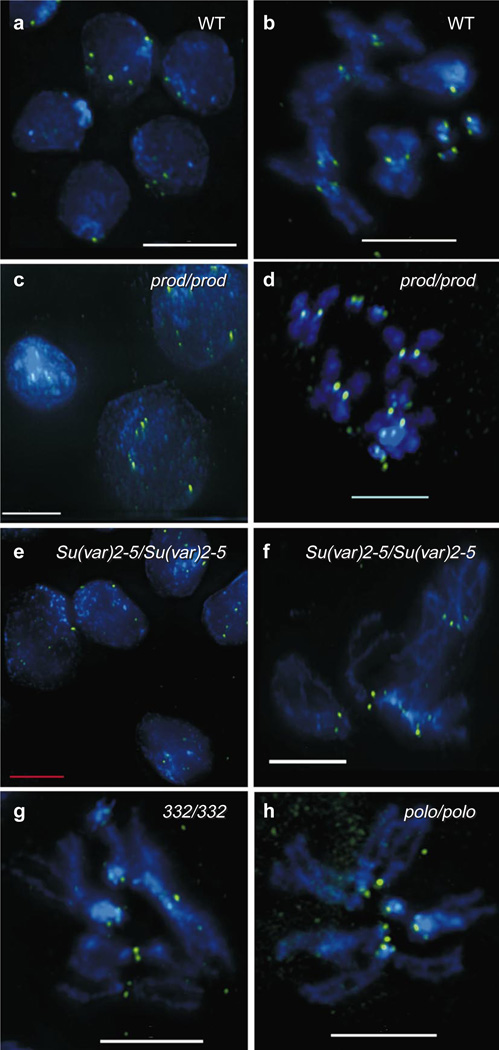
Does the spatial separation of CID chromatin, outer kinetochore proteins and centric heterochromatin proteins reflect functional independence? We examined CID localization in larval neuroblasts from animals lacking PROD, HP1, MEI-S332 or POLO kinase. In interphase nuclei and mitotic chromosomes from homozygous prod mutants, CID remained localized in the typical punctate pattern (Fig. 5c, d) observed in wild type (Fig. 5a, b), despite visible centromere region hypocondensation. Similarly, CID was localized in the typical punctate pattern in interphase nuclei from homozygous mutant Su(var)2–5 (HP1) neuroblasts (Fig. 5e).
Figure 5. CID localization is unaffected by mutations in other centromere components and proteins involved in heterochromatin structure.
CID (green) was localized by indirect immunofluorescence in larval neuroblasts from wild-type (WT), prod, Su(var)2–5, mei-S332 and polo homozygous mutant animals. a, b, CID localization in wild-type interphase and metaphase figures from a line containing the minichromosome derivative Dp31E. c, d, CID localization is unaffected by mutations in prod, even in the presence of visible centric decondensation. e, f, CID localization is unaffected in interphase cells and metaphase chromosomes of Su(var)2–5 mutants. g, CID localization is unaffected by mutations in mei-S332. h, A circular metaphase spread from a polo mutant shows that CID localization is unaffected by mutations in polo kinase. Scale bars, 5 µm.
In mutant metaphase spreads exhibiting the Su(var)2–5 telomere fusion phenotype13 CID still localized in the characteristic double-dot pattern (Fig. 5f). Furthermore, CID was also localized in the characteristic double dot pattern in homozygous mei-S332 mutant larval neuroblasts (Fig. 5g). Finally, in metaphases exhibiting circular spreads indicative of centrosome disorganization, characteristic of polo mutations, CID remained localized in characteristic double dots (Fig. 5h).Thus, the analyses of CID localization in mutant neuroblasts shows that the assembly and maintenance of centromeric chromatin in interphase and metaphase is not dependent on the presence of proteins required for normal centromere region condensation (PROD), heterochromatin structure (HP1), centric cohesion (MEI-S332), or outer kinetochore function (POLO kinase).
CID inhibition results in mislocalization of several transient kinetochore proteins and MEI-S332
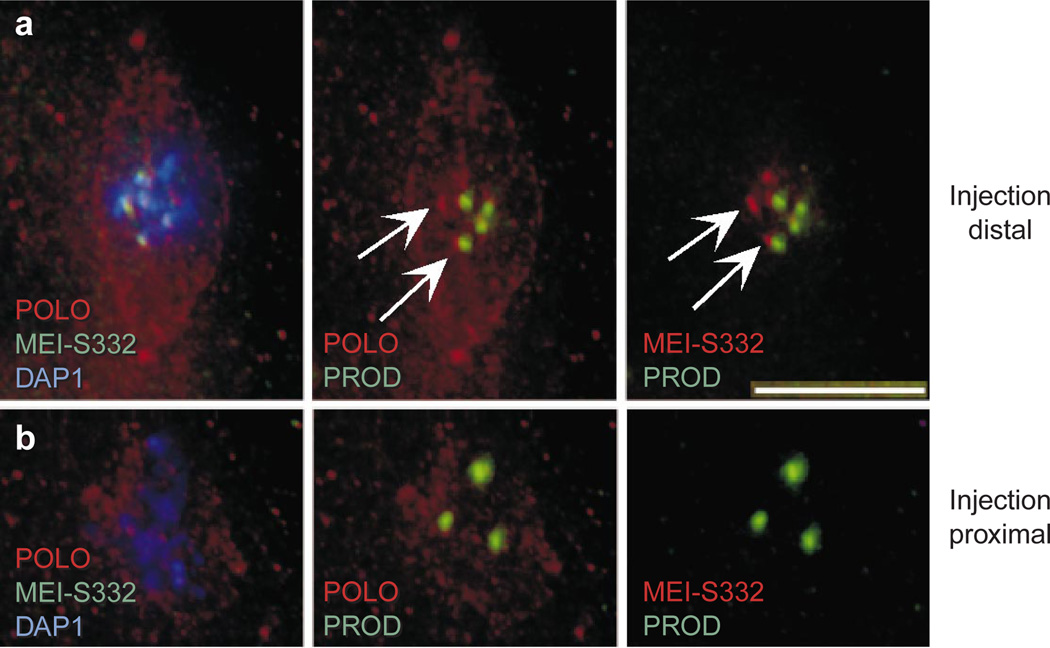
Although CID localization is not dependent on the presence of PROD, HP1, MEI-S332 or POLO kinase, the mutant analyses did not determine whether the localization of these proteins depended on CID. Therefore, we examined POLO kinase, MEI-S332 and PROD localization in embryos in which CID function was inhibited. In embryonic nuclei close to the site of injection, where high levels of CID antibody binding and the most severe mitotic defects were observed (see Fig. 2), POLO kinase localization was diffuse and apparently absent from kinetochores, as judged by counterstaining with PROD (compare Fig. 6a, b). Notably, in these same nuclei MEI-S332 was absent from the pericentromeric region (Fig. 6b), whereas PROD, a protein with sequence-specific satellite-binding properties, was still present in the pericentromeric region (Fig. 6b). Mitotic chromosomes more distal to the injection site in the same embryo showed a wild-type distribution of POLO and MEI-S332 (Fig. 6a, arrows), indicating that mislocalization was a direct result of anti-CID injection.
Figure 6. CID disruption results in mislocalization of transient kinetochore components and a sister cohesion protein.
Embryos injected with anti-CID antibody were fixed after injection and processed for immunofluorescence to determine whether localizations of other centromere region components were disrupted. a, Nucleus distal to the site of injection from the same embryo as the nucleus shown in b. Chromosomes aligned at the metaphase plate show concentrated PROD (on chromosomes 2 and 3 only), POLO and MEI-S332 localization to the kinetochore or pericentric heterochromatin (arrows), as well as POLO localization to the centrosomes, and a well-organized spindle (faint POLO staining). b, Chromosomes proximal to the injection site display chromosome misalignment and a disorganized spindle, normal PROD staining, and no MEI-S332 and POLO concentration in the centromere region. Scale bar, 5 µm.
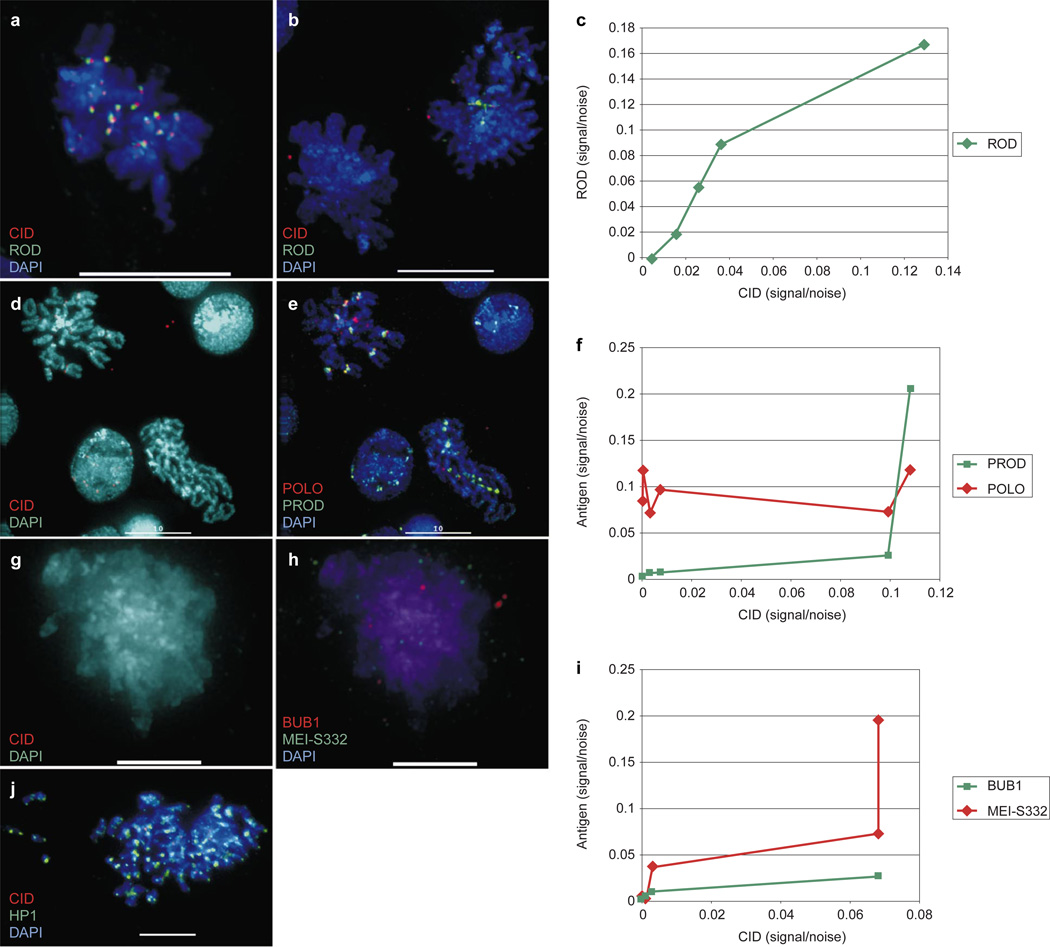
The localizations of ROD (Fig. 7a–c), CENP-meta30 outer kinetochore, CENP-E kinesin-like protein homologue (data not shown), POLO kinase (Fig. 7d–f), BUB1 (Fig. 7g–i) and MEI-S332 (Fig. 7g–i), but not PROD (Fig. 7d–f) or HP1 (Fig. 7j), were also disrupted in Kc cells displaying mitotic defects as a result of RNAi inhibition of CID expression. Quantitative deconvolution microscopy revealed that transient kinetochore component recruitment was proportional to the amount of CID present at the kinetochore (Fig. 7c, f, i), whereas PROD recruitment was independent of CID levels (Fig. 7f). Thus, CID function is required for the recruitment or maintenance of transient kinetochore components and a centric cohesion protein (MEI-S332), but is not required for the localization of PROD or HP1. These results also indicate that the pleiotropic mitotic defects observed in anti-CID injection and RNAi are likely to be caused by a failure to recruit or bind transient kinetochore components and a centric cohesion protein.
Figure 7. CID RNAi results in mislocalization of many transient kinetochore components and a sister cohesion protein.
Kc cells treated with CID dsRNA were processed for immunofluorescence to determine whether the localizations of other centromere components were disrupted. a, Control cell showing normal CID and ROD localization. b, Metaphases from RNAi-treated cells. The metaphase on the right shows a small but detectable amount of CID staining, accompanied by a decreased and delocalized amount of ROD staining; the metaphase on the left shows no detectable CID staining and no detectable ROD staining. c, Quantitative immunofluorescence shows that ROD localization depends on the amount of CID present at the kinetochore. d, Metaphase spreads exhibiting varying degrees of CID inhibition. The spread on the left has a small but detectable amount of CID, whereas the spread on the left has no detectable CID. e, Same spreads as in d; the spread with CID has detectable POLO and PROD; the spread on the right has no detectable POLO, but does have detectable, normally localized PROD. f, Quantitative immunofluorescence shows that POLO localization, but not PROD, is dependent on the amount of CID present at the kinetochore. g, h, Mitotic figure lacking detectable CID, also lacks detectable localized BUB1 and MEI-S332. i, Quantitative immunofluorescence shows that both BUB1 and MEI-S332 localization are dependent on the amount of CID present at the kinetochore. j, Mitotic figure with very low CID levels shows no disruption of telomeric or diffuse pericentric HP1 localization. Scale bars, 5 µm (g, h); 10 µm (a, b, d, e, j).
Discussion
CENP-A has been proposed to be an epigenetic mark required for determining centromere identity and thus the assembly of the kinetochore. Here we have used deconvolution microscopy and a minichromosome deletion series to show that CID is associated with the inner kinetochore and that CID localization correlates with centromere function, specifically the presence of the molecular-genetically defined centromere and neocentromere DNA.
The presence of CID on neocentromeres shows that these structurally acentric yet functional chromosomes have acquired centromeric chromatin, consistent with the presence of outer kinetochore components15,20. Furthermore, the presence of CID on neocentromeres shows that the location of centromeric chromatin is independent of sequence, and that centromeric chromatin can confer centromere identity on normally non-centromeric DNA—a state that is then propagated faithfully through replication and division.
We have also shown that CID is required for kinetochore assembly and cell-cycle progression in early Drosophila embryos and Kc tissue-culture cells. Mitotic defects were observed previously in human cells after CENP-A antibody injection31 and in the mouse CENP-A knockout8; however, our ‘live studies’ have allowed us to examine the temporal and cytological effects of CENP-A disruption in greater detail. Both antibody injection into embryos and RNAi inhibition in Kc cells resulted in several defects expected for centromere dysfunction: specifically, aberrant prometaphase congression; chromosome attachment to spindle microtubules; entry into anaphase; anaphase poleward segregation; and failure to resolve properly at telophase.
The mislocalization of ROD, BUB1, CENP-meta, POLO and MEI-S332 in nuclei displaying missegregation phenotypes shows that the defects are correlated with aberrant kinetochore structure and the recruitment of transient kinetochore proteins and other centromere region proteins. These results extend the earlier observation that the inner kinetochore protein CENP-C is mislocalized in the CENP-A knockout mouse8 to outer kinetochore proteins. Notably, the amount of outer kinetochore components present at the kinetochore was proportional to the amount of CID, suggesting that the kinetochore may be composed of a repeated substructure32.
CID disruption may decrease the number or size of functional subunits, which is sufficient to cause defects in mitosis because mitotic defects are observed in cells with decreased but visible amounts of CID. Disruption of the centromere/kinetochore substructure may be responsible for different degrees of mislocalization of outer kinetochore components, which results in the observed pleiotropy of mitotic phenotypes. Karyokinesis defects observed after CID inhibition may be the result of the failure of chromosomal passenger proteins33 to localize to the kinetochore and consequently to the spindle and midbody. On the basis of the mislocalization of several outer kinetochore components, we conclude that CID is epistatic to transient kinetochore components (Fig. 8b); these results support the hypothesis that CENP-A proteins are involved directly in the epigenetic marking of the site of kinetochore formation, and show conclusively that proper kinetochore function is required for many cell-division processes.
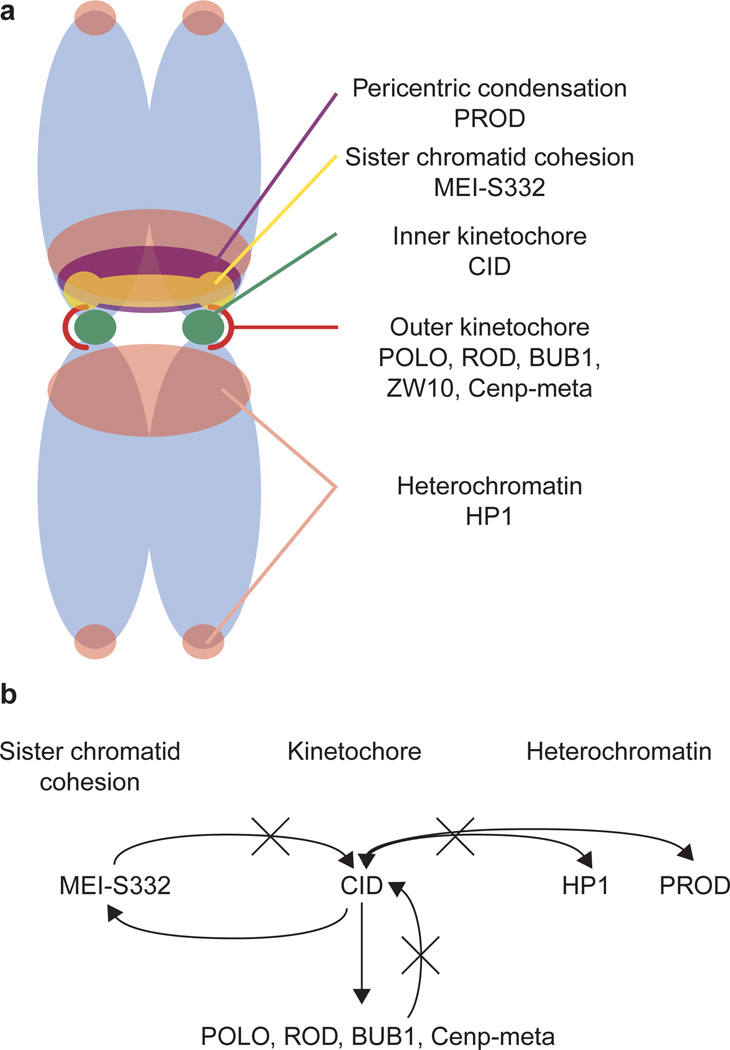
Figure 8. Structural and functional relationships within the Drosophila centromere region.
a, Summary of the spatial relationships of various centromere region and kinetochore components on metaphase chromosomes. b, Epistasis diagram depicting the functional relationships of the components shown in a. Three separate domains and pathways, which all affect chromosome inheritance, are shown: kinetochore, centric heterochromatin, and sister chromatid cohesion. Epistasis analyses show that CID is essential for recruiting all outer kinetochore proteins tested here, as well as a sister cohesion protein (MEI-S332), and that CID and two flanking heterochromatin proteins are functionally independent. Thus, CID is at or near the top of the kinetochore assembly and the MEI-S332-mediated cohesion pathways.
More complete CID disruption in embryos resulted in a severe interphase arrest phenotype, showing that CID function is also required before entry into mitosis, which is similar to one of the phenotypes observed after injection of anti-CENP-A antibodies into HeLa cells31. The use of real-time analysis allowed us to conclude unambiguously that the nuclei are arrested before entry into mitosis. The interphase arrest phenotype suggests that CID functions in interphase, where it is constitutively bound to centromeres, and that there may be another cell-cycle checkpoint that monitors kinetochore assembly and blocks entry into mitosis if this process is compromised.
This putative ‘kinetochore assembly’ checkpoint is also likely to be responsible for the delay in entering mitosis observed in nuclei just distal to the injection site. It is possible that disruption of an interaction between POLO kinase and CID is responsible for this cell-cycle arrest as POLO is required for entry into mitosis34. Clearly, further investigation is necessary to elucidate the precise pathway involved in this arrest, and to determine whether there is a checkpoint that monitors kinetochore assembly before entry into mitosis.
Genetic and protein localization studies have implicated several proteins in regulating centromere function1. Until now, it has been difficult to determine whether these proteins are involved in kinetochore formation and function, other centromere functions, or functions independent of the centromere/kinetochore. The structural and functional analyses presented here support the hypothesis that distinct spatial and functional domains exist in the centromere and adjacent regions. Previously, cytological studies in humans revealed the presence of spatially distinct protein domains in the mitotic kinetochore35.
We have extended these observations by investigating the domain structure of the kinetochore and the centromere region, including the flanking heterochromatin, and have provided data that reveal the functional separation and interdependence of these structural domains. We have found that centromeric chromatin is the central and most essential component of the centromere region, and is required for entry into mitosis and chromosome movement during mitosis, as well as for recruiting proteins to the kinetochore and flanking domains (Fig. 8). The domain organization of the centromere observed in Drosophila is similar to the situation in S. pombe, where different proteins occupy distinct subdomains within the centromere region36, and are required for separable chromosome segregation processes37.
Studies in a variety of organisms have indicated that the centromere region is a site of specialized sister cohesion, not only in meiosis I but also in mitosis38. For example, normal homologue disjunction requires that MEI-S332 functions to maintain sister chromatid cohesion in the centric regions throughout meiosis I and until anaphase of meiosis II. Although MEI-S332 is involved in sister chromatid cohesion, it is not a cohesin; moreover, its localization differs from that observed in S. cerevisiae, where cohesins are concentrated at the centromere and associated with centromeric chromatin39,40.
MEI-S332 is required for proper chromosome inheritance in Drosophila, but surprisingly the chromosome still retains kinetochore protein localization and function if MEI-S332 is eliminated. Despite the spatial and functional separation of MEI-S332 and CID, MEI-S332 is dependent on functional centromeric chromatin for its recruitment to the centromere region: it is mislocalized in anti-CID injected embryos and RNAi-treated Kc cells. CID localization is not, however, dependent on the presence of MEI-S332. Therefore, CID is epistatic to MEI-S332 in the pathway responsible for the assembly and/or maintenance of this physically and functionally distinct centromere region domain in mitosis (Fig. 8b). The relationship between CID, kinetochore function and MEI-S332-mediated cohesion warrants further genetic and biochemical analyses. It will be particularly interesting to determine the significance of the consistent asymmetric positioning of MEI-S332 to only one side of the CID chromatin, as well as its impact on CID during meiosis, where mutant phenotypes are more severe50,54.
The role of heterochromatin in centromere function
Heterochromatin encodes several inheritance functions, including homologue pairing in meiosis I, sister chromatid cohesion and interactions with anti-poleward forces41–43. The conserved location of centromeres in heterochromatin suggests that heterochromatin proteins, such as PROD and HP1, may be required for establishing or maintaining centromeres. We have shown that both PROD and HP1 are not detectable in CID chromatin (Fig. 8a) and that prod and Su(var)2–5 mutations do not affect the localization of CID. Furthermore, neither PROD nor HP1 localization is affected in anti-CID-injected embryos or RNAi-treated Kc cells (Fig. 8b).
We conclude that PROD and HP1 function in the pericentromeric regions to promote normal condensation and chromosome segregation—processes distinct from the centromere/kinetochore. Although the kinetochore is typically embedded in large blocks of heterochromatin, we have provided evidence that it may be structurally and functionally distinct from the closely juxtaposed pericentromeric or centric heterochromatin.
If centromere chromatin structure is distinct from centric heterochromatin, why are centromeres embedded in heterochromatin in almost all multicellular eukaryotes? Perhaps the flanking heterochromatin does provide an environment that is necessary for the formation of a centromere-specific higher order chromatin structure. In addition, although PROD and HP1 are not necessary for CID localization, they may encode functions redundant with other heterochromatic proteins that establish or maintain proper kinetochore structure. It will be interesting to determine whether protein localization and mutant analyses with other centric heterochromatin proteins are consistent with our results for prod and Su(var)2–5 (HP1).
We have shown that Drosophila CENP-A/CID is required for kinetochore formation and mitotic function, cell-cycle progression and recruiting transient kinetochore components and a sister chromatid cohesion protein. In contrast, CID and proteins that function in the pericentric heterochromatin are physically and functionally independent. Our results support the hypothesis that CENP-A proteins are central to many mitotic processes, and may be a component of the epigenetic mark responsible for centromere identity and function. Future studies should investigate what mechanism is responsible for loading CENP-A specifically into CENP-A chromatin in a replication-independent manner44,53, as this process may be the key to understanding maintenance of the epigenetic mark and centromere identity.
Methods
Cell culture
Drosophila S2 and Kc cells were grown in DES media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco), 100 µg ml−1 penicillin and 100 µg ml−1 streptomycin.
Fly culture
We maintained all derivatives of the free duplication/minichromosome Dp(1:f)1187 (referred to as Dps)21 used in this study as inbred stocks.Mutant stocks were obtained from the Bloomington stock center, except for the Su(var) 2–5 allele, which was isolated in a screen for mutations that affect TDAPEV (K. Donaldson et al., unpublished data).Mutants were maintained over balancer chromosomes marked with Act5C–GFP45.We identified homozygous mutant larvae used for neuroblast preparations by selecting for larvae that did not express GFP.
Antibody production and purification
Full-length CID was cloned into pQE31 (Qiagen) by introducing a 5′ BamHI and a 3′ KpnI site using polymerase chain reaction, and clones were verified by sequencing. Protein was produced in M15 cells and purified according to the manufacturer’s instructions, isolated from a 15% SDS–PAGE gel, and used to induce antibodies in chickens (Aves Labs). Antibodies were affinity purified against full-length CID by standard methods.
Western analysis
We prepared total nuclear protein from 0–12-h embryos by solubilizing purified nuclei in 8 M urea. Proteins were separated on 15% SDS–PAGE gels, processed for western blot using standard protocols and detected using anti-chicken horseradish peroxidase (1:5000, Chemicon).
Antibody injection
Affinity-purified chicken anti-CID (1.2 mg ml−1) was labelled with tetramethyl-rhodamine using the FluoReporter kit (Molecular Probes) according to the manufacturer’s instructions, except that sodium azide was omitted from the desalting column. We performed injections according to standard protocols46, except that we visualized embryos using a Deltavision workstation. For single-colour time-lapse microscopy, three images spaced 1-µm apart in the z axis were acquired every 15 s; for two-colour time-lapse, three images of each wavelength were acquired every 23 s. For post-injection staining, embryos were injected and allowed to develop for 15–30 min at room temperature then fixed and processed for immunofluorescence as described below. For quantification of mitotic defects, we observed all nuclei in a movie for two mitotic division cycles after injection.
RNA interference
RNAi was performed as described47 using the full-length CID open reading frame, except that fresh dsRNA was added every 24 h for 4 d. Cells were monitored every 24 h after RNA addition by immunofluorescence for reduction in CID protein levels, which generally took 4–6 d (probably necessary to dilute this constitutive chromatin component), and varied greatly in a given cell population. We stained RNAi-treated cells for H3 and phosphorylated H3 (PH3) as a control for nonspecific inhibition, and found no differences between treated and untreated cells.
Cytological preparations
S2 cells were prepared using a modification of a published protocol13. S2 cells were arrested with colcemid at a concentration of 0.5 µg ml−1 for 1.5–2 h. All simultaneous localizations were performed with and without colcemid, and no differences were in 0.5% (w/v) sodium citrate at a concentration of 2 × 105 cells per ml for 7 min. We put 500 µl of this cell suspension in a single-chamber Cytospin funnel and spun it for 10 min at 900 r.p.m. (90g) on high acceleration in a Shandon Cytospin 3. The cells were fixed in 4% paraformaldehyde for 5 min, blocked in PBS + 0.1% Triton X-100 + 1% dry nonfat milk for 30 min to 1 h. For tubulin staining of RNAi treated cells, we omitted the colcemid and sodium citrate treatments and fixed the cells with 4% paraformaldehyde, 0.05% gluteraldehyde and 10 µM taxol for 7 min. For BUB1 staining of RNAi treated cells, colcemid was added to the culture 2 h before slide preparation. Slides were then processed for immunofluorescence as described below.
Larval brains were dissected in PBS and neuroblast squashes were prepared as described48. For the Dp staining experiment, only brains shown in Fig. 1e were incubated in 3 µg ml−1 colcemid in PBS for 1.5 h before squashing. Brains shown in Figs 1e (γ238) and 5d were incubated in 0.02 N NaOH for 20 s after fixation in formaldehyde instead of acetic acid. We froze slides in liquid nitrogen, removed the coverslips and immersed them in PBS + 0.1% Triton X-100, before transferring them to PBS-T-M (PBS + 0.1% Triton, 1% non-fat dry milk) to block for 1 h.
Embryos were prepared for immunofluorescence as described49, except that embryos were fixed in 4% paraformaldehyde/heptane for 10 min, and the vitellin membranes were removed by hand with a tungsten needle. Embryos were then processed for immunofluorescence as described below.
Immunofluorescence
Antibodies were used at the following dilutions: chicken anti-CID (1:1,000 for western, 1:500 for embryos, 1:500 for S2 cells, 1:100 for brains and 1.2 mg ml−1 for injection), rabbit anti-CID6 (1:500 for brains, 1:2,500 for S2 cells), rabbit anti-PROD10 (1:5,000 for S2 cells), guinea-pig anti-MEI-S332 (ref. 50; 1:1,000 for S2 cells), mouse anti-POLO51 (1:50 for S2 cells), rabbit anti ZW-1015 (1:500 for S2 cells), rabbit anti-BUB1 (C. Sunkel, personal communication; 1:1,000 for S2 cells), rabbit anti-ROD16 (1:1,000 for S2 cells), mouse anti-tubulin (Sigma) and mouse anti-HP1 (ref. 52; 1:50 for S2 cells).
Blocked slides were incubated with diluted primary antibody(s) either at room temperature for 1–2 h or overnight at 4 °C, and then washed three times for 5 min in PBS-T-M before being incubated with the appropriate secondary antibody for 1 h at room temperature. We used the following secondary antibodies (Jackson ImmunoResearch): donkey anti-rabbit fluorescein isothiocyanate (FITC), donkey anti-rabbit Cy5, donkey anti-mouse FITC, donkey anti-chicken Cy3, donkey anti-mouse Cy5, and donkey anti-guinea-pig FITC. All secondary antibodies were used at 1:100 dilution unless otherwise specified. Slides were washed three times for 5 min in PBST (PBS + 0.1% Triton), mounted in Vectashield (Vector Laboratories) containing 1µg ml−1 4′,6-diamidino-2-phenylindole (DAPI) and visualized as described below. For Dp staining experiments, CID localization was scored in a minimum of 20 cells from at least 4 different brains.
Microscopy and image analysis
All images were captured using an Applied Precision Deltavision Workstation. The registration of all multifluor localizations was verified by imaging 1-µm fluorescent beads saturated with four distinct fluorochromes (Molecular Probes).We collected images as a stack of 0.1-µm increments in the z axis, and deconvolved them using the conservative algorithm with 10 iterations. Deconvolved stacked images were viewed using the Quick Projection option. Volume models were made using the model3D option of SoftWoRx, and all models presented were constructed directly from the data presented. Quantitative measurement of signal to noise ratios (S/N) in RNAi-treated cells (Fig. 7) was made by summing the total pixel intensity for each antigen in three dimensions in a box including the mitotic figure (total signal, ts), determining the signal level in discreet foci using a polygon-building algorithm as described44 (signal, s), and S/N was determined by s/ts – s. Each S/N value was plotted relative to the CID S/N for the same cell using an x–y scatter plot (Microsoft Excel). Measurements were performed for six cells of varying degrees of CID inhibition for each antigen reported; however, HP1 was omitted from this analysis owing to the diffuse nature of the Hp1 distribution. Initial adjustments were made in SoftWoRx, then the image was imported to Iris Showcase, and finally to Adobe Photoshop.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank K. Hari, J. Cordeiro and W. Sullivan for help with antibody injection, and K. Maggert, K. Sullivan and members of the Karpen lab for guidance and critical comments on the manuscript. We are grateful to B. Sullivan, B. Sullivan, S. Henikoff, C. Sunkel, T. Orr-Weaver, T. Torok, R. Karess and B. Williams for reagents, and M. Baker and The Salk Institute Sequencing Facility for analyses. M.B. is supported by an CMG training grant, and this project was funded by a grant from the NIH.
Footnotes
Supplementary Information is available on Nature Cell Biology’s website (http://cellbio.nature.com).
References
- 1.Dobie KW, Hari KL, Maggert KA, Karpen GH. Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- 2.Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl Acad. Sci. USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 5.Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 6.Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl Acad. Sci. USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 8.Howman EV, et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 10.Torok T, Harvie PD, Buratovich M, Bryant PJ. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. Genes Dev. 1997;11:213–225. doi: 10.1101/gad.11.2.213. [DOI] [PubMed] [Google Scholar]
- 11.Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 12.Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 13.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 14.Basu J, et al. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nature Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 16.Scaerou F, et al. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J. Cell Sci. 1999;112:3757–3768. doi: 10.1242/jcs.112.21.3757. [DOI] [PubMed] [Google Scholar]
- 17.Wordeman L, Steuer ER, Sheetz MP, Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J. Cell Biol. 1991;114:285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- 19.Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- 20.Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy TD, Karpen GH. Localization of centromere function in a Drosophila minichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X, Wahlstrom J, Karpen G. Molecular structure of a functional Drosophila centromere. Cell. 1997;91:1007–1019. doi: 10.1016/s0092-8674(00)80491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- 24.LeBlanc HN, Tang TT, Wu JS, Orr-Weaver TL. The mitotic centromeric protein MEI-S332 and its role in sister- chromatid cohesion. Chromosoma. 1999;108:401–411. doi: 10.1007/s004120050392. [DOI] [PubMed] [Google Scholar]
- 25.Lopez J, Karpen G, Orr-Weaver T. Sister-chromatid cohesion via MEI-S332 and kinetochore assembly are separable functions of the Drosophila centromere. Curr. Biol. 2000;10:997–1000. doi: 10.1016/s0960-9822(00)00650-3. [DOI] [PubMed] [Google Scholar]
- 26.Platero JS, Csink AK, Quintanilla A, Henikoff S. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- 28.Saffery R, et al. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 29.Ekwall K, et al. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 30.Yucel JK, et al. CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J. Cell Biol. 2000;150:1–11. doi: 10.1083/jcb.150.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Figueroa J, Saffrich R, Ansorge W, Valdivia M. Microinjection of antibodies to centromere protein CENP-A arrests cells in interphase but does not prevent mitosis. Chromosoma. 1998;107:397–405. doi: 10.1007/s004120050323. [DOI] [PubMed] [Google Scholar]
- 32.Zinkowski RP, Meyne J, Brinkley BR. The centromere–kinetochore complex: a repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams RR, et al. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- 34.Karaiskou A, Jessus C, Brassac T, Ozon R. Phosphatase 2A and polo kinase, two antagonistic regulators of cdc25 activation and MPF auto-amplification. J. Cell Sci. 1999;112:3747–3756. doi: 10.1242/jcs.112.21.3747. [DOI] [PubMed] [Google Scholar]
- 35.Pluta AF, Cooke CA, Earnshaw WC. Structure of the human centromere at metaphase. Trends Biochem Sci. 1990;15:181–185. doi: 10.1016/0968-0004(90)90158-8. [DOI] [PubMed] [Google Scholar]
- 36.Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 37.Hahnenberger KM, Carbon J, Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol. Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieder CL, Cole R. Chromatid cohesion during mitosis: lessons from meiosis. J. Cell Sci. 1999;112:2607–2613. doi: 10.1242/jcs.112.16.2607. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 40.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 41.Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 42.Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 43.Murphy TD, Karpen GH. Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- 44.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casso D, Ramirez-Weber FA, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 1999;88:229–232. doi: 10.1016/s0925-4773(99)00174-4. erratum ibid. 91, 451–454 (2000) [DOI] [PubMed] [Google Scholar]
- 46.Sharp DJ, et al. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clemens JC, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonaccorsi S, Giansanti MG, Gatti M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nature Cell Biol. 2000;2:54–56. doi: 10.1038/71378. [DOI] [PubMed] [Google Scholar]
- 49.Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- 50.Tang TTL, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llamazares S, et al. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 52.Kellum R, Raff JW, Alberts BM. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J. Cell Sci. 1995;108:1407–1418. doi: 10.1242/jcs.108.4.1407. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan BA, Karpen GH. Centromere identity in Drosophila is not determiend in vivo by replication timing. J. Cell. Biol. doi: 10.1083/jcb.200103001. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeBlanc HN, Tang TT, Wu JS, Orr-Weaver TL. The mitotic centromeric protein MEI-S332 and its role in sister-chromatid cohesion. Chromosoma. 1999;108:401–411. doi: 10.1007/s004120050392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.