Abstract
Autophagy is one of the newest and fastest emerging research areas in biomedical life sciences. Autophagosomes, large double-membrane vesicles enclosing cytoplasmic components targeted for degradation, are the hallmark of this catabolic pathway. The origin of the lipid bilayers composing these transport carriers has been the central enigma of the field since the discovery of autophagy. A series of recent studies has implicated several cellular organelles as the possible source of the autophagosomal membranes, if anything further clouding our view. In this compendium, we will discuss these apparently contradictory results and briefly emphasize the relevance of determining the lipid source used for autophagy for future translational research, for example in drug discovery programs.
Autophagy as a gatekeeper of cellular functions
The degradation of damaged and excess organelles as well as the elimination of invading pathogens is essential to maintain cell homeostasis. Autophagy is the principal catabolic pathway allowing the cell to survive the stress of these and other intrinsic and extrinsic insults. Because of its ability to rapidly eliminate unwanted structures, this conserved eukaryotic pathway plays a central role in a multitude of physiological processes including type II programmed cell death, development, cellular remodelling and differentiation [1-3]. In addition, it plays a protective role against aging, tumorigenesis, neurodegeneration and infections [4-7]. As a consequence of its crucial role in cell and organism physiology, impaired autophagy is correlated with various severe pathologies including cardiovascular and autoimmune diseases, neuro- and myo-degenerative disorders, and malignancies [8-10].
Autophagy in five stages
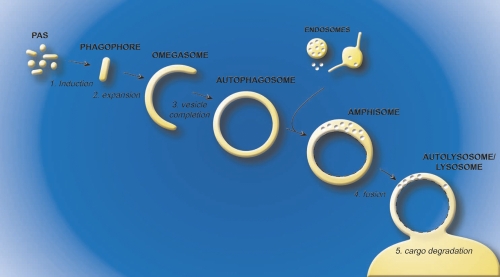
During autophagy, structures targeted for degradation are sequestered into large double-membrane vesicles called autophagosomes. Two characteristics make autophagosomes a unique type of cellular transport carrier. First, the cargo is surrounded by two lipid bilayers and second, these giant vesicles generally have an average diameter of approximately 700 nm, which can further expand to accommodate large structures such as cellular organelles and bacteria [11-13]. Autophagosome biogenesis and consumption can be divided in five discrete steps: induction, expansion, vesicle completion, fusion and cargo degradation (Figure 1) [11, 12]. The initial event upon autophagy induction is the formation of a membranous cistern called the phagophore or isolation membrane [14-16]. This compartment appears to be generated from what has been defined in yeast as the phagophore assembly site or pre-autophagosomal structure [11, 12, 17-19], a putative early autophagosomal precursor structure that is formed by the sequential association of at least a subset of the Atg (autophagy-related) proteins, proteins specifically involved in autophagy. The subsequent expansion of the phagophore through the acquisition of extra lipids permits the engulfment of the intracellular material targeted for destruction (Figure 1). The double-membrane vesicle is completed when the inner and outer bilayer fuse to form two distinct membranes, one inside the other. Then, complete autophagosomes first fuse with endosomal structures to form amphisomes, an event that appears not to occur in yeast, and then with the mammalian lysosome or the yeast and plant vacuole, allowing the degradation of the inner vesicle and its cargo by acid hydrolases residing in these lytic compartments (Figure 1). The basic metabolites generated from this catabolic processing of biological macromolecules are finally transported in the cytoplasm, where they are reused as either a source of energy or as building blocks for new proteins and lipids. To define the membranes of the autophagy pathway shown in Figure 1, we propose a working definition based on which Atg proteins characterize the different intermediate structures of autophagy (Figure 2).
Figure 1. Model for autophagosome biogenesis and cargo degradation.
The process of autophagy can be divided in five steps. The induction (1) is elicited by the formation of the phagophore (or isolation membrane), which then expands (2) around the material targeted for degradation. The omegasome probably represents a phagophore expansion intermediate. The closure of the growing phagophore (3) leads to the formation of a complete autophagosome. The autophagosome then fuses with endosomal structures to become an amphisome, an organelle specific to high eukaryotic cells, where the sequestered material starts to be degraded. Subsequently, the amphisome fuses (4) with lysosomes (vacuoles in plants and yeast) to generate autolysosomes, in the interior of which resident hydrolases break down the internal membrane of autophagosomes (dashed lines) and the cargo into basic metabolites (5). These metabolites are finally transported in the cytoplasm where they are reused as either a source of energy or building blocks for new proteins and lipids. Abbreviations: PAS, phagophore assembly site.
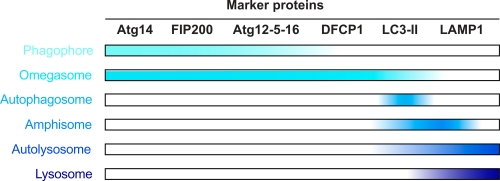
Figure 2. A working definition of the membrane carriers mediating autophagy.
A defined order for the Atg (autophagy-related) protein recruitment to the phagophore assembly site has been established in yeast [18] and recent results in mammalian cells suggest that this hierarchy is conserved [17]. Based on the available experimental data, it is possible to use a few proteins to begin to molecularly define the various autophagosomal intermediates. The distribution of the following marker proteins is shown: Atg14L is a subunit of the phosphatidylinositol-3-kinase complex involved in autophagy [60]. FIP200 associates with ULK1/2, Atg13, Atg101 into the ULK1/2 complex [61]. DFCP-1 (double FYVE-containing protein 1) could represent the marker protein that allows us to distinguish omegasomes from phagophores [30]. Atg12, Atg5 and Atg16 form a complex, e.g. Atg12-5-16, which predominantly localizes to the phagophore, but Atg16L may have an even earlier role in autophagy [41]. Lipidated LC3, e.g. LC3-II, and Lamp-1 (lysosomal-associated membrane protein 1) are well-characterized marker proteins of the late compartments of autophagy. Note that LC3-II is present in the interior of the amphisome/autophagosome and sensitive to lysosomal hydrolyases while Lamp-1 is an integral membrane protein and resistant to these hydrolyases. Abbreviations: Atg, autophagy-related; DFCP1, double FYVE-containing protein 1; LAMP1, lysosomal-associated membrane protein 1.
The autophagosomal membranes and their promiscuous origin
During the last decade, an immense effort has been invested to try to understand how autophagosomes are formed. One of the key questions in the field that still lacks a clear answer is the identity of the source of the membranes composing these large transport carriers (Figure 3) [20, 21]. One major difficulty has been that phagophores as well as autophagosomes can be considered unique organelles because they do not contain any marker proteins of other subcellular compartments [19, 22-24]. Recently, several groups have approached this question using cutting-edge microscopy techniques combined with biochemistry approaches. The results of their work are controversial because they point to different subcellular origins of the lipid bilayers composing the phagophores and/or the autophagosomes.
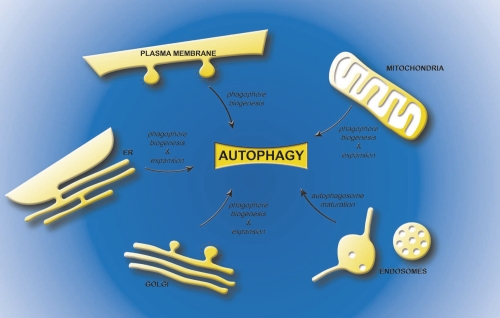
Figure 3. Possible sources of the lipids bilayers for the different intermediates of autophagy.
The plasma membrane, the endoplasmic reticulum, the mitochondria and the Golgi have been proposed to contribute to phagophore biogenesis. In addition, the endoplasmic reticulum, Golgi and mitochondria, could also provide the membranes necessary for the phagophore expansion, while endosomal compartments are necessary for the maturation of autophagosomes into amphisomes. Abbreviations: ER, endoplasmic reticulum.
The endoplasmic reticulum
Originally, the first organelle proposed as the source of autophagosomal membranes has been the endoplasmic reticulum, through morphological studies performed in the 60's [22, 25]. These observations were subsequently supported by immuno-ultrastructural observations made by Bill Dunn in 1990, in which integral membrane proteins of the rough endoplasmic reticulum were detected in both the inner and outer membranes of these vesicles [26]. More recently, using electron tomography, two groups have confirmed and emphasised these pioneering observations by revealing the existence of a physical connection between the endoplasmic reticulum and the forming autophagosomes [27, 28]. In the study of Hayashi-Nishino and co-workers, preparations from cells expressing (or not) the Atg4BC74A construct, which allows the accumulation of unsealed autophagosomes [29], were submitted to a 3-dimensional (3D) reconstruction. This analysis revealed that the endoplasmic reticulum and the growing phagophore are intimately associated, and form an endoplasmic reticulum-isolation membrane complex, suggesting that the nascent autophagosome is a subdomain of the endoplasmic reticulum. The most interesting observation was that the rough endoplasmic reticulum is connected to the outer as well as the inner membrane of the phagophore through a single point of contact, supporting the notion that lipids could be supplied via direct transfer at the sites of membrane contact. Ylä-Anttila and co-workers obtained similar findings in a different cell type (Table 1) but they have reported the existence of several points of contact between these two organelles [28]. This discrepancy could be due to either the different experimental systems used or the fact that the 3D membrane profile models are interpreted and hand drawn by a person, and variations can emerge if the same electron microscopy images are processed into a tomogram by different people.
Table 1. An overview of organelles, tissues and type of autophagy induction used in different studies on membrane origin of autophagosome.
| Organelle implicated in autophagy | Tissue | Autophagy induction | Starvation period | Reference |
|---|---|---|---|---|
| Endoplasmic reticulum | Primary cells isolated from rat livers | Fasted animals | Overnight | [22] |
| Endoplasmic reticulum | Primary cells isolated from mice renal proximal convoluted tubules and hepatic parenchymal cells | Fasted animals | Overnight | [25] |
| Endoplasmic reticulum | Primary cells from rat livers | i) Fasted animal ii) Amino acid starvation of isolated livers |
i) 18-24h | [26] |
| Endoplasmic reticulum | NIH 3T3 mouse embryonic fibroblasts expressing (or not) the Atg4BC74A construct | Amino acid, serum and glucose starvation | 1h | [27] |
| Endoplasmic reticulum | i) Mouse embryonic stem cells ii) Normal rat kidney (NRK) cells iii) Rat renal proximal tubular (NRK-52E) cells |
Amino acid starvation | 1h | [28] |
| Mitochondria | NRK cells | i) Amino acid and serum starvation ii) Serum starvation |
1h | [39] |
| Plasma membrane | Human cervical cancer (HeLa) cells | Amino acid and serum starvation | 6h | [41] |
| Plasma membrane | Human cervical cancer (Hela) cells | i) Amino acid and serum starvation ii) 1 μg/ml rapamycin |
i) 1h to 4h ii) 1h |
[42] |
| Plasma membrane/Golgi | Yeast Saccharomyces cerevisiae | Nitrogen starvation | 0.5h to 2h | [43] |
| Golgi | Yeast Saccharomyces cerevisiae | Nitrogen starvation | 4h | [44] |
| Golgi | Rat livers | Fasted animals | 24h | [48] |
| Golgi | Yeast Saccharomyces cerevisiae | Nitrogen starvation | [49] | |
| Golgi/endosomal compartments | Yeast Saccharomyces cerevisiae | Rich medium | - | [19, 46] |
| trans-Golgi network/endosomal compartments | Human embryonic kidney 293 (HEK293) cells | Nutrient starvation | 2h | [47] |
Abbreviations: HEK293, human embryonic kidney 293; NRK, normal rat kidney.
Phosphatidylinositol-3-phosphate, a phosphoinositide essential for autophagosome formation, is enriched in specific endoplasmic reticulum subdomains under autophagy-inducing conditions [30]. This finding has been corroborated by a new report revealing that Atg14L, a subunit of the phosphatidylinositol-3-kinase complex involved in autophagy, is associated with the endoplasmic reticulum surface [31]. It has also been shown that cup-shaped structures called omegasomes emerge from these phosphatidylinositol-3-phosphate-rich subdomains [30]. The fact that these compartments are positive for components of the autophagy machinery, such as Ulk1/Atg1, Atg14L, Atg16L, LC3/Atg8 and two mammalian homologues of yeast Atg18, WIPI1 and WIPI2, provides evidence that omegasomes are nascent autophagosomes [17, 30-32]. Double FYVE-containing protein 1 (DFCP1), an endoplasmic reticulum resident protein, is associated with omegasomes [30] and was found to be present in the autophagosomal membranes connected to the endoplasmic reticulum in one electron tomography study, strengthening the functional link between autophagy and the endoplasmic reticulum [27]. Are, then, phagophores and omegasomes the same structure? Currently, in the absence of a detailed molecular and mechanistic characterization, the answer to this question is just a matter of opinion and definition. We favor, however, a model where omegasomes represent expanding phagophores (Figure 1). In the absence of ATG5, phagophores can be observed by electron microscopy [16], but LC3 and DFCP1 are not recruited to these membrane cisternae [16, 17] suggesting that these two proteins are required for a step subsequent to the phagophore formation, very likely its expansion. As a result, DFCP1 and possibly LC3 could be bona fide marker proteins to differentiate the omegasome from the phagophores (Figure 2).
It has also been revealed that Rab1/Ypt1, a small GTPase mediating membrane transport in the early steps of the secretory pathway, is also involved in autophagy [33-36]. It remains to be established, however, whether Rab1/Ypt1 is regulating lipid bilayer delivery from the endoplasmic reticulum to the nascent autophagosomes or is present on membranes other than that from the endoplasmic reticulum during autophagy. All together, these data corroborate the original idea that autophagosomes emerge from the endoplasmic reticulum. Importantly, 30% of the autophagosome precursors observed in the 3D tomographic studies were not part of the endoplasmic reticulum-isolation membrane complex, raising the possibility that there may be at least one other membrane source for autophagy [27]. A careful analysis of the autophagosome membranes not in the endoplasmic reticulum-isolation membrane complex is required to determine if, for example, they contain DFCP1. It is interesting to note that yeast does not possesses a DFCP1 homologue and that, in this organism, autophagosomes do not appear to form in proximity to the endoplasmic reticulum vacuole but rather close to the vacuole [37, 38].
Mitochondria
In 2010, the group of Lippincott-Schwartz proposed the outer membrane of the mitochondria as the main source of the autophagosomal lipid bilayers [39]. Their findings were obtained from extensive fluorescent microscopy analyses in cells stably expressing CFP-LC3 and in most cases overexpressed mitochondrial outer membrane probes. Under amino acid starvation conditions (designed to induce autophagy), these two sets of marker proteins were found to co-localize on both mitochondria and autophagosomes suggesting that the membranes of this carrier and those of mitochondria are linked. The idea that there is a potential direct physical connection between these two organelles was reinforced by the visualization of growing autophagosomes in close proximity to mitochondria using live-cell imaging technology and electron microscopy. The model that the authors proposed is that there is lipid transfer between these two organelles, with the mitochondria supplying the forming autophagosomes with newly synthesized phospholipids, in particular the phosphatidylethanolamine required for LC3 lipidation [11, 12, 40]. While the co-localization of their mitochondrial marker proteins with LC3 indicates that mitochondria are involved in phagophore expansion, their experiments have not clarified whether the mitochondrial lipids are also necessary for generation of the phagophore. In the same study the authors also showed that the connection between endoplasmic reticulum and mitochondria is crucial because in its absence, starvation-induced autophagosomes are not formed [39]. This observation has led them to suggest that the endoplasmic reticulum contribution and the mitochondria could be equally important as membrane providers for autophagy. Another possible interpretation, however, is that disruption of endoplasmic reticulum homeostasis could affect the contribution of mitochondria (or an organelle connected to it) to phagophore biogenesis and/or expansion. Recently, it has been shown that Salmonella-containing autophagosomal structures are positive for DFCP1 and associated with the endoplasmic reticulum but negative for the mitochondrial marker protein used by the Lippincott-Schwartz group suggesting that mitochondrial lipids may not be involved in the biogenesis of all autophagosomes [33].
The plasma membrane
In parallel, the laboratory of David Rubinsztein has reported that lipids of the plasma membrane directly contribute to forming autophagosomes [41]. In particular, their study shows that the clathrin heavy chain, an element of the endocytic vesicle coat, binds to Atg16L1, a component of the Atg autophagy machinery. This association appears to be crucial for the generation of an early autophagosomal precursor, possibly a phagophore or an omegasome, because the formation of autophagosomes is reduced when the interaction between Atg16L1 and the clathrin heavy chain is disrupted [41]. Additional investigations from the same group have emphasized this initial discovery by revealing that maturation of the Atg16L1-positive early autophagosomal precursors requires their homotypic fusion through the action of the plasma membrane-soluble N-ethylmaleimide-sensitive-factor attachment receptor (SNARE) protein VAMP7 and its interacting partners [42]. The involvement of plasma membrane SNAREs in the early steps of autophagy has also been highlighted by work performed in yeast [43]. Additional support for the plasma membrane in autophagosome biogenesis has also indirectly provided through the discovery that components of the exocyst, a tethering complex that, in concert with SNAREs, mediates the fusion of post-Golgi vesicles with the plasma membrane, associates with nascent autophagosomes and is essential for nutrient starvation-induced autophagy [44, 45].
The Golgi
These last two studies have also raised the idea of the Golgi being a potential source for autophagosomal membranes because the exocyst complex is present at the trans-Golgi network as well as on post-Golgi vesicles [44, 45]. Also, the work on yeast revealed that plasma membranes SNAREs regulate the organization of Atg9-positive tubulovesicular membrane organelles [44], Interestingly, Atg9, a transmembrane Atg protein essential for autophagy, localizes to the trans-Golgi network, post-Golgi and endosomal compartments [19, 46, 47] and in yeast plays a central role in generating the phagophore assembly site [19]. In the 90's, the Golgi was implicated in autophagosome biogenesis because the growing extremities of the phagophores as well as complete autophagosomes were decorated with lectins that recognize glycans exclusively present in post-Golgi membranes [48]. This concept has recently been reinforced by work in yeast [49], where it has also been shown that large multi-subunit Golgi tethering complexes, lobe B of the conserved oligomeric Golgi complex and subunits of TRAPPIII, a complex homologous to the TRAPPI and TRAPPII oligomers, are essential for autophagy [34, 50].
Can all these membrane sources be reconciled in a single model?
While studies on the membrane origin of autophagosomes have started to provide some answers about the molecular mechanism and machinery, they have also created some confusion regarding the source of the lipids forming the membrane carriers mediating autophagy.
This apparent discrepancy between the conclusions reached by the different laboratories could be in part due to different experimental approaches and techniques used in the various laboratories. More importantly, the different contributions could vary depending on the tissues and conditions used to trigger autophagy (Table 1), with cells able to derive the membranes from the most suitable or expendable reservoirs.
This hypothesis could explain the reported contradictory results as different studies used different cell types, organisms and diverse conditions to induce starvation (Table 1). Thus, in a tissue with a defined function, in response to a specific stress stimulus, autophagy would be supplied with membranes from the most optimal reservoir: an organelle that could guarantee the delivery of a large amount of lipids, but ideally would not adversely affect the specific functions of the tissue. From a cursory look at the current available data, one might conclude that fasting animals utilize endoplasmic reticulum while nitrogen-starved yeast use Golgi (Table 1). However, accurate comparative studies are needed to determine whether, for example, NRK cells starved of amino acids indeed use the endoplasmic reticulum as the source for autophagosomal membranes [28] and if they turn to mitochondria when autophagy is induced by either serum, or serum and amino acid, deprivation [39]. Along the same lines, it would be important to study the various marker proteins used to implicate the different organelles in autophagy [26, 39, 41, 42] in parallel in the same experimental setup.
Another possibility that should not be discarded a priori is that autophagosomes could be mosaics of membranes derived from more than one organelle. For example, the phagophore could originate from one compartment and the additional lipid bilayers required for its expansion could be acquired from another source (Figure 3). For example, the plasma membrane and Atg9-positive membranes could contribute to the formation of the phagophore while the endoplasmic reticulum, mitochondria and/or Golgi might be necessary for its expansion. In this regard, it is important to note that the endosomal system provides the membranes required for the last steps of autophagy, i.e. amphisome and autolysosome formation (Figure 3) [51]. The advantage to the cell of having a spectrum of membrane sources to choose from is the availability of a large supply of lipids to sustain the progression of autophagy; autophagosomes are huge carriers and a multitude of them are produced upon induction of this degradative pathway. One could imagine that a single intracellular organelle could not provide enough membranes, especially during prolonged period of starvation or stress.
Finally, it still remains to be determined whether the different organelles implicated so far in autophagosome biogenesis, contribute to non-selective bulk autophagy or to selective forms of autophagy such as mitophagy (selective degradation of mitochondria), or pexophagy (selective degradation of peroxisomes). In this regard, the observation that the endoplasmic reticulum is found connected to both the inside and the outside membrane of the phagophore [26-28] could suggest that the endoplasmic reticulum found in the interior of the closing phagophore is not undergoing the general process of bulk autophagy but rather is specifically degraded through a seleletive type of autophagy known as endoplasmic reticulumphagy or reticulophagy [52, 53]. A similar explanation could be applicable to the other organelles acting in the phagophore and autophagosome biogenesis.
Is this hypothesis, implicating multiple membrane sources for autophagosomes, something reconcilable at the molecular mechanistc level? Potentially yes. Apart from the transmembrane protein Atg9, the core Atg machinery is composed of soluble proteins that transiently associate with membranes. Consequently, the cell could select the membrane source by targeting the signal and/or the molecule initiating the assembly of the Atg machinery to the organelle of choice.
Autophagosomal membranes and drug therapies
Experimental evidence indicates that autophagosome biogenesis is probably a very complex process on different levels, including its regulation in response to the different cellular and environmental cues, and in the choice of membrane sources.
Is there any therapeutic value in determining, or at the very least understanding, the origin of the autophagosomal membranes? We believe this discovery will not have a significant therapeutic value per se but it will be a fundamental step forward in the investigation of autophagosome biogenesis. However, defects in autophagy underlie various diseases, and the delivery of lipid bilayers from their source to the site of autophagosome formation is a highly regulated process that involves several of the proteins that have been shown to be essential for the progression of autophagy and these could be the target for drugs modulating autophagy [54]. It is also worth bearing in mind that the task of assigning a function to a protein is generally better facilitated when the step of a pathway in which it participates is defined.
A good example is the transmembrane protein Atg9. Because of its intrinsic association with lipid bilayers, it has been postulated that Atg9 could be involved is supplying constituents of the membranes necessary to the biogenesis of autophagosomes. This concept is supported by the finding that a change in Atg9 trafficking is directly associated with autophagy induction [19, 47], probably because of its key contribution to the phagophore assembly site/phagophore formation [19]. Not surprisingly, Atg9 transport is controlled by signaling cascades [55]. While Atg9 itself has not been to date associated to particular diseases, alterations of factors regulating the trafficking of this protein such as the phosphatidylinositol-3-kinase complex involved in autophagy [47, 56] are the direct cause of tumorigenesis [57, 58]. In this context, the tumor-suppressor Bif-1 has also been linked to Atg9 [59]. These observations support the possibility that specific illnesses could be the phenomenological manifestation of a misregulation of lipid bilayer flux during autophagy. As a result, the factors modulating these pathways would be the optimal targets for drugs aimed to restore normal membrane supply and consequently proper progression of autophagy.
Conclusions and future directions
Since the discovery of autophagy, the membrane origin of the autophagosomes has been the subject of intense debate in the field. The recent studies employing advanced technologies have confirmed and extended the pioneering ultrastructural observations and have provided some insights on the membrane origin of these unique carriers. The diverse conclusions of the recent work, however, have not given an unequivocal answer yet, but rather they have raised new questions that now need to be addressed. The work on this phenomenon has just started.
Acknowledgments
The authors wish to thank Marc van Peski for the preparation of the figures. Fulvio Reggiori is supported by the Netherlands Organization for Health Research and Development (ZonMW-VIDI-917.76.329), by the Netherlands Organization for Scientific Research (Chemical Sciences, ECHO grant-700.59.003, and Earth and Life Sciences, Open Program grant-821.02.017). Sharon Tooze is supported by Cancer Research UK.
Abbreviations
- Atg
autophagy-related
- DFCP1
double FYVE-containing protein 1
- SNARE
soluble N-ethylmaleimide-sensitive-factor attachment receptor
- 3D
3-dimensional
Competing interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/3/25
Contributor Information
Sharon A. Tooze, Email: sharon.tooze@cancer.org.uk.
Fulvio Reggiori, Email: F.Reggiori@umcutrecht.nl.
References
- 1.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–77. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–49. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–12. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–7. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 15 Nov 2011
- 12.Yoshimori T, Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr Opin Cell Biol. 2008;20:401–7. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 13.Eskelinen EL. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 14.Militello RD, Colombo MI. A membrane is born: origin of the autophagosomal compartment. Curr Mol Med. 2011;11:197–203. doi: 10.2174/156652411795243441. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 15.Eskelinen EL, Reggiorri F, Baba M, Kovács AL, Seglen PO. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy. 2011;7:935–56. doi: 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 16.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 17.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–76. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 18.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 19.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reggiori F, Tooze SA. The EmERgence of autophagosomes. Dev Cell. 2009;17:747–8. doi: 10.1016/j.devcel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–5. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 22.Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733. [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 23.Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–24. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 24.Yokota S. Formation of autophagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate. Eur J Cell Biol. 1993;61:67–80. [PubMed] [Google Scholar]
- 25.Ericsson JL. Studies on induced cellular autophagy. I. Electron microscopy of cells with in vivo labelled lysosomes. Exp Cell Res. 1969;55:95–106. doi: 10.1016/0014-4827(69)90462-5. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 26.Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1923–33. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 27.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Kerstin Radtke and Michel Desjardins 23 Dec 2009, Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 28.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Daniel Klionsky 01 Dec 2009
- 29.Fujita N, Havashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–9. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 31.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 32.Polson HE, de Lartique J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–22. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, Welch MD, Ktistakis NT, Kim PK, Klionsky DJ, Brumell JH. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 34.Lynch-Day M.A, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–6. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, Brown S, O'Kane CJ, Rubinsztein DC. alpha-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190:1023–37. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Vojo Deretic 04 Oct 2010, Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 36.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–61. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 37.Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–73. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 16Evaluated by Eric Lau and Ze'ev Ronai 01 Jun 2010, Yi Xiang and Yanzhuang Wang 01 Jul 2010, Maria Markaki and Nektarios Tavernarakis 21 Jul 2010, Roy Gross 21 Oct 2010, Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 40.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 20 Oct 2010
- 42.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–17. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Thierry Galli 09 Sep 2011, Vojo Deretic 18 Oct 2011, Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 43.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Sascha Martens 28 Jul 2011, Kerstin Radtke and Michel Desjardins 04 Oct 2011
- 44.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–69. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 45.Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, Camonis JH, Yeaman C, Levine B, White MA. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–67. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 13Evaluated by Sascha Martens 17 Feb 2011, Jianglan Liu and Wei Guo 23 Feb 2011, Raphael Valdivia 24 Feb 2011, Muriel Mari and Fulvio Reggiori 17 May 2011
- 46.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 47.Young AR, Chan EY, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto A, Masaki R, Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J Histochem Cytochem. 1990;38:573–80. doi: 10.1177/38.4.2319125. [DOI] [PubMed] [Google Scholar]
- 49.van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi Is Required for the Expansion of the Autophagosomal Phagophore in Yeast Saccharomyces cerevisiae. Mol Biol Cell. 21:2270–84. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–14. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–8. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 52.Kincaid MM, Cooper AA. ERADicate ER stress or die trying. Antioxid Redox Signal. 2007;9:2373–87. doi: 10.1089/ars.2007.1817. [DOI] [PubMed] [Google Scholar]
- 53.Kondratyev M, Avezov E, Shenkman M, Groisman B, Lederkremer GZ. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp Cell Res. 2007;313:3395–407. doi: 10.1016/j.yexcr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 55.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2009;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/S1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 57.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 58.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 11Evaluated by Harald Stenmark 22 Oct 2007, Daniel Klionsky 07 Nov 2007, Muriel Mari and Fulvio Reggiori 16 Nov 2011
- 59.Takahashi Y, Meyerkord CL, Wang HG. BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1. Autophagy. 2008;4:121–4. doi: 10.4161/auto.5265. [DOI] [PubMed] [Google Scholar]
- 60.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–9. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]