Abstract
Optical tweezers have become one of the primary weapons in the arsenal of biophysicists, and have revolutionized the new field of single-molecule biophysics. Today’s techniques allow high-resolution experiments on biological macromolecules that were mere pipe dreams only a decade ago.
On a microscopic scale, the living world is a chaotic mix of biochemistry in a complex environment whose temperature tends to lie between the freezing and boiling points of water, the main ingredient of life. Brownian motion inside living cells constantly jostles biomolecular components such as proteins, lipids, carbohydrates and nucleic acids. Conventional light microscopy, which has a resolution limited by optical diffraction, is typically unable to image the tiny nanometre-scale motions of biomolecules. The weak non-covalent bonds that hold biomolecules (and entire cells) together exert forces that are typically in the piconewton range. To study the physics and chemistry of life, and thus to begin to understand its mechanisms, we require experimental methods of measuring nanometre-length distances and piconewton-scale forces.
The advent of the laser-based gradient-force optical trap, or ‘optical tweezers’, invented by Arthur Ashkin and co-workers at Bell labs nearly 25 years ago1, provided scientists with a tool that is ideally suited for biophysical studies at the molecular level. By selecting a wavelength in the near-infrared region of 800–1,100 nm, where light is poorly absorbed by most living matter2, optical tweezers can be used to grasp, capture and manipulate micrometre-scale objects non-invasively and with exquisite precision. The loads exerted by optical traps fall conveniently into the piconewton range, and are therefore perfectly suited to studying the forces between and within biomolecules. Combined with light sensors that can monitor the displacements of trapped objects accurately down to the subnanometre level3, optical tweezers have revolutionized the nascent field of single-molecule biophysics, making it now possible to study the processes of life at the level of individual molecules.
An optical trap exploits the forces derived from the radiation pressure produced by a high-power laser. Tightly focusing a laser beam to a diffraction-limited spot creates a steep three-dimensional light gradient in the immediate vicinity of the focus, as shown in Fig. 1a. A tiny dielectric object such as a living cell or micrometre-sized glass or plastic sphere located near the focus will experience a force that is proportional to the gradient of the light. This force tends to draw the object towards the focus, whereas the better-known scattering force tends to push the object along the direction of the laser beam. Using a microscope objective lens of high numerical aperture (typically 1.0–1.4) makes it possible to generate an optical gradient that is sufficiently steep to overcome the otherwise destabilizing effects of the scattering force, thus resulting in a stable trapping zone.
Figure 1.
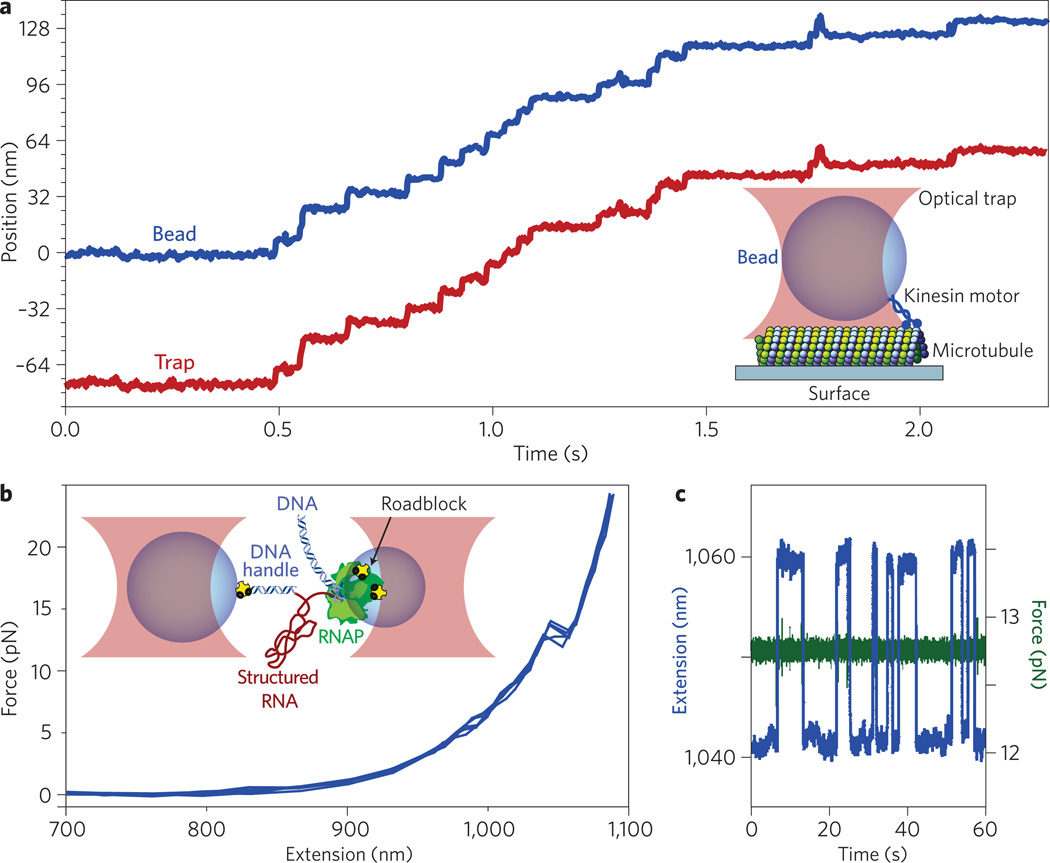
Biological applications of optical tweezers to kinesin motor stepping and RNA folding. a, Record of motion for a single kinesin motor under force-clamped conditions, displaying discrete, 8 nm steps (blue trace) as it walks along a microtubule (inset, not to scale). The trap position is servoed under computer control to maintain a fixed distance behind the bead, thereby imposing a load of a few piconewtons in a direction that hinders movement (red trace). b,c, Unfolding of a structured RNA molecule (red, inset, not to scale) using a dual-beam optical trap arrangement (pink), producing out-of-equilibrium transitions (‘rips’) as the structure unfolds under a force ramp (b) or reversible, thermally driven fluctuations in extension when clamped near equilibrium (c). To exert forces on the ends of the RNA molecule, it is hybridized at one end to a DNA ‘handle’ (blue), which is chemically (yellow) linked to the left bead (blue). The RNA emerges at its other end as a transcript from RNAP (green), which is chemically attached to the right bead (blue). The RNAP molecule is transcriptionally stalled at a roadblock (yellow) placed on the DNA template (blue).
Gradient traps essentially function as microscale three-dimensional springs that constrain fluid-immersed particles such that they undergo Brownian motion within a submicrometre-scale harmonic potential. The ability of optical tweezers to grasp and manipulate intact cells makes them useful for selecting individuals from among a mixed population or measuring the motility of swimming cells such as sperm or bacteria. In addition, traps can probe the rheology of cellular components such as the lipid membranes of eukaryotic cells or their cytoskeletons. Perhaps the most revealing applications were first realized in the early 1990s, when optical tweezers were used to measure the detailed motions of motor proteins such as kinesin4 and myosin5. Motor proteins are enzymes powered by adenosine triphosphate, which drives the subcellular movements responsible for processes such as organelle transport, cell and chromosomal division, and muscle contraction. Combining optical traps with nanometry revealed the nanometre-scale steps taken by individual motor proteins. Since then, thanks to a great many technical improvements, the spatial resolution of optical tweezers has continued to improve to a point where it is now possible to detect the subnanometre motions of individual proteins at room temperature in real time. For example, it is now possible to resolve the 3.4 Å steps taken by single molecules of RNA polymerase as they move along DNA from one base pair to the next, transcribing the genetic code6.
One advantage of optical tweezers over alternative approaches such as magnetic tweezers is that, in addition to providing better control over the associated forces, they are capable of altering the trap position at high (kilohertz) frequencies by moving the beam using acousto-optic deflectors, electro-optic deflectors or mirrors. Fast switching makes it possible, for example, to synthesize multiple independent traps by time-sharing7, or to probe multiple conformational states within a biological system. One recent study determined the binding states of kinesin heads to the microtubule by rapidly reversing the trap every 14 ms, thus alternating between applying hindering and assisting loads8.
Exerting controlled forces on single biomolecules typically requires the molecule or molecular system of interest to be attached at two points. One end of the nanometre-scale molecule (or system) is chemically attached to a small micrometre-scale bead whose size is comparable to the diffraction-limited focal spot of the optical tweezers. The bead, which is captured and held in an optical trap, acts as a convenient ‘handle’ for exerting optically controlled forces. The other end of the molecule (or system) is usually attached to a fixed substrate (Fig. 1a) or a second bead held in a separate optical trap (Fig. 1b). In an assay for kinesin-driven motility, a bead carrying a single motor protein is first trapped in a buffer containing adenosine triphosphate (the fuel), then brought into contact with a stationary microtubule bound to a glass surface, as shown in Fig. 1a. The kinesin motor attaches to the microtubule filament and steps unidirectionally along it, drawing the bead from the trap and building up a force that resists further motion of the motor. Optical instruments that are capable of monitoring the displacements of the bead in real time and adjusting the trap position (or the microscope stage) accordingly can be used to enhance this simple assay using computer feedback. The optics can be controlled either to maintain the bead at some fixed location (a position clamp) or to allow movement while maintaining a fixed distance between the bead and the trap centre (a force clamp). Position clamps are particularly useful for resolving forces at high resolution, such as the stall forces of motors, whereas force clamps are useful for resolving displacements at high resolution, such as single motor steps.
Optical traps can also exert forces while single molecules move or undergo structural changes, thus assisting or impeding molecular processes by adding or removing mechanical energy. For a simple two-state system such as the highly cooperative folding and unfolding of certain nucleic acid molecules, the load applied by an optical trap has the effect of tilting the energy landscape to favour either the folded or unfolded state. A two-state system is equally likely to be found in either state when clamped at a particular threshold force (Fig. 1a). Thermal fluctuations cause the system to transition between the two states at a rate that is exponentially sensitive to the height of the energy barrier. Performing optical force clamp experiments near equilibrium or unfolding molecules away from equilibrium at controlled rates makes it possible to derive the underlying energy landscape and the kinetics of molecular folding processes, as demonstrated by studies of hairpins formed in RNA9 or single-stranded DNA10. Such analyses require that the molecular extension of the molecule represents a good reaction coordinate, in a physical chemical sense, and that this coordinate corresponds well to structural changes. In practice, this requirement can sometimes be met by examining, piecewise, the folding of subdomains. Other approaches include pulling the system along a variety of coordinates, for example with a two-dimensional force clamp11, or by combining optical trapping with fluorescence or Förster resonance energy transfer measurements to monitor the internal motions occuring within the molecule12.
A major advance in biophysics has been the development of low-drift trapping instrumentation (Fig. 1b), which is capable of resolving ångström-scale displacements6,13,14 — around 10,000 times smaller than the wavelength of the trapping light. Such ultrahigh-resolution instruments enable the study of biosystems right down to the atomic level. A number of challenges needed to be met when designing such instruments, including the significant reduction of noise contributions arising from temperature changes, laser-pointing instabilities, ambient vibrations, and light or electronic fluctuations. Another reason for the improvement in performance is that some ultrastable instruments optically ‘levitate’ the molecular system away from any surfaces that drift. Others remove drift altogether by servoing out the stage motions14. Furthermore, the spatial resolution of a single trap can be improved by correlating the motions of two beads in a dual-beam trap15. Another important innovation has been the invention of the ‘passive force clamp’16, which can maintain a constant force over short distances (~50–100 nm), without any need for active feedback, thereby increasing data bandwidths considerably and eliminating all noise contributions associated with changing the trap position.
Clever modifications to optical tweezers have emerged as the field of optical manipulation in biology has matured. Two notable examples are time-shared traps and the optical torque wrench. Time-sharing traps produce multiple traps from a single beam by scanning along a set of predetermined positions more quickly than the trapped objects can respond (that is, faster than the Brownian roll-off frequency, which is typically in the kilohertz range). Such traps can be moved, created or turned off independently. An analogous approach enables the trapping of objects of arbitrary shape. One study involved using a ‘keyhole’ trap shape comprising a conventional point trap and a line trap (made by rapidly scanning the laser beam along a single axis) to trap microtubules along their long axes for studying depolymerization phenomena17. Multiple, independent traps — indeed, almost any desired distribution of laser light in the specimen plane — may also be produced in a versatile way by wavefront synthesis using computer-addressable spatial light modulators, allowing the realization of holographic optical tweezers18. Holographic traps have not yet had a major impact in biological applications, perhaps because of their relatively low light throughput, but that situation may change in the future.
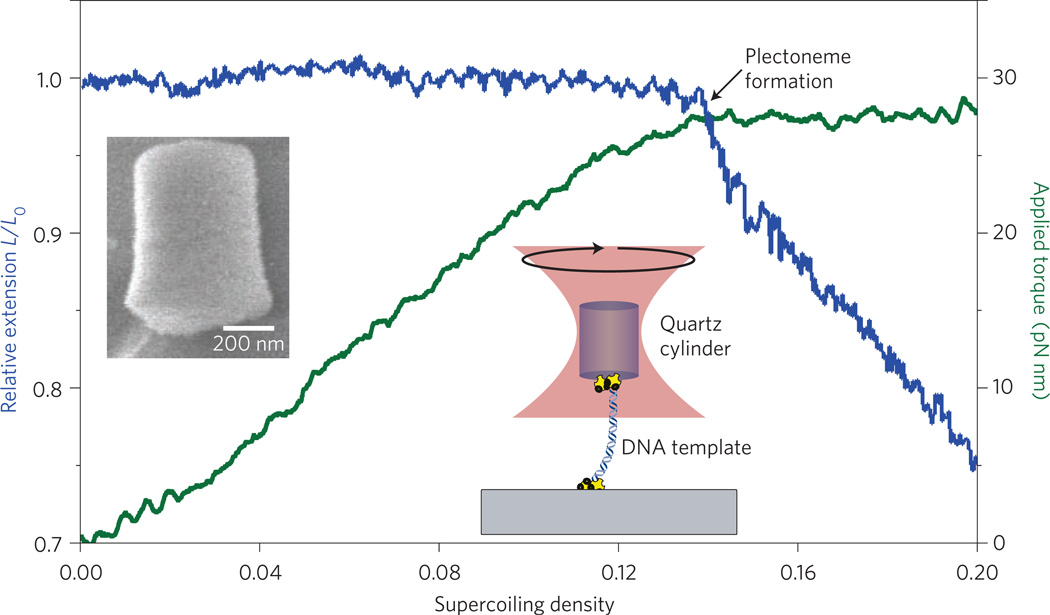
Another potentially important development is the ‘optical torque wrench’ (OTW), which is the rotational analogue of an optical force clamp19. An OTW can be used to apply a constant torque of more than 100 pN nm to a micrometre-scale birefringent object. The object may exhibit intrinsic birefringence, such as a nanofabricated cylinder made of quartz, or form birefringence, such as for an oblate spheroid made from polystyrene. The trap exerts a rotational force by using an electro-optic modulator to spin the axial orientation of the polarized light introduced to the sample. The instrument continuously monitors the difference in circular polarization between the light entering and leaving the trapped specimen. Owing to the conservation of angular momentum, this difference reports the change in angular momentum imparted to the sample, which equals the applied torque. A computer-feedback circuit is used to maintain this polarization difference at some pre-set level, thereby keeping the torque constant. The OTW offers a particularly promising way to study any biomolecules that impart torque or twist (for example, rotary biomotors, DNA topoisomerases, helicases and gyrases), or enzymes that translocate along helical paths (for example, DNA-based replication and transcription enzymes). An OTW can also facilitate studies of the nanomechanical properties of filaments subjected to twist (such as DNA, RNA, microtubules and actin filaments; Fig. 2). Work with the OTW is only just beginning.
Figure 2.
Twisting DNA with an optical torque wrench. Inset, left: scanning electron micrograph of a nanofabricated quartz cylinder. Inset, right: schematic of the experimental set-up (not to scale), in which a DNA molecule is tethered to the cylinder at one end and to a glass surface at the other. Here, the DNA is stretched by a force of 3 pN and twisted at a constant rate of 0.5 turns per second. The relative extension of the DNA (blue trace) and the applied torque (green trace) are plotted as a function of the supercoiling density, which indicates the degree of twist introduced. When the supercoiling density is around 0.14, the coiled DNA undergoes a phase transition from a twisted to a plectonemic form, which is indicated by the plateau in the applied torque and the monotonic decrease in relative extension.
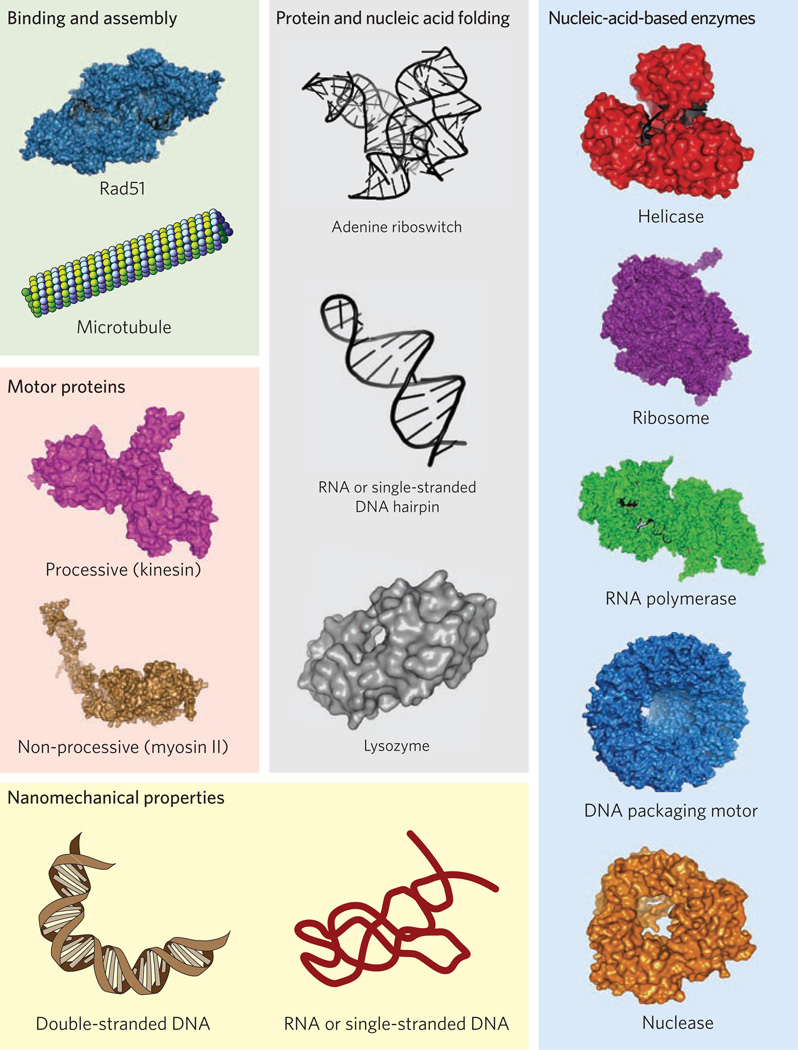
Over the past two decades, optical traps have made fundamental contributions to our detailed understanding of the mechanochemistry of molecular motors (Fig. 3). For example, experiments based on optical tweezers have been able to resolve the 8.2 nm steps taken by kinesin on microtubules4 and have demonstrated that such motors advance processively over very long distances (hundreds of steps)20. Other notable successes of optical traps include studies of the motor-like properties of the bacterial version of RNA polymerase (RNAP)21, the unique mechanisms of the eukaryotic version of RNAP (RNAP II)22, the DNA packaging motor found in the base of bacteriophages (bacterial viruses)23, the motor proteins myosin5 and dynein24, a wide variety of nucleic-acid-based enzymes that are ubiquitous in biology25,26, and the motions of ribosome, a giant macromolecular machine composed of proteins and nucleic acids27 (Fig. 3).
Figure 3.
Examples of the diverse protein and nucleic acid systems that have been studied using optical tweezers, ranging in complexity from simple hairpins, formed in RNA or double-stranded DNA, to the bacterial ribosome, a macromolecular machine comprised of over 50 protein subunits and three structured RNA molecules (not to scale). The systems have been grouped into categories (boxes).
Optical tweezers have also greatly improved our basic understanding of the nanomechanical properties of biological polymers such as nucleic acids and polypeptides28,29 (Fig. 3). Double-stranded DNA — the repository of genetic information — is known to be a relatively stiff, uncomplicated polymer whose force–extension behaviour is well-described by a worm-like chain model28 with a persistence length of ~50 nm. Similar investigations have explored the nanomechanical properties of single-stranded DNA and RNA, which are substantially different from their double-stranded counterparts. Inside cells, RNA molecules and polypeptides generally fold themselves into complex shapes. For example, some genes are controlled by the folding of short pieces of RNA formed from the initial, untranslated region of DNA located upstream of the protein-coding regions. Such RNA regulatory elements are known as riboswitches. Parts of riboswitches fold into specific forms, called aptamers, which are able to bind specific ligands (typically small metabolites) and thus monitor the health of the cell. Depending on the formation and binding state of its aptamer portion, a given riboswitch can influence the transcription, translation or splicing of downstream genes. Optical traps, in conjunction with new methods of analysis (see below), have made it possible to study riboswitch regulation in considerable detail and to reconstruct the complete energy landscapes for aptamer folding, including various structural intermediates29. Optical traps also make it possible to study protein and polypeptide folding, although the forces required to fully unfold proteins are typically larger than for nucleic acids and sometimes in excess of 50 pN, making atomic force microscopy a better option for certain types of protein-folding experiments. Successful studies using optical tweezers have already been carried out on giant proteins such as titin30, which maintains the integrity of muscle cells during contraction, and even down to tiny polypeptides such as isolated protein domains from phage T4 lysozyme31.
Optical trapping is a powerful and versatile technique that has had an enormous impact in single-molecule biophysics. When combined with other single-molecule approaches, its advantages can be leveraged to even greater effect. Traps that marry optical trapping with single-molecule fluorescence12,13 or Förster resonance energy transfer have the potential to open up entire new avenues of discovery, as demonstrated by two recent studies examining different aspects of homologous recombination in DNA32,33. Further instrument development is certain to come, which is made all the more challenging by the fact that the light entering a trap is typically 15 orders of magnitude more intense than that emerging from a single trapped fluorophore in the sample. Moreover, the intense infrared light of optical tweezers has a propensity to photobleach such dyes, often through a two-photon effect. Several experimental remedies have attempted to reduce photobleaching, including the judicious choice of dyes, the use of special anti-oxidants, spatially separating the fluorescent molecule from the trapping laser light32, and rapidly switching between the infrared trapping laser and the visible laser used to excite fluorescence13,34. Work continues on this important front.
The field of optical trapping continues to grow tremendously. As the technique becomes increasingly accessible, more and more biophysicists are using it to tackle fundamental questions in the life sciences down to the single-molecule level. We expect this trend to continue with the development of many more powerful experiments that employ state-of-the-art technologies. Optical tweezers and single-molecule biophysics are truly in their prime.
Acknowledgements
S.M.B. acknowledges support from grant GM51453 from the National Institutes of Health.
Contributor Information
Furqan M. Fazal, Department of Applied Physics, Stanford University, Stanford, California 94305, USA.
Steven M. Block, Department of Applied Physics and the Department of Biology, Stanford University, Stanford, California 94305, USA. sblock@stanford.edu
References
- 1.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Opt. Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 2.Svoboda K, Block SM. Ann. Rev. Biophys. Biomol. Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 3.Neuman KC, Block SM. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 5.Finer JT, Simmons RM, Spudich JA. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 6.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher K, Block SM. Meth. Enzymol. 1998;298:460–489. doi: 10.1016/s0076-6879(98)98040-5. [DOI] [PubMed] [Google Scholar]
- 8.Guydosh NR, Block SM. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 10.Woodside MT, et al. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Block SM, Asbury CL, Shaevitz JW, Lang MJ. Proc. Natl Acad. Sci. USA. 2003;100:2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Nature Meth. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comstock MJ, Ha T, Chemla YR. Nature Meth. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter AR, et al. Appl. Opt. 2007;46:421–427. doi: 10.1364/ao.46.000421. [DOI] [PubMed] [Google Scholar]
- 15.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Proc. Natl Acad. Sci. USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Phys. Rev. Lett. 2005;95 doi: 10.1103/PhysRevLett.95.208102. 208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerssemakers JW, et al. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 18.Grier DG. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 19.La Porta A, Wang MD. Phys. Rev. Lett. 2004;92 doi: 10.1103/PhysRevLett.92.190801. 190801. [DOI] [PubMed] [Google Scholar]
- 20.Asbury CL, Fehr AN, Block SM. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson MH, Landick R, Block SM. Mol. Cell. 2011;41:249–262. doi: 10.1016/j.molcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galburt EA, et al. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 23.Smith DE, et al. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 24.Sakakibara H, Kojima H, Sakai Y, Katayama E, Oiwa K. Nature. 1999;400:586–590. doi: 10.1038/23066. [DOI] [PubMed] [Google Scholar]
- 25.Dumont S, et al. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Biophys. J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen JD, et al. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang MD, Yin H, Landick R, Gelles J, Block SM. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 31.Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. Nature. 2010;465:637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hohng S, et al. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Mameren J, et al. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brau RR, Tarsa PB, Ferrer JM, Lee P, Lang MJ. Biophys. J. 2006;91:1069–1077. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]