Abstract
Our visual experience is initiated when the visual pigment in our retinal photoreceptors absorbs photons of light energy and initiates a cascade of intracellular events that lead to closure of cyclic-nucleotide-gated channels in the cell membrane. The resulting change in membrane potential leads in turn to reductions in the amount of neurotransmitter release from both rod and cone synaptic terminals. To measure how the light-evoked change in photoreceptor membrane potential leads to downstream activity in the retina, scientists have made electrophysiological recordings from retinal slice preparations for decades1,2. In the past these slices have been cut manually with a razor blade on retinal tissue that is attached to filter paper; in recent years another method of slicing has been developed whereby retinal tissue is embedded in low gelling temperature agar and sliced in cool solution with a vibrating microtome3,4. This preparation produces retinal slices with less surface damage and very robust light-evoked responses. Here we document how this procedure can be done under infrared light to avoid bleaching the visual pigment.
Protocol
1. Preparing Electrodes
Prepare the recording and reference electrodes by dipping them both in 1M NaCl and driving 15V between them using a 1 Hz sine wave for ~ 20 s.
Make the recording electrodes using a micropipette puller (Sutter Instruments P-97). For patch-pipettes use a borosilicate glass with an outer diameter of 1.2 mm and an inner diameter of 0.69 mm (Sutter Instruments, Cat#B120-69-10). Measure the series resistance through the tip of the electrode. Based on the size of the target cells choose an pipette tip diameter; for cell bodies ~ 20 μm in diameter use ~ 5 MΩ for cell bodies ~ 5 μm in diameter use ~ 12 MΩ.
Prepare ground/reference electrodes. Using a syringe fill a small section polyethylene tubing (Intramedic, PE205) with 1 mM NaCl that includes 3.5% Agar (Sigma; Cat# A7002) in 1M NaCl. Place this agar bridge over the Ag/AgCl reference electrode.
2. Preparing Solutions
External bath solution, Ames' media containing Bicarbonate (1 L): 1 bottle of Ames' Medium (Sigma; Cat# A1420), 1.9 g of NaHCO3, 7 mL of Penicillin-Streptomycin (Sigma; Cat# P0781) to prevent bacterial growth. Adjust osmolarity to ~ 280 mOsm.
Slicing solution, Ames' media containing the pH buffer HEPES (1 L): 1 bottle of Ames' Medium, 0.887 g of NaCl, 2.38 g of HEPES, 7 mL of Penicillin-Streptomycin. Adjust pH to ~ 7.4 using 1M NaOH and/or 1M HCl. Adjust osmolarity to ~ 280 mOsm.
Potassium-Aspartate (K-Asp) pipette internal solution (in mM): 125 K-Aspartate, 10 KCl, 10 HEPES, 5 NMG-HEDTA, 0.5 CaCl2, 1 ATP-Mg, 0.2 GTP-Mg; pH is adjusted to ~ 7.3 with NMG-OH, and osmolarity is adjusted to ~ 280 mOsm.
Agar backstop for stabilizing low gelling temperature agarose containing the retina (5% by weight), 7.5 g of Agar (Sigma; Cat# A7002) melted in 150 mL of dH2O.
3. Day of Experiment
Thaw ~ 1.5 mL of K-Asp Internal Solution and the fluorophore of your choice from freezer. Dilute the fluorophore to the appropriate concentration, which should be determined empirically.
Pour Ames' media containing NaHCO3 (~ 200 mL) into the light-tight storage container and then place into the water bath (30-32°C) and equilibrate with 5% CO2/95% O2 gas
Fill bottle with ~ 110 mL of Ames' media containing HEPES, place on ice and equilibrate with 100% O2.
Place the remaining Ames' media containing NaHCO3 in a heated reservoir on top of the Faraday cage, and equilibrate with 5% CO2/95% O2 gas. Parafilm may be used to trap the gas in the space above the solution.
Prepare the low gelling temperature agarose (3%) by adding 0.75 g (Sigma; Cat#A0701) to 25 mL of Ames' media with HEPES and heat the solution at ~ 115°C to melt the agarose. Stir until the solution clear without bubbles, and store in a water bath (~ 42°C).
Fill a recording pipette with the K-Asp internal solution and check the tip resistance.
4. Begin Experiment
Euthanize the mouse and quickly remove the eyes using curved scissors. Infrared goggles should be used for all procedures to prevent bleaching of the visual pigment.
Place the eyes on a piece of filter paper to prevent them from rolling, and using a thin double sided blade, make a slit in the cornea while holding the eye with the forceps.
As soon the eyeball is punctured, transfer they eye into a dish filled with Ames' media.
Using the slit in the cornea as an entry point, small scissors are used to cut away the remaining portions of the cornea, after which the lens and vitreous can be removed. Be careful that the retina remains attached to the eyecup, and store the eyecup in darkness at 32° C in Ames' media equilibrated with 5% CO2/95% O2.
5. Slicing
Hemisect one of the eyecups and dissociate the retina from the retinal pigment epithelium. Remove the edges of the retina to allow the tissue to lie flat.
Use a small glass Pasteur pipette with a latex bulb to fill a 35 mm Petri dish with melted agar, and drag the forceps across through the agar to test its consistency.
- When you can begin to see the agar solidify:
- Transfer the retina to the top of the agar.
- Use a twisted piece of Kimwipe to absorb the excess solution around the retina.
- Place 2-3 drops of Agar directly onto the retina and quickly place a plastic cylinder over the retina to form a wall around it. Then drip additional melted agar down the sides of the plastic cylinder while centering the retina within the agar.
- Transfer the Petri dish to an ice water bath to cool.
After ~ 45 seconds remove the tissue from the ice water bath and use a plunger to extrude the agar block out of the plastic cylinder, pushing from the side farthest from the retina.
Use the one-sided razor blade to cut out a small block of agar containing the retina and superglue the block into place against the agar backstop on the specimen disc.
Transfer the specimen disc to the vibrating microtome tray and pour ice cold Ames HEPES into the tray allowing the block with retina to be fully submersed.
Retinal slices with a thickness of ~ 200 μm can be cut at the maximum blade vibrating rate and a forward blade speed set such that it takes 4 to 5 seconds to cut through the retina on each slice.
Use a No. 11 scalpel blade to remove each usable slice one at a time
Good slices containing retina are placed into recording chambers and are weighted down with slice anchors with nylon threads crossing them. The nylon threads hold down the agar surrounding the retina, leaving the retina unobstructed for recordings.
Transfer the recording chamber to the stage of the upright microscope, close the curtain and turn on the infrared light source to view the slice on an infrared-sensitive camera.
6. Recording
Once a good retinal slice (with intact photoreceptors and the appropriate cutting angle) is identified begin perfusing the retina at a rate greater than 5 mL/min with heated Ames' media that is heated to 35 37°C and equilibrated with 5% CO2/95% O2.
Some surface damage may be apparent due to the slicing process. This usually thin layer of cells can be removed using a pipette with a broken tip, and gently sucking away the dead cell bodies. This will typically reveal live cells underneath. Identify a healthy cell body whose position is located in the strata of interest.
Fill a micropipette with the KAsp internal solution and apply positive (outward) pressure through the tip of the pipette, and lower the tip of the pipette into the recording chamber and down to the level of the cell using a micromanipulator.
Move the pipette tip such that it dimples the cell membrane, and apply gentle negative (inward) pressure to make a high resistance seal. Whole-cell mode can be achieved by applying negative pressure to the electrode to break into the cell.
Stimulating the retinal tissue using LEDs can then evoke responses to light.
Photograph the cell from which the recording was made by evoking fluorescence from the fluorophore in the pipette internal solution.
7. Representative Results
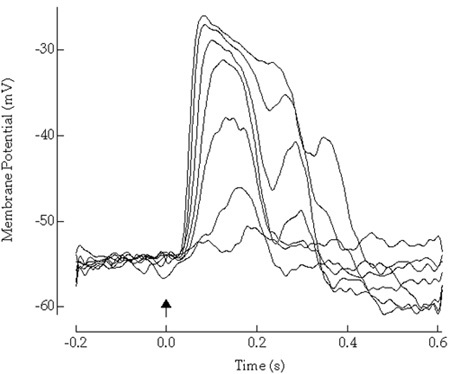 Figure 1. Here, light-evoked potentials of a dark-adapted retinal neuron to flashes of light of increasing strength are shown.
Figure 1. Here, light-evoked potentials of a dark-adapted retinal neuron to flashes of light of increasing strength are shown.
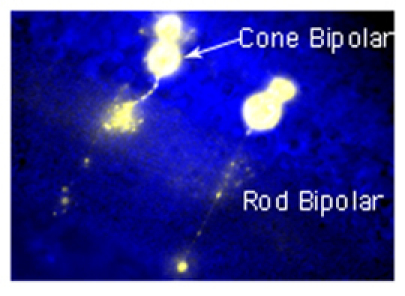 Figure 2. Fluorescence picture from a retinal slice where cells have been filled with the fluorophore, Lucifer Yellow. Evoked fluorescence shows the morphology of cells from which patch recordings were made.
Figure 2. Fluorescence picture from a retinal slice where cells have been filled with the fluorophore, Lucifer Yellow. Evoked fluorescence shows the morphology of cells from which patch recordings were made.
Discussion
Slice recording from vertebrate retinas have been made for decades1,2, and have been quite successful in providing a deeper understanding of how the retinal circuitry encodes incident light. The advantages of cutting with a vibrating microtome rather than directly with a razor blade is that the retinal slices incur less damage at the surface. With a suction pipette it is easy to remove this damage and target healthy cells just below the surface that retain their connectivity in the retinal circuit, where most of the cell types have been identified5,6, and that display robust responses to light. Thus patch clamp recordings from dark-adapted retinal slices can allow an investigator to determine the roles played by identified cell classes in neural computations.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by NIH Grant EY17606 (APS).
References
- Weblin F. Transmission along and between rods in the tiger salamander retina. J Physiol. 1978;280:449–470. doi: 10.1113/jphysiol.1978.sp012394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM. Synaptic connections among retina neurons in living slices. J Neurosci Methods. 1987;20:139–149. doi: 10.1016/0165-0270(87)90046-x. [DOI] [PubMed] [Google Scholar]
- Rieke F. Temporal contrast adaptation in salamander bipolar cell. J Neurosci. 2001;21:9445–9454. doi: 10.1523/JNEUROSCI.21-23-09445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong-Gold C, & Rieke, F Bandpass filtering at the rod to second-order cell synapse in salamander (Ambystoma tigrinum) retina. J Neurosci. 2003;23:3796–3806. doi: 10.1523/JNEUROSCI.23-09-03796.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Wassle H. Parallel processing in the mammalian retina. Nat Rev Neurosci. 2004;5:747–757. doi: 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]


