Abstract
Possible sources of variation in non-polar secondary metabolites of Portieria hornemannii, sampled from two distinct regions in the Philippines (Batanes and Visayas), resulting from different life-history stages, presence of cryptic species, and/or spatiotemporal factors, were investigated. PCA analyses demonstrated secondary metabolite variation between, as well as within, five cryptic Batanes species. Intraspecific variation was even more pronounced in the three cryptic Visayas species, which included samples from six sites. Neither species groupings, nor spatial or temporal based patterns, were observed in the PCA analysis, however, intraspecific variation in secondary metabolites was detected between life-history stages. Male gametophytes (102 metabolites detected) were strongly discriminated from the two other stages, whilst female gametophyte (202 metabolites detected) and tetrasporophyte (106 metabolites detected) samples were partially discriminated. These results suggest that life-history driven variations, and possibly other microscale factors, may influence the variation within Portieria species.
Keywords: Portieria, secondary metabolite, variation, cryptic species, life-history stages
1. Introduction
Natural products, and in particular secondary metabolites, have been the focus of study in many marine macroalgae. Seaweeds interact with their environment utilizing a rich variety of secondary metabolites [1–4]. These chemical compounds have no explicit role in the internal metabolism of the organisms [1,5,6] but serve as defense mechanisms against grazers, competitors, fouling organisms and pathogens [7–9]. The compounds are localized in specialized cells, have relatively low molecular weights (<3000 Daltons), are structurally diverse, often halogenated, and exist in low abundance (often <1% of total carbon) [1,2,6,10–12].
Secondary metabolites have drawn wide attention because of their pharmaceutical potentials, chemotaxonomic and ecological importance [13–16]. Macroalgae produce a wide range of compounds such as terpenes, phenols, fatty acids, lipopeptides, amides, alkaloids, terpenoids, lactones, pyrroles and steroids [9,15,17]. Red seaweeds belonging to the family Rhizophyllidaceae have been found to be especially rich in secondary metabolites, harboring a variety of halogenated monoterpenes. The tropical, Indo-Pacific genus Portieria (synonyms Chondrococcus and Desmia), is a prolific source of these halogenated compounds [11,12,18–24], about 80 of which have been isolated (Table S1). Among the most interesting of these compounds is halomon, a monoterpene with anti-tumor properties, isolated from samples collected in Batanes, Philippines [22,23]. However, the inconsistent availability of this compound from natural populations prevented further drug development.
Previous studies have mainly focused on discovery and pharmaceutical potentials of chemical compounds in P. hornemannii. Only a few studies have addressed inter- or intraspecific variation in secondary metabolite composition. Puglisi and Paul [25] tested the carbon/nutrient hypothesis, which postulates that the secondary metabolites produced by a certain alga are dependent on the nutrient availability. They found that variation of ochtodene concentrations in P. hornemannii cannot be attributed to nitrogen and phosphorus availability but suggested instead that light was a contributing factor. Matlock et al. [24] demonstrated strong site-to-site differences, variation within populations, and limited evidence for temporal variation in apakaochtodene levels. Some authors have emphasized the necessity to genotype the organisms in order to better understand the mechanisms that regulate the production of specific natural products [5,26–28].We found evidence for a large number of cryptic species within Portieria in the Philippines [29], but the effect of intraspecific genetic variation in relation to secondary metabolites has not been tested to date.
This study aims to understand if non-polar secondary metabolites of Portieria plants vary quantitatively or qualitatively between cryptic species and among life stage within species. We employed metabolite fingerprinting and multivariate analyses of chromatograms of extracts of samples collected in the Philippines. The large number of compounds produced by Portieria, the lack of commercial standards and the scarcity of compound identification resources (mass spectral databases) dedicated to marine secondary metabolites makes metabolite fingerprinting the most feasible analytical method to define variation between cryptic species and among life-history stages within species. In contrast to target analysis, metabolite fingerprinting does not separate individual metabolites, but instead compares spectra of whole extracts using multivariate statistics [30]. Metabolite fingerprinting is particularly useful for rapid classification of samples [31]. Our specific aims were to evaluate: (1) if there is variation of non-polar secondary metabolites between the gametophyte and tetrasporophyte life-history stages of Portieria, (2) if metabolite variation is due to the presence of phylogenetically distinct cryptic species, and (3) if patterns of variation are observed on a geographical and temporal level.
2. Results and Discussion
2.1. Cryptic Diversity
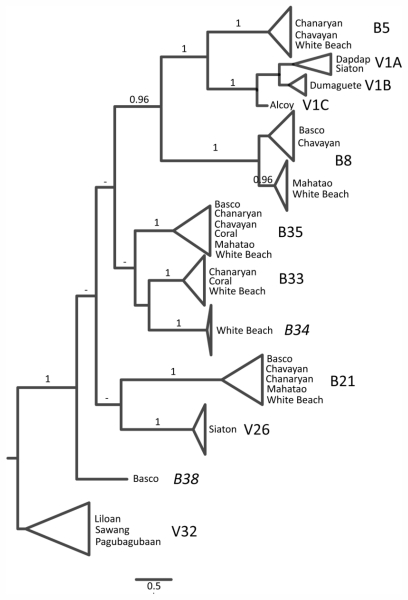
Phylogenetic analyses of 152 cox2–3 sequences revealed 12 clades of closely related sequences, preceded by long well-supported branches. Seven lineages include specimens from Batanes and five clades are restricted to the Visayas (Figure 1). These clades will further be regarded as species. Details on cryptic diversity and species delimitation are reported in Payo [29].
Figure 1.
Phylogenetic tree reconstructed using Bayesian inference based on the mitochondrial cox2–3 spacer of Portieria specimens collected from Batanes (B) and Visayas (V) Islands in the Philippines. Branch support (posterior probabilities) ≥ 0.5 are indicated at the branches. The eleven clades represent cryptic lineages, likely equivalent to species. Species B34 and B38 were not included in the chemical analysis.
2.2. Identification of Compounds in Non-Polar Extracts
Owing to the lack of standard compounds and public repositories of mass spectral data specific to marine metabolites, the identity of the compounds was investigated only by comparing the mass spectra and the Kovats retention indices to those deposited in publicly available databases (MassBase, Pherobase, Lucero Library and the free limited NIST database in the MS Search Program and AMDIS). Identifications were based on a minimum of 80% similarity of mass fragments. Using a retention index calculator [32], plant volatiles with the nearest Kovats Index (KI) to that of the compounds in the samples were retrieved. The KI used for compound identification is based on a DB5 capillary column similar to the non-polar, low bleed HP-5MS column used for GC/MS in this study. The Lucero KI based identification was verified with the mass spectra and KIs listed in Pherobase. Comparison with mass spectra of compounds found in Portieria in the literature [11,12,33] did not reveal any positive identification. Examination of mass fragments did not show any similarity of components with that of halomon’s mass spectral fragments as reported by Egorin et al. [33].
2.3. Metabolite Fingerprinting
2.3.1. Variation Between Life Stages
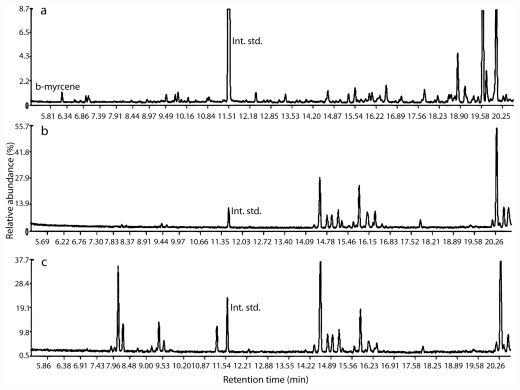
Plants used for this study belonged to a single species V1B (Figure 1) and were collected on the same day from a single site in Bantayan, Dumaguete. A total of 202 non-polar compounds were detected from gametophytes and tetrasporophyte samples by GC-MS (Table S2). Using an external standard and the databases, the first peak of each sample (RT = 6.18 min) was confirmed as β–myrcene (Figure 2a). Of the remaining compounds, only 11% (22) were identified using NIST and MassBase. Based on the NIST and MassBase identifications, five compounds were monoterpenes. Examination of the patterns of parent ions showed ten halogenated compounds containing one Cl or one Br. Two hundred compounds had a KI ranging 993–1969. Some peaks with succeeding retention times shared the same KI but had a different mass spectra composition. NIST and MassBase did not share the same identification of peaks except for β-myrcene.
Figure 2.
Portion of GC-MS total ion chromatograms of non-polar extracts of Portieria samples: (a) male gametophyte from Bantayan (Dumaguete), V1; (b) White Beach, Batanes, B21; (c) Liloan, V32. β-myrcene, a precursor of many halogenated monoterpenes, was detected only in the Bantayan specimens. Naphthalene is used as an internal standard.
KI for the last two peaks were not computed since it occurred later than the last alkane standard (C20 at 31.226 min) that was used for KI calculation. We also explored the potential of the Lucero library and Pherobase to identify compounds that Portieria might have in common with land plants. The Lucero library retrieved the nearest KIs of plant volatiles. Some of these identifications were verified at Pherobase, but mass spectra composition of the unidentified peaks was not similar to the spectra of the compound name retrieved from the database. Furthermore, we also tried to retrieve compounds with a certain KI and examined the corresponding spectra. β-Myrcene has a KI of 993.543 in this study. Pherobase listed 991 and 994 as KIs for β-myrcene in a DB5 column. Nine compounds listed in the database have a KI of 993.
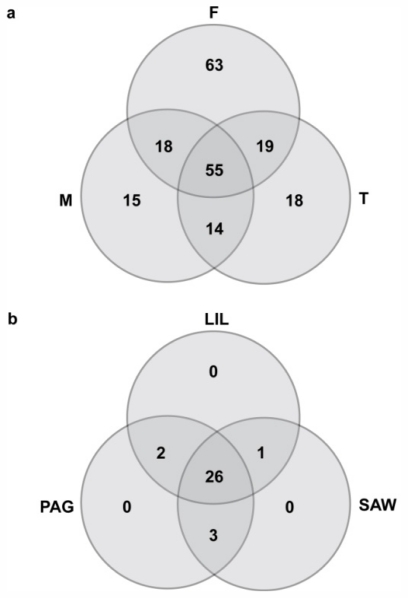
The Lucero library lists KI 993 for β-myrcene. With the large number of metabolites retrieved, this method of verification proved cumbersome especially since Pherobase currently does not allow automated mass spectral and KI comparison. Of the 202 compounds detected, 155 compounds were present in the female samples, 102 in male plants and 106 in tetrasporophytes (Figure 3a). There were 55 compounds that were shared by all life stages. Sixty-three compounds were found exclusively in female gametophyte extracts, 15 in male gametophyte extracts, and 18 in tetrasporophyte extracts. There were 14 compounds common to male and tetrasporophyte extracts. The male and female gametophyte shared 18 compounds while female and tetrasporophyte shared 19 compounds. A precursor compound of the monoterpenes, myrcene, was detected in all of the life stages but not in all replicates.
Figure 3.
Frequency distribution indicating number of shared and unique non-polar secondary metabolites: (a) within life-history stage of species V1B from Bantayan, female (F), male (M) and tetrasporophyte (T); (b) among sites of species V32 collected in Sawang (SAW), Liloan (LIL) and Pagubagubaan (PAG).
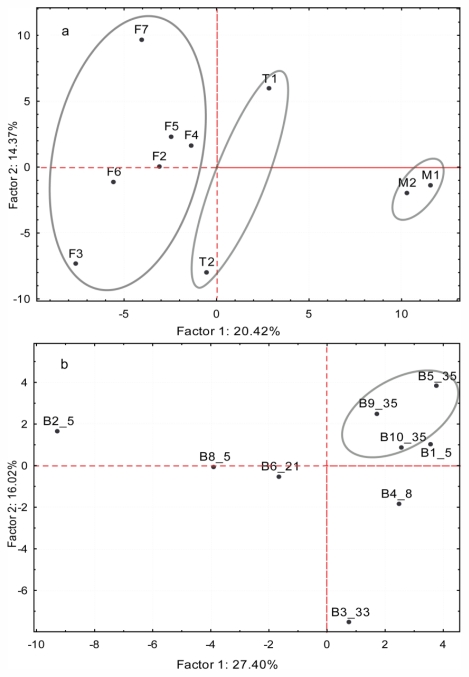
We applied a principal component analysis (PCA) on the standardized relative abundances to determine if metabolites differ among life stages (Figure 4a). The matrix contained 202 compounds (active variables) and ten samples (active cases: six female gametophytes, two male gametophytes, and two tetrasporophytes). All samples were collected on the same day from a single site, and belong to a single species. Figure 4a shows the projection of the cases on the factor plane. Factor 1 and factor 2 accounts for 20.42% and 14.37% of the variation, respectively. Factor 1 clearly discriminated the male gametophytes from the others, whilst the female gametophytes and tetrasporophytes, although separated, were more closely aligned. Factor 2 does not discriminate between life stages but points towards additional variation within life stages.
Figure 4.
Principal component analysis of GC-MS standardized relative abundance datasets which includes the compounds detected in a 67-min run of Portieria extracts. (a) Male gametophyte (M1-2) samples are clearly discriminated occurring at the positive end of the plane while female gametophyte (F2-7) and tetrasporophyte samples (T1-2) are only partially discriminated by Factor 1; (b) Batanes dataset includes five cryptic species. The clustering of B35 replicates and the scattered pattern of B5 suggest variation in component compounds exists between species but at the same time suggested that variation within species can occur.
2.3.2. Variation Between Cryptic Species
A total of 67 compounds were detected from nine samples (three technical replicates) by GC-MS (Table S3). These samples belonged to the five species B8, B5, B21, B33 and B35, as determined by DNA sequence analysis. A sample consists of pooled extract of eight individuals belonging to one species as verified by DNA analysis. β-Myrcene was not detected in any of the samples (Figure 2b). Unfortunately the three databases used (NIST, MassBase, Lucero library) did not give identical identifications of compounds. NIST identified four compounds appearing at different times as 2,2-dimethyl-3-hexanone. Comparison of mass spectra with MassBase resulted in the retrieval of straight and branched alkanes and alkenes. Compounds with retention times from 8.11 to 30.69 min had retention indices from 1065 to 1965. No KIs were computed for the rest of the compounds. Comparison of computed KIs with those in the Lucero library indicated compounds from these samples contained monoterpenes, sesquiterpenes, esters, fatty acids and alcohols. NIST, MassBase and the Lucero databases did not share the same identification of compounds. Visual examination of parent ions showed only two compounds to be halogenated containing either one Cl or one Br.
The number of compounds detected varied between the five species (Table 1) and among biological replicates. Species B35 had the highest number of compounds detected (60/67), species B33 contained 51/67 compounds detected, species B8 and B5 both had 57/67 compounds detected and species B21 had the least number of compounds (49/67). There were 21 compounds that were detected in all nine samples, while 33 compounds were present in all five species.
Table 1.
Frequency of compounds that are unique to or common to several species of Portieria found in Batanes. Values in parenthesis indicate total number of compounds found in a species, while 1 indicates presence and 0 absence of a compound.
| B21 (49) | B33 (51) | B35 (60) | B8 (57) | B5 (57) | Frequency (67) |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 1 | 0 | 1 |
| 0 | 0 | 1 | 1 | 0 | 3 |
| 0 | 0 | 1 | 1 | 1 | 3 |
| 0 | 1 | 0 | 0 | 0 | 1 |
| 0 | 1 | 0 | 1 | 0 | 2 |
| 0 | 1 | 1 | 1 | 0 | 1 |
| 0 | 1 | 1 | 1 | 1 | 7 |
| 1 | 0 | 1 | 0 | 1 | 3 |
| 1 | 0 | 1 | 1 | 0 | 2 |
| 1 | 0 | 1 | 1 | 1 | 4 |
| 1 | 1 | 0 | 0 | 1 | 2 |
| 1 | 1 | 0 | 1 | 1 | 1 |
| 1 | 1 | 1 | 0 | 1 | 4 |
| 1 | 1 | 1 | 1 | 1 | 33 |
To detect distinct groupings, we conducted a PCA based on the standardized relative abundances of the nine samples using the 67 compounds as variables (Figure 4b). The first principal axis accounted for 27.4% of the variance while the second 16.0%. Plotted scores from the PCA did not show distinct interspecific clusters. The largest variation was displayed by specimens belonging to species B5.
2.3.3. Spatial and Temporal Variation
Six sites in the Visayas (Sawang, Pagubagubaan, Dapdap, Siaton, Liloan and Bantayan (Dumaguete); n = 44) were sampled from 2007 to 2009 and investigated using Spectconnect to determine if seasonality or geography explained the variation in secondary metabolites. A total of 33 compounds were detected from these samples (Table S4), however, β-myrcene was not detected (Figure 2c). As in previous datasets, the three databases (NIST, MassBase, Lucero library) used did not give identical identifications of compounds. Comparison of mass spectra with MassBase resulted in the retrieval of eleven compounds, most likely straight and branched alkanes and alkenes. Many of the peaks detected eluted later than the standard, icosane (C20), hence were not included. Comparison of computed RIs with those in Lucero library retrieved the following compound classes: monoterpenes, sesquiterpenes, esters and fatty acids. NIST library identified two compounds, 3-hexanone, 2,2-dimethyl- and disilane. No halogenated compounds were detected.
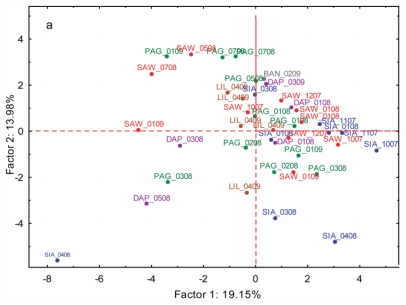
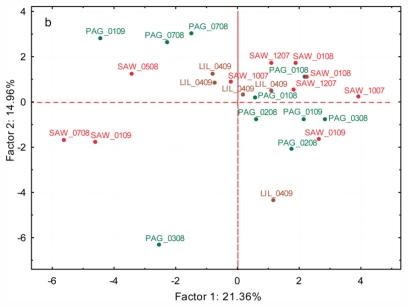
PCA was conducted using the standardized relative abundance file generated by Spectconnect (n = 44) using the 33 compounds as variables (Figure 5a). The first and second PCs accounted for 17.87% and 17.78% of the variance, respectively. No clear groupings could be deduced based on species, sampling period, or sampling site. Most of the samples clumped except for one Siaton and two Bantayan samples, which were clearly separated from the rest. A PCA excluding these three samples slightly spread the rest of the samples but likewise did not show any recognizable clusters (Figure 5a). A separate PCA analysis was conducted on samples belonging to the same species, V32, from Sawang, Pagubagubaan, and Liloan (Figure 5b). The first and second PCs accounted for 21.36% and 14.96% of the variance, respectively. Again, no clusters based on site or period of collection were detected.
Figure 5.
Principal component analysis of GC-MS standardized relative abundance datasets for detecting possible spatial and temporal patterns in: (a) the Visayas species; (b) the Visayas samples belonging to species V32. Letters indicate sampling site (DAP: Dapdap; PAG: Pagubagubaan; SAW: Sawang; SIA: Siaton) and numbers indicate month and year of sampling.
2.4. Patterns of Secondary Metabolite Distribution
Distribution of secondary metabolites in marine macroalgae exhibit macroscale and microscale patterns [34]. Macroscale patterns include global patterns within or across algal taxa, patterns within a specific habitat, and patterns correlated with biotic and abiotic factors. Microscale patterns are related to molecular and biochemical processes within an algal thallus, the spatial distribution of compounds within an algal thallus and temporal responses (short- and long-term responses). In Portieria neither macro- nor microscale patterns of secondary metabolite variation have been fully explored.
Although three decades of studies have been dedicated to the isolation of new compounds only two have investigated the variation of these, specifically ochtodene and apakaochtodene A and B [24,25]. However, a possible drawback related to these studies is the fact that individual compounds are not mutually independent but are linked via biological pathways as is the case for β-myrcene, which is the likely precursor of many monoterpenes [35], and whose presence influences the presence or absence of other halogenated monoterpene components. Hence in order to understand general or specific metabolite patterns, knowledge of the presence and absence of progenitor compounds is essential. The present study has successfully described the variation of the non-polar secondary metabolites of Portieria on a macroscale level using metabolite fingerprinting.
Reports on intraspecific variation of secondary metabolites in macroalgae have been based on variations within individuals, between individuals of a population and among geographically isolated populations [36,37]. Our results suggest substantial intraspecific variation among the life-history stages of Portieria. Male gametophytes form a clearly distinct group, while a partial discrimination was detected between female gametophytes and tetrasporophytes (Figure 4a). Furthermore, our results show that female gametophytes are chemically richer in terms of the number of secondary metabolites (Figure 3a), possibly providing protection to the carposporophyte from grazers. Many of the compounds detected appear to occur exclusively to a certain life-history stage while a relatively small group of compounds is shared among life-history stages. Similarly, male gametophytes of Asparagopsis armata were found to have lower concentration of the secondary metabolite bromoform compared to female gametophytes [37]. It has also been observed that the carposporophyte phase of A. armata was the least consumed life-history phase by the seahare, Aplysia parvula [37]. Likewise, in the brown alga Dictyota menstrualis, female gametophytes were found to produce more diterpenes compared to the other life-history stages [38].
Comparative analyses of the five cryptic species from Batanes (B5, B21, B33, B35, and B8) showed variation between, as well as within species (Figure 4b). Intraspecific variation is even more pronounced in the Visayas dataset (Figure 5). Neither species specific nor spatio-temporal patterns were evident.
In addition, a separate PCA including only specimens of species V32 (Figure 3b) did not elucidate patterns based on collection site or period. The results suggest that life history related variations combined with microscale factors have a large influence on the variation within Portieria species. Pelletreau and Targett [34] pointed out that the existence of inducible and activated defenses complicates the search for universal patterns of secondary metabolites and continues to highlight the importance of localized phenomena. We conducted a preliminary grazing experiment in outdoor flow-through aquaria to assess the influence of the herbivore (Aplysia) on the production of metabolites in Portieria. However, these efforts were beset by two factors: death of plants due to high water temperature and continuous development of microscopic Aplysia larvae on field collected plants undergoing acclimatization. Future grazing experiments will attempt to eliminate these factors. In addition more extensive sampling should confirm the patterns observed during the present study.
Finally, there was difficulty in achieving unequivocal identification of compounds in this study unveiling the need for mass spectral and KI databases dedicated solely to marine secondary metabolites. Such repositories will assist in the identification of previously reported compounds for investigations mainly focused on rapid detection and understanding of chemical patterns in marine organisms. In doing so, future studies will not just be focused in isolating and naming novel compounds as had been the trend in Portieria but will also seek to answer questions of biological or ecological relevance. Furthermore, the presence of cryptic species in Portieria highlights the need to genotype organisms ensuring that correct species identification is correlated with metabolite fingerprint, enabling more targeted approaches in future studies.
3. Experimental Section
3.1. Collection and Storage
Field samplings were performed at different periods from several sites in the Philippines (Figure S1). Plants were collected by snorkeling and SCUBA, diving depths ranging from 1–20 m. For secondary metabolite characterization of the different life-history stages of Portieria, plants were collected randomly from Bantayan near Dumaguete (Figure S1c) on December 29, 2009. To allow both chemical characterization and life-history stage verification plants were blotted dry and stored over silica.
Plants for secondary metabolites analysis between species were collected from four sites in Batanes (Basco, White Beach, Coral, and in Chavayan) (Figure S1a,b) on April 23–26, 2009. Plants were preserved in 95% ethanol in the field and stored at −23 °C afterwards. Sampling for seasonal and spatial comparison of secondary metabolites were conducted from six sites in the Central Visayas (Dapdap, Siquijor, Siquijor; Pagubagubaan, San Juan, Siquijor; Sawang, San Juan Siquijor; Bantayan, Dumaguete, Negros Oriental; Siaton, Negros Oriental; Liloan, Santander, Cebu (Figure S1a,c) from 2007 to 2009. For the first four sites, plants were collected on a monthly basis when present and when abundant enough to afford a good amount for extraction. For sites close to the Silliman Marine Laboratory, plants were immediately packed and stored at −23 °C upon arrival. When immediate freezing was not possible, plants were preserved in 95% ethanol in the field and stored at −23 °C. Frozen plants were rinsed in freshwater or in ethanol and blotted dry.
As Portieria cannot be distinguished morphologically, species identification was based on DNA sequence data (see below). For DNA characterization, thallus clippings were collected from specimens and stored in silica while in the field.
3.2. Extraction
3.2.1. Life-History Stages Comparison
Portieria has a tri-phasic isomorphic life history which includes free-living haploid male and female gametophytes, a diploid carposporophyte attached to and dependent on the female gametophyte, and a free-living diploid tetrasporophyte. Silica-dried specimens were examined under the microscope and segregated into different life-history stages (male gametophytes, female gametophytes, and tetrasporophytes). Biological replicates, each about 1 gram dry weight, were prepared for every life-history stage. Tissues were ground using mortar and pestle, dissolved in 6 mL dichloromethane (DCM) containing 50 μg·mL−1 naphthalene, and vortexed for a few seconds. The DCM extracts were allowed to stand at room temperature for 24 h in screw-cap tubes. The extracts were decanted and transferred to air tight screw-cap tubes and stored at −23 °C. The tissues were steeped in DCM for another 24 h. First and second extracts of a sample were pooled and filtered using Whatman GF/C. The combined extracts were loaded onto pre-conditioned SPE tubes containing 10 g of silica (Silicycle SiliaFlash® G60, 60–200 μm, 60 Å). Non-polar compounds were eluted using 12 mL hexane. The eluate was concentrated to 0.5 mL under N2 gas. Three 100 μL replicates were taken from this stock and used as technical replicates in the GC-MS analysis.
3.2.2. Species Comparison
The samples were extracted individually. Tissue was ground using a mortar and pestle and extracted twice in 6 mL DCM overnight. Extracts were pooled, loaded onto 2 g silica and eluted using 6 mL of hexane. Eluates of eight samples belonging to the same species, approximately weighing 10–26 g, were pooled and evaporated down to 100 μL. To assess consistency in the GC-MS analyses, three technical replicates were prepared for each sample.
HPLC solvents for extraction and chromatography (DCM and n-hexane) were sourced from Mallinckrodt or HiperSolv Chromanorm). Naphthalene (Fisher) and β-myrcene (Sigma) were used as internal and external standards, respectively.
3.2.3. Seasonal and Spatial Comparison
For seasonal analysis, plant material was ground using mortar and pestle. Twenty grams of ground material was extracted in 20 mL DCM for 24 h at room temperature. The material was extracted a second time in 20 mL DCM and the extracts were pooled. To measure the amount of green oil that was generated, one of the two pooled extracts was evaporated to dryness using nitrogen. The oil was weighed and redissolved in 20 mL DCM. The other extract was evaporated down to 20 mL. Both extracts were loaded in SPE columns containing 10 g of silica (Silicycle SiliaFlash® G60, 60–200 μm, 60 Å). Non-polar compounds were eluted from the column with 20 mL hexane and again reduced to 100 μL. A separate aliquot of ground plant tissue was used to determine dry mass.
3.3. Phylogenetic Analysis
Assignment of the sampled specimens to cryptic species was based on phylogenetic analysis of the mitochondrial cox2–3 spacer region. The phylogeny includes specimens collected from five sites (Basco, Chanaryan, Coral, Mahatao, Chavayan) in Batanes and six sites (Dapdap, Siquijor; Sawang, San Juan, Siquijor; Pagubagubaan, San Juan, Siquijor; Malo, Siaton; Bantayan, Dumaguete; Liloan, Santander) in the Visayas, Philippines. Species delineation follows the rational outlined in Payo et al. [29]. DNA extraction, PCR amplification, sequencing and sequence alignment were performed as described in Payo et al. [29].The alignment of 152 Portieria sequences was 345 bp long. Bayesian inference (BI) of phylogeny was performed using MrBayes v3.1.2 [39] under a GTR + I + Γ model as determined by jModelTest [40]. BI analyses consisted of two parallel runs of three incrementally heated chains and one cold chain each, and 3 million generations with sampling every 1000 generations. A burnin value of 750 was determined using TRACER V1.4 [41].
3.4. Metabolite Analysis, Data Processing and Multivariate Analysis
3.4.1. GC-MS Analysis
GC-MS analyses of the non-polar eluates were performed using an Agilent 6890 gas chromatograph and Agilent 6973 mass selective detector. Sample injection volume was 1 μL. Split injection with a split ratio of 20:1 was used. The carrier gas was helium with a total flow rate of 72.7 mL·min−1 and 26.20 psi column head pressure. Compounds were separated using a 30 m × 0.25 mm HP-5 MS non-polar capillary column (Hewlett-Packard, 5% phenyl methyl siloxane, 0.25 μm thickness) for a run time of 67 min. under the following oven temperature program: 50 °C initial held for 2 min, then increased at a rate of 5 °C·min−1 to 300 °C, held for 15 min. The spectrometers were run in electron-impact mode with ionization energy of 70 eV and an ion source temperature ranging 230–250 °C. The scan range was set to detect masses 50–500 amu.
3.4.2. GC-MS Data Processing
The software AMDIS [42] was used for peak identification and deconvolution of the chromatogram. This method calculates and retrieves pure (background-free) mass spectra from raw GC-MS data files based from the parameters indicated by the user. The following parameters were used for all of the analyses: medium shape requirement, medium sensitivity, and medium resolution. The ELU files generated from AMDIS were submitted for analysis using Spectconnect [43] to generate matrices of component peaks (relative abundance, retention time, integrated signal and base peak).
3.4.3. Compound Identification
In the absence of pure standards of compounds previously isolated from Portieria (except for β-myrcene), we used KI’s, freely available mass spectral databases to determine identity of component peaks, and literature. We computed the Kovat’s or retention index of each peak using the retention times of alkane standards C8–C20 and the Retention Index Calculator [32]. The retention index is derived from the interpolation (usually logarithmic), relating the adjusted retention volume (time) of the sample component to the adjusted retention volumes (times) of two standards eluted before and after the peak of a sample component (IUPAC definition). The calculated KI was automatically compared to the KI’s of compounds stored in the built-in library of the calculator. Mass spectra of component peaks were compared to compounds retrieved from MassBank [44], Pherobase [45] and the limited version of the NIST library in the MS Search Program and AMDIS. Mass spectra of components were inspected for similarity with those of the characteristic mass signals of components published in [11,12,33].
3.4.4. Data Standardization, Metabolite Fingerprinting and Multivariate Analyses
Relative abundance matrices generated from the Spectconnect analyses were used for statistical analyses. A matrix contains relative abundance values of all detected components across all samples included in the analysis. The data were standardized by obtaining a ratio between the relative abundance of a component per replicate (or sample) and the total of all components per replicate (or sample). The ratios obtained among the technical replicates were subsequently averaged. Principal component analysis was performed using Statistica 7.0 (StatSoft Inc., Tulsa, OK, USA) on each dataset to detect any pattern or groupings based from the variables which are the component compounds.
4. Conclusions
Portieria species in the Philippines are a rich source of secondary metabolites. This diversity in secondary metabolites amounts to at least 302 various compounds. The majority of which are exotic, remain undescribed and therefore are not available in natural product databases. Our study demonstrates that metabolic fingerprinting presents a practical approach to disclose intra- and interspecific patterns of secondary metabolites. Variation in secondary metabolites occurs between, as well as within, Portieria species. Preliminary results, based on a relatively small sample size, demonstrate that extensive intraspecific variation in secondary metabolites occurs between life-history stages. Female gametophytes (202 compounds) are chemically richer compared to that of the males (102) and tetrasporophytes (106). No spatio-temporal patterns were evident among the datasets. These results suggest that life-history driven variations and possibly other microscale factors may have an important influence on the variation of secondary metabolites within Portieria species. More exhaustive sampling is needed to confirm the life-stage specific metabolic fingerprints. In addition, to determine whether the intraspecific variation of the analyzed part of non-polar metabolome remains lower than the interspecific variation, it will be necessary to expand the taxon sampling across different cryptic species.
Supplementary
Table S1.
List of compounds isolated from Portieria (syn. Desmia and Chondrococcus) hornemannii, based on previous studies.
| Reference | Cpd # | Name | Mol. Formula | Description | Specimen Source |
|---|---|---|---|---|---|
| [18] | 1 | Myrcene | C10H16 | acyclic halogenated monoterpene | Amami Is., Japan |
| 2 | 7-chloro-myrcene | C10H15Cl | " | " | |
| 3 | 7-bromo-myrcene | C10H15Br | " | " | |
| 4 | (Z)-10-bromo-myrcene | C10H15Br | " | " | |
| 5 | (E)-10-bromo-myrcene | C10H15Br | " | " | |
| 6 | (Z)-10-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 7 | (E)-10-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 8 | 3-chloro-7, (Z)-10-dibromo-myrcene | C10H13Br2Cl | " | " | |
| 9 | (Z)-10-chloro-3,7-dibromo-myrcene | C10H13Br2Cl | " | " | |
| 10 | 3-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 11 | 7-bromo-10-chloro-myrcene | C10H14BrCl | " | " | |
| 12 | - | C10H15Br | cyclic halogenated monoterpene | " | |
| [19] | 1 | chondrocole A | C10H14BrClO | " | Hawaii |
| 2 | chondrocole B | C10H14BrClO | " | Hawaii | |
| [46] | 3 | - | C10H15Cl3Br2 | acyclic halogenated monoterpene | Black Point, Oahu, Hawaii |
| 4 | - | C10H16Cl3Br | " | " | |
| 5 | - | C10H15Cl2Br | " | " | |
| 6 | - | C10H16Cl2Br2 | " | Halona Blowhole Is, Oahu, Hawaii | |
| 10 | - | C10H16Cl2Br2 | acyclic halogenated monoterpene | " | |
| 11 | 6-bromo-2-chloromyrcene | C10H15ClBr | " | Black Point, Oahu, Hawaii | |
| 12 | chondrocole c | C10H14Br2O | cyclic halogenated monoterpene | " | |
| 13 | - | C10H15Cl3Br2 | acyclic halogenated monoterpene | Hawaii | |
| 14 | Z-3-bromomethylene-7-methyl-1,6-octadiene | C10H16ClBr2 | " | Halona Blowhole Is., Oahu, Hawaii | |
| 15 | - | C10H14Br2Cl2 | cyclic halogenated monoterpene | " | |
| [47] | 1 | myrcene | C10H16 | acyclic halogenated monoterpene | Kada Coast, Wakayama Pref., Japan |
| 2 | 7-chloro-myrcene | C10H15Cl | " | " | |
| 3 | 7-bromo-myrcene | C10H15Br | " | " | |
| 4 | 3,7-dichloro-myrcene | C10H14Cl2 | " | " | |
| 5 | (Z)-10-bromo-3-methoxy-α-myrcene | C11H17OBr | " | " | |
| 6 | (E)-10-bromo-3-methoxy-α-myrcene | C11H17OBr | " | " | |
| 7 | 3-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 8 | (Z)-10-bromo-1-methoxy-myrcene | C11H17OBr | " | " | |
| 9 | (E)-10-bromo-1-methoxy-myrcene | C11H17OBr | " | " | |
| [48] | (−)-3-bromomethyl-3-chloro-7-methyl-1,6-octadiene | C10H16BrCl | " | Trincomalee (Foul Point) Sri Lanka | |
| [49] | 2-(1-chloro-2-hydroxyethyl)-4,4-dimethylcyclohexa-2, 5-dienone: a precursor of 4,5-dimethylbenzo [b] furan | - | - | - | |
| [20] | 1 | (2Z,6E)-1,8-dichloro-3-chloromethyl-7-methylocta-2,6-diene | C10H15Cl3 | acyclic halogenated monoterpene | Rib Reef, Great Barrier Reef, Australia |
| 2 | (E)-1,2-dibromo-3-chloromethylene-7-methyloct-6-ene | C10H15Br2Cl | " | " | |
| 3 | (Z)-1-chloro-3-chloromethyl-7-methylocta-2,6-diene | C10H16Cl2 | " | " | |
| 4 | (Z)-1,6-dichloro-3-chloromethyl-7-methylocta-2,7-diene | C10H15Cl3 | " | " | |
| 5 | (2R*,3(8)E,4S*,6R*)-6-bromo-2-chloro-1,4-oxido-3,(8)-ochtodene | C10H14BrClO | epimeric bicyclic monoterpene | " | |
| 6 | (2S*,3(8)E,4S*,6R*)-6-bromo-2-chloro-1,4-oxido-3,(8)-ochtodene | C10H14BrClO | " | " | |
| [50] | (Z)-3-bromo-8-chloro-6-chloromethyl-2-methylocta-1,6-diene (9,6-hydroxymethyl-2-methylocta-2,8-dien-6-ol | - | - | - | |
| [21] | 1 | (2Z)-6-bromo-3-chloromethyl-1,7-dichloro-7-methylocta-2-ene | C10H16Cl3Br | acyclic halogenated monoterpene | Nelly Bay, Magnetic Island, Queensland, Australia |
| 2 | (2Z,6E)-3-chloromethyl-1-chloroocta-2,6-dien-8-al | C10H14OCl2 | " | " | |
| 3 | 3-methoxymethyl-6-methoxyl-7-methylocta-1,7(10)-dien-3-ol | C12H22 O3 | " | " | |
| 4 | (2Z,6S)-3-chloromethyl-1-methylocta-2,7(10)-dien-6-ol | C11H19 O2Cl | " | " | |
| 5 | (2Z,6S)-3-chloromethyl-6-methylocta-2,7(10)-dien-1-ol | C11H19 O2Cl | " | " | |
| [22] | 1 | halomon/6(R)-bromo-3(S)-(bromomethyl)-7-methyl-2,3,7-trichloro-1-octene | C10H15Br2Cl3 | acyclic halogenated monoterpene | Chanaryan, Batan Is., Batanes, Philippines |
| 2 | - | C10H13Cl | cyclic halogenated monoterpene | Banilad, Bacong, Negros Oriental, Philippines | |
| [23] | 2 | isohalomon | C10H15Br2Cl3 | acyclic halogenated monoterpene; isomeric with halomon | Chanaryan, Batan Is., Batanes, Philippines |
| 3 | - | C10H14BrCl3 | acyclic halogenated monoterpene | " | |
| 4 | - | C10H14Br2Cl2 | " | " | |
| 5 | - | C10H14Br3Cl | cyclic halogenated monoterpene | " | |
| 6 | - | C10H14BrCl3 | " | " | |
| 7 | - | C10H13Cl3 | " | " | |
| 8 | - | C10H15BrCl2 | acyclic halogenated monoterpene | " | |
| 9 | - | C10H16Br2Cl2 | " | " | |
| 10 | - | C10H16BrCl | " | Halona Blowhole Is, Oahu, Hawaii | |
| 11 | - | C10H15BrCl | " | " | |
| 12 | - | C10H14BrCl | cyclic halogenated monoterpene | " | |
| [8] | 1 | apakaochtodene A/6(S*)-bromo-1,4(S*),8(R*)-trichloro-2(Z)-ochtodene | C10H14Cl3Br | cyclic halogenated monoterpene | Apaka Point Beach, Guam |
| 2 | apakaochtodene B/6(S)-bromo-1,4(S), 8(R*)-trichloro-2(E)-ochtodene | C10H14Cl3Br | cyclic halogenated monoterpene | Gun Beach; Double Reef; Pago Bay; Guam | |
| [11] | 1 | myrcene/7-methyl-3-methylene-1,6-octadiene | C10 H16O | acyclic non-halogenated monoterpene | microplantlet culture, Double Reef NW Guam |
| 2 | 10E-bromomyrcene/E-3-bromomethylene-7-methyl-1,6-octadiene | C10H15Br | " | " | |
| 3 | 10Z-bromomyrcene/Z-3-bromomethylene-7-methyl-1,6-octadiene | C10H15Br | " | " | |
| 4 | 10E-bromo-3-chloro-α-myrcene/E-3-bromomethylene-6-chloro-1,7-octadiene | C10H14BrCl | " | " | |
| 5 | - | C10H14Br2 | " | " | |
| 6 | apakaochtodene B/6(S)-bromo-1,4(S), 8(R*)-trichloro-2(E)-ochtodene | C10H14BrCl3 | cyclic halogenated monoterpene | " | |
| 7 | - | C10 H16O | non-halogenated monoterpene | " | |
| 8 | bromomyrcene isomer | C10H15Br | halogenated monoterpene | " | |
| 9 | 7-chloromyrcene/2-chloro-3-methylene-7-methyl-1,6-octadiene | C10H15Cl | " | " | |
| 10 | C10H14Cl | " | " | ||
| 11 | chloromyrcene derivative | C10H16Cl | " | " | |
| 12 | bromomyrcene isomer | C10H15Br | " | " | |
| 13 | chloromyrcene derivative | C10H14Cl | " | " | |
| 14 | - | C10H18Cl2 | " | " | |
| 15 | - | C15H18 O4 | sesquiterpene | " | |
| [13] | 5 | 1,2-dibromoochtoda-3(8),5-dien-4-one | C22H12Br2O | cyclic halogenated monoterpene | Cape Zampa, Okinawa, Japan |
| 6 | 1-bromo-2-chloroochtoda-3(8),5-dien-4-one | C10H12BrClO | " | " | |
| 7 | 1,2-dichloroochtoda-3(8),5-dien-4-one | C10H12Cl2O | " | Gushichan coast, Okinawa, Japan | |
| 8 | (1Z)-1-bromoochtoda-1,3(8),5-trien-4-one | C10H11BrO | " | Cape Zampa, Okinawa, Japan | |
| 9 | (1Z)-1-chloroochtoda-1,3(8),5-trien-4-one | C10H11ClO | " | " | |
| [12] | 1 | halomon/6(R)-bromo-3(S)-(bromomethyl)-7-methyl-2,3,7-trichloro-1-octene | C10H15Br2Cl3 | acyclic halogenated monoterpene | Tolagniaro, Fort Dauphin, Madagascar |
| 2 | - | C10H13ClBr | " | " | |
| 3 | - | C10H14Cl2Br | " | " | |
| 4 | C10H15Br2Cl3 | " | " |
Table S2.
Compound peaks detected from GC-MS analysis of samples used for characterization of the life history stages of P. hornemannii. Identifications based on comparison of either Kovat’s Indices (KI) of compounds retrieved from the Retention Index Calculator [37] or mass spectral comparison of compounds retrieved from NIST or MassBase. Asterisk (*) indicates compounds with parent ions showing halogenated mass spectral patterns.
| # | Sample | Base Peak | KI | KI Based ID | Mass Spectra Based ID | |||
|---|---|---|---|---|---|---|---|---|
| RT (min) | Nearest KI | Lucero et al. [37] | NIST | MassBase | ||||
| 1 | F6_2 | 93 | 6.1863 | 994 | 993/994 | beta-myrcene/6-methyl-5-hepten-2-ol | b-myrcene | b-myrcene |
| 2 | T1_1 | 104 | 6.5874 | 1008 | 1007 | α-phellandrene | - | - |
| 3 | T1_2 | 104 | 6.5909 | 1008 | 1007 | α-phellandrene | - | - |
| 4 | M1_1 | 91 | 6.7685 | 1016 | 1017 | α-terpinene | Benzene, tert-butyl- | 4-methylacetophenone |
| 5 | F3_3 | 79 | 6.9501 | 1023 | 1024 | r-cymene | - | - |
| 6 | M1_2 | 79 | 6.9532 | 1023 | 1024 | r-cymene | - | - |
| 7 | M1_3 | 91 | 7.017 | 1026 | 1025 | p-cymene | carbonic acid | 1-phenylpropan-2-one |
| 8 | M2_2 | 91 | 7.021 | 1026 | 1025 | p-cymene | - | protopine |
| 9 | F3_2 | 91 | 7.0387 | 1027 | 1025 | p-cymene | - | N-Methyl-Npropagylbenzylamine |
| 10 | T2_1 | 57 | 8.0381 | 1063 | 1062 | y-terpinene | - | - |
| 11 | M2_2 | 132 | 8.8894 | 1089 | 1089 | p-mentha-2,4(8)diene | - | - |
| 12 | T2_1 | 117 | 9.5127 | 1110 | 1099 | linalool | - | - |
| 13 | F4_3 | 117 | 9.8874 | 1124 | 1123 | chrysanthenone | - | - |
| 14 | M1_3 | 117 | 9.9855 | 1128 | 1127 | α-campholenal | - | - |
| 15 | M2_2 | 91 | 10.2042 | 1136 | 1127 | α-campholenal | - | - |
| 16 | F2_2 | 91 | 10.2132 | 1136 | 1127 | α-campholenal | - | Dimethirimol |
| 17 | T2_1 | 119 | 10.4565 | 1145 | 1145 | camphor | - | Benzimidazole |
| 18 | M1_1 | 68 | 10.8826 | 1160 | 1158 | isobomeol | - | - |
| 19 | F7_3 | 68 | 10.8845 | 1160 | 1158 | isobomeol | - | - |
| 20 | T2_1 | 68 | 10.8866 | 1160 | 1158 | isobomeol | - | - |
| 21 | F3_3 | 68 | 10.8873 | 1160 | 1158 | isobomeol | - | - |
| 22 | F4_1 | 117 | 11.3242 | 1174 | 1171 | ethyl-benzoate | - | - |
| 23 | F4_1 | 50 | 11.5177 | 1180 | 1180 | m-cymen-8-ol | - | - |
| 24 | T2_2 | 127 | 11.5317 | 1180 | 1180 | m-cymen-8-ol | - | - |
| 25 | T2_3 | 134 | 11.7619 | 1188 | 1187 | p-cymen-8-ol | - | - |
| 26 | M2_2 | 105 | 12.0654 | 1197 | 1196 | methylchavicol | - | 3-cyanopyridine |
| 27 | F3_2 | 57 | 12.1827 | 1200 | 1203 | n-decanal | 3-Hexanone | - |
| 28 | F7_3 | 113 | 12.3674 | 1207 | 1207 | verbenone | - | - |
| 29 | M1_2 | 113 | 12.3831 | 1208 | 1207 | verbenone | - | - |
| 30 | T1_3 | 91 | 12.3874 | 1208 | 1207 | verbenone | - | - |
| 31 | M2_2 | 93 | 12.6294 | 1217 | 1217 | trans-carveol | - | - |
| 32 | M1_2 | 69 | 12.7588 | 1222 | 1219 | trans-carveol | - | - |
| 33 | M1_1 | 55 | 12.8397 | 1225 | 1229 | nerol | - | - |
| 34 | F7_3 | 67 | 13.1203 | 1236 | 1236 | thymol methyl ether | - | - |
| 35 | M1_1 | 79 | 13.1266 | 1236 | 1236 | thymol methyl ether | - | - |
| 36 | F5_2 | 93 | 13.127 | 1236 | 1236 | thymol methyl ether | - | Beta-pinene; y-eudesmol |
| 37 | M2_2 | 93 | 13.32 | 1243 | 1243 | carvone | - | - |
| 38 | T1_1 | 93 | 13.327 | 1243 | 1243 | carvone | 2-chloropropionyl chloride | - |
| 39 | T2_1 | 93 | 13.3305 | 1243 | 1243 | carvone | - | - |
| 40 *(Br) | F6_2 | 69 | 13.6916 | 1256 | 1256 | geraniol | - | - |
| 41 | M1_1 | 91 | 13.6953 | 1256 | 1256 | geraniol | - | |
| 42 | F2_1 | 69 | 13.6967 | 1256 | 1256 | geraniol | - | |
| 43 | F5_1 | 69 | 13.6975 | 1256 | 1256 | geraniol | - | - |
| 44 | M2_1 | 91 | 13.9837 | 1266 | 1263 | (E)-2-decenal | - | N-Methyl-Npropagylbenzylamine |
| 45 | F7_3 | 119 | 14.131 | 1271 | 1271 | geranial | - | - |
| 46 | F2_3 | 119 | 14.1354 | 1271 | 1271 | geranial | - | - |
| 47 | F3_2 | 69 | 14.4252 | 1281 | 1282 | a-terpinen-7-al | - | - |
| 48 | M2_3 | 69 | 14.4368 | 1281 | 1282 | a-terpinen-7-al | - | - |
| 49 | F5_1 | 91 | 14.6745 | 1289 | 1289 | p-cymen-7-ol | - | - |
| 50 | T2_1 | 91 | 14.68 | 1289 | 1289 | p-cymen-7-ol | - | - |
| 51 | F6_3 | 149 | 14.9157 | 1297 | 1297 | perilla alcohol | - | Benzoylcholine |
| 52 * (Cl) | F6_3 | 131 | 15.0641 | 1302 | 1302 | trans-ascaridole | - | 2-chloro-1-phenyl-2-butene |
| 53 | F6_1 | 166 | 15.3344 | 1313 | 1313 | 2E,4E-decadienal | - | 2-chloro-1-phenyl-2-butene |
| 54 | F6_3 | 81 | 15.5368 | 1320 | 1313 | 2E,4E-decadienal | - | - |
| 55 | M2_2 | 81 | 15.5381 | 1321 | 1313 | 2E,4E-decadienal | - | - |
| 56 | T1_2 | 81 | 15.5481 | 1321 | 1313 | 2E,4E-decadienal | - | - |
| 57 | M2_2 | 67 | 15.6269 | 1324 | 1313 | 2E,4E-decadienal | - | 1-Adamantanamine |
| 58 | M1_2 | 67 | 15.6354 | 1324 | 1313 | 2E,4E-decadienal | - | - |
| 59 | F3_1 | 57 | 15.7447 | 1329 | 1339 | d-elemene | - | - |
| 60 | F2_1 | 57 | 15.7649 | 1329 | 1339 | d-elemene | - | - |
| 61 | M2_2 | 91 | 15.8827 | 1334 | 1339 | d-elemene | - | - |
| 62 | M1_2 | 91 | 15.8885 | 1334 | 1339 | d-elemene | - | - |
| 63 | M2_2 | 139 | 16.5157 | 1357 | 1357 | eugenol | - | - |
| 64 | F6_1 | 115 | 16.5278 | 1358 | 1357 | eugenol | - | - |
| 65 | M2_3 | 166 | 16.5392 | 1358 | 1357 | eugenol | - | - |
| 66 | M2_2 | 91 | 17.02 | 1375 | 1373 | a-ylangene | - | - |
| 67 | M2_3 | 91 | 17.0254 | 1376 | 1373 | a-ylangene | - | - |
| 68 | M1_2 | 91 | 17.0311 | 1376 | 1373 | a-ylangene | - | - |
| 69 | T2_1 | 127 | 17.403 | 1389 | 1389 | isolongifolene | - | Imidazole-4-acetate |
| 70 | T2_1 | 57 | 17.7476 | 1400 | 1399 | 1,7-di-epi-a-cedrene | - | - |
| 71 | M2_2 | 79 | 17.7625 | 1401 | 1402 | methyleugenol | - | - |
| 72 | M2_3 | 127 | 17.7782 | 1402 | 1402 | methyleugenol | - | - |
| 73 | T2_2 | 67 | 18.1041 | 1415 | 1415 | cis-a-bergarnotene | - | - |
| 74 | T1_3 | 79 | 18.18 | 1418 | 1419 | β-caryophyllene | - | - |
| 75 | F6_1 | 79 | 18.1866 | 1419 | 1419 | β-caryophyllene | - | - |
| 76 | T1_1 | 79 | 18.1928 | 1419 | 1419 | β-caryophyllene | Pyridine | - |
| 77 | F5_1 | 79 | 18.1934 | 1419 | 1419 | β-caryophyllene | - | camphene |
| 78 | M1_2 | 79 | 18.1939 | 1419 | 1419 | β-caryophyllene | - | - |
| 79 * (Br) | F7_2 | 212 | 18.3193 | 1424 | 1420 | β-caroyophyllene | - | - |
| 80 | F6_2 | 210 | 18.4564 | 1429 | 1420 | β-caroyophyllene | - | - |
| 81 | M2_3 | 131 | 18.4673 | 1430 | 1420 | β-caroyophyllene | - | - |
| 82 | M1_1 | 67 | 18.5462 | 1433 | 1435 | trans-a-bergamotene | - | - |
| 83 | M1_2 | 67 | 18.5503 | 1433 | 1435 | trans-a-bergamotene | - | - |
| 84 | F2_2 | 133 | 18.6084 | 1435 | 1435 | trans-a-bergamotene | - | - |
| 85 | T2_2 | 69 | 18.6925 | 1439 | 1438 | trans-a-bergamotene | - | - |
| 86 | F3_3 | 69 | 18.6982 | 1439 | 1440 | a-guaiene | - | - |
| 87 | M2_3 | 69 | 18.7018 | 1439 | 1440 | a-guaiene | - | - |
| 88 | M1_1 | 91 | 18.7475 | 1441 | 1440 | a-guaiene | - | - |
| 89 | F4_2 | 69 | 18.8243 | 1444 | 1440 | a-guaiene | - | - |
| 90 | F7_2 | 69 | 18.8273 | 1444 | 1440 | a-guaiene | - | - |
| 91 | T1_3 | 69 | 18.8306 | 1444 | 1440 | a-guaiene | - | - |
| 92 | F2_2 | 210 | 19.1226 | 1456 | 1455 | a-humulene | - | - |
| 93 | F3_3 | 133 | 19.3339 | 1464 | 1464 | a-acoradiene | - | |
| 94 | T2_2 | 105 | 19.4843 | 1469 | 1469 | drima-7,9(11)-diene | - | - |
| 95 | T2_1 | 105 | 19.4852 | 1469 | 1469 | drima-7,9(11)-diene | - | - |
| 96 | F7_2 | 131 | 19.4915 | 1470 | 1469 | drima-7,9(11)-diene | - | - |
| 97 | F5_1 | 91 | 19.6255 | 1475 | 1474 | b-cadinene | - | - |
| 98 | F6_3 | 91 | 19.7468 | 1479 | 1479 | y-curcurnene | - | |
| 99 | F2_1 | 91 | 19.7581 | 1480 | 1480 | germacrene D | - | - |
| 100 | F4_1 | 153 | 20.0567 | 1491 | 1491 | cis-β-guaiene | - | - |
| 101 | F4_1 | 153 | 20.0567 | 1491 | 1491 | cis-β-guaiene | - | - |
| 102 | F3_2 | 71 | 20.2111 | 1496 | 1495 | a-zingiberene | - | |
| 103 | F7_2 | 135 | 20.3621 | 1502 | 1499 | a-muurolene | - | - |
| 104 | F5_1 | 91 | 20.3727 | 1503 | 1506 | d-selinene | - | - |
| 105 | T2_1 | 91 | 20.3766 | 1503 | 1506 | d-selinene | - | - |
| 106 | F2_2 | 91 | 20.3829 | 1503 | 1506 | d-selinene | -- | - |
| 107 | F6_1 | 205 | 20.6221 | 1513 | 1513 | g-cadinene | - | - |
| 108 | F4_3 | 205 | 20.6237 | 1513 | 1513 | g-cadinene | - | - |
| 109 | F7_2 | 205 | 20.6252 | 1513 | 1513 | g-cadinene | - | 5-bromo-4-methoxyoct-1-ene |
| 110 | T2_1 | 205 | 20.6278 | 1513 | 1513 | g-cadinene | p-Benzoquinone | - |
| 111 | F3_2 | 133 | 20.6781 | 1516 | 1514 | sesquicineole | - | - |
| 112 | F3_3 | 133 | 20.6803 | 1516 | 1514 | sesquicineole | - | - |
| 113 | F6_3 | 133 | 20.685 | 1516 | 1514 | sesquicineole | - | - |
| 114 | F5_1 | 69 | 20.8293 | 1522 | 1523 | d-cadinene | - | -- |
| 115 | F5_1 | 69 | 20.9154 | 1526 | 1525 | eugenyl acetate | - | - |
| 116 | F6_3 | 69 | 20.9158 | 1526 | 1525 | eugenyl acetate | - | - |
| 117 | F7_2 | 121 | 20.9204 | 1526 | 1525 | eugenyl acetate | - | - |
| 118 | T1_3 | 69 | 20.9227 | 1526 | 1525 | eugenyl acetate | - | - |
| 119 | F7_2 | 167 | 21.1103 | 1534 | 1533 | cadina-1,4-diene1 | - | - |
| 120 | T2_2 | 91 | 21.1498 | 1536 | 1538 | a-cadinene | - | - |
| 121 | F6_1 | 167 | 21.43 | 1547 | 1548 | elemol | - | - |
| 122 | F7_2 | 167 | 21.4346 | 1547 | 1548 | elemol | - | - |
| 123 | T1_3 | 69 | 21.467 | 1549 | 1549 | elemol | - | -- |
| 124 | F3_2 | 69 | 21.4709 | 1549 | 1549 | elemol | - | - |
| 125 | F7_2 | 91 | 21.598 | 1554 | 1552 | elemicin | - | - |
| 126 | F6_2 | 91 | 21.6852 | 1557 | 1557 | germacrene B | - | - |
| 127 | F5_1 | 91 | 21.6859 | 1558 | 1557 | germacrene B | - | - |
| 128 | F6_3 | 91 | 21.6869 | 1558 | 1557 | germacrene B | - | - |
| 129 | M1_1 | 91 | 21.6895 | 1558 | 1557 | germacrene B | - | - |
| 130 | F6_3 | 67 | 21.7268 | 1559 | 1557 | germacrene B | - | - |
| 131 | F7_2 | 133 | 21.8253 | 1563 | 1564 | β-calacorene | - | l-Asparagine |
| 132 | T1_1 | 91 | 21.8405 | 1564 | 1564 | β-calacorene | - | - |
| 133 | F7_2 | 167 | 22.0012 | 1570 | 1574 | prenopsan-8-ol | - | - |
| 134 | T1_3 | 213 | 22.018 | 1571 | 1574 | prenopsan-8-ol | - | - |
| 135 | F6_3 | 132 | 22.0226 | 1571 | 1574 | prenopsan-8-ol | - | - |
| 136 | F6_3 | 153 | 22.0226 | 1571 | 1574 | prenopsan-8-ol | - | - |
| 137 | M1_1 | 91 | 22.3956 | 1586 | 1585 | gleenol | - | - |
| 138 | F2_2 | 133 | 22.5362 | 1591 | 1590 | viridflorol | - | - |
| 139 | F6_3 | 71 | 22.7179 | 1598 | 1590 | viridflorol | - | - |
| 140 | F3_2 | 71 | 22.7193 | 1598 | 1590 | viridflorol | - | - |
| 141 | F2_3 | 135 | 22.722 | 1598 | 1590 | viridflorol | - | - |
| 142 | F3_3 | 57 | 22.7547 | 1600 | 1607 | b-oplopenone | - | - |
| 143 | F3_2 | 57 | 22.7603 | 1600 | 1607 | b-oplopenone | - | - |
| 144 | F3_3 | 57 | 22.761 | 1600 | 1607 | b-oplopenone | - | - |
| 145 | F6_3 | 67 | 22.8782 | 1605 | 1607 | b-oplopenone | - | - |
| 146 | T2_2 | 68 | 23.0541 | 1613 | 1612 | tetradecanal | - | - |
| 147 | F4_3 | 148 | 23.2435 | 1622 | 1623 | silphiperfol-6-en-5-one | - | - |
| 148 | F3_1 | 135 | 23.2485 | 1622 | 1623 | silphiperfol-6-en-5-one | - | - |
| 149 | F5_3 | 91 | 23.52 | 1634 | 1633 | y-eudesmol | - | - |
| 150 | F4_3 | 91 | 23.5226 | 1634 | 1633 | y-eudesmol | - | - |
| 151 | F7_2 | 68 | 23.5261 | 1634 | 1633 | y-eudesmol | - | - |
| 152 | F7_2 | 68 | 23.5261 | 1634 | 1633 | y-eudesmol | - | -- |
| 153 | T2_2 | 68 | 23.5278 | 1634 | 1633 | y-eudesmol | - | - |
| 154 | F6_1 | 167 | 23.5309 | 1634 | 1633 | y-eudesmol | - | |
| 155 | F6_2 | 67 | 23.7239 | 1643 | 1642 | cubenol | - | allylcy-anide |
| 156 | T1_3 | 91 | 23.8325 | 1647 | 1646 | a-muurolol | - | - |
| 157 | F7_2 | 132 | 23.8759 | 1649 | 1646 | a-muurolol | - | - |
| 158 | F7_2 | 132 | 23.9988 | 1654 | 1653 | a-cadinol | - | - |
| 159 | F6_2 | 149 | 24.3377 | 1669 | 1668 | bulnesol | - | - |
| 160 | F7_2 | 149 | 24.3428 | 1669 | 1668 | bulnesol | - | - |
| 161 | F6_3 | 167 | 24.6091 | 1680 | 1682 | a-bisabolol | - | - |
| 162 | F6_1 | 57 | 24.6252 | 1681 | 1682 | a-bisabolol | - | - |
| 163 | F7_2 | 69 | 24.8113 | 1689 | 1686 | 8-cedren-13-ol | - | - |
| 164 | F7_2 | 69 | 24.8207 | 1689 | 1686 | 8-cedren-13-ol | - | - |
| 165 | F2_3 | 67 | 24.9282 | 1693 | 1686 | 8-cedren-13-ol | - | - |
| 166 | F6_1 | 57 | 25.0854 | 1700 | 1686 | 8-cedren-13-ol | - | Heptadecane |
| 167 | F7_2 | 91 | 25.3006 | 1710 | 1686 | 8-cedren-13-ol | - | - |
| 168 | F3_2 | 71 | 25.3224 | 1711 | 1735 | oplopanone | - | - |
| 169 | F3_3 | 68 | 25.4814 | 1718 | 1735 | oplopanone | - | - |
| 170 | F6_3 | 67 | 25.7278 | 1730 | 1735 | oplopanone | - | - |
| 171 | F7_2 | 103 | 25.7292 | 1730 | 1735 | oplopanone | - | - |
| 172 | F2_1 | 67 | 25.7318 | 1730 | 1735 | oplopanone | - | - |
| 173 | F2_3 | 133 | 25.8305 | 1735 | 1735 | oplopanone | - | - |
| 174 | F3_1 | 117 | 25.8366 | 1735 | 1735 | oplopanone | - | - |
| 175 | F7_2 | 167 | 26.3223 | 1757 | 1761 | benzyl-benzoate | - | - |
| 176 | T2_1 | 167 | 26.5838 | 1769 | 1761 | benzyl-benzoate | - | 6-Methylmer-captopurine |
| 177 | F6_2 | 117 | 26.7333 | 1775 | 1789 | 8-a-acetoxyelemol | - | - |
| 178 | T2_2 | 103 | 26.8111 | 1779 | 1789 | 8-a-acetoxyelemol | - | - |
| 179 | F3_1 | 67 | 26.8577 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 180 | F6_1 | 67 | 26.8599 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 181 | F3_2 | 67 | 26.8671 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 182 | T2_1 | 67 | 26.8697 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 183 * (Cl2) | F6_1 | 103 | 26.8838 | 1782 | 1789 | 8-a-acetoxyelemol | - | - |
| 184 | F3_1 | 103 | 26.8852 | 1782 | 1789 | 8-a-acetoxyelemol | - | - |
| 185 * (Br) | F6_1 | 67 | 27.4286 | 1806 | 1798 | nootkatone | - | - |
| 186 * (Br) | F7_2 | 67 | 27.4342 | 1806 | 1798 | nootkatone | - | - |
| 187 | F2_1 | 67 | 27.4383 | 1807 | 1798 | nootkatone | - | - |
| 188 | F6_3 | 103 | 27.4961 | 1810 | 1798 | nootkatone | - | - |
| 189 | F6_1 | 67 | 27.5012 | 1810 | 1798 | nootkatone | - | - |
| 190 | F6_2 | 67 | 27.5676 | 1813 | 1827 | isopropyl tetradecanoate | - | - |
| 191 | F6_2 | 69 | 28.0351 | 1836 | 1827 | isopropyl tetradecanoate | - | - |
| 192 | F3_2 | 69 | 28.0387 | 1836 | 1827 | isopropyl tetradecanoate | - | - |
| 193 | F6_1 | 91 | 28.3153 | 1849 | 1867 | flourensiadiol | - | - |
| 194 | F3_2 | 149 | 28.735 | 1869 | 1867 | flourensiadiol | - | - |
| 195 * (ClBr) | F6_2 | 69 | 29.2918 | 1895 | 1878 | hexadecanol | - | - |
| 196 | F5_3 | 67 | 29.4766 | 1903 | 1927 | methyl hexadecanoate | - | - |
| 197 | T2_1 | 67 | 29.4889 | 1904 | 1927 | methyl hexadecanoate | - | - |
| 198 | F7_2 | 91 | 30.7705 | 1968 | 1999 | eicosane | - | - |
| 199 | T2_2 | 91 | 30.7753 | 1969 | 1999 | eicosane | - | - |
| 200 | F3_2 | 91 | 30.7855 | 1969 | 1999 | eicosane | - | - |
| 201 | T2_2 | 64 | 31.6475 | - | - | - | - | - |
| 202 | T2_2 | 129 | 38.661 | - | - | - | - | - |
Table S3.
Compound peaks detected from GC-MS analysis of samples for evaluation of non-polar secondary metabolite patterns of 5 cryptic species of P. hornemanni found in Batanes, Philippines. Identifications based on comparison of either Kovat’s Indices (KI) of compounds retrieved from the Retention Index Calculator [37] or mass spectral comparison of compounds retrieved from NIST or MassBase. Asterisk (*) indicates compounds with parent ions showing halogenated mass spectral patterns.
| # | Sample | Base Peak | KI | KI Based ID | Mass Spectra Based ID | |||
|---|---|---|---|---|---|---|---|---|
| RT (min) | Nearest KI | Lucero et al. [37] | NIST | Massbase | ||||
| 1 | B4B | 57 | 8.1122 | 1065 | 1062 | y-terpinene | - | - |
| 2 | B4C | 69 | 8.7386 | 1085 | 1085 | artemisia-alcohol | - | - |
| 3 | B4C | 57 | 9.1807 | 1098 | 1097 | linalool | - | - |
| 4 | B4B | 71 | 9.1841 | 1098 | 1097 | linalool | - | - |
| 5 | B6C | 57 | 9.4047 | 1106 | 1099 | linalool | 3-hexanone, 2,2-dimethyl- | 2-methylbutane |
| 6 | B4C | 69 | 11.2515 | 1172 | 1171 | ethyl-benzoate | - | - |
| 7- * (Cl) | B3A | 69 | 11.2560 | 1172 | 1171 | ethyl-benzoate | - | - |
| 8 | B4A | 57 | 14.3547 | 1279 | 1277 | trans-carvone-oxide | - | 2,2-dimethylbutane |
| 9 | B4B | 57 | 14.3593 | 1279 | 1277 | trans-carvone-oxide | - | 3-ethylhexane |
| 10 | B3B | 57 | 14.5560 | 1285 | 1286 | borneol-acetate | - | |
| 11 | B3A | 69 | 15.2678 | 1310 | 1306 | undecanal | - | trans-4-octene |
| 12 | B3A | 57 | 15.6497 | 1325 | 1314 | 2E,4E-decadienal | - | |
| 13 | B6B | 57 | 15.6518 | 1325 | 1314 | 2E,4E-decadienal | - | |
| 14 | B4A | 71 | 16.5884 | 1360 | 1357 | eugenol | - | 2-methylpentane |
| 15 | B6C | 57 | 17.8154 | 1403 | 1403 | italicene | - | 2,2-dimethylbutane |
| 16 | B3A | 57 | 17.8289 | 1404 | 1403 | italicene | - | |
| 17 | B10 | 71 | 20.306 | 1500 | 1499 | a-muurolene | - | Pentacosane |
| 18 | B3A | 57 | 20.4025 | 1504 | 1506 | d-selinene | - | tripropylamine |
| 19 | B4A | 191 | 20.715 | 1517 | 1514 | sesquicineole | - | |
| 20 | B6C | 57 | 21.0008 | 1529 | 1532 | cadina-1,4-diene | - | |
| 21- * (Br) | B3B | 57 | 21.0024 | 1529 | 1532 | cadina-1,4-diene | - | 1-octene |
| 22 | B4A | 69 | 21.5566 | 1552 | 1552 | elemicin | - | |
| 23 | B4A | 57 | 25.1819 | 1704 | 1686 | 8-cedren-13-ol | - | |
| 24 | B4B | 71 | 25.4142 | 1715 | 1735 | oplopanone | - | Hexacosane |
| 25 | B4B | 69 | 26.0498 | 1745 | 1747 | 6S-7R-bisabolone | - | 10-methyl-1-dodecanol |
| 26 | B4A | 57 | 27.4031 | 1805 | 1798 | nootkatone | - | 4-methyl-1-pentene |
| 27 | B3B | 57 | 27.9515 | 1832 | 1827 | isopropyl-tetradecanoate | - | 2,4-dimethylpentane |
| 28 | B3B | 71 | 29.9932 | 1930 | 1927 | methyl-hexadecanoate | - | docosane |
| 29 | B6B | 57 | 30.4182 | 1951 | 1927 | methyl-hexadecanoate | - | - |
| 30 | B10 | 69 | 30.5987 | 1960 | 1927 | methyl-hexadecanoate | - | - |
| 31 | B4C | 73 | 30.6949 | 1965 | 1999 | eicosane | - | - |
| 32 | B6B | 71 | 32.0267 | - | - | 3-hexanone, 2,2-dimethyl- | 2,2-dimethylbutane | |
| 33 | B3B | 71 | 32.0458 | - | - | - | 2,2-dimethylbutane | |
| 34 | B6B | 69 | 34.1452 | - | - | - | docosane | |
| 35 | B3B | 69 | 34.7128 | - | - | - | - | |
| 36 | B4C | 69 | 34.7233 | - | - | - | - | |
| 37 | B6C | 71 | 35.0713 | - | - | - | - | |
| 38 | B3C | 71 | 36.7162 | - | - | - | 3-ethylpentane | |
| 39 | B9B | 395 | 38.2697 | - | - | - | - | |
| 40 | B6C | 71 | 38.8875 | - | - | - | 3-ethylhexane | |
| 41 | B6B | 71 | 39.0147 | - | - | - | - | |
| 42 | B4C | 57 | 39.4792 | - | - | - | 3-ethylhexane | |
| 43 | B6B | 57 | 39.4822 | - | - | - | 3,3-dimethyl-1-butene | |
| 44 | B4C | 71 | 39.6266 | - | - | - | 3-ethylpentane | |
| 45 | B3C | 71 | 39.7884 | - | - | - | - | |
| 46 | B6B | 57 | 40.4997 | - | - | 3-hexanone, 2,2-dimethyl- | 2-methylbutane | |
| 47 | B6C | 57 | 40.7684 | - | - | - | 3-ethylpentane | |
| 48 | B3C | 71 | 40.7722 | - | - | - | - | |
| 49 | B4C | 57 | 43.09 | - | - | - | 3-ethylpentane | |
| 50 | B6B | 57 | 43.0982 | - | - | - | 2,2,4,6,6-pentamethylheptane | |
| 51 | B6C | 85 | 43.2865 | - | - | - | 2-methylnonane | |
| 52 | B6B | 71 | 43.3628 | - | - | - | 3-ethylpentane | |
| 53 | B3C | 71 | 43.3691 | - | - | - | - | |
| 54 | B3B | 85 | 43.7727 | - | - | - | 3,3-dimethyl-1-butene | |
| 55 | B4B | 85 | 44.3901 | - | - | - | Amitrole | |
| 56 | B6B | 71 | 44.3953 | - | - | - | 3-aminopropiononitrile | |
| 57 | B4A | 71 | 46.2638 | - | - | - | - | |
| 58 | B3C | 71 | 46.8143 | - | - | - | 2,3-dimethylbutane | |
| 59 | B6B | 71 | 46.822 | - | - | 3-hexanone, 2,2-dimethyl- | - | |
| 60 | B6C | 71 | 47.3544 | - | - | - | - | |
| 61 | B4A | 71 | 47.7467 | - | - | - | 3-ethylypentane | |
| 62 | B3A | 57 | 47.7529 | - | - | - | 3,3-dimethyl-1-butene | |
| 63 | B4A | 71 | 48.0072 | - | - | - | - | |
| 64 | B6B | 71 | 48.0159 | - | - | - | - | |
| 65 | B6C | 71 | 48.0169 | - | - | - | - | |
| 66 | B4A | 71 | 48.0321 | - | - | - | - | |
| 67 | B4B | 71 | 50.7731 | - | - | - | - | |
Table S4.
Compound peaks detected from GC-MS analysis of samples used for evaluation of temporal and spatial patterns of non-polar metabolites of three cryptic species of P. hornemanni found in the Visayas. Identifications based on comparison of either Kovat’s Indices (KI) of compounds retrieved from the Retention Index Calculator [37] or mass spectral comparison of compounds retrieved from NIST or MassBase. Asterisk (*) indicates compounds with parent ions showing halogenated mass spectral patterns.
| # | Sample | Base Peak | KI | KI Based ID | Mass Spectra Based ID | ||
|---|---|---|---|---|---|---|---|
| RT | Nearest KI | Lucero et al. [37] | Massbase | ||||
| 1 | 3A | 57 | 8.1118 | 1065 | 1062 | y-terpinene | undecane |
| 2 | 15A | 57 | 14.556 | 1285 | 1286 | borneol acetate | 1-nonene |
| 3 | 9A | 57 | 14.556 | 1285 | 1286 | borneol acetate | pentacosane |
| 4 | 29A | 57 | 20.3128 | 1500 | 1499 | a-muurolene | tricosane |
| 5 | 15B | 71 | 20.5458 | 1510 | 1509 | β-bisabolene | 3-ethylpentane |
| 6 | 15A | 57 | 25.186 | 1705 | 1686 | 8-cedren-13-ol | - |
| 7 | 34A | 57 | 25.4206 | 1716 | 1735 | oplopanone | - |
| 8 | 15B | 71 | 26.6674 | 1772 | 1761 | benzyl-benzoate | 2-methylbutane |
| 9 | 15A | 57 | 27.8534 | 1827 | 1827 | isopropyl tetradecanoate | |
| 10 | 44BB | 71 | 30.0015 | 1930 | 1927 | methyl hexadecanoate | docosane |
| 11 | 8B | 57 | 30.4069 | 1950 | 1927 | methyl hexadecanoate | |
| 12 | 15B | 71 | 31.6111 | - | - | - | - |
| 13 | 15A | 57 | 31.8326 | - | - | - | - |
| 14 | 15B | 57 | 31.8342 | - | - | - | 1,4-dimethylhexane |
| 15 | 44BA | 71 | 34.1535 | - | - | - | docosane |
| 16 | 15B | 71 | 34.2967 | - | - | - | 3-ethylpentane |
| 17 | 15A | 57 | 34.76 | - | - | - | 1-octene |
| 18 | 15A | 71 | 35.4496 | - | - | - | 2-methylbutane |
| 19 | 15B | 57 | 35.5757 | - | - | - | - |
| 20 | 15B | 57 | 35.5757 | - | - | - | 2,2-dimethylbutane |
| 21 | 44BA | 71 | 37.9407 | - | - | - | docosane |
| 22 | 15B | 410 | 38.2743 | - | - | - | - |
| 23 | 15A | 71 | 39.3484 | - | - | - | 3-ethylhexane |
| 24 | 15A | 71 | 40.0877 | - | - | - | 2,2-dimethylbutane |
| 25 | 15A | 71 | 40.0981 | - | - | - | beta-Aminopropionitrile |
| 26 | 15B | 71 | 42.5358 | - | - | - | 2-methylbutane |
| 27 | 15B | 57 | 43.9626 | - | - | - | 4-methyloctane |
| 28 | 44C | 71 | 44.6454 | - | - | - | - |
| 29 | 15A | 57 | 45.2255 | - | - | - | 2,2-dimethylbutane |
| 30 | 15A | 71 | 47.3529 | - | - | - | - |
| 31 | 15B | 57 | 47.3534 | - | - | - | - |
| 32 | 15B | 57 | 47.3581 | - | - | - | 2,4-dimethylpentane |
| 33 | 15A | 71 | 47.6237 | - | - | - | 2,4-dimethylhexane |
Map of sampling sites. (a) Map of the Philippines indicating location of Batanes and Visayas; (b) Sampling sites in Batan and Sabtang Islands in Batanes; (c) Sampling sites in Siquijor, Negros, and Cebu Islands in the Visayas).
Acknowledgments
The authors are grateful to Abner Bucol, Zacharias Generoso and Jacinta Lucañas of IEMS-Silliman University for field assistance, Batanes MENRO Francis Domingo for field and dive assistance, Frederik Lynen (Department of Organic Chemistry, UGent) and co-workers for the helpful suggestions and access to GC-MS and HPLC instruments during the exploratory period of the chemical analysis. Special gratitude goes to Elie Verleyen and Wim Vyverman (Protistology and Aquatic Ecology, UGent) for allowing us to use the GC-MS equipment (FWO Grant 1516906N) and Renaat Dasseville for the valuable technical assistance. Funding for this research was provided by the Flemish Interuniversity Council (VLIR) as part of the PhD Grant to D.A. Payo.
References
- 1.Williams D.H., Stone M.J., Hauck P.R., Rahman S.K. Why are secondary metabolites (natural products) biosynthesized? J. Nat. Prod. 1989;52:1189–1208. doi: 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- 2.Hay M.E., Fenical W. Chemical ecology and marine biodiversity: Insights and products from the sea. Oceanography. 1996;9:10–20. [Google Scholar]
- 3.Wise M.L. Monoterpene biosynthesis in marine algae. Phycologia. 2003;42:370–377. [Google Scholar]
- 4.Bhat S.V., Nagasampagi B.A., Sivakumar M. Chemistry of Natural Products. Narosa Publishing House; New Delhi, India: 2005. [Google Scholar]
- 5.Bourgaud F., Gravot A., Milesi S., Gontier E. Production of plant secondary metabolites: A historical perspective. Plant Sci. 2001;161:839–851. [Google Scholar]
- 6.Gribble G.W. The diversity of naturally produced organohalogens. Chemosphere. 2003;52:289–297. doi: 10.1016/S0045-6535(03)00207-8. [DOI] [PubMed] [Google Scholar]
- 7.Dworjanyn S.A., de Nys R., Steinberg P.D. Localisation and surface quantification of secondary metabolites in the red alga Delisea pulchra. Mar. Biol. 1999;133:727–736. [Google Scholar]
- 8.Gunatilaka A.A.L., Paul V.J., Park P.U., Puglisi M.P., Gitler A.D., Eggleston D.S., Haltiwanger R.C., Kingston D.G.I. Apakaochtodenes a and b: Two tetrahalogenated monoterpenes from the red marine alga Portieria hornemannii. J. Nat. Prod. 1999;62:1376–1378. doi: 10.1021/np9901128. [DOI] [PubMed] [Google Scholar]
- 9.Kladi M., Vagias C., Roussis V. Volatile halogenated metabolites from marine red algae. Phytochem. Rev. 2004;3:337–366. [Google Scholar]
- 10.Fenical W. Halogenation in the Rhodophyta: A review. J. Phycol. 1975;11:245–259. [Google Scholar]
- 11.Barahona L.F., Rorrer G.L. Isolation of halogenated monoterpenes from bioreactor-cultured microplantlets of the macrophytic red algae Ochtodes secundiramea and Portieria hornemannii. J. Nat. Prod. 2003;66:743–751. doi: 10.1021/np0206007. [DOI] [PubMed] [Google Scholar]
- 12.Andrianasolo E.H., France D., Cornell-Kennon S., Gerwick W.H. DNA methyl transferase inhibiting halogenated monoterpenes from the Madagascar red marine alga Portieria hornemannii. J. Nat. Prod. 2006;69:576–579. doi: 10.1021/np0503956. [DOI] [PubMed] [Google Scholar]
- 13.Kuniyoshi M., Oshiro N., Miono T., Higa T. Halogenated monoterpenes having a cyclohexadienone from the red alga Portieria hornemanni. J. Chin. Chem. Soc. 2003;50:167–170. [Google Scholar]
- 14.Smit A.J. Medicinal and pharmaceutical uses of seaweed natural products: A review. J. Appl. Phycol. 2004;16:245–262. [Google Scholar]
- 15.Kamenarska Z., Ivanova A., Stancheva R., Stoyneva M., Stefanov K., Dimitrova-Konaklieva S., Popov S. Volatile compounds from some Black Sea red algae and their chemotaxonomic application. Bot. Mar. 2006;49:47–56. [Google Scholar]
- 16.Blunt J.W., Copp B.R., Hu W.P., Munro M.H.G., Northcote P.T., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2007;24:31–86. doi: 10.1039/b603047p. [DOI] [PubMed] [Google Scholar]
- 17.Bhadury P., Wright P.C. Exploitation of marine algae: Biogenic compounds for potential antifouling applications. Planta. 2004;219:561–578. doi: 10.1007/s00425-004-1307-5. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa N., Naya Y., Enomoto S. New halogenated monoterpenes from Desmia (Chondrococcus) hornemanni. Chem. Lett. 1974;3:1333–1336. [Google Scholar]
- 19.Burreson B.J., Woolard F.X., Moore R.E. Chondrocole a and b, two halogenated dimethylhexahydrobenzofurans from the red alga Chondrococcus hornemannii (Mertens) Schmitz. Tetrahedron Lett. 1975;16:2155–2158. [Google Scholar]
- 20.Coll J.C., Wright A.D. Tropical marine algae.1. New halogenated monoterpenes from Chondrococcus hornemannii (Rhodophyta, Gigartinales, Rhizophyllidaceae) Aust. J. Chem. 1987;40:1893–1900. [Google Scholar]
- 21.Wright A.D., Konig G.M., Sticher O., de Nys R. Five new monoterpenes from the marine red alga Portieria hornemannii. Tetrahedron. 1991;47:5717–5724. [Google Scholar]
- 22.Fuller R.W., Cardellina J.H., Kato Y., Brinen L.S., Clardy J., Snader K.M., Boyd M.R. A pentahalogenated monoterpene from the red alga Portieria hornemannii produces a novel cytotoxicity profile against a diverse panel of human tumor cell lines. J. Med. Chem. 1992;35:3007–3011. doi: 10.1021/jm00094a012. [DOI] [PubMed] [Google Scholar]
- 23.Fuller R.W., Cardellina J.H., Jurek J., Scheuer P.J., Alvaradolindner B., McGuire M., Gray G.N., Steiner J.R., Clardy J., Menez E., et al. Isolation and structure activity features of halomon-related antitumor monoterpenes from the red alga Portieria hornemannii. J. Med. Chem. 1994;37:4407–4411. doi: 10.1021/jm00051a019. [DOI] [PubMed] [Google Scholar]
- 24.Matlock D.B., Ginsburg D.W., Paul V.J. Spatial variability in secondary metabolite production by the tropical red alga Portieria hornemannii. Hydrobiologia. 1999;399:267–273. [Google Scholar]
- 25.Puglisi M.P., Paul V.J. Intraspecific variation in the red alga Portieria hornemannii: Monoterpene concentrations are not influenced by nitrogen or phosphorus enrichment. Mar. Biol. 1997;128:161–170. [Google Scholar]
- 26.Miller K., Alvarez B., Battershill C., Northcote P., Parthasarathy H. Genetic, morphological, and chemical divergence in the sponge genus Latrunculia (Porifera: Demospongiae) from New Zealand. Mar. Biol. 2001;139:235–250. [Google Scholar]
- 27.Pietra F. Biodiversity and Natural Product Diversity. Vol. 21. Pergamon Trento; Italy: 2002. p. 351. [Google Scholar]
- 28.McGovern T.M., Hellberg M.E. Cryptic species, cryptic endosymbionts, and geographical variation in chemical defences in the bryozoan Bugula neritina. Mol. Ecol. 2003;12:1207–1215. doi: 10.1046/j.1365-294x.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- 29.Payo D.A. Ph.D. Thesis. Ghent University; Gent, Belgium: 2011. Diversity of the Marine Red Alga Portieria in the Philippines, an Integrative Approach. [Google Scholar]
- 30.Fiehn O., Kind T. Metabolite Profiling in Blood Plasma. In: Weckwerth W., editor. Methods in Molecular Biology, Metabolomics: Methods and Protocols. Vol. 358. Humana Press; Totowa, NJ, USA: 2006. pp. 3–17. [DOI] [PubMed] [Google Scholar]
- 31.Fiehn O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 32.Lucero M., Estell R., Tellez M., Frederickson E. A retention index calculator simplifies identification of plant volatile organic compounds. Phytochem. Anal. 2009;20:378–384. doi: 10.1002/pca.1137. [DOI] [PubMed] [Google Scholar]
- 33.Egorin M.J., Rosen D.M., Benjamin S.E., Callery P.S., Sentz D.L., Eiseman J.L. In vitro metabolism by mouse and human liver preparations of halomon, an antitumor halogenated monoterpene. Cancer Chemother. Pharmacol. 1997;41:9–14. doi: 10.1007/s002800050701. [DOI] [PubMed] [Google Scholar]
- 34.Pelletreau K.N., Targett N.M. New perspectives for addressing patterns of secondary metabolites in marine macroalgae. Algal Chem. Ecol. 2008:121–146. [Google Scholar]
- 35.Wise M.L., Rorrer G.L., Polzin J.J., Croteau R. Biosynthesis of marine natural products: Isolation and characterization of a myrcene synthase from cultured tissues of the marine red alga Ochtodes secundiramea. Arch. Biochem. Biophys. 2002;400:125–132. doi: 10.1006/abbi.2002.2780. [DOI] [PubMed] [Google Scholar]
- 36.Thornber C., Stachowicz J.J., Gaines S. Tissue type matters: Selective herbivory on different life history stages of an isomorphic alga. Ecology. 2006;87:2255–2263. doi: 10.1890/0012-9658(2006)87[2255:ttmsho]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Verges A., Paul N.A., Steinberg P.D. Sex and life-history stage alter herbivore responses to a chemically defended red alga. Ecology. 2008;89:1334–1343. doi: 10.1890/07-0248.1. [DOI] [PubMed] [Google Scholar]
- 38.Cavalcanti D.N., de Oliveira M.A.R., De-Paula J.C., Barbosa L.S., Fogel T., Pinto M.A., de Palmer Paixão I.C.N., Teixeira V.L. Variability of a diterpene with potential anti-HIV activity isolated from the Brazilian brown alga Dictyota menstrualis. J. Appl. Phycol. 2010;5:1–4. [Google Scholar]
- 39.Huelsenbeck J.P., Ronquist F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 40.Posada D. Jmodeltest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 41.Rambaut A., Drummond A.J. Tracer v1.4. [(accessed on 11 October 2007)]. Available online: http://beast.bio.ed.ac.uk/Tracer.
- 42.AMDIS. [(accessed on 7 March 2010).]. Available online: http://www.amdis.net.
- 43.Styczynski M.P., Moxley J.F., Tong L.V., Walther J.L., Jensen K.L., Stephanopoulos G.N. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. [(accessed on 23 February 2010)];Anal. Chem. 2007 79:966–973. doi: 10.1021/ac0614846. Available online: http://spectconnect.mit.edu. [DOI] [PubMed] [Google Scholar]
- 44.Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K., et al. Massbank: A public repository for sharing mass spectral data for life sciences. [(accessed on 18 March 2010)];J. Mass Spectrometry. 2010 45:703–714. doi: 10.1002/jms.1777. Available online: http://www.massbank.jp. [DOI] [PubMed] [Google Scholar]
- 45.The Pherobase: Database of Pheromones and Semiochemicals. [(accessed on 11 August 2010).]. Available online: http://www.pherobase.com.
- 46.Burreson B.J., Woolard F.X., Moore R.E. Evidence for the biogenesis of halogenated myrcenes from the red alga Chondrococcus hornemanni. Chem. Lett. 1975;4:1111–1114. [Google Scholar]
- 47.Ichikawa N., Naya Y., Enomoto S. Halogenated monoterpene derivatives from Desmia (Chondrococcus) japonicus. Proc. Jpn. Acad. 1975;51:562–565. [Google Scholar]
- 48.Woolard F.X., Moore R.E., Mahendran M., Sivapalan A. (−)-3-bromomethyl-3-chloro-7- methyl-1 6-octadiene from Sri Lankan Chondrococcus hornemanni. Phytochemistry. 1976;15:1069–1070. [Google Scholar]
- 49.Higa T. 2-(1-chloro-2-hydroxyethyl)-4,4-dimethylcyclohexa-2,5-dienone: A precursor of 4, 5-dimethylbenzo [β] furan from the red alga Desmia hornemanni. Tetrahedron Lett. 1985;26:2335–2336. [Google Scholar]
- 50.Coll J.C., Wright A.D. Tropical marine-algae. 6. New monoterpenes from several collections of Chondrococcus hornemannii (Rhodophyta, Gigartinales, Rhizophyllidaceae) Aust. J. Chem. 1989;42:1983–1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
List of compounds isolated from Portieria (syn. Desmia and Chondrococcus) hornemannii, based on previous studies.
| Reference | Cpd # | Name | Mol. Formula | Description | Specimen Source |
|---|---|---|---|---|---|
| [18] | 1 | Myrcene | C10H16 | acyclic halogenated monoterpene | Amami Is., Japan |
| 2 | 7-chloro-myrcene | C10H15Cl | " | " | |
| 3 | 7-bromo-myrcene | C10H15Br | " | " | |
| 4 | (Z)-10-bromo-myrcene | C10H15Br | " | " | |
| 5 | (E)-10-bromo-myrcene | C10H15Br | " | " | |
| 6 | (Z)-10-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 7 | (E)-10-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 8 | 3-chloro-7, (Z)-10-dibromo-myrcene | C10H13Br2Cl | " | " | |
| 9 | (Z)-10-chloro-3,7-dibromo-myrcene | C10H13Br2Cl | " | " | |
| 10 | 3-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 11 | 7-bromo-10-chloro-myrcene | C10H14BrCl | " | " | |
| 12 | - | C10H15Br | cyclic halogenated monoterpene | " | |
| [19] | 1 | chondrocole A | C10H14BrClO | " | Hawaii |
| 2 | chondrocole B | C10H14BrClO | " | Hawaii | |
| [46] | 3 | - | C10H15Cl3Br2 | acyclic halogenated monoterpene | Black Point, Oahu, Hawaii |
| 4 | - | C10H16Cl3Br | " | " | |
| 5 | - | C10H15Cl2Br | " | " | |
| 6 | - | C10H16Cl2Br2 | " | Halona Blowhole Is, Oahu, Hawaii | |
| 10 | - | C10H16Cl2Br2 | acyclic halogenated monoterpene | " | |
| 11 | 6-bromo-2-chloromyrcene | C10H15ClBr | " | Black Point, Oahu, Hawaii | |
| 12 | chondrocole c | C10H14Br2O | cyclic halogenated monoterpene | " | |
| 13 | - | C10H15Cl3Br2 | acyclic halogenated monoterpene | Hawaii | |
| 14 | Z-3-bromomethylene-7-methyl-1,6-octadiene | C10H16ClBr2 | " | Halona Blowhole Is., Oahu, Hawaii | |
| 15 | - | C10H14Br2Cl2 | cyclic halogenated monoterpene | " | |
| [47] | 1 | myrcene | C10H16 | acyclic halogenated monoterpene | Kada Coast, Wakayama Pref., Japan |
| 2 | 7-chloro-myrcene | C10H15Cl | " | " | |
| 3 | 7-bromo-myrcene | C10H15Br | " | " | |
| 4 | 3,7-dichloro-myrcene | C10H14Cl2 | " | " | |
| 5 | (Z)-10-bromo-3-methoxy-α-myrcene | C11H17OBr | " | " | |
| 6 | (E)-10-bromo-3-methoxy-α-myrcene | C11H17OBr | " | " | |
| 7 | 3-bromo-7-chloro-myrcene | C10H14BrCl | " | " | |
| 8 | (Z)-10-bromo-1-methoxy-myrcene | C11H17OBr | " | " | |
| 9 | (E)-10-bromo-1-methoxy-myrcene | C11H17OBr | " | " | |
| [48] | (−)-3-bromomethyl-3-chloro-7-methyl-1,6-octadiene | C10H16BrCl | " | Trincomalee (Foul Point) Sri Lanka | |
| [49] | 2-(1-chloro-2-hydroxyethyl)-4,4-dimethylcyclohexa-2, 5-dienone: a precursor of 4,5-dimethylbenzo [b] furan | - | - | - | |
| [20] | 1 | (2Z,6E)-1,8-dichloro-3-chloromethyl-7-methylocta-2,6-diene | C10H15Cl3 | acyclic halogenated monoterpene | Rib Reef, Great Barrier Reef, Australia |
| 2 | (E)-1,2-dibromo-3-chloromethylene-7-methyloct-6-ene | C10H15Br2Cl | " | " | |
| 3 | (Z)-1-chloro-3-chloromethyl-7-methylocta-2,6-diene | C10H16Cl2 | " | " | |
| 4 | (Z)-1,6-dichloro-3-chloromethyl-7-methylocta-2,7-diene | C10H15Cl3 | " | " | |
| 5 | (2R*,3(8)E,4S*,6R*)-6-bromo-2-chloro-1,4-oxido-3,(8)-ochtodene | C10H14BrClO | epimeric bicyclic monoterpene | " | |
| 6 | (2S*,3(8)E,4S*,6R*)-6-bromo-2-chloro-1,4-oxido-3,(8)-ochtodene | C10H14BrClO | " | " | |
| [50] | (Z)-3-bromo-8-chloro-6-chloromethyl-2-methylocta-1,6-diene (9,6-hydroxymethyl-2-methylocta-2,8-dien-6-ol | - | - | - | |
| [21] | 1 | (2Z)-6-bromo-3-chloromethyl-1,7-dichloro-7-methylocta-2-ene | C10H16Cl3Br | acyclic halogenated monoterpene | Nelly Bay, Magnetic Island, Queensland, Australia |
| 2 | (2Z,6E)-3-chloromethyl-1-chloroocta-2,6-dien-8-al | C10H14OCl2 | " | " | |
| 3 | 3-methoxymethyl-6-methoxyl-7-methylocta-1,7(10)-dien-3-ol | C12H22 O3 | " | " | |
| 4 | (2Z,6S)-3-chloromethyl-1-methylocta-2,7(10)-dien-6-ol | C11H19 O2Cl | " | " | |
| 5 | (2Z,6S)-3-chloromethyl-6-methylocta-2,7(10)-dien-1-ol | C11H19 O2Cl | " | " | |
| [22] | 1 | halomon/6(R)-bromo-3(S)-(bromomethyl)-7-methyl-2,3,7-trichloro-1-octene | C10H15Br2Cl3 | acyclic halogenated monoterpene | Chanaryan, Batan Is., Batanes, Philippines |
| 2 | - | C10H13Cl | cyclic halogenated monoterpene | Banilad, Bacong, Negros Oriental, Philippines | |
| [23] | 2 | isohalomon | C10H15Br2Cl3 | acyclic halogenated monoterpene; isomeric with halomon | Chanaryan, Batan Is., Batanes, Philippines |
| 3 | - | C10H14BrCl3 | acyclic halogenated monoterpene | " | |
| 4 | - | C10H14Br2Cl2 | " | " | |
| 5 | - | C10H14Br3Cl | cyclic halogenated monoterpene | " | |
| 6 | - | C10H14BrCl3 | " | " | |
| 7 | - | C10H13Cl3 | " | " | |
| 8 | - | C10H15BrCl2 | acyclic halogenated monoterpene | " | |
| 9 | - | C10H16Br2Cl2 | " | " | |
| 10 | - | C10H16BrCl | " | Halona Blowhole Is, Oahu, Hawaii | |
| 11 | - | C10H15BrCl | " | " | |
| 12 | - | C10H14BrCl | cyclic halogenated monoterpene | " | |
| [8] | 1 | apakaochtodene A/6(S*)-bromo-1,4(S*),8(R*)-trichloro-2(Z)-ochtodene | C10H14Cl3Br | cyclic halogenated monoterpene | Apaka Point Beach, Guam |
| 2 | apakaochtodene B/6(S)-bromo-1,4(S), 8(R*)-trichloro-2(E)-ochtodene | C10H14Cl3Br | cyclic halogenated monoterpene | Gun Beach; Double Reef; Pago Bay; Guam | |
| [11] | 1 | myrcene/7-methyl-3-methylene-1,6-octadiene | C10 H16O | acyclic non-halogenated monoterpene | microplantlet culture, Double Reef NW Guam |
| 2 | 10E-bromomyrcene/E-3-bromomethylene-7-methyl-1,6-octadiene | C10H15Br | " | " | |
| 3 | 10Z-bromomyrcene/Z-3-bromomethylene-7-methyl-1,6-octadiene | C10H15Br | " | " | |
| 4 | 10E-bromo-3-chloro-α-myrcene/E-3-bromomethylene-6-chloro-1,7-octadiene | C10H14BrCl | " | " | |
| 5 | - | C10H14Br2 | " | " | |
| 6 | apakaochtodene B/6(S)-bromo-1,4(S), 8(R*)-trichloro-2(E)-ochtodene | C10H14BrCl3 | cyclic halogenated monoterpene | " | |
| 7 | - | C10 H16O | non-halogenated monoterpene | " | |
| 8 | bromomyrcene isomer | C10H15Br | halogenated monoterpene | " | |
| 9 | 7-chloromyrcene/2-chloro-3-methylene-7-methyl-1,6-octadiene | C10H15Cl | " | " | |
| 10 | C10H14Cl | " | " | ||
| 11 | chloromyrcene derivative | C10H16Cl | " | " | |
| 12 | bromomyrcene isomer | C10H15Br | " | " | |
| 13 | chloromyrcene derivative | C10H14Cl | " | " | |
| 14 | - | C10H18Cl2 | " | " | |
| 15 | - | C15H18 O4 | sesquiterpene | " | |
| [13] | 5 | 1,2-dibromoochtoda-3(8),5-dien-4-one | C22H12Br2O | cyclic halogenated monoterpene | Cape Zampa, Okinawa, Japan |
| 6 | 1-bromo-2-chloroochtoda-3(8),5-dien-4-one | C10H12BrClO | " | " | |
| 7 | 1,2-dichloroochtoda-3(8),5-dien-4-one | C10H12Cl2O | " | Gushichan coast, Okinawa, Japan | |
| 8 | (1Z)-1-bromoochtoda-1,3(8),5-trien-4-one | C10H11BrO | " | Cape Zampa, Okinawa, Japan | |
| 9 | (1Z)-1-chloroochtoda-1,3(8),5-trien-4-one | C10H11ClO | " | " | |
| [12] | 1 | halomon/6(R)-bromo-3(S)-(bromomethyl)-7-methyl-2,3,7-trichloro-1-octene | C10H15Br2Cl3 | acyclic halogenated monoterpene | Tolagniaro, Fort Dauphin, Madagascar |
| 2 | - | C10H13ClBr | " | " | |
| 3 | - | C10H14Cl2Br | " | " | |
| 4 | C10H15Br2Cl3 | " | " |
Table S2.
Compound peaks detected from GC-MS analysis of samples used for characterization of the life history stages of P. hornemannii. Identifications based on comparison of either Kovat’s Indices (KI) of compounds retrieved from the Retention Index Calculator [37] or mass spectral comparison of compounds retrieved from NIST or MassBase. Asterisk (*) indicates compounds with parent ions showing halogenated mass spectral patterns.
| # | Sample | Base Peak | KI | KI Based ID | Mass Spectra Based ID | |||
|---|---|---|---|---|---|---|---|---|
| RT (min) | Nearest KI | Lucero et al. [37] | NIST | MassBase | ||||
| 1 | F6_2 | 93 | 6.1863 | 994 | 993/994 | beta-myrcene/6-methyl-5-hepten-2-ol | b-myrcene | b-myrcene |
| 2 | T1_1 | 104 | 6.5874 | 1008 | 1007 | α-phellandrene | - | - |
| 3 | T1_2 | 104 | 6.5909 | 1008 | 1007 | α-phellandrene | - | - |
| 4 | M1_1 | 91 | 6.7685 | 1016 | 1017 | α-terpinene | Benzene, tert-butyl- | 4-methylacetophenone |
| 5 | F3_3 | 79 | 6.9501 | 1023 | 1024 | r-cymene | - | - |
| 6 | M1_2 | 79 | 6.9532 | 1023 | 1024 | r-cymene | - | - |
| 7 | M1_3 | 91 | 7.017 | 1026 | 1025 | p-cymene | carbonic acid | 1-phenylpropan-2-one |
| 8 | M2_2 | 91 | 7.021 | 1026 | 1025 | p-cymene | - | protopine |
| 9 | F3_2 | 91 | 7.0387 | 1027 | 1025 | p-cymene | - | N-Methyl-Npropagylbenzylamine |
| 10 | T2_1 | 57 | 8.0381 | 1063 | 1062 | y-terpinene | - | - |
| 11 | M2_2 | 132 | 8.8894 | 1089 | 1089 | p-mentha-2,4(8)diene | - | - |
| 12 | T2_1 | 117 | 9.5127 | 1110 | 1099 | linalool | - | - |
| 13 | F4_3 | 117 | 9.8874 | 1124 | 1123 | chrysanthenone | - | - |
| 14 | M1_3 | 117 | 9.9855 | 1128 | 1127 | α-campholenal | - | - |
| 15 | M2_2 | 91 | 10.2042 | 1136 | 1127 | α-campholenal | - | - |
| 16 | F2_2 | 91 | 10.2132 | 1136 | 1127 | α-campholenal | - | Dimethirimol |
| 17 | T2_1 | 119 | 10.4565 | 1145 | 1145 | camphor | - | Benzimidazole |
| 18 | M1_1 | 68 | 10.8826 | 1160 | 1158 | isobomeol | - | - |
| 19 | F7_3 | 68 | 10.8845 | 1160 | 1158 | isobomeol | - | - |
| 20 | T2_1 | 68 | 10.8866 | 1160 | 1158 | isobomeol | - | - |
| 21 | F3_3 | 68 | 10.8873 | 1160 | 1158 | isobomeol | - | - |
| 22 | F4_1 | 117 | 11.3242 | 1174 | 1171 | ethyl-benzoate | - | - |
| 23 | F4_1 | 50 | 11.5177 | 1180 | 1180 | m-cymen-8-ol | - | - |
| 24 | T2_2 | 127 | 11.5317 | 1180 | 1180 | m-cymen-8-ol | - | - |
| 25 | T2_3 | 134 | 11.7619 | 1188 | 1187 | p-cymen-8-ol | - | - |
| 26 | M2_2 | 105 | 12.0654 | 1197 | 1196 | methylchavicol | - | 3-cyanopyridine |
| 27 | F3_2 | 57 | 12.1827 | 1200 | 1203 | n-decanal | 3-Hexanone | - |
| 28 | F7_3 | 113 | 12.3674 | 1207 | 1207 | verbenone | - | - |
| 29 | M1_2 | 113 | 12.3831 | 1208 | 1207 | verbenone | - | - |
| 30 | T1_3 | 91 | 12.3874 | 1208 | 1207 | verbenone | - | - |
| 31 | M2_2 | 93 | 12.6294 | 1217 | 1217 | trans-carveol | - | - |
| 32 | M1_2 | 69 | 12.7588 | 1222 | 1219 | trans-carveol | - | - |
| 33 | M1_1 | 55 | 12.8397 | 1225 | 1229 | nerol | - | - |
| 34 | F7_3 | 67 | 13.1203 | 1236 | 1236 | thymol methyl ether | - | - |
| 35 | M1_1 | 79 | 13.1266 | 1236 | 1236 | thymol methyl ether | - | - |
| 36 | F5_2 | 93 | 13.127 | 1236 | 1236 | thymol methyl ether | - | Beta-pinene; y-eudesmol |
| 37 | M2_2 | 93 | 13.32 | 1243 | 1243 | carvone | - | - |
| 38 | T1_1 | 93 | 13.327 | 1243 | 1243 | carvone | 2-chloropropionyl chloride | - |
| 39 | T2_1 | 93 | 13.3305 | 1243 | 1243 | carvone | - | - |
| 40 *(Br) | F6_2 | 69 | 13.6916 | 1256 | 1256 | geraniol | - | - |
| 41 | M1_1 | 91 | 13.6953 | 1256 | 1256 | geraniol | - | |
| 42 | F2_1 | 69 | 13.6967 | 1256 | 1256 | geraniol | - | |
| 43 | F5_1 | 69 | 13.6975 | 1256 | 1256 | geraniol | - | - |
| 44 | M2_1 | 91 | 13.9837 | 1266 | 1263 | (E)-2-decenal | - | N-Methyl-Npropagylbenzylamine |
| 45 | F7_3 | 119 | 14.131 | 1271 | 1271 | geranial | - | - |
| 46 | F2_3 | 119 | 14.1354 | 1271 | 1271 | geranial | - | - |
| 47 | F3_2 | 69 | 14.4252 | 1281 | 1282 | a-terpinen-7-al | - | - |
| 48 | M2_3 | 69 | 14.4368 | 1281 | 1282 | a-terpinen-7-al | - | - |
| 49 | F5_1 | 91 | 14.6745 | 1289 | 1289 | p-cymen-7-ol | - | - |
| 50 | T2_1 | 91 | 14.68 | 1289 | 1289 | p-cymen-7-ol | - | - |
| 51 | F6_3 | 149 | 14.9157 | 1297 | 1297 | perilla alcohol | - | Benzoylcholine |
| 52 * (Cl) | F6_3 | 131 | 15.0641 | 1302 | 1302 | trans-ascaridole | - | 2-chloro-1-phenyl-2-butene |
| 53 | F6_1 | 166 | 15.3344 | 1313 | 1313 | 2E,4E-decadienal | - | 2-chloro-1-phenyl-2-butene |
| 54 | F6_3 | 81 | 15.5368 | 1320 | 1313 | 2E,4E-decadienal | - | - |
| 55 | M2_2 | 81 | 15.5381 | 1321 | 1313 | 2E,4E-decadienal | - | - |
| 56 | T1_2 | 81 | 15.5481 | 1321 | 1313 | 2E,4E-decadienal | - | - |
| 57 | M2_2 | 67 | 15.6269 | 1324 | 1313 | 2E,4E-decadienal | - | 1-Adamantanamine |
| 58 | M1_2 | 67 | 15.6354 | 1324 | 1313 | 2E,4E-decadienal | - | - |
| 59 | F3_1 | 57 | 15.7447 | 1329 | 1339 | d-elemene | - | - |
| 60 | F2_1 | 57 | 15.7649 | 1329 | 1339 | d-elemene | - | - |
| 61 | M2_2 | 91 | 15.8827 | 1334 | 1339 | d-elemene | - | - |
| 62 | M1_2 | 91 | 15.8885 | 1334 | 1339 | d-elemene | - | - |
| 63 | M2_2 | 139 | 16.5157 | 1357 | 1357 | eugenol | - | - |
| 64 | F6_1 | 115 | 16.5278 | 1358 | 1357 | eugenol | - | - |
| 65 | M2_3 | 166 | 16.5392 | 1358 | 1357 | eugenol | - | - |
| 66 | M2_2 | 91 | 17.02 | 1375 | 1373 | a-ylangene | - | - |
| 67 | M2_3 | 91 | 17.0254 | 1376 | 1373 | a-ylangene | - | - |
| 68 | M1_2 | 91 | 17.0311 | 1376 | 1373 | a-ylangene | - | - |
| 69 | T2_1 | 127 | 17.403 | 1389 | 1389 | isolongifolene | - | Imidazole-4-acetate |
| 70 | T2_1 | 57 | 17.7476 | 1400 | 1399 | 1,7-di-epi-a-cedrene | - | - |
| 71 | M2_2 | 79 | 17.7625 | 1401 | 1402 | methyleugenol | - | - |
| 72 | M2_3 | 127 | 17.7782 | 1402 | 1402 | methyleugenol | - | - |
| 73 | T2_2 | 67 | 18.1041 | 1415 | 1415 | cis-a-bergarnotene | - | - |
| 74 | T1_3 | 79 | 18.18 | 1418 | 1419 | β-caryophyllene | - | - |
| 75 | F6_1 | 79 | 18.1866 | 1419 | 1419 | β-caryophyllene | - | - |
| 76 | T1_1 | 79 | 18.1928 | 1419 | 1419 | β-caryophyllene | Pyridine | - |
| 77 | F5_1 | 79 | 18.1934 | 1419 | 1419 | β-caryophyllene | - | camphene |
| 78 | M1_2 | 79 | 18.1939 | 1419 | 1419 | β-caryophyllene | - | - |
| 79 * (Br) | F7_2 | 212 | 18.3193 | 1424 | 1420 | β-caroyophyllene | - | - |
| 80 | F6_2 | 210 | 18.4564 | 1429 | 1420 | β-caroyophyllene | - | - |
| 81 | M2_3 | 131 | 18.4673 | 1430 | 1420 | β-caroyophyllene | - | - |
| 82 | M1_1 | 67 | 18.5462 | 1433 | 1435 | trans-a-bergamotene | - | - |
| 83 | M1_2 | 67 | 18.5503 | 1433 | 1435 | trans-a-bergamotene | - | - |
| 84 | F2_2 | 133 | 18.6084 | 1435 | 1435 | trans-a-bergamotene | - | - |
| 85 | T2_2 | 69 | 18.6925 | 1439 | 1438 | trans-a-bergamotene | - | - |
| 86 | F3_3 | 69 | 18.6982 | 1439 | 1440 | a-guaiene | - | - |
| 87 | M2_3 | 69 | 18.7018 | 1439 | 1440 | a-guaiene | - | - |
| 88 | M1_1 | 91 | 18.7475 | 1441 | 1440 | a-guaiene | - | - |
| 89 | F4_2 | 69 | 18.8243 | 1444 | 1440 | a-guaiene | - | - |
| 90 | F7_2 | 69 | 18.8273 | 1444 | 1440 | a-guaiene | - | - |
| 91 | T1_3 | 69 | 18.8306 | 1444 | 1440 | a-guaiene | - | - |
| 92 | F2_2 | 210 | 19.1226 | 1456 | 1455 | a-humulene | - | - |
| 93 | F3_3 | 133 | 19.3339 | 1464 | 1464 | a-acoradiene | - | |
| 94 | T2_2 | 105 | 19.4843 | 1469 | 1469 | drima-7,9(11)-diene | - | - |
| 95 | T2_1 | 105 | 19.4852 | 1469 | 1469 | drima-7,9(11)-diene | - | - |
| 96 | F7_2 | 131 | 19.4915 | 1470 | 1469 | drima-7,9(11)-diene | - | - |
| 97 | F5_1 | 91 | 19.6255 | 1475 | 1474 | b-cadinene | - | - |
| 98 | F6_3 | 91 | 19.7468 | 1479 | 1479 | y-curcurnene | - | |
| 99 | F2_1 | 91 | 19.7581 | 1480 | 1480 | germacrene D | - | - |
| 100 | F4_1 | 153 | 20.0567 | 1491 | 1491 | cis-β-guaiene | - | - |
| 101 | F4_1 | 153 | 20.0567 | 1491 | 1491 | cis-β-guaiene | - | - |
| 102 | F3_2 | 71 | 20.2111 | 1496 | 1495 | a-zingiberene | - | |
| 103 | F7_2 | 135 | 20.3621 | 1502 | 1499 | a-muurolene | - | - |
| 104 | F5_1 | 91 | 20.3727 | 1503 | 1506 | d-selinene | - | - |
| 105 | T2_1 | 91 | 20.3766 | 1503 | 1506 | d-selinene | - | - |
| 106 | F2_2 | 91 | 20.3829 | 1503 | 1506 | d-selinene | -- | - |
| 107 | F6_1 | 205 | 20.6221 | 1513 | 1513 | g-cadinene | - | - |
| 108 | F4_3 | 205 | 20.6237 | 1513 | 1513 | g-cadinene | - | - |
| 109 | F7_2 | 205 | 20.6252 | 1513 | 1513 | g-cadinene | - | 5-bromo-4-methoxyoct-1-ene |
| 110 | T2_1 | 205 | 20.6278 | 1513 | 1513 | g-cadinene | p-Benzoquinone | - |
| 111 | F3_2 | 133 | 20.6781 | 1516 | 1514 | sesquicineole | - | - |
| 112 | F3_3 | 133 | 20.6803 | 1516 | 1514 | sesquicineole | - | - |
| 113 | F6_3 | 133 | 20.685 | 1516 | 1514 | sesquicineole | - | - |
| 114 | F5_1 | 69 | 20.8293 | 1522 | 1523 | d-cadinene | - | -- |
| 115 | F5_1 | 69 | 20.9154 | 1526 | 1525 | eugenyl acetate | - | - |
| 116 | F6_3 | 69 | 20.9158 | 1526 | 1525 | eugenyl acetate | - | - |
| 117 | F7_2 | 121 | 20.9204 | 1526 | 1525 | eugenyl acetate | - | - |
| 118 | T1_3 | 69 | 20.9227 | 1526 | 1525 | eugenyl acetate | - | - |
| 119 | F7_2 | 167 | 21.1103 | 1534 | 1533 | cadina-1,4-diene1 | - | - |
| 120 | T2_2 | 91 | 21.1498 | 1536 | 1538 | a-cadinene | - | - |
| 121 | F6_1 | 167 | 21.43 | 1547 | 1548 | elemol | - | - |
| 122 | F7_2 | 167 | 21.4346 | 1547 | 1548 | elemol | - | - |
| 123 | T1_3 | 69 | 21.467 | 1549 | 1549 | elemol | - | -- |
| 124 | F3_2 | 69 | 21.4709 | 1549 | 1549 | elemol | - | - |
| 125 | F7_2 | 91 | 21.598 | 1554 | 1552 | elemicin | - | - |
| 126 | F6_2 | 91 | 21.6852 | 1557 | 1557 | germacrene B | - | - |
| 127 | F5_1 | 91 | 21.6859 | 1558 | 1557 | germacrene B | - | - |
| 128 | F6_3 | 91 | 21.6869 | 1558 | 1557 | germacrene B | - | - |
| 129 | M1_1 | 91 | 21.6895 | 1558 | 1557 | germacrene B | - | - |
| 130 | F6_3 | 67 | 21.7268 | 1559 | 1557 | germacrene B | - | - |
| 131 | F7_2 | 133 | 21.8253 | 1563 | 1564 | β-calacorene | - | l-Asparagine |
| 132 | T1_1 | 91 | 21.8405 | 1564 | 1564 | β-calacorene | - | - |
| 133 | F7_2 | 167 | 22.0012 | 1570 | 1574 | prenopsan-8-ol | - | - |
| 134 | T1_3 | 213 | 22.018 | 1571 | 1574 | prenopsan-8-ol | - | - |
| 135 | F6_3 | 132 | 22.0226 | 1571 | 1574 | prenopsan-8-ol | - | - |
| 136 | F6_3 | 153 | 22.0226 | 1571 | 1574 | prenopsan-8-ol | - | - |
| 137 | M1_1 | 91 | 22.3956 | 1586 | 1585 | gleenol | - | - |
| 138 | F2_2 | 133 | 22.5362 | 1591 | 1590 | viridflorol | - | - |
| 139 | F6_3 | 71 | 22.7179 | 1598 | 1590 | viridflorol | - | - |
| 140 | F3_2 | 71 | 22.7193 | 1598 | 1590 | viridflorol | - | - |
| 141 | F2_3 | 135 | 22.722 | 1598 | 1590 | viridflorol | - | - |
| 142 | F3_3 | 57 | 22.7547 | 1600 | 1607 | b-oplopenone | - | - |
| 143 | F3_2 | 57 | 22.7603 | 1600 | 1607 | b-oplopenone | - | - |
| 144 | F3_3 | 57 | 22.761 | 1600 | 1607 | b-oplopenone | - | - |
| 145 | F6_3 | 67 | 22.8782 | 1605 | 1607 | b-oplopenone | - | - |
| 146 | T2_2 | 68 | 23.0541 | 1613 | 1612 | tetradecanal | - | - |
| 147 | F4_3 | 148 | 23.2435 | 1622 | 1623 | silphiperfol-6-en-5-one | - | - |
| 148 | F3_1 | 135 | 23.2485 | 1622 | 1623 | silphiperfol-6-en-5-one | - | - |
| 149 | F5_3 | 91 | 23.52 | 1634 | 1633 | y-eudesmol | - | - |
| 150 | F4_3 | 91 | 23.5226 | 1634 | 1633 | y-eudesmol | - | - |
| 151 | F7_2 | 68 | 23.5261 | 1634 | 1633 | y-eudesmol | - | - |
| 152 | F7_2 | 68 | 23.5261 | 1634 | 1633 | y-eudesmol | - | -- |
| 153 | T2_2 | 68 | 23.5278 | 1634 | 1633 | y-eudesmol | - | - |
| 154 | F6_1 | 167 | 23.5309 | 1634 | 1633 | y-eudesmol | - | |
| 155 | F6_2 | 67 | 23.7239 | 1643 | 1642 | cubenol | - | allylcy-anide |
| 156 | T1_3 | 91 | 23.8325 | 1647 | 1646 | a-muurolol | - | - |
| 157 | F7_2 | 132 | 23.8759 | 1649 | 1646 | a-muurolol | - | - |
| 158 | F7_2 | 132 | 23.9988 | 1654 | 1653 | a-cadinol | - | - |
| 159 | F6_2 | 149 | 24.3377 | 1669 | 1668 | bulnesol | - | - |
| 160 | F7_2 | 149 | 24.3428 | 1669 | 1668 | bulnesol | - | - |
| 161 | F6_3 | 167 | 24.6091 | 1680 | 1682 | a-bisabolol | - | - |
| 162 | F6_1 | 57 | 24.6252 | 1681 | 1682 | a-bisabolol | - | - |
| 163 | F7_2 | 69 | 24.8113 | 1689 | 1686 | 8-cedren-13-ol | - | - |
| 164 | F7_2 | 69 | 24.8207 | 1689 | 1686 | 8-cedren-13-ol | - | - |
| 165 | F2_3 | 67 | 24.9282 | 1693 | 1686 | 8-cedren-13-ol | - | - |
| 166 | F6_1 | 57 | 25.0854 | 1700 | 1686 | 8-cedren-13-ol | - | Heptadecane |
| 167 | F7_2 | 91 | 25.3006 | 1710 | 1686 | 8-cedren-13-ol | - | - |
| 168 | F3_2 | 71 | 25.3224 | 1711 | 1735 | oplopanone | - | - |
| 169 | F3_3 | 68 | 25.4814 | 1718 | 1735 | oplopanone | - | - |
| 170 | F6_3 | 67 | 25.7278 | 1730 | 1735 | oplopanone | - | - |
| 171 | F7_2 | 103 | 25.7292 | 1730 | 1735 | oplopanone | - | - |
| 172 | F2_1 | 67 | 25.7318 | 1730 | 1735 | oplopanone | - | - |
| 173 | F2_3 | 133 | 25.8305 | 1735 | 1735 | oplopanone | - | - |
| 174 | F3_1 | 117 | 25.8366 | 1735 | 1735 | oplopanone | - | - |
| 175 | F7_2 | 167 | 26.3223 | 1757 | 1761 | benzyl-benzoate | - | - |
| 176 | T2_1 | 167 | 26.5838 | 1769 | 1761 | benzyl-benzoate | - | 6-Methylmer-captopurine |
| 177 | F6_2 | 117 | 26.7333 | 1775 | 1789 | 8-a-acetoxyelemol | - | - |
| 178 | T2_2 | 103 | 26.8111 | 1779 | 1789 | 8-a-acetoxyelemol | - | - |
| 179 | F3_1 | 67 | 26.8577 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 180 | F6_1 | 67 | 26.8599 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 181 | F3_2 | 67 | 26.8671 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 182 | T2_1 | 67 | 26.8697 | 1781 | 1789 | 8-a-acetoxyelemol | - | - |
| 183 * (Cl2) | F6_1 | 103 | 26.8838 | 1782 | 1789 | 8-a-acetoxyelemol | - | - |
| 184 | F3_1 | 103 | 26.8852 | 1782 | 1789 | 8-a-acetoxyelemol | - | - |
| 185 * (Br) | F6_1 | 67 | 27.4286 | 1806 | 1798 | nootkatone | - | - |
| 186 * (Br) | F7_2 | 67 | 27.4342 | 1806 | 1798 | nootkatone | - | - |
| 187 | F2_1 | 67 | 27.4383 | 1807 | 1798 | nootkatone | - | - |
| 188 | F6_3 | 103 | 27.4961 | 1810 | 1798 | nootkatone | - | - |
| 189 | F6_1 | 67 | 27.5012 | 1810 | 1798 | nootkatone | - | - |
| 190 | F6_2 | 67 | 27.5676 | 1813 | 1827 | isopropyl tetradecanoate | - | - |
| 191 | F6_2 | 69 | 28.0351 | 1836 | 1827 | isopropyl tetradecanoate | - | - |
| 192 | F3_2 | 69 | 28.0387 | 1836 | 1827 | isopropyl tetradecanoate | - | - |
| 193 | F6_1 | 91 | 28.3153 | 1849 | 1867 | flourensiadiol | - | - |
| 194 | F3_2 | 149 | 28.735 | 1869 | 1867 | flourensiadiol | - | - |
| 195 * (ClBr) | F6_2 | 69 | 29.2918 | 1895 | 1878 | hexadecanol | - | - |
| 196 | F5_3 | 67 | 29.4766 | 1903 | 1927 | methyl hexadecanoate | - | - |
| 197 | T2_1 | 67 | 29.4889 | 1904 | 1927 | methyl hexadecanoate | - | - |
| 198 | F7_2 | 91 | 30.7705 | 1968 | 1999 | eicosane | - | - |
| 199 | T2_2 | 91 | 30.7753 | 1969 | 1999 | eicosane | - | - |
| 200 | F3_2 | 91 | 30.7855 | 1969 | 1999 | eicosane | - | - |
| 201 | T2_2 | 64 | 31.6475 | - | - | - | - | - |
| 202 | T2_2 | 129 | 38.661 | - | - | - | - | - |
Table S3.
Compound peaks detected from GC-MS analysis of samples for evaluation of non-polar secondary metabolite patterns of 5 cryptic species of P. hornemanni found in Batanes, Philippines. Identifications based on comparison of either Kovat’s Indices (KI) of compounds retrieved from the Retention Index Calculator [37] or mass spectral comparison of compounds retrieved from NIST or MassBase. Asterisk (*) indicates compounds with parent ions showing halogenated mass spectral patterns.
| # | Sample | Base Peak | KI | KI Based ID | Mass Spectra Based ID | |||
|---|---|---|---|---|---|---|---|---|
| RT (min) | Nearest KI | Lucero et al. [37] | NIST | Massbase | ||||
| 1 | B4B | 57 | 8.1122 | 1065 | 1062 | y-terpinene | - | - |
| 2 | B4C | 69 | 8.7386 | 1085 | 1085 | artemisia-alcohol | - | - |
| 3 | B4C | 57 | 9.1807 | 1098 | 1097 | linalool | - | - |
| 4 | B4B | 71 | 9.1841 | 1098 | 1097 | linalool | - | - |
| 5 | B6C | 57 | 9.4047 | 1106 | 1099 | linalool | 3-hexanone, 2,2-dimethyl- | 2-methylbutane |
| 6 | B4C | 69 | 11.2515 | 1172 | 1171 | ethyl-benzoate | - | - |
| 7- * (Cl) | B3A | 69 | 11.2560 | 1172 | 1171 | ethyl-benzoate | - | - |
| 8 | B4A | 57 | 14.3547 | 1279 | 1277 | trans-carvone-oxide | - | 2,2-dimethylbutane |
| 9 | B4B | 57 | 14.3593 | 1279 | 1277 | trans-carvone-oxide | - | 3-ethylhexane |
| 10 | B3B | 57 | 14.5560 | 1285 | 1286 | borneol-acetate | - | |
| 11 | B3A | 69 | 15.2678 | 1310 | 1306 | undecanal | - | trans-4-octene |
| 12 | B3A | 57 | 15.6497 | 1325 | 1314 | 2E,4E-decadienal | - | |
| 13 | B6B | 57 | 15.6518 | 1325 | 1314 | 2E,4E-decadienal | - | |
| 14 | B4A | 71 | 16.5884 | 1360 | 1357 | eugenol | - | 2-methylpentane |
| 15 | B6C | 57 | 17.8154 | 1403 | 1403 | italicene | - | 2,2-dimethylbutane |
| 16 | B3A | 57 | 17.8289 | 1404 | 1403 | italicene | - | |
| 17 | B10 | 71 | 20.306 | 1500 | 1499 | a-muurolene | - | Pentacosane |
| 18 | B3A | 57 | 20.4025 | 1504 | 1506 | d-selinene | - | tripropylamine |
| 19 | B4A | 191 | 20.715 | 1517 | 1514 | sesquicineole | - | |
| 20 | B6C | 57 | 21.0008 | 1529 | 1532 | cadina-1,4-diene | - | |
| 21- * (Br) | B3B | 57 | 21.0024 | 1529 | 1532 | cadina-1,4-diene | - | 1-octene |
| 22 | B4A | 69 | 21.5566 | 1552 | 1552 | elemicin | - | |
| 23 | B4A | 57 | 25.1819 | 1704 | 1686 | 8-cedren-13-ol | - | |
| 24 | B4B | 71 | 25.4142 | 1715 | 1735 | oplopanone | - | Hexacosane |
| 25 | B4B | 69 | 26.0498 | 1745 | 1747 | 6S-7R-bisabolone | - | 10-methyl-1-dodecanol |
| 26 | B4A | 57 | 27.4031 | 1805 | 1798 | nootkatone | - | 4-methyl-1-pentene |
| 27 | B3B | 57 | 27.9515 | 1832 | 1827 | isopropyl-tetradecanoate | - | 2,4-dimethylpentane |
| 28 | B3B | 71 | 29.9932 | 1930 | 1927 | methyl-hexadecanoate | - | docosane |
| 29 | B6B | 57 | 30.4182 | 1951 | 1927 | methyl-hexadecanoate | - | - |
| 30 | B10 | 69 | 30.5987 | 1960 | 1927 | methyl-hexadecanoate | - | - |
| 31 | B4C | 73 | 30.6949 | 1965 | 1999 | eicosane | - | - |
| 32 | B6B | 71 | 32.0267 | - | - | 3-hexanone, 2,2-dimethyl- | 2,2-dimethylbutane | |
| 33 | B3B | 71 | 32.0458 | - | - | - | 2,2-dimethylbutane | |
| 34 | B6B | 69 | 34.1452 | - | - | - | docosane | |
| 35 | B3B | 69 | 34.7128 | - | - | - | - | |
| 36 | B4C | 69 | 34.7233 | - | - | - | - | |
| 37 | B6C | 71 | 35.0713 | - | - | - | - | |
| 38 | B3C | 71 | 36.7162 | - | - | - | 3-ethylpentane | |
| 39 | B9B | 395 | 38.2697 | - | - | - | - | |
| 40 | B6C | 71 | 38.8875 | - | - | - | 3-ethylhexane | |
| 41 | B6B | 71 | 39.0147 | - | - | - | - | |
| 42 | B4C | 57 | 39.4792 | - | - | - | 3-ethylhexane | |
| 43 | B6B | 57 | 39.4822 | - | - | - | 3,3-dimethyl-1-butene | |
| 44 | B4C | 71 | 39.6266 | - | - | - | 3-ethylpentane | |
| 45 | B3C | 71 | 39.7884 | - | - | - | - | |
| 46 | B6B | 57 | 40.4997 | - | - | 3-hexanone, 2,2-dimethyl- | 2-methylbutane | |
| 47 | B6C | 57 | 40.7684 | - | - | - | 3-ethylpentane | |
| 48 | B3C | 71 | 40.7722 | - | - | - | - | |
| 49 | B4C | 57 | 43.09 | - | - | - | 3-ethylpentane | |
| 50 | B6B | 57 | 43.0982 | - | - | - | 2,2,4,6,6-pentamethylheptane | |
| 51 | B6C | 85 | 43.2865 | - | - | - | 2-methylnonane | |
| 52 | B6B | 71 | 43.3628 | - | - | - | 3-ethylpentane | |
| 53 | B3C | 71 | 43.3691 | - | - | - | - | |
| 54 | B3B | 85 | 43.7727 | - | - | - | 3,3-dimethyl-1-butene | |
| 55 | B4B | 85 | 44.3901 | - | - | - | Amitrole | |
| 56 | B6B | 71 | 44.3953 | - | - | - | 3-aminopropiononitrile | |
| 57 | B4A | 71 | 46.2638 | - | - | - | - | |
| 58 | B3C | 71 | 46.8143 | - | - | - | 2,3-dimethylbutane | |
| 59 | B6B | 71 | 46.822 | - | - | 3-hexanone, 2,2-dimethyl- | - | |
| 60 | B6C | 71 | 47.3544 | - | - | - | - | |
| 61 | B4A | 71 | 47.7467 | - | - | - | 3-ethylypentane | |
| 62 | B3A | 57 | 47.7529 | - | - | - | 3,3-dimethyl-1-butene | |
| 63 | B4A | 71 | 48.0072 | - | - | - | - | |
| 64 | B6B | 71 | 48.0159 | - | - | - | - | |
| 65 | B6C | 71 | 48.0169 | - | - | - | - | |
| 66 | B4A | 71 | 48.0321 | - | - | - | - | |
| 67 | B4B | 71 | 50.7731 | - | - | - | - | |
Table S4.
Compound peaks detected from GC-MS analysis of samples used for evaluation of temporal and spatial patterns of non-polar metabolites of three cryptic species of P. hornemanni found in the Visayas. Identifications based on comparison of either Kovat’s Indices (KI) of compounds retrieved from the Retention Index Calculator [37] or mass spectral comparison of compounds retrieved from NIST or MassBase. Asterisk (*) indicates compounds with parent ions showing halogenated mass spectral patterns.
| # | Sample | Base Peak | KI | KI Based ID | Mass Spectra Based ID | ||
|---|---|---|---|---|---|---|---|
| RT | Nearest KI | Lucero et al. [37] | Massbase | ||||
| 1 | 3A | 57 | 8.1118 | 1065 | 1062 | y-terpinene | undecane |
| 2 | 15A | 57 | 14.556 | 1285 | 1286 | borneol acetate | 1-nonene |
| 3 | 9A | 57 | 14.556 | 1285 | 1286 | borneol acetate | pentacosane |
| 4 | 29A | 57 | 20.3128 | 1500 | 1499 | a-muurolene | tricosane |
| 5 | 15B | 71 | 20.5458 | 1510 | 1509 | β-bisabolene | 3-ethylpentane |
| 6 | 15A | 57 | 25.186 | 1705 | 1686 | 8-cedren-13-ol | - |
| 7 | 34A | 57 | 25.4206 | 1716 | 1735 | oplopanone | - |
| 8 | 15B | 71 | 26.6674 | 1772 | 1761 | benzyl-benzoate | 2-methylbutane |
| 9 | 15A | 57 | 27.8534 | 1827 | 1827 | isopropyl tetradecanoate | |
| 10 | 44BB | 71 | 30.0015 | 1930 | 1927 | methyl hexadecanoate | docosane |
| 11 | 8B | 57 | 30.4069 | 1950 | 1927 | methyl hexadecanoate | |
| 12 | 15B | 71 | 31.6111 | - | - | - | - |
| 13 | 15A | 57 | 31.8326 | - | - | - | - |
| 14 | 15B | 57 | 31.8342 | - | - | - | 1,4-dimethylhexane |
| 15 | 44BA | 71 | 34.1535 | - | - | - | docosane |
| 16 | 15B | 71 | 34.2967 | - | - | - | 3-ethylpentane |
| 17 | 15A | 57 | 34.76 | - | - | - | 1-octene |
| 18 | 15A | 71 | 35.4496 | - | - | - | 2-methylbutane |
| 19 | 15B | 57 | 35.5757 | - | - | - | - |
| 20 | 15B | 57 | 35.5757 | - | - | - | 2,2-dimethylbutane |
| 21 | 44BA | 71 | 37.9407 | - | - | - | docosane |
| 22 | 15B | 410 | 38.2743 | - | - | - | - |
| 23 | 15A | 71 | 39.3484 | - | - | - | 3-ethylhexane |
| 24 | 15A | 71 | 40.0877 | - | - | - | 2,2-dimethylbutane |
| 25 | 15A | 71 | 40.0981 | - | - | - | beta-Aminopropionitrile |
| 26 | 15B | 71 | 42.5358 | - | - | - | 2-methylbutane |
| 27 | 15B | 57 | 43.9626 | - | - | - | 4-methyloctane |
| 28 | 44C | 71 | 44.6454 | - | - | - | - |
| 29 | 15A | 57 | 45.2255 | - | - | - | 2,2-dimethylbutane |
| 30 | 15A | 71 | 47.3529 | - | - | - | - |
| 31 | 15B | 57 | 47.3534 | - | - | - | - |
| 32 | 15B | 57 | 47.3581 | - | - | - | 2,4-dimethylpentane |
| 33 | 15A | 71 | 47.6237 | - | - | - | 2,4-dimethylhexane |
Map of sampling sites. (a) Map of the Philippines indicating location of Batanes and Visayas; (b) Sampling sites in Batan and Sabtang Islands in Batanes; (c) Sampling sites in Siquijor, Negros, and Cebu Islands in the Visayas).