Abstract
Heart failure (HF) is a common disease that has been attributed, in part, to deprivation of cardiac energy. As a result, the interplay between metabolism and ATP production is fundamental in determining the mechanisms driving the disease progression. Due to its central role in energy production, metabolism, calcium homeostasis, and oxidative stress the mitochondrion has been suggested to play a pivotal role in the progression of the heart to failure. Nevertheless, the mitochondrion’s specific role(s) and the proteins contributing to the development and progression of HF are not entirely clear. Thus, changes in mitochondrial proteomic make-up during HF have garnered great interest. With the continued development of advanced tools for assessing proteomic make-up, characterization of mitochondrial proteomic changes during disease states such as HF, are being realized. These studies have begun to identify potential biomarkers of disease progression as well as protein targets that may provide an avenue for therapeutic intervention. The goal of this review is to highlight some of the changes in mitochondrial proteomic make-up that are associated with the development of HF in an effort to identify target axes and candidate proteins contributing to the disease development. Results from a number of different HF models will be evaluated to gain insight into some of the similarities and differences in mitochondrial proteomic alterations associated with morphological and functional changes that result from the disease.
Keywords: Mitochondria, Proteomics, Heart Failure
INTRODUCTION
Mitochondrial dysfunction is a hallmark of numerous diseases, including heart failure (HF). Deprivation of cardiac energy and ATP production are fundamental contributors to the development of HF(26). A number of studies have focused on the contribution of the mitochondrion in the progression of HF, due to its central role in energy production, metabolism, calcium homeostasis, and oxidative stress, all of which may play a role in the pathogenesis. Nevertheless, it remains unclear as to the exact mechanisms accounting for progression of the heart into failure and the nature of the mitochondrion’s contribution. Because HF can result from a number of pathogenic stimuli, numerous models with distinct etiologies have been utilized for determining the mechanisms contributing to disease progression. Many of the proposed mechanisms incorporate the mitochondrion in the pathogenic sequelae (7). With the development of advanced proteomic tools and in light of the dynamic nature of the mitochondrion, proteomic changes associated with HF are being examined. Determination of mitochondrial proteomic alterations associated with HF is necessary for biomarkers elucidation and for identification of potential protein targets for therapeutic intervention. This review will highlight some of the changes in mitochondrial proteomic make-up associated with HF development. Mitochondrial proteomic changes in a number of different HF models as well as those resulting from pathologies contributing to HF, such as diabetes mellitus will be explored (see Table 1 for a summary).
Table 1.
Changes in mitochondrial proteomes in various heart failure studies.
| SPECIES | MODEL | METHODOLOGY | UPREGULATED | DOWN-REGULATED | REFERENCE |
|---|---|---|---|---|---|
| Human | End-stage | 2D-PAGE, LC-MS/MS | Not identified | PDH, VDAC1, Antiox, TCA, ETC | #37 |
| Human | End-stage | 2D-PAGE, PDQUEST | mtCK | MDH | #19, #20 |
| Rat | TAC | Label-free, LC-MS/MS | PDH, TCA, ETC (I, II, V) | FAO, ETC (I, III, IV, V) | #7 |
| Rat | Spontaneously-hypertensive | iTRAQ, MuDPIT MS/MS | Hsp, PDH, Ketone TCA, Apoptosis, Antiox, ETC (I, III, IV) | FAO, ETC (II, V), TCA, mtCK | #17 |
| Rat | Spontaneously-hypertensive | 2D-DIGE, MALDI MS/MS | FAO, TCA, ETC (I, II) | FAO, PDH, ETC (V), VDAC1 | #16, #24 |
| Rat | ACF | 1D-BN-PAGE | --- | ETC (SSM I, III, IV, V) | #36 |
| Dog | DHF, CRT | 2D-DIGE, ESI-MS/MS, BN-PAGE | ETC (, II, III, V), PC, PDH, ADH, Antiox, PHB, VDAC3 | Cyto c, VDAC2 | #2 |
| Rat | Coronary ligation | 2D-PAGE, MALDI-TOF-MS | PDH, TCA | FAO, ETC (I, III, IV, V) | #39 |
| Rabbit | Anthracycline | 2D-DIGE, MALDI-TOF/TOF-MS | ETC (IV, V), PDH | ETC (I), mtCK, VDAC1, ANT1, Antiox | #33 |
| Mouse | Desmin null | 2D-PAGE, MALDI-TOF-MS | Ketone, acetate, ETC (I), VDAC2, Hsp, Ubi1 | Mal/Asp shuttle, Ketone, PDH | #12 |
| Rat | STZ | 2D-PAGE, MALDI-TOF-MS | FAO, Antiox | ETC (I, III) mtCK, VDAC, Hsp | #35 |
| Rat | STZ | 2D-PAGE, MALDI-TOF-MS | FAO, PDH Hsp, ETC (V) |
ETC (I), TCA, ALDH2 | #14 |
| Rat | STZ | iTRAQ, MudPIT MS/MS | FAO, TCA ETC (V) |
ETC (I-V), TCA, mtCK, Hsp, ALDH2, Antiox | #18 |
| Mice | OVE26 | 2D-PAGE, MALDI-TOF-MS | FAO, ETC (V), TCA, PHB | EF-Tu | #32 |
| Mouse | Akita | Label-free, LC-MS/MS | FAO, ETC (V) | TCA, ETC (I, IV) | #6 |
| Mouse | STZ | 2D-DIGE, TRAQ, MudPIT, LC-MALDI TOF/TOF MS |
ETC (SSM: I III, IV, V), TCA (SSM) | FAO, ETC (IFM: I, IV, V), TCA (IFM), Hsp (IFM) | #5 |
| Mouse | Db/db | iTRAQ, LC-MALDI TOF/TOF MS | FAO (SSM, IFM), ETC (IFM: I, III, IV, V), TCA (SSM, IFM) | FAO (IFM), ETC (SSM: I, III, IV V), Hsp (SSM), PRDX5 (SSM) | #10 |
| Mouse | Db/db | 2D-PAGE, LC-ESI-MS/MS, MALDI-TOF/TOF MS | SCOT (Nitration) | --- | #40 |
| Mouse | ALS | 2D-PAGE, MADI-TOF MS | SCOT (Nitration) mtCK (Nitration) PRDX3 (Nitration) |
--- | #34 |
PDH, pyruvatedehydrogenase; VDAC, voltage dependent anion channel; Antiox, antioxidant; TCA, tricarboxylic acid cycle; ETC, electron transport chain; mtCK, mitochondrial creatinekinase; MDH, malatedehydrogenase; FAO, fatty acid oxidation; Hsp, heat shock protein; ADH, aldehydedehydrogenase; PC, pyruvatecarboxylase; Cytoc, cytochromec; ANT, adenine nucleotide translocator; Ubi1, ubiquitin-like 1; ALDH, aldehydedehydrogenase; PHB, prohibitin; EF-Tu, Elongation factor Tu; SCOT, succinyl-CoA:3-oxoacid CoA transferase; PRDX, peroxiredoxin; SSM, subsarcolemmal mitochondria; IFM, interfibrillar mitochondria.
MITOCHONDRIA PROTEOME DYNAMICS AND MAKE-UP
Mitochondria comprise approximately 35–40% of cardiomyocyte volume (17). Not surprisingly, the mitochondria’s proteomeis highly dynamic during pathological stimuli. With the use of genomic data, it is estimated that the mitochondrial proteome contains approximately 1500 distinct proteins, some of which may be isoforms resulting from various post-translational modifications (PTM) (8, 21). Using a set of complimentary approaches that included in-depth protein mass spectrometry, microscopy, and machine learning, a compendium has been constructed consisting of 1098 genes and their associated proteins products across 14 different mouse tissues (27). The compendium, entitled MitoCarta, is estimated to be over 85% complete and is currently the most comprehensive proteomic characterization of the mitochondrion (27). The mitochondrion possesses its own genome which encodes for 13 proteins which are transcribed and translated within the organelle (4). Thus, the vast majority of mitochondrial proteins (>99%) are encoded by the nuclear genome and require import into the mitochondrion by a complex process of coordinated import machinery (9). The mitochondrion is composed of several subcompartments including the outer mitochondrial membrane (OMM), intermembrane space (IMS), inner mitochondrial membrane (IMM), and matrix. With the increased interest in mitochondrial contribution to disease states, an ever-expanding set of proteomic tools have been developed for assessing proteome make-up as well as identifying PTM status. The details of these methodologies will not be addressed at this time, but are the subject of a number of reviews focusing on strengths, weaknesses, and the appropriateness of the various techniques for the evaluation of a given protein target or a specific submitochondrial locale (matrix, OMM, IMS, IMM)(3, 11, 13, 22, 23, 29, 38).
MITOCHONDRIAL PROTEOMIC CHANGES IN HEART FAILURE
The Failing Human Heart
To gain insight into mitochondrial proteomic alterations associated with human HF, biopsies from patients undergoing coronary artery bypass surgery prior to as well as following revascularization, and patients with end-stage HF undergoing heart transplantation were examined (37). Bypass surgery patients were classified as having reversibly dysfunctional myocardium (RDM) following surgery, which was reflected by improved cardiac function by at least one full wall motion score, or irreversibly dysfunctional myocardium (IRDM) following surgery indicating lack of functional recovery. Two-dimensional gel electrophoresis (2D-PAGE) followed by protein identification via LC-MS/MS revealed differential responses in the cardiac phenotypes relative to reference control tissue. Among the proteins identified as being down-regulated during end-stage HF were six mitochondrial enzymes involved in substrate metabolism and energy generation. In addition, voltage dependent anion-selective protein 1 (VDAC1) as well as superoxide dismutase 2 (MnSOD) were also decreased, suggesting reduced superoxide scavenging potential may be associated with end-stage HF in the human heart. Similarly, down-regulation of six mitochondrial enzymes in IRDM was observed which included electron transport chain (ETC) complex subunits (proteins from complex I), components of the pyruvatedehydrogenase (PDH) complex, and a tricarboxylic acid (TCA) cycle intermediate. Preserved levels of mitochondrial enzymes were observed in RDM, suggesting that mitochondrial proteomic make-up is associated with recovery of function following revascularization.
Proteomic alterations associated with dilated cardiomyopathy in the human heart were examined in a set of early studies in which right atrial tissue from end-stage HF patients obtained during heart transplantation was compared to left ventricular tissue from coronary bypass patients with normal ejection fractions (19, 20, 28). Using 2D-PAGE and computer-assisted analyses, these authors observed a decrease in malatedehydrogenase and an increase in mitochondrial creatine kinase suggesting that dilated cardiomyopathy is associated with effects on cardiac substrate utilization and the creatine phosphate shuttle.
Animal Models of Heart Failure
A number of different animal models of HF have been utilized to determine mechanisms of disease progression, including the potential contribution of the mitochondrion. Examination and description of some of these models has been reported previously (1, 15, 25). Amongst the most explored animal models of HF is the pressure-overload model which includes hypertension-induced hypertrophy leading to failure. Using a transverse aortic constriction model, HF in rats was examined at 20 weeks (7). Mitochondrial proteomes were characterized by LC-MS/MS using a label-free analysis methodology. A reduction in fatty-acid oxidation (FAO) enzymes was observed while components of the PDH complex as well as TCA cycle enzymes were upregulated. ETC complex protein contents showed a differential response with the majority (proteins from complexes I, III, IV, and V) displaying significantly lower abundances, while ten ETC proteins (proteins from complexes I, II, and V) displaying significantly greater abundances. The authors suggest that the general decrease in FAO proteins in the mitochondrion may contribute to a reduction in FAO rates during HF, whereas the differential regulation of ETC proteins may only partially explain respiratory impairments observed in the failing heart.
The spontaneously hypertensive rat (SHR) is widely utilized to model progressive hypertensive cardiac hypertrophy. Using iTRAQ labeling coupled with MuDPIT MS/MS, Jüllig and colleagues characterized the mitochondrial proteome of 18 month old male SHR. Among the increased proteins observed were mitochondrial heat shock proteins (hsp60, hsp10, mthsp70), PDH complex subunits, proapoptotic factors, proteins putatively involved in permeability transition (cytochromec, cyclophilin D), reactive oxygen species (ROS) production and handling proteins (GPx1, MAO A), and ETC complex subunits (proteins from complexes I, III, and IV). In contrast, several ETC complex subunits (proteins from complexes II and V), mitochondrial creatine kinase, and proteins involved in FAO were significantly reduced. The authors conclude that metabolic remodeling associated with depressed FAO, increased glycolytic flux, and inefficient respiration in the failing heart may underlie the pathological progression into failure by reducing membrane stability, enhancing apoptotic susceptibility, and compromising energy transfer to extra mitochondrial sources. Similar results were obtained in a series of studies examining SHR at earlier time points of one and four months (16, 24). The expression of an enzyme involved in long chain FAO (LCFA) was decreased at the prehypertensive stage (one month) while the protein expression of an enzyme involved in short chain FAO (SCFA) was increased at a later stage (four months), which the authors suggest may be a compensatory response to long chain FAO dysfunction. Similar to Jüllig et al., the authors observed a differential dysregulation of ETC complex components though both sets of studies observed similar decreases in complex V protein contents suggesting a similar mechanism of decreased ATP synthesis. Perhaps the most interesting finding in these studies was the observation that different isoforms of the same protein, trifunctional enzyme alpha subunit were differentially expressed in SHR. Additionally, mitochondrial proteome changes seemed to be most dynamic at one month of age in the SHR, prior to hypertension development.
Mitochondrial subpopulation protein contents in a rat volume overload model caused by aortocaval fistula (ACF) were examined by Ulasova et al. (36). Using one-dimensional blue native PAGE (BN-PAGE), the authors reported a decrease in respiratory chain complexes I, III, IV, and V subunit protein composition in subsarcolemmal mitochondria (SSM) of ACF hearts compared to sham controls, with no impact on interfibrillar mitochondria (IFM) respiratory chain complexes. Changes in SSM respiratory complexes of ACF animals were also associated with decreased SSM respiration rates, suggesting that SSM and IFM are differentially affected by ACF which may impart specific effects on mitochondria located in different subcellular spatial locales.
Pacing-induced HF models are used to explore the influence of conduction abnormalities that result in discoordinated contraction which can contribute to the progression of the heart into failure. Cardiac resynchronization therapy (CRT) is a clinical treatment for patients with regional conduction delays, and has been shown to improve heart function and enhance survival. Using a dog model of either dyssynchronous HF (six weeks pacing at 200 bpm) or a CRT protocol (three weeks dyssynchrony followed by three weeks of CRT), Agnetti and colleagues determined mitochondrial proteome changes using 2D-PAGE, ESI MS/MS, and BN-PAGE(2). CRT altered the expression of a number of mitochondrial proteins including increases in ETC complex subunits (proteins from complexes I, II, III, V), proteins involved in metabolic pathways supplying Krebs cycle intermediates (pyruvate carboxylase, PDH, aldehydedehydrogenase), proteins that participate in protein synthesis and import (prohibitin 2), and ROS scavenging enzymes (thioredoxin-dependent peroxidasereductase). Of greatest interest was the observation that CRT influenced the PTM (phosphorylation) status of complex V subunits which was associated with an increase in the amount of mature intact F0F1 complex. The authors suggest that CRT alters cellular proteins involved in energy metabolism which may be important for enhanced systolic work performance in the failing heart.
Using a rat model of left anterior descending coronary artery ligation Wang et al. performed proteomic analyses on isolated mitochondria through 2D-PAGE followed by MALDI-TOF-MS.(39). Eight weeks following coronary ligation, eight mitochondrial proteins were upregulated following the protocol, including glycolytic enzymes, PDH complex subunits, and TCA cycle enzymes. In contrast, 19 mitochondrial proteins were significantly down-regulated following ligation, including proteins involved in FAO and oxidative phosphorylation (proteins from complexes I, III, IV, V). The results are in agreement with other studies providing proteome evidence for a metabolic switch from FAO to glycolysis in HF. Similar conclusions were drawn by Sterba et al who examined proteomic changes following ten weeks of exposure in rabbits to the anticancer anthracycline, daunorubicin, which has been linked to dilated cardiomyopathy and HF(33). Significant changes in mitochondrial proteins included increased expression of ETC complex subunits (proteins from complexes IV, V) and PDH subunits. In contrast, two ETC complex I subunits as well as mitochondrial creatinekinase, VDAC1, ANT1, and MnSOD were down-regulated following daunorubicin exposure.
Examining a desmin null mouse model of heart failure, Fountoulakis et al. determined changes in the mitochondrial proteome using 2D-PAGE and MALDI-TOF-MS (12). The authors report significant increases in the expression of proteins participating in ketone and acetate catabolism, ETC complex subunits (proteins from complex I), VDAC2, mthsp70 and ubiquitin-like 1. In contrast, proteins involved in the malate/aspartate shuttle, ketone utilization, and PDH protein subunits were significantly decreased. Though a number of observed changes were not statistically significant, the authors suggest a high percentage of similar protein changes (> 90 %) in the desmin null mouse model relative to other HF models.
MITOCHONDRIA PROTEOMIC CHANGES INDIABETES MELLITUS
Conditions such as diabetes mellitus can precede the development of HF, and the mitochondrion has been implicated in the pathogenesis. Alterations in the proteomic make-up of mitochondria have been observed in a number of different type 1 and type 2 diabetic models. Using a streptozotocin (STZ) model to induce type 1 diabetes mellitus, mitochondrial proteome changes in rats one and four weeks post injection were examined (35). 2D-PAGE coupled with MALDI-TOF-MS revealed 30 altered proteins within the diabetic mitochondria including elevated levels of FAO proteins, ETC complex proteins (proteins from complex I) and heat shock proteins (hsp60, mthsp70). Similarly, Hamblin et al. examined the proteomic profile of cardiac tissue from STZ-treated rats eight weeks following exposure through 2D-PAGE and MALDI-TOF-MS (14). In agreement with other studies, numerous FAO proteins were upregulated, while antioxidant defense proteins were down-regulated suggesting that alterations in proteins involved in substrate utilization and prooxidant/antioxidant milieu are associated with the diabetic heart. iTRAQ proteomic labeling coupled with MuDPIT MS/MS was utilized to examine STZ-treated rat cardiac mitochondria four months post diabetes mellitus onset (18). In agreement with other studies, proteomic dysregulation of LCFA enzymes and antioxidant defense proteins were prevalent. Specifically, LCFA enzymes were upregulated in the diabetic mitochondria in conjunction with decreased catabolic SCFA enzymes, which may be related to diabetes-associated lipotoxicity. Additionally, antioxidant proteins (ALDH2, GPx1) and heat shock proteins (hsp60, hsp10) were decreased in diabetic mitochondria suggesting diminished ROS scavenging potential and dysregulation of protein folding and degradation.
Using the OVE26 genetic type 1 diabetic model, proteomic changes in whole heart tissue from four month old mice were analyzed through 2D-PAGE and MALDI-TOF-MS(32). Twelve mitochondrial proteins were identified with all but one being upregulated in the diabetic heart. In agreement with other studies, several FAO protein contents were increased suggesting proteomic alterations in response to changes in metabolic fuel sources. Bugger et al. examined mitochondrial proteomes in 12 week old type 1 diabetic Akita mice through mitochondrial subcompartment selective enrichment techniques capable of separating the mitochondrial membranous and matrix fractions using a label-free approach(6). Several proteins involved in FAO were increased, while several TCA cycle proteins were decreased. In addition, ETC complex proteins were differentially altered with an increase in an ATP synthase subunit (complex V) as well as decreases in complex I and IV subunits. Proteomic profiling of spatially-distinct mitochondrial subpopulations was examined in cardiac mitochondrial subpopulations of STZ-treated mice five weeks following exposure. Using multiple proteomic techniques, including 2D-DIGE and iTRAQ coupled with MALDI-TOF-MS, the authors determined that the IFM exhibit a greater dysfunctional proteomic profile as compared to the SSM, following diabetic insult(5). Interestingly, FAO proteins were significantly decreased in the diabetic IFM. Similarly, ETC complex subunits (proteins from complexes I, IV, V), TCA cycle intermediates, and amitochondrial heat shock protein involved in import (mthsp70) were decreased in the diabetic IFM subpopulation with less of an effect on the SSM. Further, PTMs, including oxidations, were more prevalent in the diabetic IFM. These authors suggest that proteome remodeling in the diabetic heart may be subject to subcellular spatial influence. Further, loss of mthsp70 may precipitate nuclear-encoded protein import dysregulation and contribute to changes in proteome dynamics. Indeed, nuclear-encoded mitochondrial protein import into the diabetic IFM was also diminished.
Similar proteomic analyses have been conducted in the type 2 diabetic heart using a db/db mouse model. Proteomic make-up of mitochondrial subpopulations from 18 week old db/db mice was examined using iTRAQ labeling followed by MALDI-TOF-MS(10). In contrast with the type 1 diabetic model, SSM proteomes were most largely impacted as a result of the db/db phenotype. Numerous FAO proteins were significantly elevated in the db/db SSM while IFM FAO proteins showed a mixed response with several being upregulated and several being down-regulated. Further, numerous ETC proteins were significantly decreased in the diabetic SSM (proteins from complexes I, III, IV, V). Similar to the type 1 studies, these authors observed a significant decrease in mthsp70 in the diabetic SSM, also potentially implicating mitochondrial protein import deficiencies within this subpopulation.
Wang et al. explored the impact of protein nitration on four month old db/db mice by performing 2D-DIGE followed by LC-ESI-MS/MS(40). The results indicated that 18 proteins within cardiac mitochondria were nitration targets including succinyl-CoA:3-oxoacid CoA transferase (SCOT), an essential enzyme involved in ketoacidosis. Further, enhanced nitration of Tyr4 and Tyr76 on SCOT in the diabetic mitochondria was directly linked to decreased enzymatic activity of the protein suggesting that nitration of the tyrosine residues is a contributing factor for SCOT inactivation. Similar finding were reported in an alloxan-susceptible mouse model of type 1 diabetes mellitus in which the authors concluded that nitration of mitochondrial proteins may precipitate oxidative stress and dysfunctional mitochondria (34).
CONCLUSIONS
Taken together these studies suggest that the observed changes in mitochondrial proteomic make-up during HF are somewhat model, species, timing, and proteomic methodology specific. Nevertheless, some common features are apparent. For example, proteins involved in substrate utilization (i.e. FAO, glycolysis) seem to be impacted (i.e. upregulated, down-regulated) reflective of a functional change in a specific metabolic fuel (i.e. fatty acid, glucose, ketones). That is, as a particular substrate is favored, proteins involved in the metabolism are influenced, though the switch in fuel sources may ultimately underlie an inefficient use of a given metabolic fuel. Proteins involved in either stress response or in the regulation of prooxidant/antioxidant milieu, are impacted during HF. These findings suggest that ROS and/or oxidative stress play a role in the mechanisms driving the heart to failure and are associated with changes in mitochondrial proteins involved in these processes. ETC complex proteins are differentially regulated in a number of HF models. It is unclear why this response occurs though it has been suggested that upregulation of some ETC components may reflect a compensatory response by the mitochondrion. Interestingly, Rosca and colleagues observed a decrease in respirasomes (supercomplexes consisting of complex I, III, and IV subunits) in a canine model of coronary microembolization-induced HF (30). These authors suggest that the mitochondrial defect in this particular HF model resides in the assembly and function of the respirasome rather than in the individual complex. Decreases in ETC complex activities and/or respirasome formation could ultimately influence ATP production and may underlie contractile abnormalities as a result of insufficient ATP supply for contractile demands. Further, dysfunctional ETC complexes resulting from gain or loss of a particular protein subunit may contribute to enhanced ROS formation, depending on the ETC site and the complex being affected. Finally, though only touched upon, mitochondrial protein PTMs contribute to the diversity of the proteome (3). PTMs may account for altered proteome dynamics though the functional significance depends on the dynamic range of the effect on activity, and the extent of the modification (13).
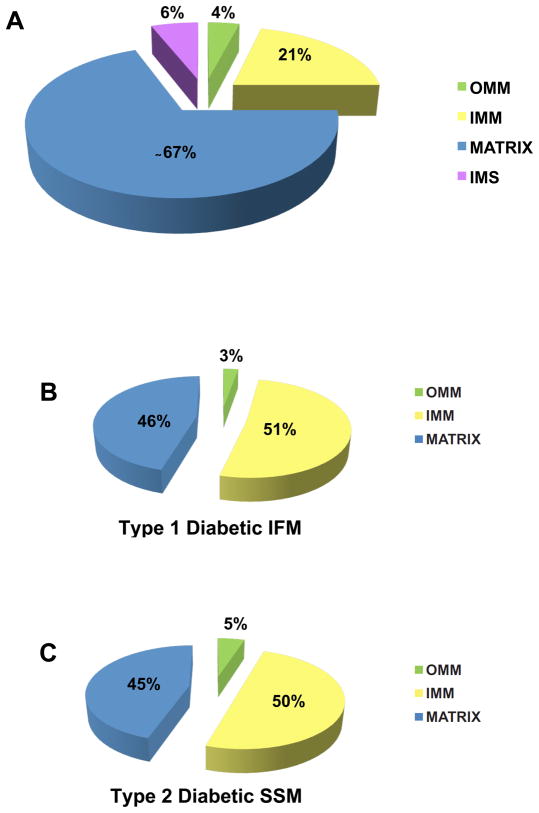
Of particular interest is the observation that proteins of the IMM, an important locus containing the ETC complexes and ATP synthesis machinery, as well as protein trafficking constituents, are specifically impacted in diabetic mitochondria. Proteins localized within the matrix represent the majority of the proteins or approximately 67% of all mitochondrial proteins (11, 31). In contrast, proteins located within the OMM, IMS, and IMM have been calculated to represent approximately 4%, 6%, and 21% of all mitochondrial proteins, respectively (Figure 1A) (11, 31). However, proteomic analyses of mitochondrial subpopulations in both type 1 and type 2 diabetes mellitus (5, 10) revealed that approximately 50% of the proteins identified as being significantly changed in both the type 1 diabetic IFM (Figure 1B) and type 2 diabetic SSM (Figure 1C), were IMM proteins. Interestingly, these two subpopulations of mitochondria were also the most functionally affected mitochondrial subpopulations for each given pathological state, as evidenced by greater dysfunction in the type 1 IFM, and type 2 SSM, respectively(5, 10). In contrast, matrix proteins, which account for a much greater percentage of the mitochondrial proteome accounted for only approximately 45% of proteins affected (Figure 1B and 1C). These results suggest that proteins within the IMM are particularly susceptible in both type 1 and type 2 diabetes mellitus in spite of the fact that less proteins reside in the IMM as compared to the matrix (21 % vs. 67% of total mitochondrial proteins). Indeed, IMM-associated mitochondrial functions including proton pump gradient, ETC respiration, ATP synthesis, and protein import appear to be impacted in many of the models being examined indicating that proteomic changes in this particular submitochondrial locale are associated with dysfunctional mitochondrial processes. These findings suggest that the environment and/or the organization of these proteins within the IMM may place them at an increased risk for proteomic alteration during diabetes mellitus and potentially other pathological states such as HF.
Figure 1. Alterations in spatially-distinct mitochondrial proteomes during diabetes mellitus.
(A) Breakdown in approximate percentages of protein contents in various mitochondrial subcompartments. Data are adapted from Schnaitman et al. J. Cell Biol. 38:158–175, 1968 and Distler et al. Proteomics. 8:4066–4082, 2008. (B) Breakdown in approximate percentages of protein contents changing in the type 1 diabetic heart IFM. (C) Breakdown in approximate percentages of protein contents changing in the type 2 diabetic heart SSM. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane; IMS, intermembrane space.
FUTURE DIRECTIONS
It is clear that the mitochondrial proteome is influenced during HF, reflecting the dynamic nature of the organelle. Nevertheless, the functional relevance of many of the reported proteomic changes is not entirely clear. With the growing interest in understanding the mitochondrion’s contribution to diseases, including HF, advancements in proteomic technologies should help to define the functional basis for many of these observed protein changes. The results of these studies will be critical for biomarker identification of disease progression and aid in the development of therapeutic interventions for specific protein targets designed to treat the failing heart.
Acknowledgments
This work was supported by National Institutes of Health Award # DP2DK083095 (J. M. H.) from the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK). Walter Baseler is a recipient of an American Heart Association Predoctoral Fellowship (#10PRE3420006). ErinneDabkowski is a recipient of an American Heart Association Predoctoral Fellowship (#0815406D).
References
- 1.Abel ED, Doenst T. Mitochondrial adaptations to physiological vs. pathological cardiac hypertrophy. Cardiovasc Res. 2011;90:234–242. doi: 10.1093/cvr/cvr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnetti G, Kaludercic N, Kane LA, Elliott ST, Guo Y, Chakir K, Samantapudi D, Paolocci N, Tomaselli GF, Kass DA, Van Eyk JE. Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ Cardiovasc Genet. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnetti G, Kane LA, Guarnieri C, Caldarera CM, Van Eyk JE. Proteomic technologies in the study of kinases: novel tools for the investigation of PKC in the heart. Pharmacol Res. 2007;55:511–522. doi: 10.1016/j.phrs.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 5.Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, Razunguzwa TT, Hollander JM. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol. 2011;300:R186–200. doi: 10.1152/ajpregu.00423.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, Ganesan B, Weimer BC, Abel ED. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes. 2009;58:1986–1997. doi: 10.2337/db09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugger H, Schwarzer M, Chen D, Schrepper A, Amorim PA, Schoepe M, Nguyen TD, Mohr FW, Khalimonchuk O, Weimer BC, Doenst T. Proteomic remodelling of mitochondrial oxidative pathways in pressure overload-induced heart failure. Cardiovasc Res. 2010;85:376–384. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- 8.Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 9.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC, Hollander JM. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol. 2010;299:H529–540. doi: 10.1152/ajpheart.00267.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Distler AM, Kerner J, Hoppel CL. Proteomics of mitochondrial inner and outer membranes. Proteomics. 2008;8:4066–4082. doi: 10.1002/pmic.200800102. [DOI] [PubMed] [Google Scholar]
- 12.Fountoulakis M, Soumaka E, Rapti K, Mavroidis M, Tsangaris G, Maris A, Weisleder N, Capetanaki Y. Alterations in the heart mitochondrial proteome in a desmin null heart failure model. J Mol Cell Cardiol. 2005;38:461–474. doi: 10.1016/j.yjmcc.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Gucek M, Murphy E. What can we learn about cardioprotection from the cardiac mitochondrial proteome? Cardiovasc Res. 2010;88:211–218. doi: 10.1093/cvr/cvq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamblin M, Friedman DB, Hill S, Caprioli RM, Smith HM, Hill MF. Alterations in the diabetic myocardial proteome coupled with increased myocardial oxidative stress underlies diabetic cardiomyopathy. J Mol Cell Cardiol. 2007;42:884–895. doi: 10.1016/j.yjmcc.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasenfuss G. Animal models of human cardiovascular disease, heart failure and hypertrophy. Cardiovasc Res. 1998;39:60–76. doi: 10.1016/s0008-6363(98)00110-2. [DOI] [PubMed] [Google Scholar]
- 16.Jin X, Xia L, Wang LS, Shi JZ, Zheng Y, Chen WL, Zhang L, Liu ZG, Chen GQ, Fang NY. Differential protein expression in hypertrophic heart with and without hypertension in spontaneously hypertensive rats. Proteomics. 2006;6:1948–1956. doi: 10.1002/pmic.200500337. [DOI] [PubMed] [Google Scholar]
- 17.Jullig M, Hickey AJ, Chai CC, Skea GL, Middleditch MJ, Costa S, Choong SY, Philips AR, Cooper GJ. Is the failing heart out of fuel or a worn engine running rich? A study of mitochondria in old spontaneously hypertensive rats. Proteomics. 2008;8:2556–2572. doi: 10.1002/pmic.200700977. [DOI] [PubMed] [Google Scholar]
- 18.Jullig M, Hickey AJ, Middleditch MJ, Crossman DJ, Lee SC, Cooper GJ. Characterization of proteomic changes in cardiac mitochondria in streptozotocin-diabetic rats using iTRAQ isobaric tags. Proteomics Clin Appl. 2007;1:565–576. doi: 10.1002/prca.200600831. [DOI] [PubMed] [Google Scholar]
- 19.Knecht M, Regitz-Zagrosek V, Pleissner KP, Emig S, Jungblut P, Hildebrandt A, Fleck E. Dilated cardiomyopathy: computer-assisted analysis of endomyocardial biopsy protein patterns by two-dimensional gel electrophoresis. Eur J Clin Chem Clin Biochem. 1994;32:615–624. doi: 10.1515/cclm.1994.32.8.615. [DOI] [PubMed] [Google Scholar]
- 20.Knecht M, Regitz-Zagrosek V, Pleissner KP, Jungblut P, Steffen C, Hildebrandt A, Fleck E. Characterization of myocardial protein composition in dilated cardiomyopathy by two-dimensional gel electrophoresis. Eur Heart J. 1994;15 (Suppl D):37–44. doi: 10.1093/eurheartj/15.suppl_d.37. [DOI] [PubMed] [Google Scholar]
- 21.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427–3440. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Mathy G, Sluse FE. Mitochondrial comparative proteomics: strengths and pitfalls. Biochim Biophys Acta. 2008;1777:1072–1077. doi: 10.1016/j.bbabio.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 23.McDonald TG, Van Eyk JE. Mitochondrial proteomics. Undercover in the lipid bilayer. Basic Res Cardiol. 2003;98:219–227. doi: 10.1007/s00395-003-0417-8. [DOI] [PubMed] [Google Scholar]
- 24.Meng C, Jin X, Xia L, Shen SM, Wang XL, Cai J, Chen GQ, Wang LS, Fang NY. Alterations of mitochondrial enzymes contribute to cardiac hypertrophy before hypertension development in spontaneously hypertensive rats. J Proteome Res. 2009;8:2463–2475. doi: 10.1021/pr801059u. [DOI] [PubMed] [Google Scholar]
- 25.Monnet E, Chachques JC. Animal models of heart failure: what is new? Ann Thorac Surg. 2005;79:1445–1453. doi: 10.1016/j.athoracsur.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 27.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pleissner KP, Regitz-Zagrosek V, Weise C, Neuss M, Krudewagen B, Soding P, Buchner K, Hucho F, Hildebrandt A, Fleck E. Chamber-specific expression of human myocardial proteins detected by two-dimensional gel electrophoresis. Electrophoresis. 1995;16:841–850. doi: 10.1002/elps.11501601139. [DOI] [PubMed] [Google Scholar]
- 29.Reichert AS, Neupert W. Mitochondriomics or what makes us breathe. Trends Genet. 2004;20:555–562. doi: 10.1016/j.tig.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res. 2008;80:30–39. doi: 10.1093/cvr/cvn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnaitman C, Greenawalt JW. Enzymatic properties of the inner and outer membranes of rat liver mitochondria. J Cell Biol. 1968;38:158–175. doi: 10.1083/jcb.38.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab. 2004;287:E896–905. doi: 10.1152/ajpendo.00047.2004. [DOI] [PubMed] [Google Scholar]
- 33.Sterba M, Popelova O, Lenco J, Fucikova A, Brcakova E, Mazurova Y, Jirkovsky E, Simunek T, Adamcova M, Micuda S, Stulik J, Gersl V. Proteomic insights into chronic anthracycline cardiotoxicity. J Mol Cell Cardiol. 2011;50:849–862. doi: 10.1016/j.yjmcc.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Turko IV, Li L, Aulak KS, Stuehr DJ, Chang JY, Murad F. Protein tyrosine nitration in the mitochondria from diabetic mouse heart. Implications to dysfunctional mitochondria in diabetes. J Biol Chem. 2003;278:33972–33977. doi: 10.1074/jbc.M303734200. [DOI] [PubMed] [Google Scholar]
- 35.Turko IV, Murad F. Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem. 2003;278:35844–35849. doi: 10.1074/jbc.M303139200. [DOI] [PubMed] [Google Scholar]
- 36.Ulasova E, Gladden JD, Chen Y, Zheng J, Pat B, Bradley W, Powell P, Zmijewski JW, Zelickson BR, Ballinger SW, Darley-Usmar V, Dell’italia LJ. Loss of interstitial collagen causes structural and functional alterations of cardiomyocyte subsarcolemmal mitochondria in acute volume overload. J Mol Cell Cardiol. 2011;50:147–156. doi: 10.1016/j.yjmcc.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbonavicius S, Wiggers H, Botker HE, Nielsen TT, Kimose HH, Ostergaard M, Lindholt JS, Vorum H, Honore B. Proteomic analysis identifies mitochondrial metabolic enzymes as major discriminators between different stages of the failing human myocardium. Acta Cardiol. 2009;64:511–522. doi: 10.2143/AC.64.4.2041617. [DOI] [PubMed] [Google Scholar]
- 38.Vo TD, Palsson BO. Building the power house: recent advances in mitochondrial studies through proteomics and systems biology. Am J Physiol Cell Physiol. 2007;292:C164–177. doi: 10.1152/ajpcell.00193.2006. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Bai L, Li J, Sun C, Zhao J, Cui C, Han K, Liu Y, Zhuo X, Wang T, Liu P, Fan F, Guan Y, Ma A. Proteomic analysis of mitochondria reveals a metabolic switch from fatty acid oxidation to glycolysis in the failing heart. Sci China C Life Sci. 2009;52:1003–1010. doi: 10.1007/s11427-009-0140-2. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Peng F, Tong W, Sun H, Xu N, Liu S. The nitrated proteome in heart mitochondria of the db/db mouse model: characterization of nitrated tyrosine residues in SCOT. J Proteome Res. 2010;9:4254–4263. doi: 10.1021/pr100349g. [DOI] [PubMed] [Google Scholar]